Abstract
Next-generation sequencing of the hypervariable V3 region of the 16s rRNA gene isolated from serial stool specimens collected from 31 patients receiving allogeneic stem cell transplantation (SCT) was performed to elucidate variations in the composition of the intestinal microbiome in the course of allogeneic SCT. Metagenomic analysis was complemented by strain-specific enterococcal PCR and indirect assessment of bacterial load by liquid chromatography-tandem mass spectrometry of urinary indoxyl sulfate. At the time of admission, patients showed a predominance of commensal bacteria. After transplantation, a relative shift toward enterococci was observed, which was more pronounced under antibiotic prophylaxis and treatment of neutropenic infections. The shift was particularly prominent in patients that developed subsequently or suffered from active gastrointestinal (GI) graft-versus-host disease (GVHD). The mean proportion of enterococci in post-transplant stool specimens was 21% in patients who did not develop GI GVHD as compared with 46% in those that subsequently developed GI GVHD and 74% at the time of active GVHD. Enterococcal PCR confirmed predominance of Enterococcus faecium or both E. faecium and Enterococcus faecalis in these specimens. As a consequence of the loss of bacterial diversity, mean urinary indoxyl sulfate levels dropped from 42.5 ± 11 µmol/L to 11.8 ± 2.8 µmol/L in all post-transplant samples and to 3.5 ± 3 µmol/L in samples from patients with active GVHD. Our study reveals major microbiome shifts in the course of allogeneic SCT that occur in the period of antibiotic treatment but are more prominent in association with GI GVHD. Our data indicate early microbiome shifts and a loss of diversity of the intestinal microbiome that may affect intestinal inflammation in the setting of allogeneic SCT.
Keywords: GVHD, Microbiome, Metagenomics, Clinical allogeneic, transplantation
INTRODUCTION
Immune responses induced by gastrointestinal (GI) microflora play a key role in the pathophysiology of graft-versus- host disease (GVHD), as shown almost 40 years ago by van Bekkum et al.’s demonstration that germ-free animals do not develop GVHD [1]. However, the exact nature of these interactions in GVHD remains unclear. Early studies focused on the ability of bacterial components such as endotoxin (lipopolysaccharide) to activate macrophages that cause inflammation [2,3]. It is now clear, however, that the intestinal microbiome exerts multiple effects that can result in specific tolerance to commensal bacteria in healthy individuals, whereas a loss of microbiome diversity is associated with a number of inflammatory diseases [4,5].
Further evidence for a role of the intestinal microbiome in GVHD was suggested by protection from experimental GVHD by the prophylactic application of probiotic lactobacilli in mice [6] and by clinical data suggesting that prophylactic use of ciprofloxacin/metronidazole antibiotics may reduce GVHD [7]. More recently, 16S rRNA gene sequencing has allowed deeper analysis of intestinal bacterial diversity. In the setting of experimental stem cell transplantation (SCT), Eriguchi et al. [8] provided evidence that murine GVHD disrupts the intestinal microbiome and suggested a loss of Paneth cells and reduced antimicrobial defensins as a potential mechanism. In line with this, we analyzed intestinal biopsies from patients with acute GVHD and observed Paneth cell loss as a hallmark of GVHD also in patients [9]. Jenq et al. [9] further confirmed loss of bacterial diversity in GVHD both in experimental models and in patients receiving allogeneic SCT.
Here, we used both molecular typing of the microbiome by pyrosequencing of the hypervariable V3 region of the 16S rRNA gene and strain-specific PCR of enterococci and an indirect metabolic approach analyzing urinary indoxyl sulfate (IS), which originates from the degradation of tryptophan to indole by colonic microbiota followed by microsomal oxidation to indoxyl and sulfonation, to monitor microbiome shifts in the course of human allogeneic SCT. We identified a major loss of bacterial diversity, which may contribute to intestinal inflammation.
METHODS
This study was approved by the Ethics Committee of the University Medical Center of Regensburg. After provision of informed consent, 31 patients and 3 donors were enrolled in a prospective study to collect stool specimens before and after allogeneic SCT. Median age of the patients was 47 (±11) years. Underlying diseases were acute myelogenous leukemia (n = 14), acute lymphoblastic leukemia (n = 5), lymphoma (n = 4),myeloma and myelodysplastic syndrome (n = 3 each), chronic myelogenous leukemia (n = 1), and aplastic anemia (n = 1). Thirteen patients received grafts from HLA-identical siblings, whereas 18 received grafts from matched unrelated donors. Eleven patients received myeloablative conditioning, and 20 patients received reduced-intensity conditioning. All patients received prophylactic antibiotics (trimethoprim/sulfamethoxazole) starting from admission until day −1, followed by ciprofloxacin/metronidazole [8] from day 0 until engraftment and empiric or therapeutic antibiotics in the case of infections. GVHD symptoms were recorded weekly for the first 100 days; GI GVHD was confirmed by endoscopic biopsy.
One hundred fifty-three stool specimens were collected at a minimum of 3 different time points: before admission until the day of transplantation, at least once on days 7 to 14 (aplastic period), and on days 21 to 28 (engraftment and early GVHD period). In 18 specimens, predominantly in patients with watery diarrhea in the aplastic period, no bacterial DNA could be extracted, which reduced the number of stool specimens subjected to successful 16S rRNA gene sequencing to 135. Later sample collection on a 2-weekly basis and in relation to occurrence of GVHD was scheduled but was mainly performed on inpatients.
Stool specimens were frozen at −80°C until use. DNA was isolated by phenol-chloroform extraction, and the concentration was standardized before amplification. The hypervariable V3 region of the 16s rRNA gene was amplified as described [10] and sequenced on a 454 GS FLX sequencer (Roche Diagnostics, Basel, Switzerland) using Gene Sequencing FLX titanium chemistry, generating on average 20,000 sequence reads per specimen. Overall taxonomic composition of each stool specimen was calculated using the VITCOMIC algorithm 9 [11]. For all DNA samples, additional strain-specific PCR of the internal transcribed ribosomal spacer region was performed according to Tyrrell et al. [12]. Urinary specimens were collected on a weekly basis and frozen at −80°C. Liquid chromatography-tandem mass spectrometry analysis of IS was performed, and results were corrected to urinary creatinine as described previously [13].
For the given sample size, rigorous statistical inference is prohibited by the multitude of confounding factors that can affect microbiome readouts in addition to GVHD. These include the exact combination and dosage of antibiotics, the underlying disease, patient nutrition, diarrhea, day of stool collection, and more. Hence, to avoid over-interpretation of our data, we confined ourselves to data description and visualization.
RESULTS
16S rRNA Gene Sequencing
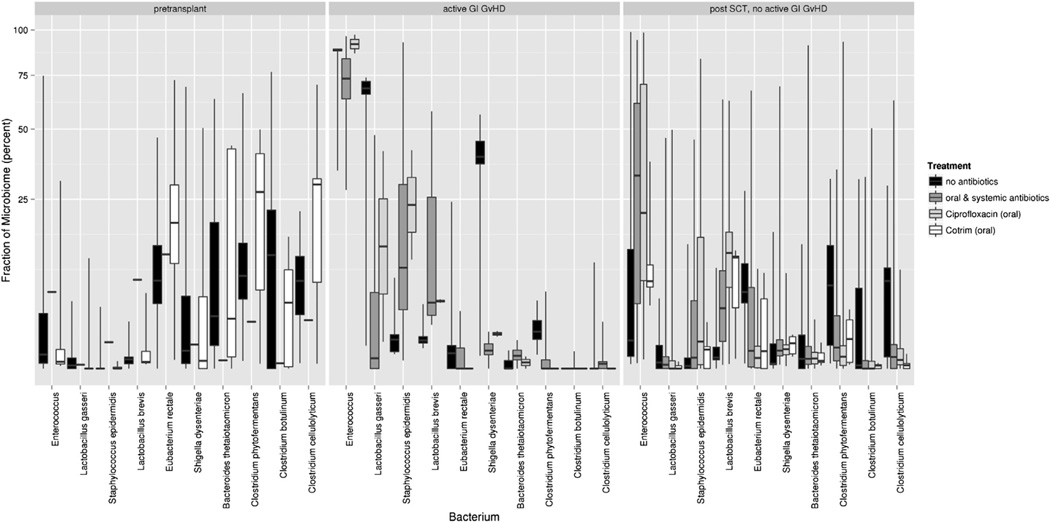
Donors showed an expected distribution of the major phyla: Actinobacteriacea were lowest with .7% ± .4% (arithmetic mean ± standard error), followed by Proteobacteriae, Bacteroidetes, and Firmicutes with 6.2% ± 2.0%, 9.1% ± 5.4%, and 81.7% ± 5.6%, respectively. Among the Firmicutes, enterococci were almost absent, whereas Eubacterium rectale (42.7% ± 16.0%), Clostridium phytofermentans (34.8% ± 20.7%), and Lactobacillus lactis (9.4% ± 1.8%) were the most dominant commensal strains. Patients at the time of admission showed a comparable distribution with some loss of the dominant commensal strains and an increase in enterococci to 6.8% ± 4.9% in previously hospitalized patients. Starting with the day of transplant, all patients showed major shifts in the microbiome with a predominant increase in the proportion of enterococci and a complementary decrease in other Firmicutes and phyla (Figure 1, Supplemental Figure 1). In active GVHD, this shift was most pronounced, with an almost complete loss of commensal Firmicutes such as Clostridia and E. rectale, whereas residual commensals were present in patients without GI GVHD. Beyond day 28, the pattern started to return to pretransplant patterns in patients without and with resolving GI GVHD but not in 2 patients who suffered from ongoing active GI GVHD.
Figure 1.
Box plots of the 10 most variable bacteria in patients pretransplant and post-transplant with and without active GVHD. Data for each bacterium are grouped according to simultaneous use of antibiotics (no antibiotics or oral ciprofloxacin/oral trimethoprim/sulfamethoxazole and oral and systemic antibiotics). Boxes show the median and lower and upper quartiles, respectively, whereas the ends of the whiskers represent the minimum and maximum of all data after removal of outliers. The fraction of microbiome shown on the y-axis was iteratively renormalized as follows: For the most prominent bacterium, the absolute fraction is shown. All reads mapping to this bacterium where removed before calculating the relative fraction of the second most abundant bacterium. For calculating the fraction of the nth most abundant bacterium, all reads of the n-1 more frequent bacteria were removed.
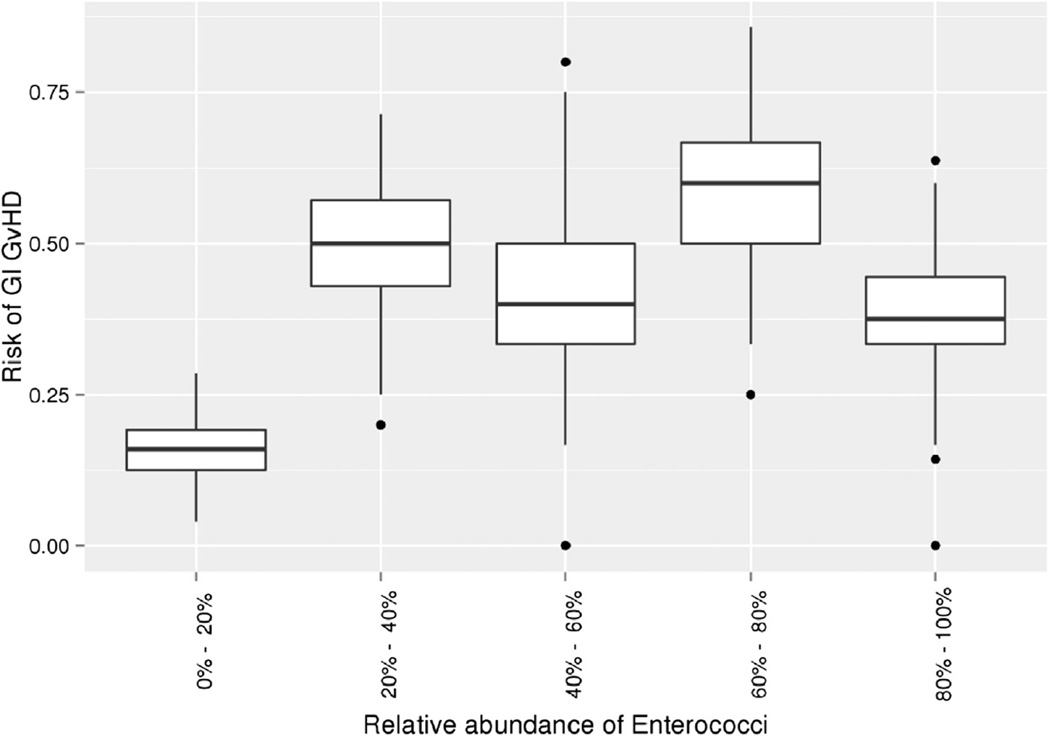
The observed changes indicated 2 factors associated with microbiome shifts. First, the use of broad-spectrum antibiotics, especially ciprofloxacin and systemic antibiotics, resulted in an increase in enterococci and a concomitant reduction in classical commensal bacteria. The second factor associated with a further increase in enterococci was GI GVHD: This was most striking when pretransplant patients were compared with patients without and with active GI GVHD as confirmed by histology. Albeit limited by a small number of samples, the abundance of enterococci in active GI GVHD was also seen in patients who did not receive antibiotics (Figure 1). The potential association of GI GVHD with enterococcal flora is also suggested by the increased frequency of GI GVHD in patients with enterococcal abundance exceeding 20% (Figure 2).
Figure 2.
Box plot of the risk of GI GVHD as a function of relative abundance of enterococci in stool specimens. Boxes show the median and lower and upper quartiles, respectively, whereas ends of the whiskers represent the 9th percentile and the 91st percentile. Outliers are shown as dots. Because the number of observations per patients varies substantially, we sampled 10,000 times randomly 2 observations per patient and averaged the number of enterococci. The distribution of risks across the 10,000 random samples is shown as box plots.
Increase in Enterococci Is Confirmed by Strain-Specific PCR
Because the applied 16s rRNA gene analysis did not distinguish between enterococcal strains, strain-specific PCR was performed to determine the contribution of both Enterococcus faecalis and Enterococcus faecium. E. faecium was significantly more frequent post-transplant, especially in patients with subsequent or active GI GVHD (Table 1). Antibiotic treatment mainly enhanced E. faecalis and to a lesser extent E. faecium(Table 2). Although enterococcal PCR does not allow absolute quantification of bacterial load, copy numbers for E. faecium were significantly increased in patients with active GI GVHD as compared with post-transplant patients without GVHD.
Table 1.
Positive Enterococcal PCR in Post-Treatment Samples in Relation to Later Occurrence or Active GI GVHD
| Post-SCT Group | No Enterococci |
E. faecalis Only |
E. faecium Only |
Both |
|---|---|---|---|---|
| Never GI GVHD | 19 (44%) | 2 (5%) | 18 (42%) | 4 (9%) |
| Subsequent GI GVHD |
3 (11%) | 3 (11%) | 8 (30%) | 13 (48%) |
| Active GI GVHD | 1 (9%) | 2 (19%) | 4 (36%) | 4 (36%) |
Table 2.
Antibiotic Treatment and Positive Enterococcal PCR
| No Enterococci |
E. faecalis Only |
E. faecium Only |
Both | |
|---|---|---|---|---|
| Pre-SCT Without antibiotics |
14 (58%) | 3 (13%) | 6 (25%) | 1 (4%) |
| With antibiotics | 4 (27%) | 4 (27%) | 5 (33%) | 2 (13%) |
| Post-SCT Without antibiotics |
8 (44%) | 0 (0%) | 7 (39%) | 3 (17%) |
| With antibiotics | 16 (25%) | 7 (11%) | 23 (36%) | 18 (28%) |
IS as an Indirect Marker of Bacterial Diversity
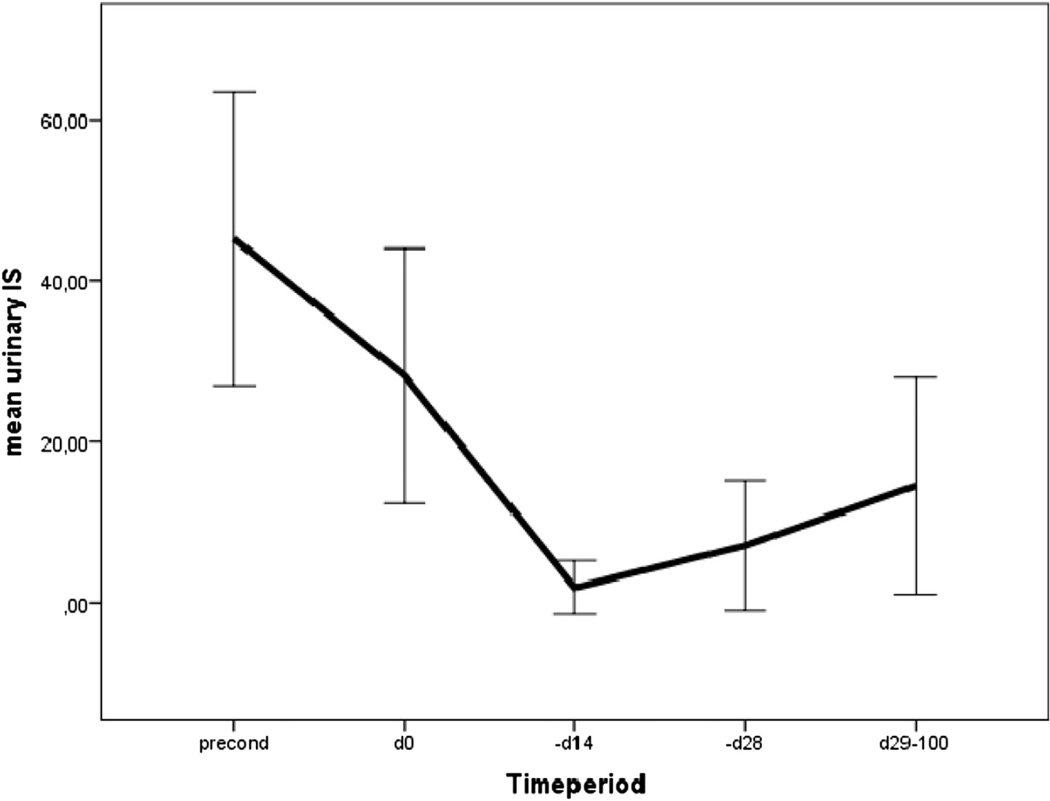
We next tested whether IS could be used as an indirect marker of bacterial load. As shown in Figure 3, urinary IS levels dropped in the period of neutropenia (which is also the period of prophylactic and therapeutic use of antibiotics) and recovered partially thereafter. Mean post-treatment levels were lower in patients with GI GVHD but not on antibiotics and in patients on antibiotics but lowest in patients with GI GVHD and under antibiotic treatment (Table 3).
Figure 3.
Time courses of urinary IS levels (µmol/L, ±2 standard errors). Significant drop of IS levels in relation to antibiotic prophylaxis (trimethoprim/sulfamethoxazole [Cotrim] until day −1, ciprofloxacin + metronidazole day −1 until engraftment [days 16 to 20]). Interaction of GVHD and antibiotic treatment is shown in Table 3.
Table 3.
Mean IS Levels in Relation to Antibiotic Treatment and GI GVHD
| Treatment | Urinary IS (±Standard Error) µmmol/L |
|---|---|
| Pre-SCT (n = 14) | 45.21 (9.10) |
| Post-SCT | |
| Without antibiotics | |
| No active GVHD (n = 19) | 28.24 (8.69) |
| With active GVHD (n = 4) | 6.97 (5.86) |
| With antibiotics | |
| No active GI GVHD (n = 60) | 6.63 (2.15) |
| With active GVHD (n = 7) | .06 (.02) |
Finally, we analyzed the correlation of IS levels with the most prominent specific microbiota. There was a negative correlation with enterococci (−.350, P < .000) and a positive correlation with colonic commensals such as E. rectale (.222, P = .018) and Clostridium phytofermentalis (.222, P = .014), whereas other bacteria such as lactobacilli failed to show any correlation.
DISCUSSION
In our study, we used 2 direct and 1 indirect approach to monitor changes in stool microbiome in the early course of allogeneic SCT. Whereas pretransplant patients had a distribution of bacteria that was quite comparable with healthy normal donors, major changes occurred during the neutropenic period and after engraftment. As shown by the comparison of 16S rRNA gene sequences in patients with and without use of antibiotics, the observed loss of commensal bacteria and the increase in enterococci in this period reflect the administration of both oral ciprofloxacin and systemic broad-spectrum antibiotics. Suppression of commensal flora by clinical use of antibiotics has been shown, and a specific mechanism for overgrowth of enterococci has been suggested [14]: Commensal bacteria are important inducers of antimicrobial peptides (AMPs) to maintain a diverse composition of the microbiome. After prolonged use of antibiotics, induction of specific AMPs such as Reg3alpha declines, which is the major peptide preventing enterococcal overgrowth [15].
In addition, our data showed additional microbiome shifts associated with GI GVHD, as enterococci were more prominent in patients with subsequent and most prominent and persistent in patients with active GI GVHD. Using an indirect metabolomic approach, we observed similar interference of both antibiotics and, to a minor extent, GI GVHD on urinary IS levels: The clear correlation of IS with commensal bacteria such as E. rectale and Clostridia is in line with the reported production of its precursor from tryptophan by colonic commensal bacteria [16].
Our study has limitations of sampling. It was difficult to extract sufficient 16S rDNA for pyrosequencing from stool specimens that had been collected from patients with watery diarrhea during the neutropenic period. A further limitation of the present study was its ability to capture only relative rather than absolute changes in abundance of enterococci in the GI tract. The existence of significant differences in absolute content of enterococci and other bacteria in the stool specimens investigated here is reflected by the fact that it took in some cases up to 9 independent PCR reactions to generate amounts of 16S rDNA sufficient for successful pyrosequencing analysis. Real-time quantitative PCR of 16S rDNA would constitute an obvious means of quantitating bacterial load but is hampered by the difficulty of normalizing 16S rDNA content to such physical measures as weight of stool specimens, which range in consistency from purely liquid to mushy, semiformed, and formed. Indirect measures, such as concentrations of serum or urine biomarkers of bacterial metabolism, may provide an alternative for estimating gut bacterial load. Currently, however, there is no serum or urine biomarker available that is specific for enterococci. Decreased levels of IS in the urine during the neutropenic period and after application of antibiotics and even more in patients with GI GVHD suggest a dramatic suppression of colonic commensals but do not reveal any information on the abundance of enterococci in the gut.
These observations may reflect to some extent the results of 2 studies reporting a disruption of the intestinal microbiome in GVHD. In the murine study by Eriguchi et al. [8], a loss of alpha-defensins as the major group of AMPs was reported in mice developing intestinal GVHD. Jenq et al. [9] tested the role of antibiotic pretreatment on both microbiome composition and the severity and outcome of GVHD. All mice that developed intestinal GVHD revealed massive changes in the intestinal microbiome. However, mice that had not received antibiotics had an overgrowth of lactobacilli, whereas ampicillin treatment resulted in a predominant enterococcal flora as observed in our patients. The murine data suggested that changes in the intestinal microbiome also directly influenced GVHD, as mice treated with ampicillin had worse survival and an increased pathology score.
However, due to the limitations of sampling and quantification, the clinical data do not allow any causal conclusion but may give rise to hypotheses worth testing. Direct analysis of the mucosal microbiome might help to overcome the limitation of insufficient quantities of bacterial DNA, particularly in watery stools, and facilitate quantification. In addition, more patients with active GI GVHD who do not receive antibiotics need to be included to dissect effects originating from antibiotics and GVHD. In an ongoing study, we are analyzing patients who have been weaned off antibiotics for several weeks before the onset of delayed acute GVHD. Preliminary results of enterococcal PCR also suggest a substantial shift to enterococci in a very high fraction of patients.
Direct contribution of enterococci to the worsening of GVHD seems possible based on the aforementioned murine data and may be caused by enterococcal epitheliolysins and other toxins that inflict epithelial damage [17,18]. However, because of the low load of bacteria in the aplastic period, it is more likely that the observed predominance of enterococci is rather an indicator of loss of bacterial diversity, as outlined in Figure 1 This loss of diversity is first induced by oral and systemic broad-spectrum antibiotics, as clearly shown by the substantial drop of IS levels. Additional contribution of GVHD to loss of diversity seems possible not only on the basis of the murine data but is also supported by our recent clinical observation on a substantial loss of Paneth cells in biopsies form patients with severe GI GVHD, which was also highly predictive for outcome [19], and might be associated with loss of AMPs. However, careful analyses of the AMPs in these intestinal biopsies are needed to demonstrate a direct impact of Paneth cell defensins on microbiome composition in GVHD patients.
Finally, the predominance of enterococci may diminish the fraction of commensals, in particular various clostridial strains considered to be important for maintaining tolerance against the intestinal microbiome by inducing regulatory T cells [20,21]. This also raises concerns regarding the practice of intestinal decontamination. Although complete decontamination may be protective against GVHD, our observation indicates that current protocols achieve incomplete rather than complete decontamination, which may be the worst scenario. Defining the role of the microbiome in development of intestinal inflammation may help to develop alternative approaches for prevention of intestinal GVHD such as the use of specific probiotics or therapeutic application of AMPs. Defining inflammatory “checkpoints” of GVHD, as described by Chakraverty et al. [22], related to the microbiome may even contribute to better dissect the deleterious effects associated with clinical GI GVHD from graft-versus-host reactions needed for optimal graft-versus-leukemia effects.
Supplementary Material
Footnotes
Financial disclosure: The authors have nothing to disclose.
Conflict of interest statement: There are no conflicts of interest to report.
SUPPLEMENTARY DATA
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.bbmt.2014.01.030.
REFERENCES
- 1.van Bekkum DW, Roodenburg J, Heidt PJ, van der Waaij D. Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. J Natl Cancer Inst. 1974;52:401–404. doi: 10.1093/jnci/52.2.401. [DOI] [PubMed] [Google Scholar]
- 2.Cooke KR, Olkiewicz K, Erickson N, Ferrara JL. The role of endotoxin and the innate immune response in the pathophysiology of acute graft versus host disease. J Endotoxin Res. 2002;8:441–448. doi: 10.1179/096805102125001046. [DOI] [PubMed] [Google Scholar]
- 3.Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 2000;95:2754–2759. [PubMed] [Google Scholar]
- 4.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerbitz A, Schultz M, Wilke A, et al. Probiotic effects on experimental graft-versus-host disease: let them eat yogurt. Blood. 2004;103:4365–4367. doi: 10.1182/blood-2003-11-3769. [DOI] [PubMed] [Google Scholar]
- 7.Beelen DW, Elmaagacli A, Muller KD, et al. Influence of intestinal bacterial decontamination using metronidazole and ciprofloxacin or ciprofloxacin alone on the development of acute graft-versus-host disease after marrow transplantation in patients with hematologic malignancies: final results and long-term follow-up of an open-label prospective randomized trial. Blood. 1999;93:3267–3275. [PubMed] [Google Scholar]
- 8.Eriguchi Y, Takashima S, Oka H, et al. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood. 2012;120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 9.Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori H, Maruyama F, Kurokawa K. VITCOMIC: visualization tool for taxonomic compositions of microbial communities based on 16S rRNA gene sequences. BMC Bioinform. 2010;11:332. doi: 10.1186/1471-2105-11-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyrrell GJ, Bethune RN, Willey B, Low DE. Species identification of enterococci via intergenic ribosomal PCR. J Clin Microbiol. 1997;35:1054–1060. doi: 10.1128/jcm.35.5.1054-1060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu W, Stevens AP, Dettmer K, et al. Quantitative profiling of tryptophan metabolites in serum, urine, and cell culture supernatants by liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2011;401:3249–3261. doi: 10.1007/s00216-011-5436-y. [DOI] [PubMed] [Google Scholar]
- 14.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinnebrew MA, Ubeda C, Zenewicz LA, et al. Bacterial flagellum stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox CR, Coburn PS, Gilmore MS. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr Protein Pept Sci. 2005;6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 18.Sava IG, Heikens E, Huebner J. Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect. 2010;16:533–540. doi: 10.1111/j.1469-0691.2010.03213.x. [DOI] [PubMed] [Google Scholar]
- 19.Levine JE, Huber E, Hammer ST, et al. Low Paneth cell numbers at onset of gastrointestinal graft-versus-host disease identify patients at high risk for nonrelapse mortality. Blood. 2013;122:1505–1509. doi: 10.1182/blood-2013-02-485813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 21.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 22.Chakraverty R, Cote D, Buchli J, et al. An inflammatory checkpoint regulates recruitment of graft-versus-host reactive T cells to peripheral tissues. J Exp Med. 2006;203:2021–2031. doi: 10.1084/jem.20060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.