Abstract
Temporal control of neuronal differentiation is critical to produce a complete and fully functional nervous system. Loss of the precise temporal control of neuronal cell fate can lead to defects in cognitive development and to disorders such as epilepsy and autism. Mechanistic target of rapamycin (mTOR) is a large serine/threonine kinase that acts as a crucial sensor of cellular homeostasis. mTOR signaling has recently emerged as a key regulator of neurogenesis. However, the mechanism by which mTOR regulates neurogenesis is poorly understood. In constrast to other functions of the pathway, ‘neurogenic mTOR pathway factors’ have not previously been identified. We have very recently used Drosophila as a model system to identify the gene unkempt as the first component of the mTOR pathway regulating neuronal differentiation. Our study demonstrates that specific adaptor proteins exist that channel mTOR signaling toward the regulation of neuronal cell fate. In this Commentary we discuss the role of mTOR signaling in neurogenesis and the significance of these findings in advancing our understanding of the mechanism by which mTOR signaling controls neuronal differentiation.
Keywords: Drosophila, D-Pax2, eye development, headcase, mTOR, neurogenesis, neuronal differentiation, photoreceptor, unkempt
The mature human brain contains around 100 billion neurons that are generated, migrate and differentiate during prenatal development. Defects in neurogenesis lead to epilepsy, autism and other common neurological disorders. To elucidate the complexity of neurogenesis we need to identify key genes and molecules that regulate the generation and differentiation of neurons. This knowledge will also lead to breakthroughs in the understanding and treatment of neurological disease.
Mechanistic target of rapamycin (mTOR) is a large serine/threonine kinase that forms 2 complexes (mTORC1 and mTORC2) that are highly conserved and regulated by upstream signals including insulin receptor (InR)/insulin like growth factor signaling, cytosolic ATP, oxygen and amino acids.1 mTOR was originally identified in budding yeast in a genetic screen for mutations conferring resistance to the immunosuppressive macrolide rapamycin.2 mTOR was subsequently shown to be a key regulator of growth control in yeast, Drosophila and mammals.29 The potency of the mTOR pathway as a regulator of growth comes from the direct control of translation and hence protein synthesis. In recent years mTOR signaling has been shown to control a number of additional fundamental cellular processes including energy metabolism, autophagy, transcription, cytoskeletal dynamics and lipid synthesis.4
Using the developing Drosophila eye as a model system, we were the first to show that the InR/mTOR pathway is a key regulator of neuronal differentiation.5-7 In this context the mTOR pathway temporally regulates the differentiation of 3 of the 8 photoreceptors and the cone cells that constitute each ommatidium (facet) of the fly eye (Fig. 1). mTOR signaling has subsequently been shown to play key roles in the genesis, migration and differentiation of neural progenitor cells in the mammalian nervous system.8-13 Inhibition of mTOR signaling in Disc1 mutant mice, a mutation associated with psychiatric illness in humans, ameliorated the behavioral phenotypes in this model.14 Furthermore, activation of mTOR signaling partially rescued the neuronal cell death phenotype caused by loss of the apical complex protein Pals1.15 The dominant genetic disorder Tuberous Sclerosis Complex (TSC), whose features include epilepsy and autism, is caused by hyperactivation of the mTOR pathway.16 mTOR signaling has also been shown to be activated in animal models of epilepsy and in human cortical dysplasia.17-19
Figure 1.
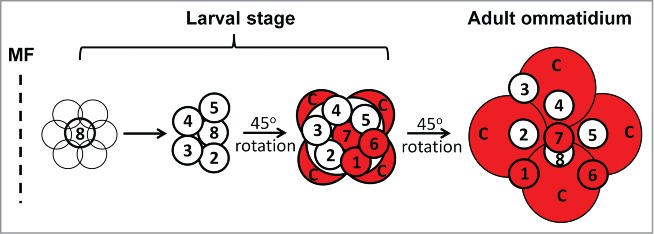
InR/mTOR signaling controls the timing of differentiation of photoreceptors 1,6,7 and cone cells. Differentiation of the 8 photoreceptor neurons and 4 non-neuronal cone cells (C) in the developing Drosophila eye begins with the differentiation of photoreceptor 8 posterior to the morphogenetic furrow (MF), followed by photoreceptors 2,5 and photoreceptors 3,4, then subsequently photoreceptors 1,6,7 and cone cells. InR/mTOR signaling, unk and hdc specifically regulates the timing of differentiation of photoreceptors 1,6,7 and cone cells (red). Anterior is to the left, dorsal is up.
mTOR signaling clearly plays a key role in neural development and neurological disease but, in contrast to other functions of this pathway, the mechanism by which mTOR regulates neurogenesis is unclear. Moreover, since none of the previously identified downstream effectors of mTOR have direct neurogenic roles, it cannot be assumed a priori that there is a specific mTOR pathway neurogenic mechanism. It is conceivable that the ability of mTOR signaling to regulate neurogenesis may be an indirect consequence of its ability to modulate protein synthesis, for example.
With these questions in mind we employed a genetic screen in Drosophila to identify putative downstream components of the mTOR pathway that regulate photoreceptor differentiation.
Through this screen we recently identified the gene unkempt (unk) as a novel negative regulator of photoreceptor differentiation acting downstream of mTORC1 (Fig. 2).20 Unk is a conserved zinc finger/RING domain protein (Fig. 3), originally identified in Drosophila,21 whose function in mammals is unknown (but see below). Unk acts together with its binding partner headcase (Hdc) to negatively regulate the differentiation of 3 of the 8 photoreceptors in the developing ommatidium, as well as non-neuronal cone cells (Fig. 1). Interestingly, neither Unk nor Hdc regulate growth and so the Unk/Hdc complex represents the point of divergence in the mTOR pathway where neuronal differentiation is uncoupled from growth control (Fig. 2).
Figure 2.
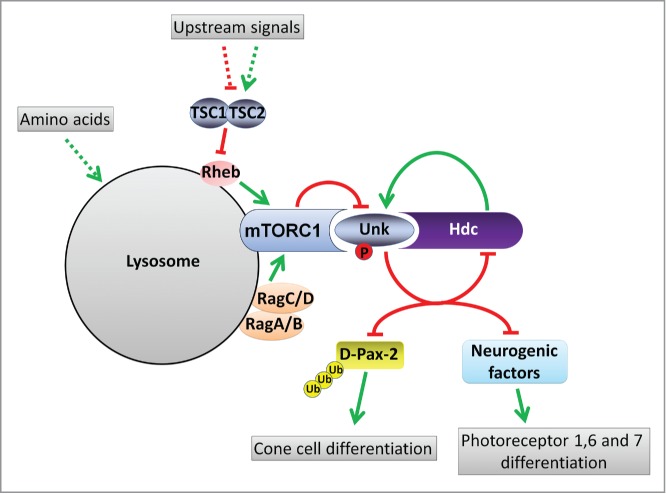
A model for the regulation of neuronal differentiation by mTOR signaling and Unk. Upstream signals regulate the TSC complex which acts to negatively regulate the small GTPase Rheb. When mTOR signaling is active the mTORC1 complex is recruited to the lysosome by the Rag proteins. We hypothesize that mTORC1 physically interacts with the Unk/Hdc complex and negatively regulates its activity by phosphorylating Unk. Unk/Hdc then negatively regulate D-Pax2 in cone cells and putative neurogenic factors in photoreceptors 1,6,7 to control their differentiation.
Figure 3.
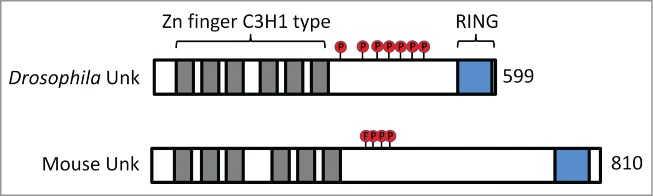
A schematic of the primary structure of Drosophila and mouse Unk proteins. See text for details.
Our work also suggests that the ability of Unk to negatively regulate cell fate comes, in part, from its interaction with the transcription factor D-Pax2. D-Pax2 is required for cone cell differentiation during Drosophila eye development.22 Unk physically interacts with D-Pax2 and negatively regulates D-Pax2 protein levels in vivo. Unk therefore directly connects mTOR signaling to a known transcriptional regulator of cell fate (Fig. 2). D-Pax2 regulates the differentiation of cone cells, but Unk and mTOR signaling also control the timing of differentiation of photoreceptors 1,6 and 7 (Fig. 1), so there must be additional factors that are regulated by Unk in these cells (Fig. 2).
Although the identity and phenotype of Unk as a critical regulator of neuronal differentiation acting downstream of mTORC1 is now established, several key mechanistic questions about the regulation and function of Unk remain: 1. What is the mechanism by which mTOR regulates Unk activity? 2. How does Unk control the levels of interacting proteins such as D-Pax2? Recently published studies have provided tantilising clues to these questions. A quantitative affinity purification/mass spectrometry study of the InR/mTOR interaction proteome in Drosophila Kc167 cells found that Unk physically interacts with the mTORC1 components mTOR, regulatory-associated protein of mTOR (raptor), Sin1 and Lst8, as well as the mTOR substrate Thor (4E-BP).23 Moreover, the interaction between mTOR, raptor, Lst8, Thor and Unk was increased upon activation of the InR pathway. This study strongly suggests that Unk, at least in this context, is a component of the mTORC1 complex. Unk has also been shown to be phosphorylated. A study that used pooled extracts from insulin stimulated and rapamycin treated Drosophila Kc167 cells identified at least 7 phosphorylated serine residues in the C-terminal region between the zinc finger and RING domains of Unk (Fig. 3).24 It is tempting to speculate that these serines may be substrates for mTOR. In support of this idea several of the phospho-serine residues identified in Unk have proline or leucine at the +1 position, consistent with the unique preference for mTOR to phosphorylate residues with proline, hydrophobic and aromatic residues at the +1 position.25 Moreover, mammalian Unk is also potentially phosphorylated by mTORC1 signaling (Fig. 3).25,26
Loss of unk causes an increase in the levels of Hdc and D-Pax-2 proteins in the Drosophila developing eye.20 Unk contains a conserved RING finger domain, characteristic of proteins involved in ubiquitin-dependent protein degradation. In accordance with this Unk has been shown to be part of a complex in human cells containing several ubiquitin conjugating enzymes.27 Moreover, studies in mammalian cells have demonstrated that Unk binds to and promotes the ubiquitination of the chromatin remodelling factor BAF60b.28 By analogy, we hypothesize that Drosophila Unk, through direct protein-protein interaction, promotes the ubiquitination and degradation of D-Pax2 and potentially other neurogenic factors (Fig. 2).
Mammalian Unk has very recently been shown to be a sequence-specific RNA binding protein.29 Unk binds several hundred different mRNA species, through the recognition of a specific motif by the zinc finger domain, and negatively regulates their translation. Unk is strongly expressed in the developing mammalian CNS and knock-down of Unk causes defects in neuronal morphology and migration. Moreover, the RNA binding function of Unk is required for the regulation of cell morphology. This study demonstrates that Unk plays a key role in regulating cell morphology and maturation during mammalian neuronal development. Whether this novel function of Unk is regulated by mTOR signaling in mammals is not known.
In conclusion, our study demonstrates that mTOR signaling is coupled to specific adaptor proteins that channel its activity toward regulating neuronal differentiation, at least in Drosophila. Future studies combining protein biochemistry with structure-function analyses will be required to fully address crucial mechanistic questions. These studies will further advance our understanding of the key role of mTOR signaling in neurogenesis and potentially provide novel insight into neurological disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The author is very grateful to Jernej Murn for communicating results prior to publication.
Funding
The original study commented on in this article was funded by King's College London and the Wellcome Trust (WT088460MA and WT089622MA) in the author's laboratory.
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006; 124:471-84; PMID:16469695; http://dx.doi.org/ 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- 2.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991; 253:905-9; PMID:1715094; http://dx.doi.org/ 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- 3.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12:21-35; PMID:21157483; http://dx.doi.org/ 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/ 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman JM, Mcneill H. Temporal control of differentiation by the insulin receptor/tor pathway in Drosophila. Cell 2004; 119:87-96; PMID:15454083; http://dx.doi.org/ 10.1016/j.cell.2004.08.028 [DOI] [PubMed] [Google Scholar]
- 6.Bateman JM, Mcneill H. Insulin/IGF signalling in neurogenesis. Cell Mol Life Sci 2006; 63:1701-5; PMID:16786222; http://dx.doi.org/ 10.1007/s00018-006-6036-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mcneill H, Craig GM, Bateman JM. Regulation of neurogenesis and epidermal growth factor receptor signaling by the insulin receptor/target of rapamycin pathway in Drosophila. Genetics 2008; 179:843-53; PMID:18505882; http://dx.doi.org/ 10.1534/genetics.107.083097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishwick KJ, Li RA, Halley P, Deng P, Storey KG. Initiation of neuronal differentiation requires PI3-kinase/TOR signalling in the vertebrate neural tube. Dev Biol 2010; 338:215-25; PMID:20004186; http://dx.doi.org/ 10.1016/j.ydbio.2009.12.001 [DOI] [PubMed] [Google Scholar]
- 9.Malagelada C, Lopez-Toledano MA, Willett RT, Jin ZH, Shelanski ML, Greene LA. RTP801/REDD1 regulates the timing of cortical neurogenesis and neuron migration. J Neurosci 2011; 31:3186-96; PMID:21368030; http://dx.doi.org/ 10.1523/JNEUROSCI.4011-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paliouras GN, Hamilton LK, Aumont A, Joppe SE, Barnabe-Heider F, Fernandes KJ. Mammalian target of rapamycin signaling is a key regulator of the transit-amplifying progenitor pool in the adult and aging forebrain. J Neurosci 2012; 32:15012-26; PMID:23100423; http://dx.doi.org/ 10.1523/JNEUROSCI.2248-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman NW, Lin TV, Zhang L, Paquelet GE, Feliciano DM, Bordey A. mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Rep 2013; 5:433-44; PMID:24139800; http://dx.doi.org/ 10.1016/j.celrep.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 12.Lafourcade CA, Lin TV, Feliciano DM, Zhang L, Hsieh LS, Bordey A. Rheb activation in subventricular zone progenitors leads to heterotopia, ectopic neuronal differentiation, and rapamycin-sensitive olfactory micronodules and dendrite hypertrophy of newborn neurons. J Neurosci 2013; 33:2419-31; PMID:23392671; http://dx.doi.org/ 10.1523/JNEUROSCI.1840-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ka M, Condorelli G, Woodgett JR, Kim WY. mTOR regulates brain morphogenesis by mediating GSK3 signaling. Development 2014; 141:4076-4086; PMID:25273085; http://dx.doi.org/ 10.1242/dev.108282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M, Li W, Huang S, Song J, Kim JY, Tian X, Kang E, Sano Y, Liu C, Balaji J, et al.. mTOR Inhibition ameliorates cognitive and affective deficits caused by Disc1 knockdown in adult-born dentate granule neurons. Neuron 2013; 77:647-654; PMID:23439118; http://dx.doi.org/ 10.1016/j.neuron.2012.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, Lehtinen MK, Sessa A, Zappaterra MW, Cho SH, Gonzalez D, Boggan B, Austin CA, Wijnholds J, Gambello MJ. et al.. The apical complex couples cell fate and cell survival to cerebral cortical development. Neuron 2010; 66:69-84; PMID:20399730; http://dx.doi.org/ 10.1016/j.neuron.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlova KA, Crino PB. The tuberous sclerosis complex. Ann N Y Acad Sci 2010; 1184:87-105; PMID:20146692; http://dx.doi.org/16912980 10.1111/j.1749-6632.2009.05117.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ljungberg MC, Bhattacharjee MB, Lu Y, Armstrong DL, Yoshor D, Swann JW, Sheldon M, D'arcangelo G. Activation of mammalian target of rapamycin in cytomegalic neurons of human cortical dysplasia. Ann Neurol 2006; 60:420-9; PMID:16912980; http://dx.doi.org/ 10.1002/ana.20949 [DOI] [PubMed] [Google Scholar]
- 18.Zeng LH, Rensing NR, Wong M. The mammalian target of rapamycin signaling pathway mediates epileptogenesis in a model of temporal lobe epilepsy. J Neurosci 2009; 29:6964-72; PMID:19474323; http://dx.doi.org/ 10.1523/JNEUROSCI.0066-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo E, Citraro R, Constanti A, De Sarro G. The mTOR signaling pathway in the brain: focus on epilepsy and epileptogenesis. Mol Neurobiol 2012; 46:662-81; PMID:22825882; http://dx.doi.org/ 10.1007/s12035-012-8314-5 [DOI] [PubMed] [Google Scholar]
- 20.Avet-Rochex A, Carvajal N, Christoforou CP, Yeung K, Maierbrugger KT, Hobbs C, Lalli G, Cagin U, Plachot C, Mcneill H. et al.. Unkempt is negatively regulated by mTOR and uncouples neuronal differentiation from growth control. PLoS Genet 2014; 10:e1004624; PMID:25210733; http://dx.doi.org/ 10.1371/journal.pgen.1004624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohler J, Weiss N, Murli S, Mohammadi S, Vani K, Vasilakis G, Song CH, Epstein A, Kuang T, English J. et al.. The embryonically active gene, unkempt, of Drosophila encodes a Cys3His finger protein. Genetics 1992; 131:377-88; PMID:1339381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev 1997; 11, 2066-78; PMID:9284046; http://dx.doi.org/ 10.1101/gad.11.16.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glatter T, Schittenhelm RB, Rinner O, Roguska K, Wepf A, Junger MA, Kohler K, Jevtov I, Choi H, Schmidt A. et al.. Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol Syst Biol 2011; 7:547; PMID:22068330; http://dx.doi.org/ 10.1038/msb.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG. et al.. PhosphoPep–a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol 2007; 3:139; PMID:17940529; http://dx.doi.org/ 10.1038/msb4100182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB. et al.. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011; 332:1317-22; PMID:21659604; http://dx.doi.org/ 10.1126/science.1199498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al.. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011; 332:1322-6; PMID:21659605; http://dx.doi.org/ 10.1126/science.1199484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S. et al.. A census of human soluble protein complexes. Cell 2012; 150:1068-81; PMID:22939629; http://dx.doi.org/ 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lores P, Visvikis O, Luna R, Lemichez E, Gacon G. The SWI/SNF protein BAF60b is ubiquitinated through a signalling process involving Rac GTPase and the RING finger protein Unkempt. FEBS J 2010; 277:1453-64; PMID:20148946; http://dx.doi.org/ 10.1111/j.1742-4658.2010.07575.x [DOI] [PubMed] [Google Scholar]
- 29.Murn J, Zarnack K, Yang YJ, Durak O, Murphy EA, Cheloufi S, Gonzalez DM, Teplova M, Curk T, Zuber J, et al.. Control of a neuronal morphology program by an RNA-binding zinc finger protein, Unkempt. Genes Dev 2015; 29:501-12; PMID:25737280; http://dx.doi.org/ 10.1101/gad.258483.115 [DOI] [PMC free article] [PubMed] [Google Scholar]


