Abstract
During development of the nervous system, it is essential to co-ordinate the processes of proliferation and differentiation. Basic helix-loop-helix transcription factors play a central role in controlling neuronal differentiation and maturation as well as being components of the combinatorial code that determines neuronal identity. We have recently shown that the ability of the proneural proteins Ngn2 and Ascl1 to drive neuronal differentiation is inhibited by cyclin dependent kinase-mediated multi-site phosphorylation. This limits downstream target promoter dwell time, thus demonstrating a direct mechanistic regulatory link between the cell cycle and differentiation machinery.Proneural proteins are key components of transcription factor cocktails that can bring about the direct reprogramming of human fibroblasts into neurons. Building on our observations demonstrating that phospho-mutant proneural proteins show an enhanced ability to drive neuronal differentiation in vivo, we see that replacing wild-type with phospho-mutant proneural proteins in fibroblast reprogramming cocktails significantly enhances the axonal outgrowth, branching and electrophysiological maturity of the neurons generated. A model is presented here that can explain the enhanced ability of dephosphorylated proneural proteins to drive neuronal differentiation, and some unanswered questions in this emerging area are highlighted.
Keywords: cell cycle, cyclin-dependent kinase, development, differentiation, phosphorylation, proneural, reprogramming, transcription factor, Xenopus
Introduction
There is a long established relationship between cell cycle lengthening and the onset of neuronal differentiation,1 but the mechanistic basis for this link has only recently been uncovered. Central to co-ordination of cell cycle and differentiation is the post-translational modification of proneural transcription factors by cyclin-dependent kinases.
The basic helix-loop-helix (bHLH) proneural factors Neurogenin2 (Ngn) and Ascl1 are master transcriptional regulators of many classes of neurons in the CNS and PNS.2 These proneural proteins, and particularly Ascl1, are also centrally important components of reprogramming transcription factor cocktails that have been used to directly convert human fibroblasts into neurons.3,4 However, these reprogramming approaches, particularly when involving fibroblasts, suffer from low efficiency and the poor maturity of the neurons generated; a more thorough understanding of how the activity of these proneural factors is controlled is required to produce mature neurons in vitro for applications such as disease modeling. Our recent work has demonstrated that multi-site phosphorylation of Ngn2 and Ascl1 plays a key role in limiting their ability to drive differentiation and maturation of neurons in vivo in developing embryos and in vitro in fibroblast reprogramming.5-7 These advances combining biochemical, in vivo and in vitro analyses would not have been possible without the use of the uniquely tractable Xenopus frog embryo system to address fundamental questions of what controls neuronal differentiation in co-ordination with cell cycle exit during development.
Proneural transcription factors control neuronal progenitor maintenance and differentiation
Ngn2 is expressed, at least at readily detectable levels, just prior to cell cycle exit in a number of classes of neural precursors, driving cell cycle exit and, subsequently, during the many aspects of differentiation. Ngn2 also has temporally distinct later roles in neuronal migration and axonal outgrowth.2,8 Ascl1/Mash1 plays a somewhat analogous role to Ngn2 in the CNS and PNS, where its upregulation drives many different processes during differentiation of selected neurons, for instance, in gaba-ergic neurons of the ventral telencephalon and nor-aderinergic neurons of the sympathetic ganglia.9-13 To fulfil these roles, Ngn2 and Ascl1 (along with other members of the proneural family) act as master regulators of batteries of downstream targets, co-ordinating multiple processes in neuronal differentiation.8,9 However, these proneural proteins also play a key early role in progenitor maintenance.9,14,15 In many regions within the developing nervous system, neurons that begin to undergo differentiation act non-cell autonomously to prevent their neighbors from differentiating via activation of Delta-Notch signaling. Briefly, proneural proteins transcriptionally upregulate the transmembrane ligand Delta. Delta binds the Notch ligand on adjacent cells, activating a downstream cascade in these neighbors that ultimately inhibits both the transcription and post-translational activity of their proneural proteins. This results in the maintenance of these adjacent cells as neuronal precursors. While this rather static view of Notch-mediated regulation of cell fate is well established for Drosophila neuroblasts,16 Notch mediated regulation of proneural proteins in vertebrates is a much more dynamic process; Kageyama's elegant work17-19 has revealed reciprocal oscillatory waves of proneural protein expression and Notch signaling that are required for neural stem cell maintenance. Conversely, the replacement of oscillatory expression with stable high proneural proteins, and stable low Notch effectors, is a prerequisite for neuronal differentiation.
Despite a basic characterization at the level of gene expression, there is still much we do not understand about how proneural proteins control the balance between proliferation and differentiation, or indeed how they can act in temporally distinct events such as during progenitor maintenance, early in differentiation around the time of cell cycle exit, and also at later stages in post-mitotic neurons controlling processes such as neuronal migration and axonal outgrowth and branching. The expression level of proneural proteins plays some part in co-ordinating these distinct roles at different times; recent work has shown that high levels of Ascl1 expression/overexpression drives cell cycle exit and differentiation after in vivo electroporation and in NS cells. However, a lower level of Ascl1 is required for neural stem cell maintenance, where it binds and supports expression of key pro-proliferative targets such as cdks, skp2 and E2F1.9,10 Moreover, the transition from progenitor maintenance to differentiation co-incides with a transition from oscillatory to stable Ascl1 expression.19 Nevertheless, whether the switch from supporting stem-ness to promoting differentiation is due solely to changes in absolute Ascl1 levels, to a change from its oscillatory to stable expression, or to changes in co-factor recruitment or other aspects of Ascl1 regulation and its context-dependent activity is currently unclear.
Multi-site phosphorylation by cdks controls the activity of proneural proteins
Against this background several years ago, building on a strong base of analysis of cell cycle control in Xenopus development,20-23 the Philpott lab set out to further understand the mechanistic links between cell cycle regulation and the function of proneural proteins during neuronal differentiation. The model system we chose to investigate first was generation of the 3 stripes of primary neurons that are the first neurons to become specified and differentiate within the neural plate in Xenopus frog embryos, a process that is directed by Ngn2.15 This system is highly sensitive to cell cycle status; enhancing cycling by increasing cdk kinase levels inhibits primary neuron differentiation while promoting cell cycle lengthening will trigger enhanced neuronal differentiation.21,22 We have used the biochemical and embryological strengths of Xenopus to characterize the mechanistic links between cell cycle and differentiation in primary neurons.
Post-translational modification of proneural proteins has previously been described as regulating aspects of their behavior. For instance, we have shown that several bHLH proneural proteins are rapidly degraded by ubiquitin-mediated proteolysis.24-26 Others have uncovered regulatory roles for phosphorylation on specific residues such as mAscl1 phosphorylation on Ser152 that regulates its ability to bind to E proteins,27 and mNgn2 phosphorylation on Tyrosine 241, that controls neuronal migration.28 However, we have found that multi-site phosphorylation on serine-proline pairs by cyclin-dependent kinases, found at highest levels in rapidly cycling cells, inhibits the ability of both Ascl1 and Ngn2 to drive neuronal differentiation; phospho-mutant forms of these proteins where all serine-proline pairs were mutated to alanine-proline (S-A mutants) showed a substantially enhanced ability to induce ectopic neurons in Xenopus5,7 (Fig. 1A). Moreover, phospho-mutant proneurals also showed increased activity in P19 embryonal carcinoma cells that respond by differentiating into neurons.5 Although in some experimental settings S-A proneural proteins are found at higher levels than their wild-type counterparts (data not shown), a difference in stability appears not to account for the enhanced activity of the phospho-mutant protein.5,7 Indeed by using chromatin immunoprecipitation we demonstrated that, even when normalizing for protein level, phospho-mutant proneural proteins bind better to their target promoters than their wild-type counterparts. Moreover, promoter binding affinity is at least semi-quantitatively dependent on the level of kinase; the more cdk kinase, the more serine-proline sites are phosphorylated, and the lower the level of promoter binding.5,7
Figure 1.
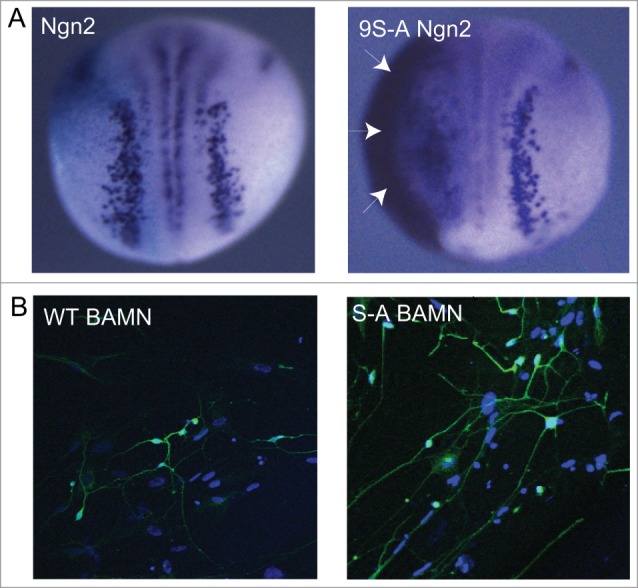
(A) Overexpression of wild-type Ngn2 induces limited ectopic neurons in the Xenopus neural plate and ectoderm on the injected side of the embryo, while 9S-A Ngn2 (phospho-mutant Ngn2) shows considerably greater activity, producing extensive ectopic neurons (arrows) on the flank of the embryo. Taken from ref.5 (B) Brn2, Ascl1, MyT1L and NeuroD together can reprogram human lung fibroblasts into neurons, stained here in green with neural β- tubulin. When wild-type Ascl1 is replaced by S-A Ascl1 (phospho-mutant Ascl1), neurons show significantly increased axonal outgrowth and branching. Taken from ref.7
Enhanced proneural binding is equally evident by ChIP on both the Delta “progenitor maintenance” promoter and promoters for “differentiation” targets such as NeuroD and MyT1.6,7 But crucially, even though enhanced binding is seen on all the promoters tested so far, “differentiation target” promoters respond to enhanced binding by the phospho-mutant proneural by a substantial increase in transcript levels (e.g. MyT1 and neural β tubulin increasing 7-fold in response to S-A Ascl1 compared to Wt Ascl1, while the Delta promoter show only a 2-fold enhancement).6,7 This means that Ngn2 and Ascl1 can activate Delta transcription and so promote progenitor maintenance while phosphorylated, as would be the case in cycling cells. However, targets driving differentiation such as NeuroD and MyT1 can only be activated effectively by a dephosphorylated proneural protein found in cells with reduced levels of cdk kinase occurring on cell cycle lengthening and exit (Fig. 2).
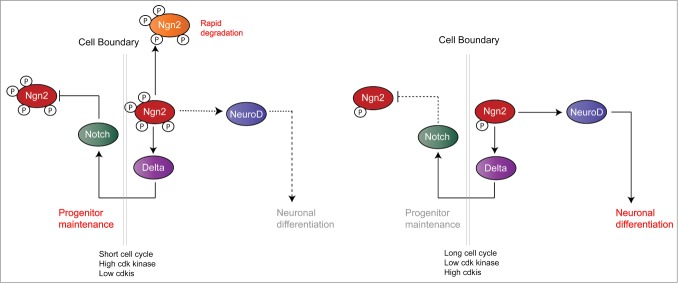
Figure 2.
Model illustrating why phosphorylated Ngn2, found when cdk kinase levels are high, favors progenitor maintenance. Conversely, dephosphorylation of Ngn2 that occurs when cdk levels drop and cdk inhibitors increase, favors differentiation. Taken from ref.7
We also have evidence that Ascl1 is highly phosphorylated in neuroblastoma, a pediatric cancer that arises from noraderinergic neurons of the developing sympathetic nervous system, and this phosphorylation inhibits its ability to drive neuronal differentiation. This finding indicates that multi-site phosphorylation of proneural proteins in response to high levels of cdks found in many cancers may contribute to their oncogenic phenotype.29
Why do distinct promoters respond differently to proneural protein phospho-status?- a model
We hypothesize that transcriptional targets respond differently to proneural protein phosphorylation status because of different epigenetic availability of the target promoters. This assumption is based on the fact that Delta is known to require little or no epigenetic modification for transcriptional activation by proneural proteins in the Xenopus neural plate, and that Delta transcript levels respond very rapidly to an increase in Ngn2 levels.6,30 In contrast, “differentiation” genes such as NeuroD respond considerably more slowly to Ngn2 expression and the NeuroD promoter has been shown to require both Swi/Snf-related nucleosome remodelling as well as p300/CBP-mediated epigenetic modification for activation.6,30,31 These observations have led us to the following model (Figs. 2 and 3), where we consider that genes driving neuronal differentiation may have epigenetically “closed” promoters, while genes driving progenitor maintenance may be epigenetically “open”.6
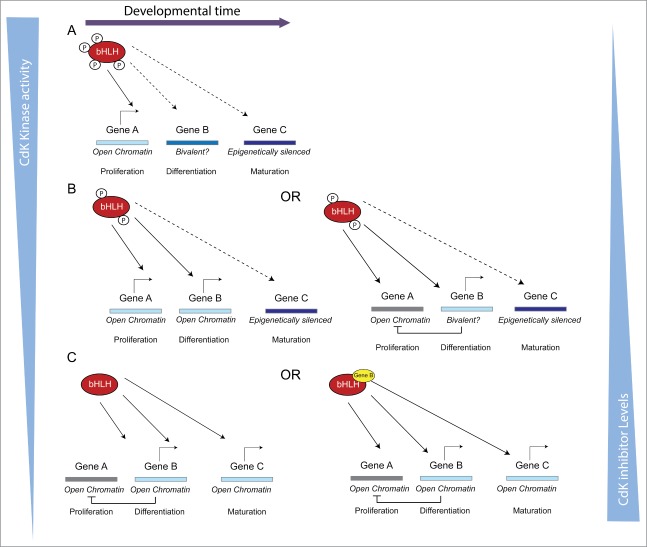
Figure 3.
Model illustrating how changing promoter dwell time by bHLH transcription factors by altering their phospho-status would have differing effects on promoter activation depending on their epigenetic availability (described in more detail in the text).
In cycling cells, levels of cdk kinases (and other proline-directed kinases such as MAP kinases) are generally high, or at least cycle rapidly between high states. These kinases phosphorylate proneural proteins in neuronal progenitors on multiple sites, limiting their ability to bind stably to promoters. Transcription factors cycle on and off promoters with a promoter dwell time estimated to be in the order multiple seconds or perhaps a few minutes (although this is an area where little quantitative data is available) e.g., ref.32,33 This is sufficient to recruit the core transcriptional machinery and activate gene expression of epigenetically available “open” promoters or progenitor maintenance targets. However, to bring about epigenetic modification and activation of “closed” promoters, transcription factors must have a sufficient dwell time per binding cycle to effectively recruit modifiers such as histone acetyl transferases and neucleosome remodellers to open chromatin to the general transcription machinery. Proneural proteins play a key role in recruiting remodellers to their multiple downstream targets.30,31 Phosphorylated proneural protein with reduced promoter binding/dwell time cannot bind “closed” targets long enough per binding cycle to allow any recruited remodellers to “open” the epigenetically unavailable chromatin of differentiation targets such as NeuroD, MyT1 and neural β-tubulin. Thus, in neural stem/progenitor cells with high cdk kinase levels, proneural proteins can drive progenitor maintenance (via the Delta/Notch oscillatory mechanism proposed by Kageyama et al18) but cannot drive neuronal differentiation.
Cell cycle lengthening alone is a trigger for neuronal differentiation.1 As the cell cycle gets longer, the levels of cyclin-dependent kinases, particularly those expressed in G1 phase such as cyclins D and E gradually drop, coinciding with a rise in cdk inhibitor levels. High levels of cdk2 and cdk1 (cdc2) are still required for key cell cycle transitions such as DNA replication and mitosis in even slowly dividing cells. However, as the G1 phase lengthens, the proportion of time spent in these states of high cdk kinase shortens relative to G1 when cdks are lower. Assuming that the phosphatases removing phosphate groups from cdk targets remain at a constant level, this would lead to a progressive decrease in the overall phosphorylation of proneural protein for extended periods as the cell cycle lengthens. This will result in enhanced target promoter binding by increasing promoter dwell time per binding cycle, and allows recruitment of the necessary epigenetic modifiers to bring about “opening” of promoters of differentiation targets.
We have also shown that Ngn2 is directly stabilized by the cdki p27Xic1,21 probably by direct binding (unpublished data). This provides a further mechanism for enhancing bHLH promoter dwell-time driven activation of downstream differentiation targets on cell cycle lengthening (Fig. 2). Once epigenetically “opened,” proneural proteins can then drive transcription of these targets to bring about differentiation and override Delta-mediated progenitor maintenance. It is also relevant that we see constitutively active Notch can post-translationally inhibit the ability of both Ngn2 and Ascl1 to drive ectopic neurogenesis in Xenopus (although the mechanism for this is currently somewhat unclear), but Notch signaling is less effective at inhibiting neurogenesis driven by phospho-mutant proneural proteins.6,7 This results in enhancement of expression of differentiation targets, and also limits the ability of Notch signaling to suppress this differentiation.
The model presented here gives a framework to understand how distinct target promoters are differentially responsive to phospho-status of proneural proteins. It may also shed light on how proneural proteins can have many potentially conflicting functions including progenitor maintenance and early and late stages of neuronal differentiation that are temporally distinct.10 Indeed, the model can be extendable to consider the different timing of expression of multiple targets and the requirement for additional co-factors for target activation, as described below (Fig. 3).
Let us consider expression of the proneural target genes A, B and C, where A is required for progenitor maintenance, B for neuronal differentiation and gene C for later stages of neuronal maturation. Gene A is epigenetically available and can be transcriptionally activated by proneural proteins whether maximally phosphorylated (PPPP-proneural) with low promoter dwell time, by partially phosphorylated proneural proteins (PP-proneural) or by fully de-phosphorylated proneural protein (deP-proneural) with maximal dwell time. Gene B is less available and perhaps shows bivalent epigenetic marks and hence is a gene poised for developmental expression after some limited epigenetic modification. This can be brought about by the enhanced promoter binding of PP-proneural but not by PPPP-proneural. PP-proneural accumulates as the cell cycle lengthens and further drives cell cycle exit. Moreover, if gene B directly promotes differentiation, it may negatively feedback to inhibit gene A expression to further slow the cell cycle and potentiate differentiation. This results in further dephosphorylation of proneural proteins to generate enough de-P proneural protein with enhanced promoter dwell time to open the “closed” enhancer/promoter of Gene C that is required for later stages of neuronal differentiation and maturation. Opening/activation of gene C promoter could be further enhanced by the product of gene B accumulating and acting with de-P proneural by binding to adjacent sites in the gene C promoter.
A number of questions remain when considering the post-translational regulation of proneural proteins by the cell cycle machinery including: is multi-site phosphorylation of proneural proteins a widespread conserved mechanism to control the ability of bHLH proteins to regulate cell differentiation and maturation in response to the kinase environment? Unpublished data in our laboratory indicates that this may indeed be the case, with several other proneural proteins displaying similar regulation. How are proneural factor protein half-life, phospho-status and oscillatory vs. stable expression integrated to give a changing transcriptional output over time? This is unclear. We do not have evidence that multi-site phosphorylation of proneural proteins affects their half-life in isolation in in vitro degradation systems (unpublished data). We do, however, see that phosphomutant proneural proteins are expressed at higher levels than their wild-type counterparts in some circumstances (unpublished data), although the reason for this has yet to be firmly established. Does multi-site phosphorylation of proneural proteins directly control recruitment of co-factors? The phosphorylated regions are either side of the bHLH and appear to be natively unstructured,34 making them prime candidates to act as adapters for co-factors that must be recruited to downstream target promoters for their activation. We are currently investigating co-factor binding and proneural protein phospho-status. Does phospho-status of proneural proteins solely affect their ability to control progenitor maintenance or differentiation or will it also affect their ability to influence neuronal subtype? We have limited preliminary evidence that this may be the case under some circumstances. However, this question may be best studied by using proneural proteins to generate mammalian neurons of defined subtype in vitro by direct reprogramming protocols (see below).
We also have evidence that high levels of phosphorylation of Ascl1 may inhibit its ability to drive neuronal differentiation in neuroblastoma, a pediatric cancer that arises from noraderinergic neurons of the developing sympathetic nervous system (unpublished data).
Enhancing neuronal maturation in vitro using phospho-mutant proneural proteins
There has been a recent explosion of work designed to generate neurons in vitro by reprogramming of other mammalian cell types including ES, iPS and fibroblasts (e.g., ref.3,35,36). In particular, much effort has been expended on producing in vitro models of neurological diseases and neurodegenerative disorders such as Parkinsons.37,38 For such “disease-in-a-dish” modeling, neurons would be ideally be generated from patient samples that recapitulate phenotypes associated with the disease, and neurons generated would also provide material for in vitro drug screening.4 Much progress has been made in producing mature neuronal cultures from ES and iPS cells, and occasionally these cells do indeed show some signs of recapitulating a neurological disease phenotype (e.g., ref.39). However, as well as the very long experimental protocols required for generation of mature neurons, there are difficulties intrinsic to this approach that may arise from trying to model diseases predominantly found in old people using nerve cells produced from ES or rejuvenated iPS cells.
These limitations have heralded the alternative approach of transcription factor-mediated direct reprogramming of fibroblasts into neurons using transcription factor cocktails that produce nerves from human fibroblasts in a matter of weeks.4,40 Proneural proteins, usually Ascl1, are an obligate component of these cocktails, while other components usually include known Ascl1 co-factors and targets such as Brn2 and MyT1L. “Induced neurons” (iN cells) expressing markers such as Map2 and neural β tubulin and showing some signs of electrophysiological maturity, such as firing of action potentials and sodium and potassium currents, are indeed generated in these reprogramming cultures.3,36-38 However iN cells are generally produced with low efficiency at a level sub-optimal both for modeling and for drug discovery applications; human fibroblast lines convert with an efficiency of generally a few percent at most, while fibroblasts from older patients show a much lower rate of conversion.40 Even more of a problem, however, is that the neurons generated from human cells tend to show poor axonal outgrowth and branching, instead resembling embryonic or foetal neurons that act as poor models for diseases of mature nerve cells.4
However, when trying to make mature neurons in vitro our recent work demonstrates that, not only must one use the correct combination of transcription factors for reprogramming, but that the post-translational control of the reprogramming factors themselves can play a part in limiting neuronal maturation; we saw that replacing WT Ascl1 and Ngn2 with their phospho-mutant counterparts in standard reprogramming protocols significantly increased neuronal maturation and axonal branching, as well as enhancing electrophysiological maturity.7 Further manipulation of the activity as well of the levels of reprogramming factors should provide further improvements in what is a promising approach to provide nerve cell both for disease modeling, and also ultimately to generate neurons for transplantation and cell replacement therapies.
Conclusions and Perspectives
Our recent work has used basic developmental biology involving a uniquely tractable model organism, Xenopus, to identify a key post-translational mode of regulation of proneural proteins, the central drivers of neuronal differentiation. We have applied this understanding of developmental regulation to enhance mammalian in vitro reprogramming protocols to generate more mature neurons for disease modeling. These studies may ultimately act as a platform for producing more mature nerve cells for transplantation therapy in the future. From our perspective, it is clear that developing and refining protocols for generation of neurons for therapeutic applications must be firmly grounded in a thorough understanding of generation of mature neurons during normal development. The future looks bright for reprogramming and regenerative medicine provided we remember this.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I am deeply indebted to members of the Philpott group whose work this commentary describes and that underpins every aspect of the model presented, and in particular to Fahad Ali, Christopher Hindley, Luke Wylie and Kevin Cheng. I also thank multiple collaborators without whom this work would not have been possible including Francois Guillemot, Marc Kirschner, Roger Barker and Rick Livesey. I also thank Fahad Ali for help with Figure preparation.
Funding
This work in the laboratory has been supported by the Medical Research Council award G0700758, a grant from the UK Neuroblastoma Society and several MRC Doctoral Training Awards, as well as support from the Rosetrees Trust, and a Cancer Research UK studentship to Christopher Hindley.
References
- 1.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol 2010; 20:233-43; PMID:20153966; http://dx.doi.org/ 10.1016/j.tcb.2010.01.006 [DOI] [PubMed] [Google Scholar]
- 2.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci 2002; 3:517-30; PMID:12094208; http://dx.doi.org/ 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- 3.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010; 463:1035-41; PMID:20107439; http://dx.doi.org/ 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang N, Ng YH, Pang ZP, Sudhof TC, Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell 2011; 9:517-25; PMID:22136927; http://dx.doi.org/ 10.1016/j.stem.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali F, Hindley C, McDowell G, Deibler R, Jones A, Kirschner M, Guillemot F, Philpott A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 2011; 138:4267-77; PMID:21852393; http://dx.doi.org/ 10.1242/dev.067900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hindley C, Ali F, McDowell G, Cheng K, Jones A, Guillemot F, Philpott A. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development 2012; 139:1718-23; PMID:22491944; http://dx.doi.org/ 10.1242/dev.077552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali FR, Cheng K, Kirwan P, Metcalfe S, Livesey FJ, Barker RA, Philpott A. The phosphorylation status of Ascl1 is a key determinant of neuronal differentiation and maturation in vivo and in vitro. Development 2014; 141:2216-24; PMID:24821983; http://dx.doi.org/ 10.1242/106377dev.106377 [DOI] [PubMed] [Google Scholar]
- 8.Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. Embo J 2007; 26:5093-108; PMID:18007592; http://dx.doi.org/ 10.1038/sj.emboj.7601923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, et al.. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev 2011; 25:930-45; PMID:21536733; http://dx.doi.org/ 10.1101/gad.627811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro DS, Guillemot F. Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle 2011; 10:4026-31; PMID:22101262; http://dx.doi.org/ 10.4161/cc.10.23.18578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci 2002; 3:517-30; PMID:12094208; http://dx.doi.org/ 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- 12.Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 1993; 75:463-76; PMID:8221886; http://dx.doi.org/ 10.1016/0092-8674(93)90381-Y [DOI] [PubMed] [Google Scholar]
- 13.Pattyn A, Guillemot F, Brunet JF. Delays in neuronal differentiation in Mash1/Ascl1 mutants. Dev Biol 2006; 295:67-75; PMID:16677628; http://dx.doi.org/ 10.1016/j.ydbio.2006.03.008 [DOI] [PubMed] [Google Scholar]
- 14.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature 1995; 375:761-6; PMID:7596407; http://dx.doi.org/ 10.1038/375761a0 [DOI] [PubMed] [Google Scholar]
- 15.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell 1996; 87:43-52; PMID:8858147; http://dx.doi.org/ 10.1016/S0092-8674(00)81321-5 [DOI] [PubMed] [Google Scholar]
- 16.Simpson P. Notch signalling in development: on equivalence groups and asymmetric developmental potential. Curr Opin Genet Dev 1997; 7:537-42; PMID:9309187; http://dx.doi.org/ 10.1016/S0959-437X(97)80083-4 [DOI] [PubMed] [Google Scholar]
- 17.Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 2008; 58:52-64; PMID:18400163; http://dx.doi.org/ 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 18.Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci 2008; 11:1247-51; PMID:18956012; http://dx.doi.org/ 10.1038/nn.2208 [DOI] [PubMed] [Google Scholar]
- 19.Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 2013; 342:1203-8; PMID:24179156; http://dx.doi.org/ 10.1126/science.1242366 [DOI] [PubMed] [Google Scholar]
- 20.Philpott A, Friend SH. E2F and its developmental regulation in Xenopus laevis. Mol Cell Biol 1994; 14:5000-9; PMID:8007993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernon AE, Devine C, Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development 2003; 130:85-92; PMID:12441293; http://dx.doi.org/ 10.1242/dev.00193 [DOI] [PubMed] [Google Scholar]
- 22.Richard-Parpaillon L, Cosgrove RA, Devine C, Vernon AE, Philpott A. G1/S phase cyclin-dependent kinase overexpression perturbs early development and delays tissue-specific differentiation in Xenopus. Development 2004; 131:2577-86; PMID:15115752; http://dx.doi.org/ 10.1242/dev.01121 [DOI] [PubMed] [Google Scholar]
- 23.Vernon AE, Movassagh M, Horan I, Wise H, Ohnuma S, Philpott A. Notch targets the Cdk inhibitor Xic1 to regulate differentiation but not the cell cycle in neurons. EMBO Rep 2006; 7:643-8; PMID:16648822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vosper JM, Fiore-Heriche CS, Horan I, Wilson K, Wise H, Philpott A. Regulation of neurogenin stability by ubiquitin-mediated proteolysis. Biochem J 2007; 407:277-84; PMID:17623011; http://dx.doi.org/ 10.1042/BJ20070064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vosper JM, McDowell GS, Hindley CJ, Fiore-Heriche CS, Kucerova R, Horan I, Philpott A. Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J Biol Chem 2009; 284:15458-68; PMID:19336407; http://dx.doi.org/ 10.1074/jbc.M809366200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roark R, Itzhaki L, Philpott A. Complex regulation controls Neurogenin3 proteolysis. Biology Open 2012; 1:1264-72; PMID:23259061; http://dx.doi.org/ 10.1242/bio.20121750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinals F, Reiriz J, Ambrosio S, Bartrons R, Rosa JL, Ventura F. BMP-2 decreases Mash1 stability by increasing Id1 expression. Embo J 2004; 23:3527-37; PMID:15318167; http://dx.doi.org/ 10.1038/sj.emboj.7600360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al.. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 2005; 48:45-62; PMID:16202708; http://dx.doi.org/ 10.1016/j.neuron.2005.08.032 [DOI] [PubMed] [Google Scholar]
- 29.Wylie LA, Hardwick LJ, Papkovskaia TD, Thiele CJ, Philpott A. Ascl1 phospho-status regulates neuronal differentiation in a Xenopus developmental model of neuroblastoma. Disease Models Mechan 2015; PMID:25786414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koyano-Nakagawa N, Wettstein D, Kintner C. Activation of Xenopus genes required for lateral inhibition and neuronal differentiation during primary neurogenesis. Mol Cell Neurosci 1999; 14:327-39; PMID:10588388; http://dx.doi.org/ 10.1006/mcne.1999.0783 [DOI] [PubMed] [Google Scholar]
- 31.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development 2005; 132:105-15; PMID:15576411; http://dx.doi.org/ 10.1242/dev.01548 [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al.. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 2014; 156:1274-85; PMID:24630727; http://dx.doi.org/ 10.1016/j.cell.2014.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 2012; 484:251-5; PMID:22498630; http://dx.doi.org/ 10.1038/nature10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDowell GS, Hindley CJ, Lippens G, Landrieu I, Philpott A. Phosphorylation in intrinsically disordered regions regulates the activity of Neurogenin2. BMC Biochem 2014; 15:24; PMID:25374254; http://dx.doi.org/ 10.1186/s12858-014-0024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfisterer U, Wood J, Nihlberg K, Hallgren O, Bjermer L, Westergren-Thorsson G, Lindvall O, Parmar M. Efficient induction of functional neurons from adult human fibroblasts. Cell Cycle 2011; 10:3311-6; PMID:21934358; http://dx.doi.org/ 10.4161/cc.10.19.17584 [DOI] [PubMed] [Google Scholar]
- 36.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al.. Induction of human neuronal cells by defined transcription factors. Nature 2011; 476:220-3; PMID:21617644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al.. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011; 476:224-7; PMID:21725324; http://dx.doi.org/ 10.1038/nature10284 [DOI] [PubMed] [Google Scholar]
- 38.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A 2011; 108:10343-8; PMID:21646515; http://dx.doi.org/ 10.1073/pnas.1105135108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livesey FJ. Human stem cell models of dementia. Human Mol Genet 2014; 23:R35-R9; PMID:24939911; http://dx.doi.org/ 10.1093/hmg/ddu302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ang CE, Wernig M. Induced neuronal reprogramming. J Compar Neurol 2014; 522:2877-86; PMID:24771471; http://dx.doi.org/ 10.1002/cne.23620 [DOI] [PMC free article] [PubMed] [Google Scholar]