Abstract
The imprinted miR379–410 cluster contains 38 microRNAs (miRNAs) that are involved in diverse neurodevelopmental processes and are important regulators of neuronal function. The implications of these miRNAs in neurological diseases have been recently recognized.In the present minireview, the current findings regarding the brain-specific functions of miR379–410 cluster miRNAs are summarized and discussed.
Keywords: DLK1-DIO3, miR-134, miR-369–3p, miR-496, miR-543, miR379–410, N-cadherin, neurogenesis, neuronal function
Introduction
MicroRNAs (miRNAs) are small, ∼22-nucleotide noncoding RNAs, which bind to sequences located mainly in the 3′-untranslated regions (3′UTRs) of their target transcripts. Upon miRNA binding, the respective target mRNAs are generally either degraded or blocked in their translation (summarized in1,2). Recent studies have implicated miRNAs in various aspects of neuronal development and function. During development, miRNAs regulate neural induction, neural stem cell expansion, neuronal differentiation and subtype specification as well as neuronal migration. Furthermore, miRNAs are critically involved in regulating neuritogenesis, neuronal function and synaptic plasticity (summarized in3,4). The first experiments that demonstrated the essential roles of miRNAs during neuronal development were based upon ablation of components of the miRNA processing machinery, i.e. Dicer or DGCR8. Since then, neural functions of single miRNAs have been identified in a number of studies. Nevertheless, many more miRNAs are expressed in the developing and/or mature brain and the functions of the majority of these brain-expressed miRNAs are still unknown.
The miR379–410 cluster is a large genomic miRNA cluster with brain-specific functions. It is located on chromosome 14 in humans and 12 in mice and is exclusively expressed from the maternal allele. It contains 38 brain-expressed miRNAs, only few of which have so far been studied concerning their brain-specific functions. This limited number of studies indicates that miR379–410 cluster miRNAs are involved in various aspects of neurodevelopment, neuronal maturation and function and that their mis-regulation might contribute to the development of neurological and neuropsychiatric diseases. In this review I will summarize the current knowledge about the miR379–410 cluster miRNA expression in the brain, its role in neurodevelopment and neuronal function and its involvement in neurological diseases (Table 1).
Table 1.
Functions and targets of miR379–410 cluster miRNAs
| Mir379–410 cluster miRNA | Brain targets | Brain-specific function | Involvement in brain disorders | Reference |
|---|---|---|---|---|
| miR-134 | Pum2, Chrdl-1, Dcx, Limk1, CREB | Promotes NPC proliferation, inhibits neuron migration, sensitizes neurons to BMP-induced neurite outgrowth, activity-induced, promotes activity-induced dendritogenesis, inhibits spine growth and density | Silencing miR-134 is neuroprotective in a mouse epilepsy model; downregulated in PBMCs from schizophrenia patients; upregulated in dorsolateral PFC Brodmann area 46 in schizophrenia | 25,28,29,46,50-52 |
| miR-154 | — | — | Downregulated in PBMCs from schizophrenia patients, Upregulated in cerebellum in Mecp2 KO mice, downregulated in glioma | 52,54,57 |
| miR-299 | — | — | Upregulated in cerebellum in Mecp2 KO mice, downregulated in glioma | 54,57 |
| miR-300 | — | — | Upregulated in cerebellum in Mecp2 KO mice | 54 |
| miR-323–3p | — | — | Downregulated in PBMCs from schizophrenia patients, Upregulated in cerebellum in Mecp2 KO mice, downregulated in glioma | 52,54,57 |
| miR-329 | — | Activity-induced, promotes activity-induced dendritogenesis | Downregulated in PBMCs from schizophrenia patients | 28,52 |
| miR-369–3p | N-cadherin | Promotes neuronal differentiation, inhibits neuronal migration | Upregulated in cerebellum in Mecp2 KO mice, downregulated in glioma | 21,54,57 |
| miR-376a | Hes5 | Promotes neuronal differentiation | Upregulated in lymphoblastoid cell lines from ASD patients, downregulated in glioma | 30,53,57 |
| miR-377 | — | — | Upregulated in cerebellum in Mecp2 KO mice | 54 |
| miR-380 | — | — | Upregulated in cerebellum in Mecp2 KO mice | 54 |
| miR-381 | — | Activity-induced, promotes activity-induced dendritogenesis | Upregulated in cerebellum in Mecp2 KO mice, downregulated in glioma | 28,54,57 |
| miR-382 | — | — | Upregulated in cerebellum in Mecp2 KO mice | 54 |
| miR-409–3p | — | — | Downregulated in PBMCs from schizophrenia patients | 52 |
| miR-410 | — | — | Downregulated in PBMCs from schizophrenia patients, upregulated in cerebellum in Mecp2 KO mice | 52,54 |
| miR-485 | SV2A | Activity-induced, inhibits spine density and maturation, inhibits spontaneous synaptic activity | Downregulated in PBMCs from schizophrenia patients | 25,26,52 |
| miR-487b | — | — | Downregulated in PBMCs from schizophrenia patients, upregulated in glioblastoma | 52,56 |
| miR-495 | — | Activity-induced | Downregulated in lymphoblastoid cell lines from ASD patients, upregulated in cerebellum in Mecp2 KO mice | 28,53,54 |
| miR-496 | N-cadherin | Promotes neuronal differentiation, inhibits neuronal migration | 21 | |
| miR-541 | — | Activity-induced | 28 | |
| miR-543 | N-cadherin | Promotes neuronal differentiation, inhibits neuronal migration | Upregulated in cerebellum in Mecp2 KO mice, downregulated in glioblastoma | 21,54,56 |
| miR-544 | — | — | Downregulated in PBMCs from schizophrenia patients, upregulated in cerebellum in Mecp2 KO mice | 52,54 |
| miR-654–5p | — | — | Downregulated in PBMCs from schizophrenia patients | 52 |
The miR379–410 Cluster is Located Within the DLK1-DIO3 Imprinted Region
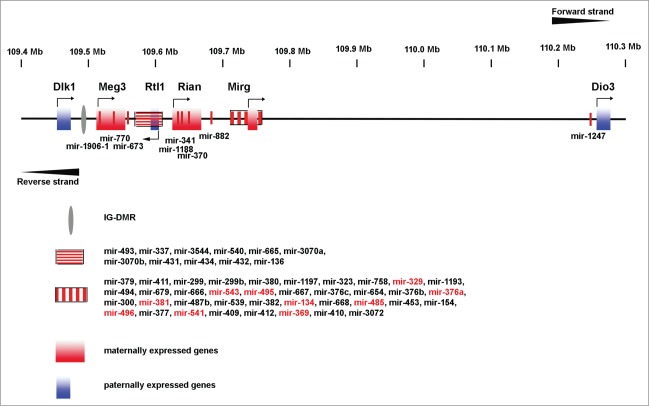
The miR379–410 cluster is located within the DLK1-DIO3 imprinted region which is situated on chromosome 14q32 in humans and on the distal part of chromosome 12 (12qF1) in mice (summarized in5; Fig. 1). In 1993 Cattanach and Rasberry provided the first evidence that genomic imprinting at this locus is important.6 They generated mice with either maternal or paternal uniparental disomy (upd) of the whole chromosome 12 or only of its distal part (Mat(Di)12; MatDp(dist12) or Pat(Di)12; PatDp(dist12)). Mat(Di)12, MatDp(dist12), Pat(Di)12 and PatDp(dist12) mice were prenatally lethal and exhibited severe developmental defects (see below and summarized in5). Since these groundbreaking experiments, a growing number of human patients have been reported with maternal or paternal upd of human chromosome 14. In humans both paternal and maternal upd for chromosome 14 (upd(14)pat and upd(14)mat) cause distinct phenotypes. Upd(14)pat (OMIM 608149) causes a severe phenotype that includes skeletal abnormalities such as a bell-shaped thorax, facial dysmorphisms and developmental delay/intellectual disability (ID). Upd(14)mat leads to a milder phenotype than upd(14)pat including short stature, hypotonia, mild facial abnormalities, precocious onset of puberty and mild developmental delay.7,8 In the mouse, upd for chromosome 12 also results in distinct phenotypes that partially overlap with those in human upd(14). Thus, paternal upd for chromosome 12 [PatDi(12)] leads to prenatal lethality, abdominal distension, skeletal defects, cardiac abnormalities and placentomegaly. [MatDi(12)] causes a phenotype that includes perinatal lethality, growth failure and placental hypoplasia.9,10
Figure 1.
Schematic overview of the DLK1-DIO3 genomic region on mouse chromosome 12 (GRCm38/mm10 Assembly). Genes, long noncoding RNAs and miRNAs are noted. MiR379–410 cluster miRNAs that have been subject to functional studies in the brain are depicted in red color. Pseudogenes and snoRNAs are not shown.
In the years following the discovery of genomic imprinting on mouse chromosome 12 and human chromosome 14 the imprinted region which was later named DLK1-DIO3 region was characterized and it was shown that it contains several coding and non-coding genes (summarized in5).
The DLK1-DIO3 region spans ∼850 kb and contains the paternally expressed genes DLK1, RTL1 and DIO3 and maternally expressed genes MEG3 (Gtl2 in mice), MEG8 (RIAN in mice) and antisense RTL1 (RTL1as). DLK1 acts as an antagonist of Notch signaling and regulates cell differentiation.11,12 RTL1 is a retrotransposon-like gene expressed in a subset of embryonic tissues as well as in the placenta and is essential for proper placental development.13,14 DIO3 is a type 3-iodothyronine deiodinase, which degrades thyroid hormone.15,16 Both MEG3 and MEG8 are noncoding RNAs.
Imprinting in this region is controlled by 2 distinct, differentially methylated regions (DMR): a primary, germline-derived DLK1-MEG3 intergenic DMR (IG-DMR) and a secondary, post-fertilization-derived MEG3-DMR. Both DMRs are hyper-methylated on the paternal allele and hypomethylated on the maternal allele (summarized in5).
The DLK1-DIO3 imprinted region also contains 53 miRNAs on the forward strand and one miRNA on the reverse strand. Most of these miRNAs were discovered by a computer-assisted approach in 2004.17,18 The 54 miRNAs are separated into 3 different clusters with miR-2392 and miR-770 being situated in the DLK1-MEG3 region, the second cluster between MEG3 and MEG8 and the third (the miR379–410 cluster) between MEG8 and DIO3.
Because miRNAs of the region are exclusively expressed from the maternal allele they are expected to be upregulated in patients with upd(14)mat and downregulated in patients with upd(14)pat as is the case for mouse [MatDi(12)] and [PatDi(12)]. In line with growing evidence that miRNAs of the miR379–410 cluster are essential regulators of neurogenesis and neuron function it seems likely that such a mis-expression of these miRNAs contributes to the development of an ID, which is seen in patients with upd(14)pat. Identified deletions in humans encompassing parts of the DLK1-DIO3 region support this hypothesis. Thus, maternal inheritance of these deletions fully phenocopied the upd(14)pat phenotype although DLK1 levels were normal. This finding suggests that the upd(14)pat phenotype (including the ID) might be caused by a loss of maternally expressed noncoding RNAs.5 However, the early lethality that occurs in the mouse models has precluded the analyses of possible impairments in brain function that might be caused by altered miRNA expression.9 A recently generated knockout mouse model may clarify this issue.19
The miRNAs of the miR379–410 cluster are highly conserved between human and mouse. Most of the pre-miRNAs of the cluster are arranged in tandem arrays of closely related sequences resulting from genomic duplications.20 Unlike the pre-miRNAs the mature sequences show a high diversity and predicted miRNA target genes as well. Because of the broad spectrum of predicted targets and the specific expression patterns of miR379–410 cluster miRNAs it has been suggested that these miRNAs might target numerous genes in specific cell types.20 In addition, we have shown recently that numerous miRNAs of the cluster target the same gene, N-cadherin, in an additive manner in the developing mouse neocortex.21
The regulation of expression of the genes and miRNAs located in the DLK1-DIO3 region is poorly studied. It is hypothesized but not yet proven that the maternally expressed genes and miRNAs of the DLK1-DIO3 region are transcribed as one giant polycistronic transcript from which individual genes and miRNAs are derived by posttranscriptional processing.22 However, this may be an oversimplification. While the genes and the few miRNAs of the region that have been studied share similar expression domains in the developing and mature brain, differences in expression do exist. Gtl2, for example, is not only expressed during development but also in the adult brain, whereas expression of miR-410 is restricted to developmental and early postnatal stages.23,24 In contrast, expression of 2 other miRNAs of the region, miR-134 and miR-485, increases postnatally.25,26 More evidence exists that, besides a giant polycistronic transcript, expression of the genes and miRNAs of the region may be driven by gene-specific transcriptional mechanisms. Thus, 7 independently maternally expressed noncoding RNA genes including Gtl2 and Mirg have recently been identified in the region on mouse chromosome 12qF1.27 Moreover, it has been shown that several miRNAs of the miR379–410 cluster but not Gtl2 undergo an activity-induced upregulation in their expression which is driven by the binding of Mef2 to a binding site ∼20 kb upstream of the miR379–410 cluster.26,28 During embryonic neocortex development miR-134 is poorly expressed in neural progenitors but is upregulated in postmitotic neurons, whereas 3 other miRNAs of the cluster, miR-369–3p, -496 and -543 are expressed in neural progenitors and neurons.21,25 MiR-134 as well as miR-369 and miR-496 are encoded within exons or introns of the noncoding RNA gene Mirg in the mouse and are therefore likely co-regulated at the transcriptional level. Expression differences may therefore be due to posttranscriptional regulation, which should be studied in the future.
miRNAs of the miR379–410 Cluster Regulate Neurogenesis and Neuronal Migration in the Mouse Embryo
Brain-enriched miRNAs are involved in all aspects of neurogenesis and fine-tune the expression of those genes that control proliferation and differentiation of neural progenitor cells (NPCs). While some miRNAs, e.g. miR-9, miR-124 and let-7, promote neuronal differentiation, others, such as miR-184, maintain NPCs in a proliferating state. In addition, brain-enriched miRNAs regulate neuronal migration (e.g.,21,29). Several studies have shown that miRNAs of the miR379–410 cluster are involved in all these processes.
The Notch pathway controls NPC expansion and cell fate specifications in the developing brain. Pathway activation maintains NPCs in a proliferating state whereas its downregulation promotes neuronal differentiation. Several components of the Notch pathway are subject to miRNA-dependent regulation, which may be important to balance Notch signaling in NPCs (summarized in3). MiR-124 targets the Notch ligand Jag1 and the Notch effector Sox9. MiR-9 targets several members of the Hes family of transcription factors in zebrafish and murine NPCs, thereby controlling the timing of NPC differentiation. A member of the miR379–410 cluster, miR-376a, targets Hes5 mRNA and positively regulates neuronal differentiation of cultured rat cortical stem cells.30 Although an interesting finding, it remains to be analyzed if such a regulation of Hes5 by miR376a occurs in vivo and is necessary for driving neurogenesis in the embryonic CNS.
Whereas miR-376a promotes neural differentiation another miRNA of the cluster, miR-541, targets synapsin I and inhibits neurite formation and neuronal differentiation when overexpressed in PC12 cells.31 However, as for miR376a, there is presently a lack of in vivo evidence.
Recent studies have indicated that miRNAs of the miR379–410 cluster are important regulators of neurogenesis and neuronal migration in the developing neocortex in vivo where they have both overlapping and antagonistic functions. As outlined above, miR-134 is poorly expressed in neural progenitors and is upregulated in postmitotic neurons. When overexpressed in neural progenitors, however, miR-134 promotes cell proliferation in vitro and in vivo. In postmitotic neurons, miR-134 negatively regulates their migration into the cortical plate by targeting Chordin-like 1 (Chrdl-1) and Doublecortin (Dcx) and sensitizes neurons to BMP-induced neurite outgrowth.29 The regulation of Dcx by miR-134 is an interesting finding as Dcx is an important effector of the Reelin pathway and controls neuronal migration. Mutations in Reelin pathway genes, e.g. the RELN, DCX or LIS1 genes, cause a neuronal migration disorder, lissencephaly, in humans. An abnormal reduction of RELN has also been observed in schizophrenia and autism (summarized in32). The Reelin pathway is fine-tuned by the action of several miRNAs that inhibit neuronal migration. Beside miR-134, other miRNAs that are not encoded within the miR379–410 cluster have been implicated in Reelin pathway regulation. Recent studies carried out in chicken hypothalamic cells and neuroblastoma cells suggest that miR-138 and miR-128 target reelin and miR-128 DCX in addition.33,34 MiR-139–5p inhibits cell migration in vitro and modulates rat brain development by targeting Lis1.35 MiR-22 and miR-124 target components of the CoREST/REST transcriptional repressor complex in the cortical wall, thereby regulating Dcx indirectly in migrating neurons.36 A mechanism involving miRNAs may explain reduced expression of Reelin pathway components in neurodevelopmental disorders and should be explored in the future.
As outlined above, miR-134 has stage-specific effects on embryonic cortical development. Also other miRNAs of the miR379–410 cluster have these stage-specific effects. We have recently found that 3 members of the cluster, miR-369–3p, -496 and -543, are expressed in NPCs and in neurons. These miRNAs target the mRNA encoding the cell adhesion protein N-cadherin in an additive manner.21 N-cadherin is widely expressed in the developing neocortex in neural progenitor cells and in neurons and has multiple functions. In the ventricular zone of the neocortex N-cadherin is an integral part of the adherens junctions and is essential for maintaining the architecture of the neocortex.37,38 In radial glia cells (RGCs), N-cadherin activates β-catenin signaling and enhances proliferation.38,39 In migrating neurons, N-cadherin is required for the motility of the cells toward the cortical plate.40,41 Although N-cadherin is widely expressed in the developing neocortex its expression levels have to be kept in a physiological range because both its under- as well as its overexpression causes defects in neuronal differentiation and migration. We have shown that the miRNAs of the miR379–410 cluster fine-tune the expression of N-cadherin and thereby ensure proper neurogenesis and neuronal migration. In contrast to miR-134 but similar to other brain-enriched miRNAs such as miR-124, they attenuate radial glia cell proliferation and enhance differentiation. In migrating neurons, they overlap in their function with miR-134 and negatively regulate neuronal migration.21 These results indicate that the miRNAs of the miR379–410 cluster not only differ in their expression patterns but also in their functions. Murine MiR-134 as well as miR-369–3p and miR-496 are encoded within exons or introns of the noncoding RNA Mirg and therefore likely share the same transcriptional regulation. Differences in expression of these miRNAs may thus be due to specific posttranscriptional mechanisms.
miRNAs of the miR379–410 Cluster Control Neuronal Function
Neuronal activity regulates the expression of numerous brain-enriched miRNAs, with important consequences for neuronal homeostasis. Whereas some miRNAs such as the CREB-dependent miR-132 and miR-212 and all miR379–410 cluster miRNAs analyzed so far, are induced by neuronal activity others, e.g., miR-124, are decreased (summarized in42).
The activity-induced miR379–410 cluster miRNAs that have been analyzed so far, i.e., miR-134, -381, -329, -495, -541 and -485, control neuronal maturation and function.26,28 For example, miR-485 targets the synaptic vesicle protein SV2A and negatively regulates spine density and maturation and reduces spontaneous synaptic activity, thereby controlling homeostatic synaptic plasticity.26 Inhibition of miR-134, -381 or -329 but not of miR-495 or -541 blocks activity-induced dendritogenesis. The miRNA of the miR379–410 cluster which has been studied in most detail, miR-134, has apparently opposing effects by, at the same time, promoting activity-induced dendritogenesis and inhibiting spine growth by targeting Pum2 and Limk1, respectively. It has been suggested that these apparently opposing functions are indeed parts of the same homeostatic network that controls the overall neuronal excitability.28 Both miR-134 and miR-485 localize to soma and dendrites in cultured hippocampal neurons and it is proposed that neuronal activity elevates miR-134 expression throughout the cell while concurrently inhibiting miR-134 locally in dendrites. Another possibility is that the upregulation of miR-134 expression upon neuronal stimulation is restricted to certain cell types. Indeed, several recent studies have shown that the activity-dependent increase of miR-134 is small in comparison to miR-132 and miR-212 and is restricted to parvalbumin, SST, glutamate decarboxylase 2 (GAD2) and Calretinin-positive interneurons. In contrast, miR-132 and miR-212 are predominantly expressed in Calcium/Calmodulin-dependent protein kinase II α (CamKIIα) positive glutamatergic neurons.43-45
Conflicting results exist concerning the regulation of spine density by miR-134. Thus the knockdown of miR-134 led to a reduction in spine density in vivo46 but not in vitro.25 The different experimental conditions used in the 2 studies might be causal for the observed differences.
It has been shown in vivo that miR-134 is a target of the transcription factor SIRT and controls long-term potentiation (LTP) and memory formation.46,47 Brain-specific SIRT1 knockout mice show defects in LTP and cortical and hippocampal dependent memory. SIRT1 is a transcription factor that, together with YY1, represses miR-134 expression by binding to 2 sites that are located ∼1 kb and 4 kb upstream of miR-134 in the genome.47 In the SIRT1 knockout mice miR-134 is upregulated which leads to decreased protein levels of the miR-134 target CREB and consequently to a downregulation of the CREB target brain-derived neurotrophic factor (BDNF). In vivo miR-134 knockdown experiments using injections of LNA-miR-134 probes into hippocampal CA1 region of SIRT1 knockout mice restored LTP and partially restored memory impairments in these mice. In agreement with cell-type-specific functions for miR-134, miR-132 and miR-212 (see above) a miRNA microarray carried out with RNA from SIRT1 knockout hippocampi did not show an obvious decrease in expression of the CREB targets miR-132 or miR-212.
It is conceivable that the subcellular localization of a certain miRNA may influence its activity and may be important for the regulation of different sets of targets, thereby increasing the complexity of miRNA-mediated regulation in neurons. Whether other miRNAs of the miR379–410 cluster exhibit a specific subcellular localization or cell-type-specific expression is not known. Recent evidence has accumulated demonstrating that miRNA function is not restricted to the cytoplasm but that instead some miRNAs can re-enter the nucleus where they may be involved in posttranscriptional gene silencing of nuclear transcripts, transcriptional gene silencing and activation and regulation of alternative splicing. Similar to other cell types miRNAs have been detected in the nuclei of NPCs and neurons.48,49 Whereas only a small fraction (5%) of neuron-expressed miRNAs is enriched in the nucleus the majority of miRNAs (including miR379–410 cluster miRNAs) seems to shuttle between the nucleus and cytoplasm in neurons. Future studies should aim at identifying possible nuclear functions of these miRNAs.
The Roles of miR379–410 Cluster miRNAs in Brain Disorders
In addition to their possible involvement in the upd(14)pat phenotype (see above) miRNAs of the miR379–410 cluster have been associated to neurodevelopmental disorders (i.e. epilepsy, schizophrenia and autism) and brain tumors (i.e., gliomas).
Silencing miR-134 has neuroprotective functions in a mouse epilepsy model. Status epilepticus (SE) results in elevated levels of mature miR-134 in vitro and in vivo.46,50 Inhibition of miR-134 by injecting miR-134 specific antagomirs into the mouse ventricle prevented kainic acid (KA)-induced neurotoxicity in hippocampal neurons. Thus, silencing miR-134 reduced spontaneous recurrent seizures and altered pathologic hallmarks of temporal lobe epilepsy, i.e. progressive neuron loss, gliosis and rearrangement of mossy fibers.
Altered levels of miR379–410 cluster miRNAs occur in human schizophrenia patients. Thus, several miRNAs of this cluster, including miR-134, miR-329, miR-409–3p, miR-487b, miR-544, miR-654–5p, miR-485–3p, miR-323–3p, miR-410 and miR-154, are significantly downregulated in peripheral blood mononuclear cells from schizophrenia patients.51 The significance of their downregulation, however, is not clear. In the brain, 2 miRNAs of the cluster, miR-134 and miR-382, were found to be upregulated in the dorsolateral prefrontal cortex Brodmann area 46 in postmortem samples of schizophrenia patients.52 In light of the known functions of miR379–410 cluster miRNAs during processes that are also affected in schizophrenia, e.g. the regulation of dendritic spine density, an altered expression of these miRNAs is of potential interest and may very well contribute to disease development. Additional studies are needed to functionally evaluate the role of miR379–410 cluster miRNAs in schizophrenia.
MiRNAs from the miR379–410 cluster were also reported to be differentially expressed between lymphoblastoid cell lines from autistic monozygotic twins and their unaffected sibling.53 Further evidence for a possible involvement of miR379–410 cluster miRNAs in the etiology of autism spectrum disorders (ASD) comes from a study that analyzed changes in miRNA expression in methyl-CpG-binding protein 2 (Mecp2) knockout cerebella.54 Mutations in the human MECP2 gene cause Rett syndrome, an autism spectrum disorder with additional severe neurological symptoms, e.g., ataxia and seizures. MiR379–410 cluster miRNAs were constantly upregulated in the mouse knockout cerebella and were transcriptionally repressed by Mecp2. Again, functional studies are needed to shed light on the roles of these miRNAs in ASD.
Several studies have investigated the miRNA expression profiles in gliomas from human patients and the mouse and have found a downregulation of miR379–410 cluster miRNAs in comparison to normal brain tissue.55-57 Lavon et al. have found that gliomas display a miRNA expression signature that is reminiscent of the profile of stem cells. Typical stem cell miRNAs such as the miR302–367 cluster miRNAs were upregulated in both gliomas and mouse NPCs. The miR379–410 cluster miRNAs were downregulated in both gliomas and normal mouse NPCs.57 This suggests that miR379–410 cluster miRNAs may represent the largest tumor suppressor miRNA cluster in the genome. Further studies should aim at confirming the role of this cluster in glioma and test whether an increase in their expression may reduce tumorigenesis.
Conclusions and Outlook
It has been convincingly shown that miRNAs of the miR379–410 cluster are important regulators of various aspects of embryonic neurogenesis, neuronal migration and function (Fig. 2, Table 1). The knowledge acquired thus far, however, may only comprise the tip of the iceberg because only few miRNAs of the cluster have been studied and the available data is limited. Concerning neurogenesis the following open questions have to be addressed in the future: (i) are other miRNAs of the cluster involved in embryonic neurogenesis and (ii) are miR379–410 cluster miRNAs also necessary for adult neurogenesis?
Figure 2.
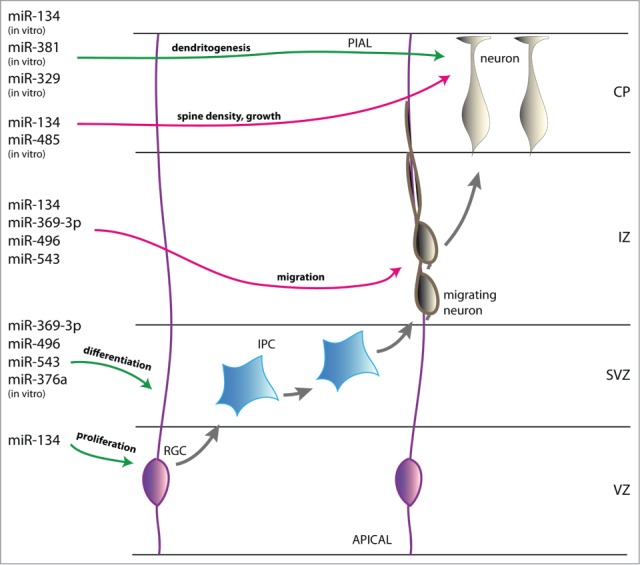
miR379–410 cluster miRNA regulation of cerebral cortex neurogenesis. Schematic represention of the developing cortical wall. During embryonic development of the cerebral cortex radial glial cells (RGCs, pink) divide asymmetrically to self-renew and generate neurons either directly or indirectly via intermediate progenitor cells (IPCs, blue). The miR379–410 cluster miRNA miR-134 promotes neural precursor cell (NPC) proliferation whereas miR-369–3p, -496, -543 and -376a induce differentiation. Newborn neurons (beige) migrate along RGC processes into the cortical plate (CP) and reach their final destination via somal translocation. Neuronal migration is negatively regulated by the miR379–410 cluster miRNAs miR-134, -369–3p, -496 and -543. After settling, neurons initiate their terminal differentiation. MiR-134 and -485 negatively regulate spine density and maturation and miR-134, -381 and -329 enhance activity-dependent dendritogenesis. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate; green arrow, activation; red arrow, repression.
Mis-expression of the miRNAs of the cluster might contribute to the pathogenesis of neurodevelopmental disorders and neuropsychiatric diseases such as ID and autism that might be caused by failures of neurogenesis, neuronal migration or neuronal homeostasis. For example, the development of an ID in patients with upd(14)pat might be caused by the downregulation of the miR379–410 cluster miRNAs that are exclusively expressed from the maternal allele. Additional studies are needed to clarify these issues. While the mouse has proven to be a suitable model system to study the brain-specific functions of the miR379–410 cluster miRNAs, the development of induced pluripotent stem cell (iPSC) models may help to enhance our knowledge about miR379–410 cluster miRNA functions in humans.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the University Medical Centre Mainz.
References
- 1. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9:102-14; PMID:18197166; http://dx.doi.org/ 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- 2. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/ 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stappert L, Roese-Koerner B, Brustle O. The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res 2014; 359:47-64; PMID:25172833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meza-Sosa KF, Pedraza-Alva G, Perez-Martinez L. microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci 2014; 8:175; PMID:25009466; http://dx.doi.org/ 10.3389/fncel.2014.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet 2008; 24:306-16; PMID:18471925; http://dx.doi.org/ 10.1016/j.tig.2008.03.011 [DOI] [PubMed] [Google Scholar]
- 6. Cattanach BM, Rasberry C. Evidence of imprinting involving the distal region of Chr 12. Mouse Genome 1993; 91:858 [Google Scholar]
- 7. Kurosawa K, Sasaki H, Sato Y, Yamanaka M, Shimizu M, Ito Y, Okuyama T, Matsuo M, Imaizumi K, Kuroki Y, et al. . Paternal UPD14 is responsible for a distinctive malformation complex. Am J Med Genet 2002; 110:268-72; PMID:12116236; http://dx.doi.org/ 10.1002/ajmg.10404 [DOI] [PubMed] [Google Scholar]
- 8. Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, Yamamori S, Kishimoto H, Nakayama M, Tanaka Y, et al. . Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 2008; 40:237-42; PMID:18176563; http://dx.doi.org/ 10.1038/ng.2007.56 [DOI] [PubMed] [Google Scholar]
- 9. Georgiades P, Watkins M, Surani MA, Ferguson-Smith AC. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 2000; 127:4719-28; PMID:11023874 [DOI] [PubMed] [Google Scholar]
- 10. Sutton VR, McAlister WH, Bertin TK, Kaffe S, Wang JC, Yano S, Shaffer LG, Lee B, Epstein CJ, Villar AJ. Skeletal defects in paternal uniparental disomy for chromosome 14 are re-capitulated in the mouse model (paternal uniparental disomy 12). Hum Genet 2003; 113:447-51; PMID:12938037; http://dx.doi.org/ 10.1007/s00439-003-0981-x [DOI] [PubMed] [Google Scholar]
- 11. Baladron V, Ruiz-Hidalgo MJ, Nueda ML, Diaz-Guerra MJ, Garcia-Ramirez JJ, Bonvini E, Gubina E, Laborda J. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res 2005; 303:343-59; PMID:15652348; http://dx.doi.org/ 10.1016/j.yexcr.2004.10.001 [DOI] [PubMed] [Google Scholar]
- 12. Nueda ML, Baladron V, Sanchez-Solana B, Ballesteros MA, Laborda J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J Mol Biol 2007; 367:1281-93; PMID:17320900; http://dx.doi.org/ 10.1016/j.jmb.2006.10.043 [DOI] [PubMed] [Google Scholar]
- 13. Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, Hino T, Suzuki-Migishima R, Kohda T, Ogura A, et al. . Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet 2008; 40:243-8; PMID:18176565; http://dx.doi.org/ 10.1038/ng.2007.51 [DOI] [PubMed] [Google Scholar]
- 14. Brandt J, Veith AM, Volff JN. A family of neofunctionalized Ty3/gypsy retrotransposon genes in mammalian genomes. Cytogenet Genome Res 2005; 110:307-17; PMID:16093683; http://dx.doi.org/ 10.1159/000084963 [DOI] [PubMed] [Google Scholar]
- 15. Hernandez A, Park JP, Lyon GJ, Mohandas TK, Germain DL. Localization of the type 3 iodothyronine deiodinase (DIO3) gene to human chromosome 14q32 and mouse chromosome 12F1. Genomics 1998; 53:119-21; PMID:9787088; http://dx.doi.org/ 10.1006/geno.1998.5505 [DOI] [PubMed] [Google Scholar]
- 16. Galton VA, Martinez E, Hernandez A, Germain EA, Bates JM, Germain DL. Pregnant rat uterus expresses high levels of the type 3 iodothyronine deiodinase. J Clin Invest 1999; 103:979-87; PMID:10194470; http://dx.doi.org/ 10.1172/JCI6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet 2003; 34:261-2; PMID:12796779; http://dx.doi.org/ 10.1038/ng1171 [DOI] [PubMed] [Google Scholar]
- 18. Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 2004; 14:1741-8; PMID:15310658; http://dx.doi.org/ 10.1101/gr.2743304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labialle S, Marty V, Bortolin-Cavaille ML, Hoareau-Osman M, Pradere JP, Valet P, Martin PG, Cavaille J. The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J 2014; 33:2216-30; PMID:25124681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kircher M, Bock C, Paulsen M. Structural conservation versus functional divergence of maternally expressed microRNAs in the Dlk1/Gtl2 imprinting region. BMC Genomics 2008; 9:346; PMID:18651963; http://dx.doi.org/ 10.1186/1471-2164-9-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rago L, Beattie R, Taylor V, Winter J. miR379-410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin. EMBO J. 2014; 33(8):906-20. http://dx.doi.org/ 10.1002/embj.201386591 Epub 2014 Mar 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tierling S, Dalbert S, Schoppenhorst S, Tsai CE, Oliger S, Ferguson-Smith AC, Paulsen M, Walter J. High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics 2006; 87:225-35; PMID:16309881; http://dx.doi.org/ 10.1016/j.ygeno.2005.09.018 [DOI] [PubMed] [Google Scholar]
- 23. da Rocha ST, Tevendale M, Knowles E, Takada S, Watkins M, Ferguson-Smith AC. Restricted co-expression of Dlk1 and the reciprocally imprinted non-coding RNA, Gtl2: implications for cis-acting control. Dev Biol 2007; 306:810-23; PMID:17449025; http://dx.doi.org/ 10.1016/j.ydbio.2007.02.043 [DOI] [PubMed] [Google Scholar]
- 24. Wheeler G, Ntounia-Fousara S, Granda B, Rathjen T, Dalmay T. Identification of new central nervous system specific mouse microRNAs. FEBS Lett 2006; 580:2195-200; PMID:16566924; http://dx.doi.org/ 10.1016/j.febslet.2006.03.019 [DOI] [PubMed] [Google Scholar]
- 25. Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature 2006; 439:283-9; PMID:16421561; http://dx.doi.org/ 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- 26. Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci U S A 2011; 108:11650-5; PMID:21697510; http://dx.doi.org/ 10.1073/pnas.1017576108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hagan JP, O'Neill BL, Stewart CL, Kozlov SV, Croce CM. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One 2009; 4:e4352; PMID:19194500; http://dx.doi.org/ 10.1371/journal.pone.0004352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J 2009; 28:697-710; PMID:19197241; http://dx.doi.org/ 10.1038/emboj.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex 2011; 21:1857-69; PMID:21228099; http://dx.doi.org/ 10.1093/cercor/bhq262 [DOI] [PubMed] [Google Scholar]
- 30. Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, Luthi-Carter R. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci 2013; 33:5127-37; PMID:23516279; http://dx.doi.org/ 10.1523/JNEUROSCI.0600-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J, Liu LH, Zhou Y, Li YP, Shao ZH, Wu YJ, Li MJ, Fan YY, Shi HJ. Effects of miR-541 on neurite outgrowth during neuronal differentiation. Cell Biochem Funct 2011; 29:279-86; PMID:21452340; http://dx.doi.org/ 10.1002/cbf.1747 [DOI] [PubMed] [Google Scholar]
- 32. Folsom TD, Fatemi SH. The involvement of Reelin in neurodevelopmental disorders. Neuropharmacology 2013; 68:122-35; PMID:22981949; http://dx.doi.org/ 10.1016/j.neuropharm.2012.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kisliouk T, Meiri N. MiR-138 promotes the migration of cultured chicken embryonic hypothalamic cells by targeting reelin. Neuroscience 2013; 238:114-24; PMID:23438760; http://dx.doi.org/ 10.1016/j.neuroscience.2013.02.020 [DOI] [PubMed] [Google Scholar]
- 34. Evangelisti C, Florian MC, Massimi I, Dominici C, Giannini G, Galardi S, Bue MC, Massalini S, McDowell HP, Messi E, et al. . MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J 2009; 23:4276-87; PMID:19713529; http://dx.doi.org/ 10.1096/fj.09-134965 [DOI] [PubMed] [Google Scholar]
- 35. Huang Y, Jiang J, Zheng G, Chen J, Lu H, Guo H, Wu C. miR-139-5p modulates cortical neuronal migration by targeting Lis1 in a rat model of focal cortical dysplasia. Int J Mol Med 2014; 33:1407-14; PMID:24647639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Volvert ML, Prevot PP, Close P, Laguesse S, Pirotte S, Hemphill J, Rogister F, Kruzy N, Sacheli R, Moonen G, et al. . MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep 2014; 7:1168-83; PMID:24794437; http://dx.doi.org/ 10.1016/j.celrep.2014.03.075 [DOI] [PubMed] [Google Scholar]
- 37. Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M. N-cadherin mediates cortical organization in the mouse brain. Dev Biol 2007; 304:22-33; PMID:17222817; http://dx.doi.org/ 10.1016/j.ydbio.2006.12.014 [DOI] [PubMed] [Google Scholar]
- 38. Zhang J, Woodhead GJ, Swaminathan SK, Noles SR, McQuinn ER, Pisarek AJ, Stocker AM, Mutch CA, Funatsu N, Chenn A. Cortical neural precursors inhibit their own differentiation via N-cadherin maintenance of beta-catenin signaling. Dev Cell 2010; 18:472-9; PMID:20230753; http://dx.doi.org/ 10.1016/j.devcel.2009.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J, Shemezis JR, McQuinn ER, Wang J, Sverdlov M, Chenn A. AKT activation by N-cadherin regulates beta-catenin signaling and neuronal differentiation during cortical development. Neural Dev 2013; 8:7; PMID:23618343; http://dx.doi.org/ 10.1186/1749-8104-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jossin Y, Cooper JA. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat Neurosci 2011; 14:697-703; PMID:21516100; http://dx.doi.org/ 10.1038/nn.2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kawauchi T, Sekine K, Shikanai M, Chihama K, Tomita K, Kubo K, Nakajima K, Nabeshima Y, Hoshino M. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron 2010; 67:588-602; PMID:20797536; http://dx.doi.org/ 10.1016/j.neuron.2010.07.007 [DOI] [PubMed] [Google Scholar]
- 42. Sim SE, Bakes J, Kaang BK. Neuronal activity-dependent regulation of microRNAs. Mol Cells 2014; 37:511-7; PMID:24957213; http://dx.doi.org/ 10.14348/molcells.2014.0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 2012; 73:35-48; PMID:22243745; http://dx.doi.org/ 10.1016/j.neuron.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, Pena JT, Tuschl T, Bramham CR. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci 2010; 31:636-45; PMID:20384810; http://dx.doi.org/ 10.1111/j.1460-9568.2010.07112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chai S, Cambronne XA, Eichhorn SW, Goodman RH. MicroRNA-134 activity in somatostatin interneurons regulates H-Ras localization by repressing the palmitoylation enzyme, DHHC9. Proc Natl Acad Sci U S A 2013; 110:17898-903; PMID:24127608; http://dx.doi.org/ 10.1073/pnas.1317528110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O'Tuathaigh C, Waddington JL, Prenter S, et al. . Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med 2012; 18:1087-94; PMID:22683779; http://dx.doi.org/ 10.1038/nm.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 2010; 466:1105-9; PMID:20622856; http://dx.doi.org/ 10.1038/nature09271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khudayberdiev SA, Zampa F, Rajman M, Schratt G. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front Mol Neurosci 2013; 6:43; PMID:24324399; http://dx.doi.org/ 10.3389/fnmol.2013.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jeffries CD, Fried HM, Perkins DO. Additional layers of gene regulatory complexity from recently discovered microRNA mechanisms. Int J Biochem Cell Biol 2010; 42:1236-42; PMID:20460095; http://dx.doi.org/ 10.1016/j.biocel.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang XM, Jia RH, Wei D, Cui WY, Jiang W. MiR-134 blockade prevents status epilepticus like-activity and is neuroprotective in cultured hippocampal neurons. Neurosci Lett 2014; 572:20-5; PMID:24810882; http://dx.doi.org/ 10.1016/j.neulet.2014.04.049 [DOI] [PubMed] [Google Scholar]
- 51. Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry 2011; 69:180-7; PMID:21111402; http://dx.doi.org/ 10.1016/j.biopsych.2010.09.030 [DOI] [PubMed] [Google Scholar]
- 52. Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry 2012; 17:827-40; PMID:21727898; http://dx.doi.org/ 10.1038/mp.2011.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med 2010; 2:23; PMID:20374639; http://dx.doi.org/ 10.1186/gm144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, Ruan X, Ruan Y, Sun YE. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A 2010; 107:18161-6; PMID:20921386; http://dx.doi.org/ 10.1073/pnas.1005595107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Henriksen M, Johnsen KB, Olesen P, Pilgaard L, Duroux M. MicroRNA expression signatures and their correlation with clinicopathological features in glioblastoma multiforme. Neuromolecular Med 2014; 16:565-77; PMID:24817689; http://dx.doi.org/ 10.1007/s12017-014-8309-7 [DOI] [PubMed] [Google Scholar]
- 56. Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One 2011; 6:e24248; PMID:21912681; http://dx.doi.org/ 10.1371/journal.pone.0024248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lavon I, Zrihan D, Granit A, Einstein O, Fainstein N, Cohen MA, Zelikovitch B, Shoshan Y, Spektor S, Reubinoff BE, et al. . Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol 2010; 12:422-33; PMID:20406893; http://dx.doi.org/ 10.1093/neuonc/nop041 [DOI] [PMC free article] [PubMed] [Google Scholar]