Abstract
Interactions between FGF and Wnt/ bcat signaling control development of the midbrain. The nature of this interaction and how these regulate patterning, growth and differentiation is less clear, as it has not been possible to temporally dissect the effects of one pathway relative to the other. We have employed pharmacological and genetic tools to probe the temporal and spatial roles of FGF and Wnt in controlling the specification of early midbrain neurons. We identify a β-catenin (bcat) independent role for GSK-3 in modulating FGF activity and hence neuronal patterning. This function is complicated by an overlap with bcat-dependent regulation of FGF signaling, through the regulation of sprouty4. Additionally we reveal how attenuation of Axin protein function can promote fluctuating levels of bcat activity that are dependent on FGF activity. This highlights the complex nature of the interactions between FGF and Wnt/ bcat and reveals that they act at multiple levels to control each others activity in the midbrain.
Keywords: axin, GSK-3, IWR-1, mesencephalon, neuron, trigeminal, Tankyrase, zebrafish, β-catenin
Abbreviations
- MTN
mesencephalic trigeminal nucleus
- nTPC
nucleus of the tract of the posterior commissure
- Wnt
wingless related integrated site
- FGF
Fibroblast growth factor
- CA-fgfr1
constitutively active fgf receptor 1
- BIO
6-bromoindirubin-3′-oxime
- IWR-1
Inhibitors of Wnt Response 1
- cat
β-catenin
- dkk1
dickkopf Wnt signaling pathway inhibitor 1
- GSK-3
glycogen synthase kinase
- A-P
anterior-posterior
- GFP
green fluorescent protein
- hpf
hours post fertilization
- dpf
days post fertilization.
Introduction
The control of neurogenesis is critical for ensuring the correct number of neurons form at the appropriate place during development and regeneration. Disruption to this process occurs in a number of congenital disorders that result in cognitive impairment.1 Signaling pathways play a critical role in controlling where and when neurons form, so are presumed to be robustly regulated, to ensure that fluctuations of any one pathway are buffered and do not perturb neurogenesis. Buffering of signaling pathway activity is achieved through regulatory interactions between pathways, that act to limit or promote activity, for instance by controlling expression of ligands or the activity of activators or repressors. The interaction between Wnt and FGF signaling in the developing midbrain is a well-explored example of this interplay between 2 critical pathways.2,3
FGF and Wnt Regulation of Neurogenesis
The isthmus arises at the midbrain-hindbrain boundary and acts as an organizer for adjacent tissues.3,4 In the midbrain, FGFs secreted from the isthmus controls the patterning of the midbrain and anterior hindbrain.5-10 At later stages, FGF activity controls the onset of neurogenic differentiation across the midbrain11,12 and acts to specify dopaminergic and serotenergic neurons in the ventral midbrain.13-15 Changes to midbrain identify in animals showing reduced levels of FGF signaling are presumed to reflect a key role of FGFs in specifying regional fate in the midbrain. Specifically, mice or zebrafish with loss of function of Fgf8 show hyoplastic midbrain and anterior hindbrain.16,17
The canonical Wnt signaling pathway activates the nuclear transcription co-factor β-catenin (bcat) and is important for multiple aspects of neuronal development. It is known to be important for regulating cell proliferation through the cell cycle regulator cyclinD1 and c-myc and perturbations to Wnt affect midbrain size.18-23 Wnt/ bcat signaling is also a regulator of neurogenesis and induces expression of Neurogenin1, Neurogenenin2 and NeuroD1.24,25 Both early specification and later differentiation of dopaminergic neurons in the ventral midbrain are regulated by Wnt/ bcat signaling.26-30
Cross-Talk between Wnt and FGF
A well-characterized regulatory feedback loop operates to maintain Wnt1 and Fgf8 expression at the isthmus.2 Manipulations of Wnt or FGF signaling in chick embryos have revealed that both pathways are required for activity of the other.31 However, it is unclear if there is a hierarchy of events in which one pathway controls the others and whether they alter the biological responses of each other. Computational modeling of the spatial expression of Wnt and FGF signaling pathway genes in mutant mice has suggested that FGF signaling is required for the maintenance of Wnt1 expression at the isthmus, but not induction.32 A number of studies reveal that Wnts and FGFs have multiple roles during midbrain development and neurogenesis, independent of their role in conferring regional patterning. In cortical neurons, bcat acts to promote neuronal differentiation and can override FGF signals that promote proliferation, by inducing expression of the pro-neurogenic factor Neurogenin1.24 Neural stem cells likewise respond differently to Wnt and FGF signaling dependent on whether both signals are present. In the presence of FGF2, bcat promotes neural stem cell proliferation; in an absence of FGF2 bcat promotes neuronal differentiation.33 Thus an interplay between Wnt and FGF regulates both neuronal progenitor proliferation, but also differentiation. The balance of these respective roles is therefore critical for orchestrating growth of the developing brain and ensuring that appropriate neuronal populations form in the correct sites.
We have previously investigated how development of early forming dorsal brain neurons are regulated by FGF and Wnt signaling.34 Using a combination of pharmacological and genetic manipulations we found that positioning and number of mesencephalic trigeminal nucleus (MTN) neurons is dependent on the level of FGF and Wnt activity. We and others have shown that FGF signaling from the isthmus controls neuronal differentiation in the midbrain, through regulation of Hairy-related genes her5 and him.8,35,36 A posterior retraction of her5 expression toward the isthmus controls where and when neuronal differentiation occurs in the midbrain.37 Using pea3 as a readout of FGF activity, we postulated this corresponds to a gradient of FGF signaling that is retracting posteriorly in the midbrain during development.34 We investigated the interaction of FGF and Wnt in controlling this process by applying inhibitors of FGF receptors (SU5402) and GSK-3 (BIO). This revealed that FGF signaling is GSK-3 dependent and that the FGF receptor inhibitor sprouty4 is regulated by Wnt signaling. It is important to understand the nature of these interactions because of their potential impact on the spatiotemporal control of neurogenesis in the brain. We have therefore investigated the role of Wnt/ bcat activity in controlling development of MTN neurons in the midbrain and how the interaction with FGF signaling affects this process.
In this work, we identify an interaction between GSK-3 and FGF intracellular signaling pathways, independent of the Wnt/ bcat pathway. Specifically we note that attenuation of Wnt/ bcat signaling or of Axin function do not alter positioning of MTN neurons, in contrast to an inhibition of GSK-3 function. Furthermore, FGF responsive genes are affected differently by bcat over-expression and manipulation of GSK-3. Intriguingly, we find that bcat-regulated genes do not respond in a linear manner to Axin protein levels, suggesting that feedback loops act to modulate the output of Wnt/ bcat during midbrain development.
Methods
Animals and embryo manipulations
All experiments were performed in accordance with UK Home Office regulations. Embryos were grown at 28.5°C as previously described.38 Lines used were AB (considered to be wildtype), masterblind (mbl),39, Tg[elavl3:gfp],40 Tg[dusp6:d2eGFP],41 Tg[hsp70l:dkk1b-gfp],42 Tg[hsp70:ca-fgfr1],43 Tg[hsp70:gal4],44 Tg[dlx5a/6a:eGFP],45 Tg[UAS:HA-β-catenin],34 Tg[TOPdGFP].46
Heat shock induction was performed by moving embryos to 37°C for 2 hours at 16.5 hours post fertilization (hpf). Pharmacological treatments were performed by applying either SU5402 (Sigma), BIO (Invitrogen) or IWR-1 (Merck) diluted in embryo medium as previously described.34 For all experimental conditions, a minimum of n = 10 embryos were used; for each individual experiment containing multiple conditions, embryos from the same clutch were used to minimise variation in developmental stage.
In situ hybridization and Immunohistochemistry
Gene expression was visualised by in situ hybridization and proteins detected by immunohistochemistry as previously described.47,48 Antibodies used were anti-HuC/D (1 : 500, Invitrogen), anti-Isl1 (1 : 200, DSHB), anti-acetylated tubulin (1 : 200, Sigma), anti-GFP (1 : 500, AMS Biotechnology), anti-HA (1 : 300, Roche). Back labeling of axons was performed by applying DiI or DiD to muscles of fixed 5 dpf larvae using a sharpened tungsten needle. Fluorescent images were acquired using a Nikon C-1 Eclipse confocal microscope and processed using Photoshop (Adobe).
Mathematical modeling
Statistical models and analyses were generated using the R programming language (R Development Core Team, 2010) as described previously.34 A minimum of 10 measurements were used for each condition in each experiment. The models used tested variables across a number of datasets and assessed batch effects for significance.
The model of Lee et al.49,50 describing Wnt signaling and bcat activity was converted into equivalent code for the Python programming language. Under the assumption that degradation of Axin (reaction 15 in the original model, described by the rate constant k15) is primarily through its ubiquitination by Tankyrase, this model was then simulated under varying values of k15 (other rates were kept as in the original model description).
Results
FGF signaling controls positioning and number or early forming midbrain neurons
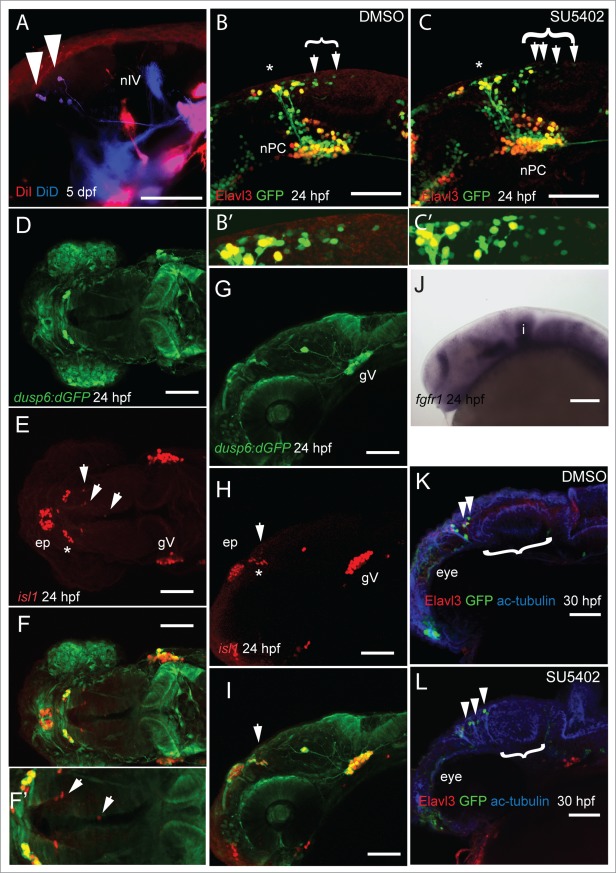
We have previously identified MTN neurons in the anterior midbrain by retrograde labeling of adductor mandibulae muscles using DiI.34 In order to ascertain whether MTN neurons also innervate other cranial muscles, we labeled lateral rectus and levator arcus palantini muscles with DiD and adductor mandibulae muscle with DiI in 5 day post fertilization (dpf) larvae. We found that MTN neurons were back labeled with both DiD and DiI, but all labeled MTN neurons were restricted to the anterior midbrain, as we have found previously (Fig. 1A). To understand why MTN neurons arise in the anterior midbrain, we used a transgenic line that labels developing neurons in the developing brain. The elavl3 gene encodes the HuC protein, a marker of differentiated neurons.40 At 24 hpf, GFP expressing (GFP+) neurons in Tg[elavl3:eGFP] embryos are observed in the dorsal brain, in the nucleus of the posterior commissure (nTPC) and in the anterior midbrain. GFP+ MTN neurons undergo differentiation in an anterior-posterior manner from 22 hpf as shown by the presence of HuC protein (Fig. 1B). Treatment with the FGF receptor antagonist, SU5402, at stages prior to MTN differentiation, leads to an increased number of MTN neurons that lie at more posterior positions along the midbrain, relative to control animals (Fig. 1C).34 This suggests that FGF signaling controls where MTN neurons will form in the midbrain during development. Isl1 is expressed by all primary neurons in the developing zebrafish brain.51 To determine whether FGF signaling is active in the anterior midbrain where MTN neurons will first differentiate, we labeled MTN neurons by an anti-Isl1 antibody and compared to GFP localization in a reporter line for FGF signaling that expresses GFP under the control of the Dusp6 promoter (Tg[dusp6:d2GFP]). Strikingly, there was no co-localization of GFP and Isl1 revealing that MTN neurons differentiate in regions devoid of FGF signaling (Fig. 1D–I). Commensurate with this, fgfr1 is not expressed in anterior midbrain cells at this stage, suggesting that cells are not able to respond to FGF signaling (Fig. 1J). We wished to know whether a temporal inhibition of FGF signaling during stages prior to MTN formation would affect the development of later-forming neuronal populations in the midbrain including the optic tectum. To investigate this, embryos were treated from 14–24 hpf with SU5402 or DMSO, then drug washed away and embryos allowed to develop until 30 hpf. In SU5402 treated embryos we observed more posteriorly located MTN neurons in the midbrain and a smaller optic tectum (Fig. 1K and L). Thus, a temporal inhibition of FGF signaling at stages when primary neurons form, leads to perturbations of midbrain neuronal architecture.
Figure 1.
For figure legend, see page 5.
Wnt/ bcat signaling interacts with FGF to direct MTN formation
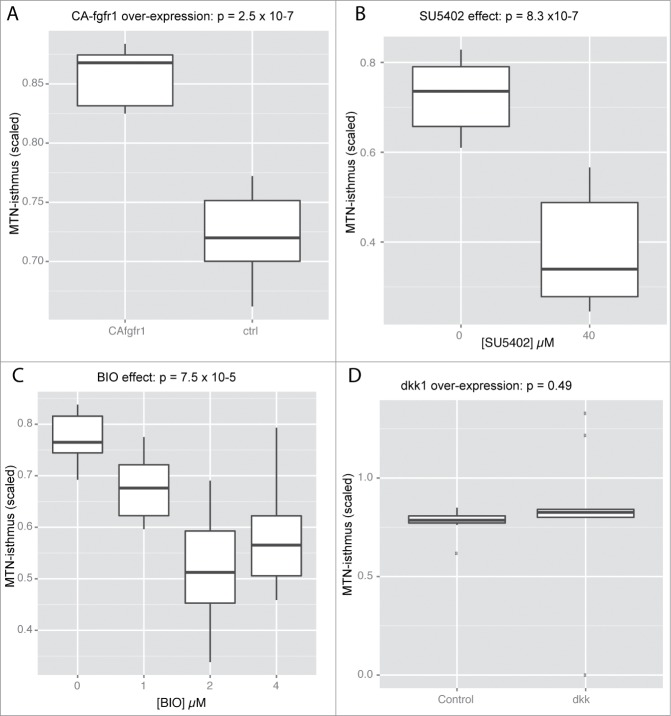
The well documented interaction between FGF and Wnt/ bcat signaling suggests that manipulations of bcat activity would also affect positioning of MTN neurons. We therefore compared how alterations of Wnt relative to FGF signaling affected the positioning of MTN neurons. MTN neuron positioning relative to the isthmus was quantified in 24 hpf embryos under the following conditions: 1) expression of a constitutively active FGF receptor (CA-Fgfr1), 2) application of SU5402, 3) expression of the Wnt antagonist dickkopf1b (dkk1), 4) application with BIO. All manipulations were performed between 14–24 hpf (SU5402, BIO) or 16.5–24 hpf (CA-Fgfr1, dkk1) as we have previously shown that MTN, but not adjacent diencephalic neurons, are sensitive to manipulations of FGF and Wnt signaling at this stage.34 The position of the most posteriorly located MTN neurons in the midbrain was measured relative to the isthmus for each condition and expressed as a ratio relative to the midbrain size to compensate for any differences in size. This revealed that over-activation of FGF resulted in an anterior shift of MTN neurons (Fig. 2A). In contrast, SU5402 treated embryos displayed posteriorly located MTN neurons relative to controls (Fig. 2B). BIO treatment caused a similar phenotype to SU5402 treatment, with MTN neurons present at more posterior positions in the midbrain (Fig. 2C). In contrast, abrogation of Wnt signaling by overexpression of dkk1 had no effect on MTN positioning (Fig. 2D). This suggests that the posterior displacement of MTN neurons observed in BIO treated embryos may reflect a role for GSK-3 in controlling neuronal differentiation along the dorsal midbrain, independent of its role in regulating Wnt signaling.
Figure 2.
MTN A-P positioning along the dorsal midbrain is regulated by FGF and GSK-3 activity, but does not require Wnt signaling. Box plots representing the minimal distance between MTN neurons and the isthmus scaled relative to midbrain size for embryos expressing CA-fgfr1 (A), exposed to 0 or 40µM SU5402 (B), exposed with 0, 1, 2, 4µM BIO (C), or expressing dkk1b (D). Significance was determined by t-test with Welch correction (A, B, D) or by a Kruksal-Wallis test (C) by comparing to embryos not expressing transgenes or treated only with DMSO carrier (0µM). N = 10 for each condition.
GSK3 and bcat differentially regulate FGF signaling in the midbrain
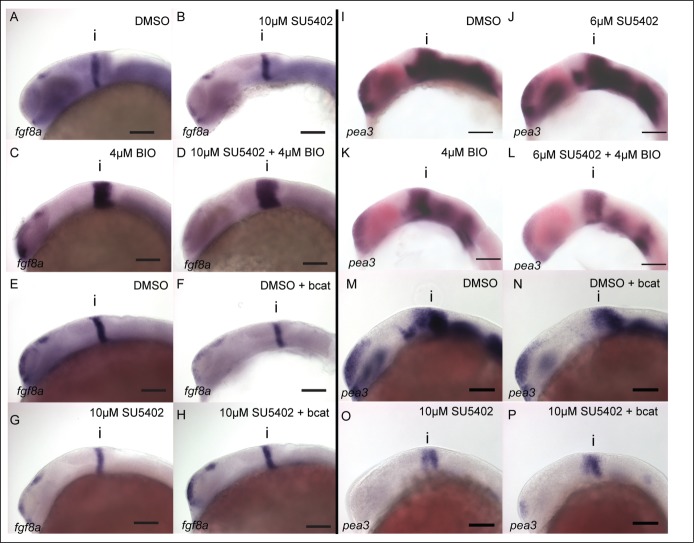
The different changes to neuronal pattering caused by altering Wnt and FGF activity suggests that they do not interact in a simple feedback loop to control neurogenesis in the midbrain. To understand the importance of GSK-3 in mediating interactions between Wnt and FGF, we compared the responses of FGF signaling genes to alterations of GSK-3 and bcat activity. Previously, we showed that BIO treatment up-regulates fgf8a expression at the isthmus. To determine whether FGF-dependent genes such as fgf8a are regulated by GSK-3 or bcat activity, the affects of GSK-3 inhibition or bcat over-expression were compared when FGF signaling was attenuated using the inhibitor SU5402. Embryos were treated with 10µM SU5402 from 14 hpf in the presence of varying doses of BIO or a stabilised HA tagged bcat. Expression of fgf8a at the isthmus was assessed at 24 hpf by in situ hybridization and found to be down-regulated by SU5402 treatment (Fig. 3A and B). In contrast, BIO causes elevated fgf8a expression compared to controls (Fig. 3C). Surprisingly, in the presence of both BIO and SU5402 fgf8a expression was higher than in embryos treated with BIO alone (Fig. 3D). Overexpression of bcat resulted in reduced fgf8a expression, unlike BIO treatment (Fig. 3E and F). In the presence of SU5402, over-expression of bcat then rescued fgf8a relative to embryos treated with SU5402 alone (Fig. 3G and H). These results were consistent between embryos (see also Fig. S1) and reveal that fgf8a responds differently to BIO treatment and bcat overexpression. However, both BIO and bcat over-expression are able to rescue fgf8a expression in the presence of SU5402.
Figure 3.
FGF activity in the midbrain is GSK-3 dependent, but is not regulated by β-catenin. Expression of fgf8a (A–H) and pea3 (I–P) was visualized by in situ hybridization in 24 hpf embryos treated with DMSO (A, E, I, M), SU5402 at 10µM or 6µM (B, G, J, O), BIO at 4µM (C, K), SU5402 and BIO together (D, L), when over-expressing bcat (F, N), when over-expressing bcat and treated with SU5402 (H, P). Isthmus (i). Scale bars: 100µm.
To show how fgf8a responses compare to a transcriptional read-out of FGF activity we assessed the response of the ETS family gene pea3 at the isthmus and midbrain signaling under different levels of FGF, GSK-3 and bcat activity. Both SU5402 and BIO treatment cause a reduction of pea3 expression (Fig. 3I–K). SU5402 was used at 6µM in combination with BIO as higher SU5402 concentrations (10µM) lead to a loss of pea3 expression, precluding quantification. In the presence of SU5402, BIO caused a further reduction of pea3 expression than when treated with BIO or SU5402 alone (Fig. 3L).
Overexpression of bcat also leads to a reduction of pea3 expression, similar to BIO (Fig. 3M and N). In contrast to BIO, over-expression of bcat in the presence of SU5402 does not further repress pea3 (Fig. 3O and P, see also Fig. S2). These differential responses of genes in the FGF signaling pathway to BIO and bcat overexpression suggests that GSK-3 affects FGF signaling in a bcat-independent manner (Table 1).
Table 1.
Summary of gene expression changes in the midbrain at 24 hpf due to alterations of Wnt or FGF activity from 14 hpf (SU5402, IWR-1, BIO) or 16.5 hpf (dkk1, βummar
| Signaling pathway manipulation | gene expression in the midbrain | |||||||
|---|---|---|---|---|---|---|---|---|
| Wnt | FGF | treatment | lef1 | wnt1 | pea3 | her5 | spry4 | fgf8 |
| ↑ | BIO | ↑ | ↓ | ↓ | ↓ | ↑ | ↑ | |
| ↑ | bcat | ↑ | ↑ | ↓ | ↓ | ↑ | ↓ | |
| ↓ | dkk1 | ↓ | ↓ | ↓ | ↓ | |||
| ↓ | IWR-1 | ↓ | ↑ | ↓ | ||||
| ↑ | CA-fgfr1 | ↑ | ↑ | ↑ | ↑ | ↓ | ||
| ↓ | SU5402 | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | |
| ↑ | ↓ | BIO + SU5402 | ↑ | ↓ | ↓ | ↓ | ↑ | ↑ |
| ↑ | ↓ | bcat + SU5402 | ↑ | ↓ | ↓ | ↓ | ||
| ↓ | ↓ | IWR-1 + SU5402 | ↓ | ↓ | ↓ | ↓ | ↓ | |
| reduced expression relative to control | ||||||||
| no change relative to control | ||||||||
| elevated expression relative to control | ||||||||
Wnt responsive genes are affected differently by changes to β-catenin and GSK-3 activity
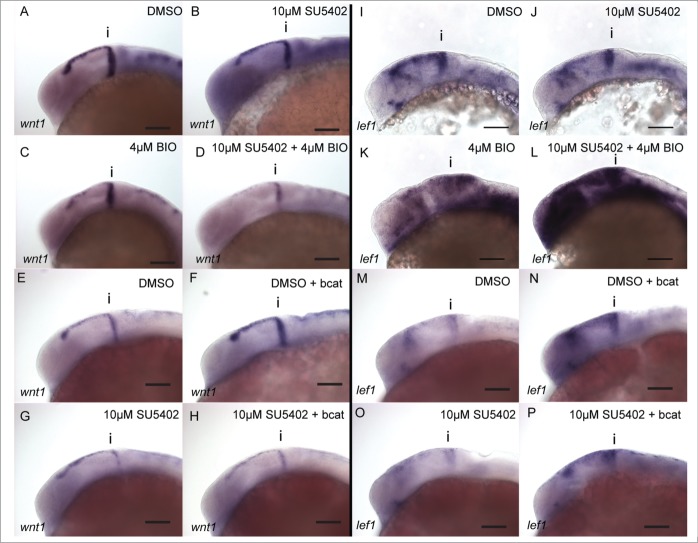
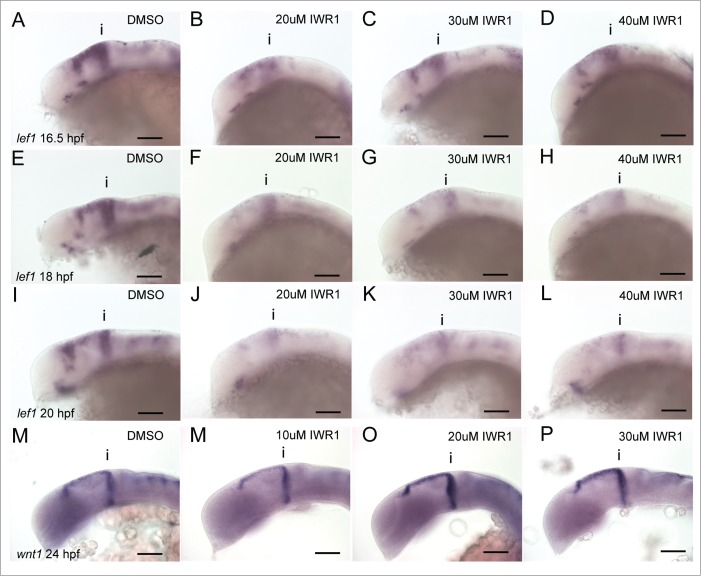
GSK-3 may also regulate Wnt signaling independently of its role in inhibiting β-catenin activity in the bcat destruction complex (bcatDC). To test this we investigated how genes in the Wnt pathway are affected by inhibiting FGF signaling in conjunction with GSK-3 inhibition compared to bcat over-expression. Application of SU5402 or BIO leads to reduced wnt1 expression (Fig. 4A–C). Application of BIO in presence of SU5402 causes a further inhibition of wnt1 (Fig. 4D). In contrast to the response of wnt1 to BIO treatment, wnt1 is up-regulated by overexpression of bcat (Fig. 4E and F). Application of SU5402 abrogates this response (Fig. 4G and H, see also Fig. S3). This reveals a differential response of wnt1 to inhibition of GSK-3 and elevated β-catenin activity, suggesting that GSK-3 has a bcat-independent function in controlling wnt1 expression.
Figure 4.
wnt1 in the midbrain is GSK-3 dependent, but is not regulated by β-catenin. Expression of wnt1 (A–H) and lef1 (I–P) was visualised by in situ hybridization in 24 hpf embryos treated with DMSO (A, E, I, M), SU5402 at 10µM (B, G, J, O), BIO at 4µM (C, K), SU5402 and BIO together (D, L), when over-expressing bcat (F, N), when over-expressing bcat and treated with SU5402 (H, P). Isthmus (i). Scale bars: 100µm.
To determine how GSK-3 inhibition or bcat over-expression can affect Wnt/ bcat activity, we used lef1 as a transcriptional readout. SU5402 treatment results in down-regulation of lef1 (Fig. 4I and J). In contrast, inhibition of GSK-3 by BIO results in a slight up-regulation of lef1 expression (Fig. 4K). In the presence of SU5402 and BIO, lef1 expression was strongly upregulated (Fig. 4L). This upregulation was much greater than in embryos treated with BIO alone, implying that GSK-3 inhibition causes a much stronger activation of bcat when FGF signaling is likewise inhibited. We then evaluated how overexpression of bcat affects lef1 expression. As predicted, bcat over-expression causes an up-regulation of lef1 (Fig. 4M and N). In the presence of SU5402, bcat overexpression can rescue lef1 expression (Fig. 4O and P, see also Fig. S4).
In summary, we found that lef1 expression is reduced by SU5402, indicating a requirement for FGF activity to promote bcat signaling. Both BIO and bcat can rescue this reduction in the presence of SU5402, suggesting bcat regulation of lef1 is not dependent on FGF activity.
Inhibition of Wnt signaling does not affect MTN neuron positioning in the midbrain
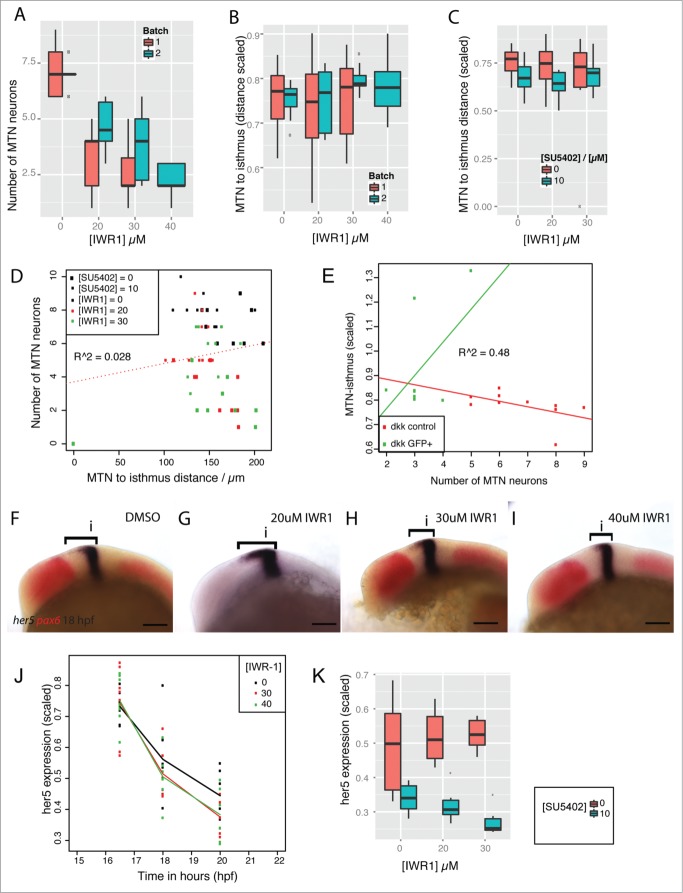
To further probe the relative roles of Wnt and FGF signaling in controlling each others activity, we focused on the role of the key regulator of the bcat destruction complex, Axin. We have previously described how IWR-1 acts in an antagonistic manner to SU5402 to control the number of MTN neurons that form.34 Inhibition of Wnt signaling (by over-expression of dkk1) does not affect MTN positioning (Fig. 2D). We therefore tested whether stabilization of Axin, through inhibition of Tankyrase, would affect MTN positioning in the midbrain. Although application of varying IWR-1 doses resulted in the formation of fewer MTN neurons (Fig. 5A) there was no effect on the distance of MTN neurons to the isthmus (Fig. 5B). A comparison of MTN neuron number relative to the MTN-isthmus distance showed no correlation when embryos were exposed to IWR-1 in the presence of SU5402 (Fig. 5C). We than asked whether changes to the MTN-isthmus distance in embryos exposed to IWR-1 and SU5402 could be explained by the application of either IWR-1 or SU5402. We find that there is no correlation bewteen MTN neuron number and distance to the isthmus in embryos exposed to both IWR-1 and SU5402 (R = 0.028, Fig. 5D). Two-way ANOVA tests revealed that IWR-1 had no significant affect (p = 0.616) on MTN neuron positioning, but SU5402 had a strong affect (p = 0.0012). Moreover, there is no interaction effect expected if IWR-1 and SU5402 are acting synergistically or antagonistically (p = 0.839). In contrast, models that describe MTN neuron number revealed that there is both a strong IWR-1 affect (p = 1.23 × 10−6, Table 2) and a strong interaction effect between IWR-1 and SU5402 (p = 0.0048, Table 2). These models indicate that although inhibition of Tankyrase results in fewer MTN neurons, this does not occur in conjunction with posterior shifts of MTN neurons observed following abrogation of FGF activity (by SU5402) or inhibition of GSK-3 (by BIO). Loss of Wnt signaling by overexpression of dkk1 likewise causes a decrease in the number of MTN neurons forming, but this does not correlate with altered positioning within the midbrain (R = 0.48, Fig. 5E). This suggests that inhibition of Wnt signaling by either dkk1 over-expression or IWR-1 application does not perturb the positioning of MTN neurons.
Figure 5.
For figure legend, see page 11.
Table 2.
Results from Poisson models describing MTN neuron numbers as a function of SU5402 at 10µM (δ([SU5402],10) and IWR-1 at 20 (δ,([IWR120], 20) or 30µM (δr 3IWR1],30). The baseline number of MTN neurons is represented by µ and interactions represented by d. Addition of IWR-1 at 20 or 30µM results in significant changes to MTN neuron number (p<0.001), but SU5402 at 10µM does not (p > 0.05). Application of both SU5402 and IWR-1 results in significant changes to MTN neuron numbers that cannot be explained by simply additive effects, but rather represent interaction effects e.g. dIWR1 = 20
& SU5402 = 10.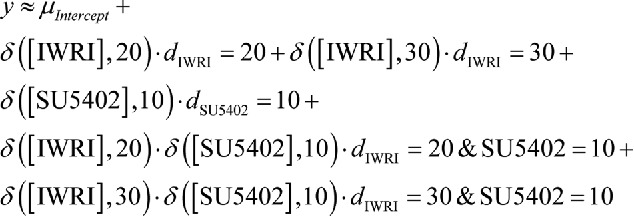
| Estimate Std. | Error | z value | Pr(>|z|) | |
|---|---|---|---|---|
| (Intercept) | 1.9617 | 0.1250 | 15.693 | < 2e-16 |
| f(IWR1)20 | −0.7916 | 0.2238 | −3.536 | 0.000406 |
| f(IWR1)30 | −1.2197 | 0.2515 | −4.850 | 1.23e-06 |
| f(SU5402)10 | 0.1316 | 0.1712 | 0.768 | 0.442269 |
| f(IWR1)20:factor(SU5402)10 | 0.5066 | 0.2832 | 1.789 | 0.073613 |
| f(IWR1)30:factor(SU5402)10 | 0.8670 | 0.3074 | 2.820 | 0.004796 |
The Hairy-related gene her5 controls the spatiotemporal onset of neurogenesis in the midbrain and hence where MTN neurons form.8,35 The expression of her5 along the A-P extent of the dorsal midbrain is controlled by GSK-3 and FGF activity.34 To determine whether her5 shows differential responses to IWR-1 over time, we measured the spatial expression of her5 at 16.5, 18 and 20 hpf after application of IWR-1 at 14 hpf. We found that no significant changes to her5 expression occurred initially after exposure to IWR-1, although by 18 hpf the her5 expression was reduced relative to control embryos (Fig. 5F–J). However, by 24 hpf, there no longer an effect of IWR-1 treatment on her5 expression in the midbrain.
Having shown that her5 is not significantly affected by IWR-1, we then tested if inhibition of Wnt signaling would affect the response of her5 to FGF activity. IWR-1 and SU5402 were applied simultaneously at 14 hpf and the spatial expression of her5 measured along the dorsal midbrain at 24 hpf. Plots of her5 expression revealed that IWR-1 did not alter the response of her5 SU5402 (Fig. 5K). We confirmed this by generating models for the response of her5 to IWR-1 and SU5402 (Table 3). These revealed that application of IWR-1 did not have a significant affect on her5 (p = 0.322). In contrast, SU5402 had a significant effect (p = 0.000282). Furthermore, IWR-1 did not significantly alter the response of her5 to SU5402 (p = 0.0763). This reveals that bcat activity is not required for the response of her5 to FGF activity and so does not control the A-P position of MTN neurons in the dorsal midbrain, unlike GSK-3.
Table 3.
Results from generalized linear models describing expression of her5 (scaled relative to midbrain size) as i) a function of SU5402 at 10µM (fSU540210) and IWR-1 at either 20 or 30µM (fIWR120, fIWR130), or ii) a function of SU5402 at 10µM relative to IWR-1 (fIWR1). Addition of IWR-1 at 20 or 30µM does not result in significant changes to her5 expression (p > 0.05), but SU5402 does (p < 0.001). Addition of 30µM IWR-1 in the presence of SU5402 has an affect on her5 expression with low significance (p < 0.1), but overall IWR-1 does not significantly alter the response of her5 to SU5402 (p = 0.076). Fit of data to models R2 = 0.67
i) her5 expression as a function of IWR-1 and SU5402 concentration.
| Estimate Std. | Error | t value | Pr(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 0.48836 | 0.02693 | 18.136 | < 2e-16 *** |
| fIWR120 | 0.03078 | 0.03942 | 0.781 | 0.440123 |
| fIWR130 | 0.03709 | 0.04113 | 0.902 | 0.373348 |
| fSU540210 | −0.14845 | 0.03808 | −3.898 | 0.000418 *** |
| fIWR120:fSU540210 | −0.05031 | 0.05697 | −0.883 | 0.383250 |
| fIWR130:fSU540210 | −0.10557 | 0.05817 | −1.815 | 0.078133. |
ii) her5 expression as a function of IWR-1 and SU5402 presence. 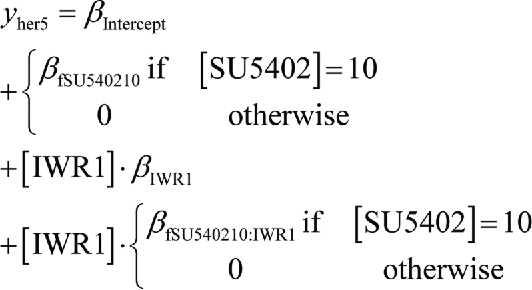
| ||||
| |
Estimate Std. |
Error |
t value |
Pr(>|t|) |
| (Intercept) | 0.489451 | 0.025566 | 19.145 | < 2e-16*** |
| fSU540210 | −0.145272 | 0.036210 | −4.012 | 0.000282*** |
| IWR1 | 0.001297 | 0.001294 | 1.003 | 0.322598 |
| fSU540210:IWR1 | −0.003342 | 0.001833 | −1.823 | 0.076349. |
bcat activity responses to IWR-1 are not linear relative to dose or time of exposure
Our observation that IWR-1 application caused a transient decrease in her5 expression led us to wonder whether it caused repression of bcat activity in a dose-dependent and uniform manner. We therefore used expression of lef1 in the midbrain as a readout of bcat activation and compared embryos treated with IWR-1 at varying doses (20, 30, 40µM) from 14 hpf after 2.5 (16.5 hpf), 4 (18 hpf) and 6 (20 hpf) hours of exposure. Intriguingly, we found that lef1 responses were affected more by application of 20µM IWR-1 than either 30 or 40 µM after 2.5 hours post application (Fig. 6A–D). At later stages this changes, until by 12 hours post application lef1 is most strongly reduced in embryos treated with the higher doses of 30 and 40µM (Fig. 6E–L). Why lef1 should be more affected by a lower, rather than higher dose of IWR-1 initially is unclear. Expression of wnt1 at the isthmus is responsive to BIO and bcat activity (Fig. 4). We found that the lowest dose of IWR-1 tested (10µM), resulted in the strongest downregulation of wnt1 expression (Fig. 6M–P). We then turned to an alternative reporter of Wnt/ bcat activity, the Tg[TOPdGFP] transgenic line, to determine if these responses of lef1 represent Wnt activity. In this transgenic line, we were unable to discriminate any differences in bcat activity when treated at different IWR-1 doses (Table 1, data not shown). This may reflect the relative insensitivity to changes in Wnt signaling levels previously reported.52 We therefore examined other readouts of bcat activity to determine how they responded to different concentrations of IWR-1 over time.
Figure 6.
Tankyrase inhibition does not lead to linear reductions of bcat activity over time. Lateral views of 16.5 (A–D), 18 (E–H), 20 hpf (I–L) embryos processed by in situ hybridization to reveal expression of lef1 expression following treatment with IWR-1 at 0, 20, 30, 40 µM IWR-1 from 14 hpf. Lateral views of 24 hpf embryos processed by in situ hybridization to reveal expression of wnt1 expression following treatment with IWR-1 at 0, 20, 30, 40 µM IWR-1 from 14 hpf. Isthmus (i). Scale bars: 100µm.
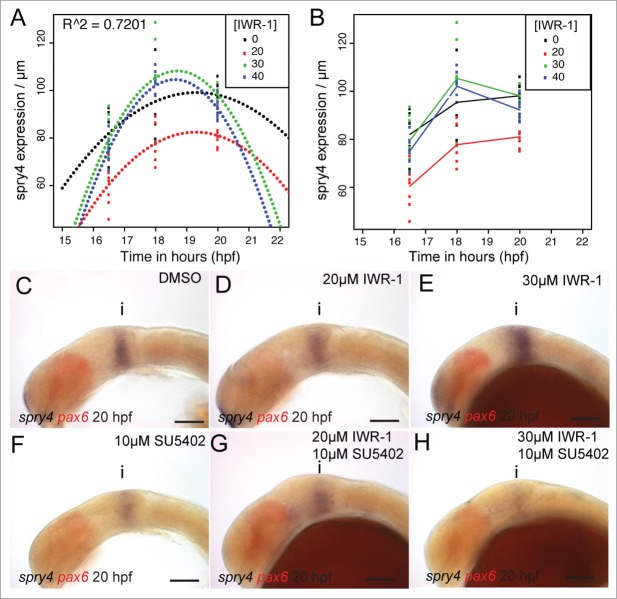
Previously, we found that the FGF receptor inhibitor, sprouty4 (spry4) shows a dual response to FGF and Wnt/ GSK-3 activity in the midbrain.34 Strikingly, we noted that spry4 is sensitive to the level of bcat activity independently from FGF activity. We therefore measured the response of spry4 to varying doses of IWR-1 from 14 hpf, to determine how similar its response was to that of lef1. As observed for lef1, expression of spry4 was initially reduced most strongly when 20µM IWR-1 was applied, compared to 30 or 40µM (Fig. 7A and B). Continued exposure to IWR-1 revealed that 20µM had the strongest effect on spry4. In contrast, higher doses led to an elevated spry4 expression 4 hours after application relative to DMSO treated control embryos. After 6 hours exposure, all doses of IWR-1 caused a reduced expression of spry4, with the lower dose showing the strongest effect. This difference in the response of spry4 to low or high doses of IWR-1 suggests that a compensatory feedback loop may operate to promote spry4 expression in response to reduced bcat activity. A good candidate for this is FGF signaling, as Wnt and FGF signaling interact to maintain the isthmus and promote each others activity.10,31 We therefore tested if abrogation of FGF activity would alter the dose-dependent response of spry4 to IWR-1, by simultaneously applying both SU5402 and IWR-1. As before, we noted that application of 20µM IWR-1 caused a more severe reduction of spry4 that 30µM (Fig. 7C–E). Likewise, we observed that 10µM SU5402 caused a reduction, but not abrogation of spry4 expression (Fig. 7F). In the presence of SU5402, the response of spry4 to different doses of IWR-1 is altered and the 30µM dose causes a stronger reduction than 20µM (Fig. 7G–H). This suggests that at higher doses of IWR-1, FGF signaling compensates for reduced bcat activity, by promoting spry4 expression. It also reveals that small reductions of bcat activity do not induce this feedback response by FGF signaling.
Figure 7.
spry4 shows a bivalent response to changes in Wnt and FGF signaling. Polynomials were fitted against the spatial expression of spry4 in the dorsal midbrain (µm) at 16.5, 18, 20 hpf following treatment with IWR-1 from 14 hpf (A) and showed a good fit (R2 = 0.7201, n = 10 for each condition). A line plot of the spry4 expression domain (scaled to midbrain size) in the same embryos (B) reveals that treatment with 20µM IWR-1 had the strongest affect (n = 10 for each condition). Lateral views of 20 hpf embryos processed by in situ hybridization to reveal spry4 (blue) and pax6 (red) expression following treatment from 14 hpf with DMSO (C), 20µM IWR-1 (D), 30µM IWR-1 (E), 10µM SU5402 (F), 20µM IWR-1 with 10µM SU5402 (G), or 30µM IWR-1 with 10µM SU5402 (H). Isthmus (i). Scale bars: 100µm (C–H).
A model of Axin inhibition predicts oscillating bcat activity
We found that Wnt/ bcat responsive genes lef1 and spry4 show a non-linear response to IWR-1 and this changes relative to the duration of exposure. One putative explanation for the differential effects of IWR-1 dosage on Wnt-target genes may therefore be due to feedback from FGF signaling to promote elevated bcat signaling. IWR-1 inhibition of Tnk reduces the rate of Axin protein degradation, a key limiting step in the activation of bcat.53,54 We used the model of Lee et al49 to examine how changing the rate at which Axin is ubiquinated by Tnk affects bcat activity. We modified the model such that the Axin degradation term was dependent on the concentration of Tnk:
Therefore, adding IWR-1 will reduce the contribution of Tnk to the rate (k15) at which Axin is degraded and so decrease this rate overall. A plot of Axin protein relative to different rates of Axin degradation reveals that at high IWR-1 doses (corresponding to low concentrations of Tnk), Axin protein accumulates (Fig. 8A). This corresponds with a decrease in free bcat. We wondered how Axin concentration (and thus Tnk activity) was related to bcat concentration in the model and observed a nonlinear response for the bcat, divided into 2 regions (Fig. 8B): a flat region corresponding to very low Axin concentrations and an inversely linear region corresponding to higher concentrations, described by the (fitted) power law equation:
Figure 8.
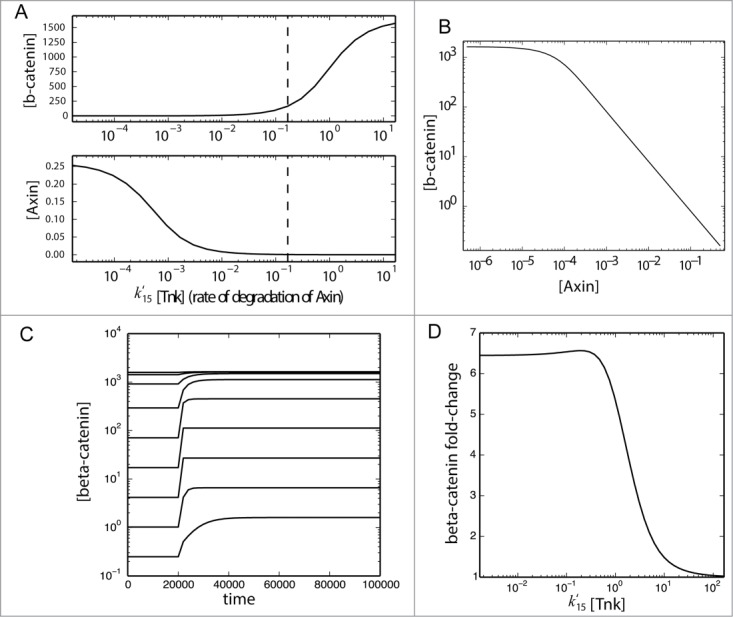
A model for bcat activity reveals differential responses to Tnk activity. The model of Lee et al.52 and Goentoro and Kirschner53 was modified such that Axin degradation (reaction 15) became linearly dependent on the concentration of Tankyrase [Tnk] with a new second-order kinetic constant k15'. The value of k15' was assumed to be equal to the value of k15 (0.167), and the basal concentration of Tankyrase was assumed to be 1 (in arbitrary units). Steady-state values of Axin and b-catenin concentration (brackets) were obtained from the simulation of the model under varying concentrations of Tankyrase (both above and below the basal concentration indicated by a dashed vertical line) and plotted as functions of the total rate of degradation of Axin (A) and as a parametric plot (B). Next, the model was run under different rates of Axin degradation (k15′[Tnk]) until it reached a steady state (s1), then stimulated with addition of Wnt protein at t=20000 and run until a steady-state was again reached (s2). These time courses were plotted to confirm a steady state was reached (C). For each value of k15'[Tnk] the fold change in the steady-state of [β-catenin] was calculated (s1/s2) and plotted (D).
This linear region corresponds to Axin concentrations measured from Xenopus eggs and used for construction of the model, hence provides the region most likely occurring in a biological context. An analysis of how free bcat concentration changes relate to induction of a Wnt/ bcat induced phenotype or induction of siamois or Xnr3 in Xenopus embryos suggested it is the fold change of free bcat concentration that dictates the response to Wnt signaling.50,55 Using the same model as above, we investigated how the concentration of bcat changes relative to different rates of Axin degradation prior to and after Wnt stimulation (Fig. 8C). As before, high rates of Axin degradation lead to high levels of bcat. We then used this result to ask how the fold change of free bcat is affected by the Axin degradation rate (Fig. 8D). Intriguingly, we note that the fold change of bcat is predicted to be relatively high when Axin degradation is high or medium, but that it precipitously drops at low rates of Axin degradation. Our interpretation of this model is that IWR-1 will reduce the levels of free bcat, by promoting elevated levels of Axin, but also allow a greater fold change in bcat levels upon Wnt stimulation.
GSK-3 regulation of MTN positioning is independent of bcat activity
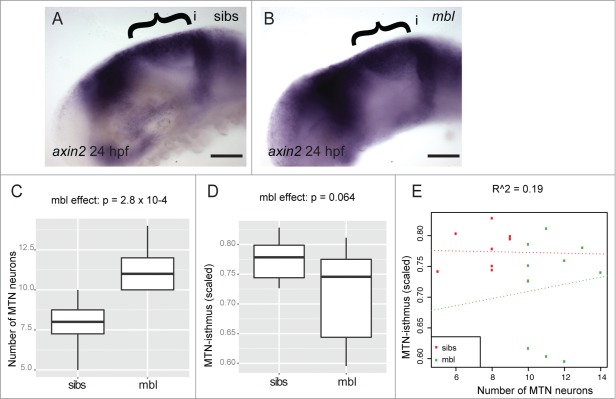
Our results reveal that BIO inhibits GSK-3 activity, leading to elevated bcat activity and an increase in the number of MTN neurons that show a more posterior location in the midbrain. In contrast, inhibition of bcat activity, through inhibition of Tankyrase enzyme or overexpression of dkk1, does not result in posteriorly located MTN neurons, but does lead to the formation of more neurons. As GSK-3 is important for regulating multiple intracellular signaling pathways, it may therefore have a bcat-independent role in controlling neurogenesis. To test if Wnt signaling can affect MTN positioning independently of GSK-3 function, we examined whether MTN development was altered in an axin1 mutant, masterblind (mbl). We observe elevated axin2 expression in mbl mutants indicative of increased bcat activity, similar to that observed after BIO treatment (Fig. 9A and B). We also note that mbl mutants have significantly more MTN neurons than in wildtype siblings, again similar to embryos treated with BIO (Fig. 9C). However, mbl mutants do not show posteriorly located neurons and there is no correlation between MTN number and positioning relative to the isthmus (Fig. 9D and E). This reveals that the increased number of MTN neurons caused by BIO treatment can be attributed to elevated bcat activity, but the posteriorly located MTN neurons observed in these embryos cannot be explained by increased bcat activity in the midbrain.
Figure 9.
Masterblind mutants possess more MTN neurons, but do not show a posterior displacement of MTN neurons. Lateral views of 24 hpf embryos processed by in situ hybridization to reveal elevated axin2 expression in midbrain (bracket) of masterblind mutants (mbl, B), relative to wildtype siblings (sibs, A). Box plot of the number of MTN neurons (C) or MTN-isthmus distance (scaled by midbrain size, D) in 24 hpf mbl mutants and sibs (n = 10 for each condition). There is a strong effect by mbl on MTN number (p = 2.8 × 10−4) but not on MTN-isthmus distance (p = 0.064). Dot plot of MTN number against MTN-isthmus distance (scaled) for mbl mutants and sibs (E). A line of best fit reveals no correlation between MTN neuron number and distance (R = 0.19, n = 10 for each condition). Isthmus (i). Scale bars: 100µM (A,B).
Discussion
We have addressed how the interplay between Wnt and FGF activity controls the spatiotemporal specification of neurons in the developing midbrain. Our findings reveal that a variety of feedback and feed-forward regulatory loops operate to control both the level and site of activity of each pathway. This has implications for where neurons form in the brain during development, but also how many neurons will form and thus affect progenitor cell populations important for later-forming neurons. We find that FGF activity controls the number and positioning of MTN neurons, whereas Wnt signaling is primarily important for controlling the number of neurons that form. Furthermore, we identify a putative role for GSK-3 in modulating FGF-target gene activity and hence neuronal positioning, independent of bcat function.
Previously we could show that FGF activity regulates the number and positioning of MTN neurons in the midbrain and that this is mediated by FGF control of her5 expression. From these findings we predicted that alterations to FGF at early stages would affect subsequent development of later neuronal populations in the midbrain. We now present evidence that this occurs, as expansion of the MTN, due to reductions to FGF activity over a defined window of time, results in a smaller optic tectum. This reveals that a disruption to FGF activity across the midbrain upsets the controlled temporal onset of differentiation, needed for the appropriate spatial formation of discrete neuronal populations. Sustaining an appropriate level of FGF activity at the isthmus is therefore crucial for controlling where and when neurons are specified in the forming midbrain. There is a large body of evidence for an auto-regulatory interaction between Wnt and FGF signaling at the isthmus during midbrain development. Perturbations to this interaction are expected to affect midbrain patterning and development. In this study we have asked whether the interaction between Wnt and FGF occurs by control of bcat activity or of GSK-3. Inhibition of axin1 function, over-expression of dkk1 or over-activation of Axin function using IWR-1 all fail to alter MTN positioning, although they do increase the number of neurons that form. In contrast, GSK-3 inhibition promotes both the formation of extra neurons and the presence of posteriorly located neurons in the midbrain (Fig. 10A). We interpret these results to suggest that GSK-3 regulates FGF signaling across the midbrain independently of bcat. It further reveals that bcat activity controls the number of MTN neurons that form, independently of FGF signaling, but dependent on GSK-3 activity (Fig. 10B). Support for this interpretation comes from a screen to identify Wnt and FGF responsive genes in the developing tail bud of zebrafish.56 Similar to our approach, LiCl and SU5402 were used as tools to manipulate each pathway. Many genes examined showed similar responses to both GSK-3 inhibition and inhibition of FGF receptor function. Furthermore, it was shown that inhibition of GSK-3 by LiCl led to phosphorylation of ERK and therefore activation of ERK signaling. This was interpreted to suggest that Wnt/ bcat signaling was controlling FGF signaling through MAPK/ ERK activity. Based on our comparisons between BIO treatment relative to bcat overexpression, we would argue that GSK-3 has a bcat independent function in controlling MAPK/ERK activity.
Figure 10.
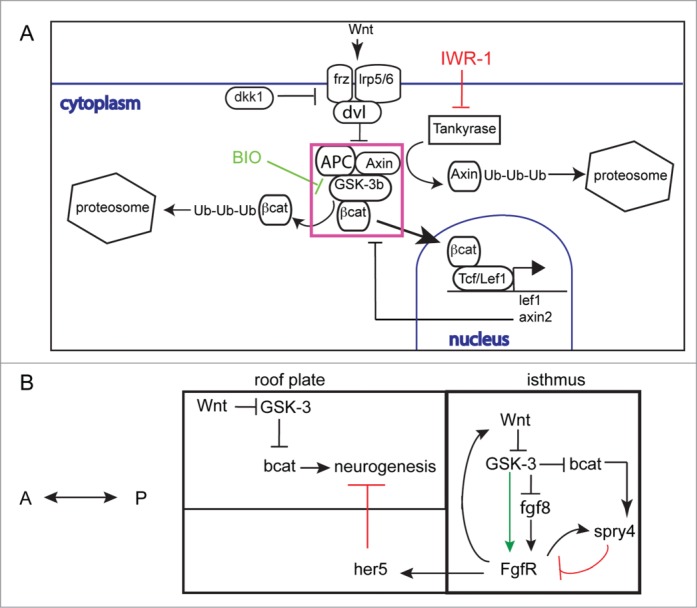
Schematic for interactions between Wnt/ bcat and FGF signaling during midbrain development (A). Secreted Wnt proteins bind frizzled (frz) and lrp5/6 cell membrane receptors complexed with dishevelled (dvl). This results in the inhibition of the ubiquitination (Ub) and subsequent degradation of bcat by the destruction complex (DC, purple box), incorporating Axin, APC and GSK-3 proteins. Free bcat concentration then rises and bcat can translocate to the nucleus to activate gene expression by binding TCF or LEF transcription factors. Inhibition of GSK-3 by BIO results in decreased bcat protein degradation. In contrast, inhibition of Tankyrase enzyme by IWR-1 prevents ubiquitination of Axin proteins and hence increased activity of the DC. A working model for Wnt-FGF interactions at the isthmus and in the midbrain during stages when MTN neurons form (B). Wnt and FGF signaling co-regulate each others activity in GSK-3 dependent and independent manners. Wnt inhibits GSK-3 activity leading to elevated bcat activity. Spry4 expression is increased in response to bcat and FGF activity (FgfR) and acts to inhibit FgfR receptors. GSK-3 is also required for FgfR activity and acts independently of bcat activity (green arrow). FGF signaling across the midbrain represses neurogenesis through activation of her5; as the her5 expression retracts posteriorly Wnt signaling acts to promote neuronal differentiation.
We found that manipulations of bcat activity altered the number of MTN neurons that formed in the midbrain. This could be due to a role of Wnt signaling in controlling cell proliferation. Alternatively, it may reflect a role of Wnts in controlling neurogenesis. Recent findings have shown that Wnt/ bcat signaling can regulate neurogenesis in the flatworm Platynereis and regulates expression of neurogenic genes Neurogenin1, Neurogenin2 and NeuroD in mouse.24,25,57 We have previously shown that MTN development in the midbrain is not regulated by neurogenin function in zebrafish suggesting that Wnt/ bcat are not directing neuronal differentiation through regulation of neurogenin genes.58 Intriguingly, the action of Wnt on neurogenesis in neural stem cells is influenced by the level of FGF activity.33 We note that in zebrafish, increasing bcat activity promotes the formation of more MTN neurons. When this is performed in the context of reduced FGF activity (by varying doses of SU5402) this effect is enhanced. This implies that in the midbrain neuronal differentiation is controlled by the action of both FGF and Wnt/ bcat signaling. When this is perturbed by enhancing or reducing bcat activity (by expressing dkk1), we observe commensurate changes to the number of neurons that form in the midbrain. We have previously shown that temporal application of BIO at stages when MTN neurons start to differentiate does not alter proliferation, but does cause an increase in MTN number.34 We suggest that this reveals a role of Wnt/ bcat signaling in controlling the process of neuronal differentiation in the midbrain.
The targeted inhibition of Wnt/ bcat activity is important for treatment of many diseases and syndromes that involve aberrant Wnt signaling.59 Tankyrase enzymes have been the focus for many such efforts, due to their important functions in degrading and so limiting Axin protein availability. The IWR and XAV939 compounds selectively inhibit Tnk, resulting in an increase in Axin protein and hence increased bcat degradation by the bcatDC.11,53 Initial reports describing the effect of IWR-1 dose on bcat activity using the SuperTopFlash assay showed that an approximate linear response occurs relative to dose. We find that in vivo, IWR-1 does not affect bcat activity in a linear manner. We note that Wnt/ bcat responsive genes lef1 and spry4 show a non-linear response to IWR-1 that changes relative to the duration of exposure. We also find that the concentration of IWR-1 alters the response over time. One intriguing observation we made is that low doses of IWR-1 initially cause a more severe inhibition of bcat responsive genes than a higher dose. Over time, higher concentrations of IWR-1 result in greater inhibition of bcat-dependent responses than lower concentrations. This change in response of bcat-responsive genes to different doses of IWR-1 suggests compensatory feedback loops modulate their response.
We have found that in the midbrain, FGF activity is required for expression of wnt1 and controls bcat activity. Likewise, bcat is able to upregulate expression of fgf8 and FGF target gene expression in the presence of SU5402. The interaction between Wnt and FGF signaling to maintain expression of their respective ligands in the isthmus is well established and occurs by a positive feedback loop.3,4 One putative explanation for the differential effects of IWR-1 dosage on Wnt-target genes may therefore be due to feedback from FGF signaling to promote elevated bcat signaling. In support of this, we note that higher doses of IWR-1 causes a rapid decrease in bcat activity and leads to increased wnt1 expression, similar to that seen by over-activation of FGF signaling. The varying response of bcat-dependent gene expression in relation to IWR-1 dose, suggests that FGF-dependent feedback loop induce an oscillatory response. Such oscillatory behavior is a prediction of models generated to explain robustness during Wnt/ bcat signaling.50,60 Our interpretation of the results we observe in vivo, is that high doses of IWR-1 promote an FGF-mediated feedback mechanism leading to an upregulation of wnt1 expression. Elevated levels of Wnt protein will promote bcatDC disassociation, until bcat-induced Axin2 levels rise sufficiently to overcome Wnt action on the bcatDC and bcat activity is then repressed. Our modification of a model for bcat activity indicates that when Axin degradation is low, the fold change is relatively insensitive to small changes in the degradation rate, but that it is far more sensitive to small changes at higher rates of degradation. This is result is analogous to previous analyses of bcat fold changes, which predicted sensitive and insensitive regions as a consequence of modifying the bcat degradation rate.50 It would thus be valuable to further determine how the concentration of free bcat respond to differing concentrations of IWR-1 concentration over time and how this corresponds to the relative concentrations of Axin1 and Axin2. Even more importantly, it will be necessary to show how bcat-activated genes respond to Tnk inhibitors in vivo, to understand how robustness in signaling pathway activity impacts on the biological outputs of drugs that aim to modulate Wnt/ bcat.
In summary, we show that components of the Wnt/ bcat pathway controls 2 aspects of midbrain development. GSK-3 acts to modulate FGF activity at the isthmus and thus controls the spatiotemporal differentiation of early neuronal populations, such as the MTN, across the midbrain. This function of GSK-3 appears to be independent to its role in regulating bcat, which acts to control the number of neurons that form. How these 2 functions of Wnt signaling are integrated is unclear. One hypothesis is that GSK-3 is required for normal activation of intracellular FGF targets, such as ERK. Thus, the interplay between Wnt/ bcat and FGF signaling occurs at a number of levels and involves a complex set of feedback loops to control their relative activity. Perturbations to any one component will lead to compensatory activity, inducing some degree of oscillatory response. In the presence of small molecular inhibitors, which do not respond dynamically to such feedback loops, oscillations will be more pronounced until they eventually reach a new equilibrium. As recently highlighted, this is relevant for understanding how cells in vivo may respond to drugs that target Wnt/ bcat activity.59 It is therefore crucial to target the most appropriate component of this pathway for effective inhibition of bcat activity with minimal interference to other important signaling pathways.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Lea Goentoro and Marc Kirschner for sharing their models for Wnt/ bcat signaling. Also, Karen Liu, Joao Peres, Holger Bielen, Corinne Houart, Geraint Thomas for constructive discussions and reagents.
References
- 1.Barkovich AJ, Millen KJ, Dobyns WB. A developmental and genetic classification for midbrain-hindbrain malformations. Brain 2009; 132:3199-230; PMID:19933510; http://dx.doi.org/ 10.1093/brain/awp247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raible F, Brand M. Divide et Impera–the midbrain-hindbrain boundary and its organizer. Trends Neurosci 2004; 27:727-34; PMID:15541513 [DOI] [PubMed] [Google Scholar]
- 3.Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci 2001; 2:99-108; PMID:11253000 [DOI] [PubMed] [Google Scholar]
- 4.Rhinn M, Brand M. The midbrain–hindbrain boundary organizer. Curr Opin Neurobiol 2001; 11:34-42; PMID:11179870 [DOI] [PubMed] [Google Scholar]
- 5.Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature 1996; 380:66-8; PMID:8598907 [DOI] [PubMed] [Google Scholar]
- 6.Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development 1999; 126:1189-200; PMID:10021338 [DOI] [PubMed] [Google Scholar]
- 7.Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 1998; 125:2381-95; PMID:9609821 [DOI] [PubMed] [Google Scholar]
- 8.Geling A, Itoh M, Tallafuss A, Chapouton P, Tannhauser B, Kuwada JY, Chitnis AB, Bally-Cuif L. bHLH transcription factor Her5 links patterning to regional inhibition of neurogenesis at the midbrain-hindbrain boundary. Development 2003; 130:1591-604; PMID:12620984 [DOI] [PubMed] [Google Scholar]
- 9.Scholpp S, Lohs C, Brand M. Engrailed and Fgf8 act synergistically to maintain the boundary between diencephalon and mesencephalon. Development 2003; 130:4881-93; PMID:12917294 [DOI] [PubMed] [Google Scholar]
- 10.Partanen J. FGF signalling pathways in development of the midbrain and anterior hindbrain. J Neurochem 2007; 101:1185-93; PMID:17326764 [DOI] [PubMed] [Google Scholar]
- 11.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al.. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009; 461:614-20; PMID:19759537; http://dx.doi.org/ 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- 12.Lahti L, Saarimaki-Vire J, Rita H, Partanen J. FGF signaling gradient maintains symmetrical proliferative divisions of midbrain neuronal progenitors. Dev Biol 2011; 349:270-82; PMID:21074523; http://dx.doi.org/ 10.1016/j.ydbio.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 13.Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell 1998; 93:755-66; PMID:9630220 [DOI] [PubMed] [Google Scholar]
- 14.Jaeger I, Arber C, Risner-Janiczek JR, Kuechler J, Pritzsche D, Chen IC, Naveenan T, Ungless MA, Li M. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development 2011; 138:4363-74; PMID:21880784; http://dx.doi.org/ 10.1242/dev.066746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosco A, Bureau C, Affaticati P, Gaspar P, Bally-Cuif L, Lillesaar C. Development of hypothalamic serotoninergic neurons requires Fgf signalling via the ETS-domain transcription factor Etv5b. Development 2013; 140:372-84; PMID:23250211; http://dx.doi.org/ 10.1242/dev.089094 [DOI] [PubMed] [Google Scholar]
- 16.Jaszai J, Reifers F, Picker A, Langenberg T, Brand M. Isthmus-to-midbrain transformation in the absence of midbrain-hindbrain organizer activity. Development 2003; 130:6611-23; PMID:14660549 [DOI] [PubMed] [Google Scholar]
- 17.Basson MA, Echevarria D, Ahn CP, Sudarov A, Joyner AL, Mason IJ, Martinez S, Martin GR. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development 2008; 135:889-98; PMID:18216176; http://dx.doi.org/ 10.1242/dev.011569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niehrs C, Acebron SP. Mitotic and mitogenic Wnt signalling. EMBO J 2012; 31:2705-13; PMID:22617425; http://dx.doi.org/ 10.1038/emboj.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paridaen JT, Danesin C, Elas AT, van de Water S, Houart C, Zivkovic D. Apc1 is required for maintenance of local brain organizers and dorsal midbrain survival. Dev Biol 2009; 331:101-12; PMID:19397905; http://dx.doi.org/ 10.1016/j.ydbio.2009.04.022 [DOI] [PubMed] [Google Scholar]
- 20.McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 1992; 69:581-95; PMID:1534034 [DOI] [PubMed] [Google Scholar]
- 21.Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 1990; 346:847-50; PMID:2202907 [DOI] [PubMed] [Google Scholar]
- 22.Rhinn M, Picker A, Brand M. Global and local mechanisms of forebrain and midbrain patterning. Curr Opin Neurobiol 2006; 16:5-12; PMID:16418000 [DOI] [PubMed] [Google Scholar]
- 23.Mulligan KA, Cheyette BN. Wnt signaling in vertebrate neural development and function. J Neuroimmune Pharmacol 2012; 7:774-87; PMID:23015196; http://dx.doi.org/ 10.1007/s11481-012-9404-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/β-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 2004; 131:2791-801; PMID:15142975 [DOI] [PubMed] [Google Scholar]
- 25.Qu Q, Sun G, Murai K, Ye P, Li W, Asuelime G, Cheung YT, Shi Y. Wnt7a regulates multiple steps of neurogenesis. Mol Cell Biol 2013; 33:2551-9; PMID:23629626; http://dx.doi.org/ 10.1128/MCB.00325-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Brown A, Ellisor D, Paul E, Hagan N, Zervas M. Dynamic temporal requirement of Wnt1 in midbrain dopamine neuron development. Development 2013; 140:1342-52; PMID:23444360; http://dx.doi.org/ 10.1242/dev.080630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castelo-Branco G, Rawal N, Arenas E. GSK-3beta inhibition/β-catenin stabilization in ventral midbrain precursors increases differentiation into dopamine neurons. J Cell Sci 2004; 117:5731-7; PMID:15522889 [DOI] [PubMed] [Google Scholar]
- 28.Joksimovic M, Yun BA, Kittappa R, Anderegg AM, Chang WW, Taketo MM, McKay RD, Awatramani RB. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat Neurosci 2009; 12:125-31; PMID:19122665; http://dx.doi.org/ 10.1038/nn.2243 [DOI] [PubMed] [Google Scholar]
- 29.Tang M, Miyamoto Y, Huang EJ. Multiple roles of β-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Development 2009; 136:2027-38; PMID:19439492; http://dx.doi.org/ 10.1242/dev.034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alves dos Santos MT, Smidt MP. En1 and Wnt signaling in midbrain dopaminergic neuronal development. Neural Dev 2011; 6:23; PMID:21569278; http://dx.doi.org/ 10.1186/1749-8104-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canning CA, Lee L, Irving C, Mason I, Jones CM. Sustained interactive Wnt and FGF signaling is required to maintain isthmic identity. Dev Biol 2007; 305:276-86; PMID:17383629 [DOI] [PubMed] [Google Scholar]
- 32.Wittmann DM, Blochl F, Trumbach D, Wurst W, Prakash N, Theis FJ. Spatial analysis of expression patterns predicts genetic interactions at the mid-hindbrain boundary. PLoS Comput Biol 2009; 5:e1000569; PMID:19936059; http://dx.doi.org/ 10.1371/journal.pcbi.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Israsena N, Hu M, Fu W, Kan L, Kessler JA. The presence of FGF2 signaling determines whether β-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Dev Biol 2004; 268:220-31; PMID:15031118 [DOI] [PubMed] [Google Scholar]
- 34.Dyer C, Blanc E, Hanisch A, Roehl H, Otto GW, Yu T, Basson MA, Knight R. A bi-modal function of Wnt signalling directs an FGF activity gradient to spatially regulate neuronal differentiation in the midbrain. Development 2014; 141:63-72; PMID:24284206; http://dx.doi.org/ 10.1242/dev.099507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geling A, Plessy C, Rastegar S, Strahle U, Bally-Cuif L. Her5 acts as a prepattern factor that blocks neurogenin1 and coe2 expression upstream of Notch to inhibit neurogenesis at the midbrain-hindbrain boundary. Development 2004; 131:1993-2006; PMID:15056616 [DOI] [PubMed] [Google Scholar]
- 36.Ninkovic J, Tallafuss A, Leucht C, Topczewski J, Tannhauser B, Solnica-Krezel L, Bally-Cuif L. Inhibition of neurogenesis at the zebrafish midbrain-hindbrain boundary by the combined and dose-dependent activity of a new hairy/E(spl) gene pair. Development 2005; 132:75-88; PMID:15590746 [DOI] [PubMed] [Google Scholar]
- 37.Tallafuss A, Bally-Cuif L. Tracing of her5 progeny in zebrafish transgenics reveals the dynamics of midbrain-hindbrain neurogenesis and maintenance. Development 2003; 130:4307-23; PMID:12900448 [DOI] [PubMed] [Google Scholar]
- 38.Westerfield M. The zebrafish book: A guide for laboratory use of the zebrafish Danio rerio. Eugene, Oregon: University of Oregon Press; 2007. [Google Scholar]
- 39.Heisenberg CP, Houart C, Take-Uchi M, Rauch GJ, Young N, Coutinho P, Masai I, Caneparo L, Concha ML, Geisler R, et al.. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes Dev 2001; 15:1427-34; PMID:11390362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, et al.. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol 2000; 227:279-93; PMID:11071755 [DOI] [PubMed] [Google Scholar]
- 41.Molina GA, Watkins SC, Tsang M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev Biol 2007; 7:62; PMID:17553162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/β-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol 2005; 15:489-500; PMID:15797017 [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez-Quevedo R, Lee Y, Poss KD, Wilkinson DG. Neuronal regulation of the spatial patterning of neurogenesis. Dev Cell 2010; 18:136-47; PMID:20152184; http://dx.doi.org/ 10.1016/j.devcel.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev 1999; 80:153-8; PMID:10072782 [DOI] [PubMed] [Google Scholar]
- 45.Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci 2000; 20:709-21; PMID:10632600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/β-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol 2002; 241:229-37; PMID:11784107 [DOI] [PubMed] [Google Scholar]
- 47.Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission 2004; ZFIN ID: ZDB-PUB-040907-1 [Google Scholar]
- 48.Nusslein-Volhard C, Dahm R. Zebrafish: A Practical Approach (The Practical Approach Series, 261). Oxford, England: Oxford University Press; 2002. [Google Scholar]
- 49.Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol 2003; 1:E10; PMID:14551908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goentoro L, Kirschner MW. Evidence that fold-change, and not absolute level, of β-catenin dictates Wnt signaling. Mol Cell 2009; 36:872-84; PMID:20005849; http://dx.doi.org/ 10.1016/j.molcel.2009.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hjorth JT, Key B. Are pioneer axons guided by regulatory gene expression domains in the zebrafish forebrain? High-resolution analysis of the patterning of the zebrafish brain during axon tract formation. Dev Biol 2001; 229:271-86; PMID:11203695 [DOI] [PubMed] [Google Scholar]
- 52.Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, Bournele D, Domenichini A, Valdivia LE, Lum L, et al.. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol 2012; 366:327-40; PMID:22546689; http://dx.doi.org/ 10.1016/j.ydbio.2012.03.023 [DOI] [PubMed] [Google Scholar]
- 53.Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al.. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 2009; 5:100-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kofahl B, Wolf J. Mathematical modelling of Wnt/β-catenin signalling. Biochem Soc Trans 2010; 38:1281-5; PMID:20863299; http://dx.doi.org/ 10.1042/BST0381281 [DOI] [PubMed] [Google Scholar]
- 55.Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Molecular cell 2009; 36:894-9.; PMID:20005851; http://dx.doi.org/ 10.1016/j.molcel.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stulberg MJ, Lin A, Zhao H, Holley SA. Crosstalk between Fgf and Wnt signaling in the zebrafish tailbud. Dev Biol 2012; 369:298-307; PMID:22796649; http://dx.doi.org/ 10.1016/j.ydbio.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demilly A, Steinmetz P, Gazave E, Marchand L, Vervoort M. Involvement of the Wnt/β-catenin pathway in neurectoderm architecture in Platynereis dumerilii. Nat Commun 2013; 4:1915; PMID:23715274; http://dx.doi.org/ 10.1038/ncomms2915 [DOI] [PubMed] [Google Scholar]
- 58.Dyer C, Linker C, Graham A, Knight R. Specification of sensory neurons occurs through diverse developmental programs functioning in the brain and spinal cord. Dev Dyn 2014; 243:1429-39; PMID:25179866; http://dx.doi.org/ 10.1002/dvdy.24184 [DOI] [PubMed] [Google Scholar]
- 59.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov 2014; 13:513-32; PMID:24981364; http://dx.doi.org/ 10.1038/nrd4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wawra C, Kuhl M, Kestler HA. Extended analyses of the Wnt/β-catenin pathway: robustness and oscillatory behaviour. FEBS Lett 2007; 581:4043-8; PMID:17678900 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.