Summary
Microbisporicin is a potent type I lantibiotic produced by the rare actinomycete M icrobispora corallina that is in preclinical trials for the treatment of infections caused by methicillin‐resistant isolates of S taphylococcus aureus (MRSA). Analysis of the gene cluster for the biosynthesis of microbisporicin, which contains two unique post‐translationally modified residues (5‐chlorotryptophan and 3, 4‐dihydroxyproline), has revealed an unusual regulatory mechanism that involves a pathway‐specific extracytoplasmic function sigma factor (MibX)/anti‐sigma factor (MibW) complex and an additional transcriptional regulator MibR. A model for the regulation of microbisporicin biosynthesis derived from transcriptional, mutational and quantitative reverse transcription polymerase chain reaction analyses suggests that MibR, which contains a C‐terminal DNA‐binding domain found in the LuxR family of transcriptional activators, functions as an essential master regulator to trigger microbisporicin production while MibX and MibW induce feed‐forward biosynthesis and producer immunity. Moreover, we demonstrate that initial expression of mib R, and thus microbisporicin production, is dependent on the ppGpp synthetase gene (relA) of M . corallina. In addition, we show that constitutive expression of either of the two positively acting regulatory genes, mib R or mib X, leads to precocious and enhanced microbisporicin production.
Introduction
Lantibiotics are ribosomally synthesised, post‐translationally modified peptides with antimicrobial activity that are produced by a number of Gram‐positive bacteria (Li and O'Sullivan, 2012). They contain lanthionine and/or methyl lanthionine bridges, which contribute to their relative resistance to proteolytic cleavage, structural rigidity and target specificity (Chatterjee et al., 2005). Only a few lantibiotic biosynthetic gene clusters derived from actinobacteria have been characterised thus far – all of them chromosomally located (Li and O'Sullivan, 2012). These clusters typically contain genes encoding the precursor peptide, enzymes responsible for a variety of posttranslational modifications, proteins involved in export and immunity, and frequently pathway‐specific regulatory proteins (Chatterjee et al., 2005; Arnison et al., 2013). Microbisporicin is a potent lantibiotic produced by Microbispora corallina (Nakajima et al., 1999); it is also made by Microbispora sp. American Type Culture Collection (ATCC) PTA5024 and known commercially as NAI‐107 (Donadio et al., 2009; Jabés et al., 2011). It is active against a wide range of Gram‐positive pathogens, including multiply drug resistant Staphylococcus aureus strains. Microbisporicin inhibits cell wall biosynthesis by binding to lipid II, the immediate precursor for peptidoglycan biosynthesis (Lazzarini et al., 2005; Münch et al., 2014). The lantibiotic contains one methyllanthionine and three lanthionine bridges, a S‐[(Z)‐2‐aminovinyl]‐D‐cysteine at its C terminus and two unique modifications, 5‐chlorotryptophan and 3, 4‐dihydroxyproline (Lazzarini et al., 2005; Castiglione et al., 2008). The absence of these latter two modifications markedly reduces the potency of the compound (Maffioli et al., 2014). Previous studies identified a cluster of 20 genes involved in microbisporicin biosynthesis, which to our knowledge, is the largest lantibiotic gene cluster identified thus far (Foulston and Bibb, 2010). It encodes a putative transcriptional activator, MibR, as well as an extracytoplasmic function (ECF) sigma factor and anti‐sigma factor pair, MibX and MibW, respectively, all involved in the regulation of microbisporicin biosynthesis. Foulston and Bibb (2011) suggested a model in which MibR acts as a master regulator to promote low levels of production of the immature, less active form of microbisporicin that lacks the unique modifications referred to earlier; this intermediate then triggers a feed‐forward mechanism mediated by the ECF‐sigma factor MibX that results in high levels of microbisporicin production. However, the direct targets of MibR and MibX remained to be confirmed experimentally, as did the signal triggering the initial activation of MibR transcription.
Antibiotic production in actinomycetes is triggered frequently by nutrient limitation (Bibb, 2005; Martín and Liras, 2012) presumably affording a selective advantage to the producing organism under starvation conditions. Guanosine tetraphosphate (ppGpp) is a key intracellular signalling molecule for sensing nutrient starvation and triggering adaptive responses in a wide range of bacteria. ppGpp induces a rapid response to amino acid starvation in Escherichia coli, Streptomyces species and other bacteria, reducing the expression of genes involved in rapid growth and often activating transcription of genes involved in specialised metabolism (Takano and Bibb, 1994; Bremer and Ehrenberg, 1995; Ochi, 2007; Gaca et al., 2015). Under conditions of nitrogen limitation, the ribosome‐bound RelA synthesises ppGpp in response to uncharged tRNAs that bind to the ribosomal A site (Cashel et al., 1996). In E. coli, ppGpp elicits transcriptional changes by interacting directly with RNA polymerase (Magnusson et al., 2005) while in Bacillus subtilis, ppGpp regulation of gene expression appears to be mediated through GTP pool levels, therefore modulating promoter activity indirectly (Krásný and Gourse, 2004). In Streptomyces coelicolor, ppGpp synthesis was shown to be required for antibiotic production under conditions of nitrogen limitation (Chakraburtty and Bibb, 1997); moreover, induction of ppGpp synthesis at levels that did not influence growth rate and under conditions of nutritional sufficiency invoked the transcription of the pathway‐specific regulatory gene actII‐orf4 and actinorhodin production (Hesketh et al., 2001).
In this study, we demonstrate the role of RelA, and presumably ppGpp synthesis, in the activation of microbisporicin biosynthesis by initially triggering the production of a precursor that subsequently induces high levels of production of the mature antibiotic. We also show that the lantibiotic can act as an extracellular signalling molecule to trigger microbisporicin production in the wider M. corallina community. We identify the individual targets of MibX and MibR, firmly establishing the complex regulatory cascade that leads to microbisporicin biosynthesis. Finally, we identify an ABC transporter that appears to confer some level of immunity to microbisporicin, and that is also required for production of the lantibiotic.
Results
gusA transcriptional fusions in S . coelicolor M1152 verify the targets of MibX and MibR
Microbisporicin production occurs in a growth phase‐dependent manner, commencing towards the end of rapid growth (Foulston, 2010). The microbisporicin biosynthetic gene cluster consists of six operons (Fig. 1A). Previous studies (Foulston and Bibb, 2011) identified an ECF‐sigma factor consensus sequence (GAACC‐N15‐GCTAC) located 8–10 nucleotides upstream of the transcriptional start sites of mibJ, mibQ, mibR, mibX and mibE, suggesting that the transcription of these genes and operons is activated directly by MibX. In contrast, the promoter region of the crucial mibABCDTUV biosynthetic operon lacks this sequence, but contains instead the motif TTGACA‐N17‐TCGACT that is likely to be recognised by the RNA polymerase holoenzyme containing the major vegetative sigma factor of M. corallina [the homologue of sigma hrdB of S. coelicolor (Brown et al., 1992; Foulston and Bibb, 2011)]. MibR, which is essential for microbisporicin production, could thus be a candidate for activating the transcription of the mibA operon in a growth phase‐dependent manner. To evaluate these bioinformatic predictions, the mibJ (299bp), mibQ (166 bp), mibR (384 bp), mibX (254 bp), mibA (244 bp) and mibE (440 bp) promoter regions were cloned separately upstream of a Streptomyces codon‐optimised β‐glucuronidase gene, gusA, in pGUS (Myronovskyi et al., 2011), and the resulting plasmids (Table 1) integrated into the ΦC31 attB sites of S. coelicolor M1152 derivatives M1598, M1594 and M1595; the last two strains carried derivatives of pIJ10257 (which integrates at the ΦBT1 attB site) in which mibR or mibX, respectively, were constitutively expressed from ermE*p, while M1598 contained just the vector pIJ10257 (Table 1).
Figure 1.

A. The microbisporicin biosynthetic gene cluster (Foulston and Bibb, 2010). Transcriptional start sites are marked by arrows and those promoter regions containing the predicted ECF‐sigma factor consensus sequence are indicated by stars; the loop downstream of mib A indicates a putative attenuator. B. β‐glucuronidase activity assays of mib promoter regions fused to gus A in S treptomyces coelicolor M1152 and derivatives containing constitutively expressed mib R (R++) or mibX (X++) were carried out after 72 h of growth. β‐Glucuronidase activity is expressed as Miller units mg−1 of protein. The filled stars indicate promoters regulated by MibX and the open star indicates the promoter regulated by MibR.
Table 1.
Plasmids used and constructed in this study
| Vector | Description | Reference |
|---|---|---|
| pUZ8002 | tra, neo, RP4 | J. Wilson and D. Figurski, personal communication |
| pIJ8600 | oriT, ΦC31 attB‐int, APRR, tipAp | Takano et al., 1995 |
| pIJ12572 | pIJ8600 + mibR | This work |
| pIJ10257 | oriT, ΦBT1 attB‐int, HYGR, ermE*p | Hong et al., 2005 |
| pIJ12574 | pIJ10257 + mibX | This work |
| pIJ12576 | pIJ10257 + mibR | This work |
| pIJ12743 | pIJ10257 + mibEF | This work |
| pIJ12750 | pIJ10257 + I‐SceI | This work |
| pIJ12738 | pKC1132 with MCS and I‐SceI site from pUC57‐Simple_SceI | Fernández‐Martínez and Bibb, 2014 |
| pGUS | oriT, ΦC31 attB‐int, APRR, promoterless codon‐optimised gusA | Myronovskyi et al., 2011 |
| pIJ12579 | pGUS + mibA promoter region | This work |
| pIJ12580 | pGUS + mibE promoter region | This work |
| pIJ12581 | pGUS + mibJ promoter region | This work |
| pIJ12582 | pGUS + mibQ promoter region | This work |
| pIJ12583 | pGUS + mibR whole (including p1 and p2) promoter region | This work |
| pIJ12584 | pGUS + mibX promoter region | This work |
| pIJ12586 | pGUS + mibRp2 promoter region | This work |
APR, apramycin; HYG, hygromycin B.
Only very low levels of gusA expression were detected from the six mib promoters in the absence of mibR and mibX (Fig. 1B). However, constitutive expression of mibX resulted in marked induction of GusA activity from the mibJ, mibQ, mibR, mibX and mibE promoters, all of which contain the ECF‐sigma factor consensus sequence, but not from the mibA promoter fusion in which the ECF‐sigma factor motif is absent (Fig. 1B, induction indicated by black stars). In contrast, induction of the mibA promoter was only detected in the strain containing the constitutively expressed mibR (Fig. 1B, indicated by the white star). Thus, in S. coelicolor, MibR activates transcription of the mibA operon whereas MibX activates transcription of the other five genes and operons in the microbisporicin biosynthetic cluster.
Analysis of the mibR promoter region reveals a second promoter independent of MibX
The studies described earlier demonstrated that the ECF‐like mibR promoter (mibRp1) is indeed dependent on the ECF‐sigma factor MibX for its activation. However, earlier S1 nuclease protection analyses had suggested that there was a second mibR promoter located further upstream (Foulston and Bibb, 2011) that might also be involved in the activating mibR expression. In an attempt to further characterise this putative promoter, reverse transcription polymerase chain reaction (RT‐PCR) experiments were carried out using RNA extracted from M. corallina 16, 24, 40 and 48 h after inoculation of production medium using a series of paired and nested oligonucleotide primers covering the sequence upstream of mibRp1 (Table S1). This revealed the presence of a second putative promoter element, mibRp2, located 449 nt–261 nt upstream of the mibR coding sequence that was active 24 h after inoculation, well before transcriptional read‐through from mibQ and microbisporicin production was observed (Fig. 2). The putative transcriptional start site of mibRp2 was determined using 5′ extension RACE (Fig. S1) and shown to lie 439–440 nt upstream of the mibR GTG start codon, inside the mibQ coding sequence. Inspection of the nucleotide sequence preceding this putative transcriptional start site failed to reveal any striking similarity to known promoter sequences, including the canonical ECF‐sigma factor recognition motif and consequently it would not be predicted to be activated by MibX.
Figure 2.
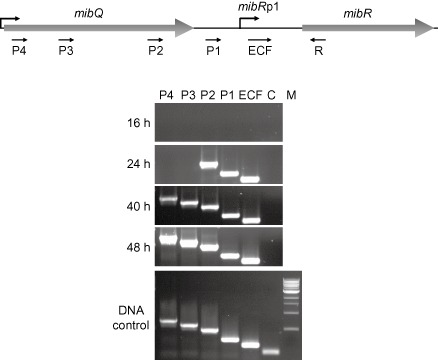
Top: Schematic representation of mib Q, mib R and their intergenic region. Bottom: Forward oligonucleotides P4, P3, P2, P1 and ECF were used in pairwise combinations with reverse oligonucleotide R in RT‐PCR reactions to identify the approximate location of the promoter upstream of mib Rp1. RNA was isolated from M icrobispora corallina grown in VSPA liquid medium at the time points indicated. Genomic DNA was used as a control to confirm the authenticity of each oligonucleotide pair. C represents amplification from an internal mib R fragment used as a negative control to verify the absence of DNA from the RNA samples. M, the 1Kb NEB ladder (NEB).
Transcriptional activation of mibRp2 is relA‐dependent
Previous studies had implicated ppGpp, produced by the ribosome‐bound ppGpp synthetase (RelA), as an intracellular signalling molecule for the initiation of antibiotic production in actinomycetes (Takano and Bibb, 1994; Martínez‐Costa et al., 1998; Ochi, 2007). Moreover, deletion of relA in S. coelicolor abolished the production of both actinorhodin and the undecylprodiginine complex of compounds under conditions of nitrogen limitation (Chakraburtty and Bibb, 1997), while induction of ppGpp synthesis at levels that did not impair growth activated actinorhodin gene transcription (Hesketh et al., 2001). To determine whether transcription from mibRp2 was dependent on ppGpp, a 478 bp fragment containing mibRp2 (corresponding to nt sequence 5245–5723 of GenBank accession HM536998.1) was cloned in pGUS generating pIJ12586, which was then introduced into the ΦC31 attB sites of S. coelicolor M145 (relA +) and S. coelicolor M571 (ΔrelA), and GusA activity assayed throughout growth in nitrogen‐limited Supplemented liquid Minimal Medium (SMM) (Fig. 3A). The results demonstrate that transcription from mibRp2 in S. coelicolor under these growth conditions is RelA‐ (and presumably ppGpp) dependent.
Figure 3.
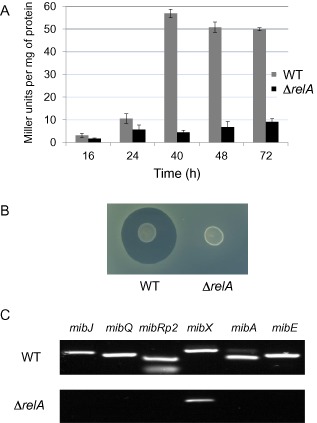
A. mib Rp2‐derived β‐glucuronidase activity in S treptomyces coelicolor M145 and M571 (ΔrelA). β‐Glucuronidase activity is given in Miller units mg−1 of protein. B. Anti‐microbial activity of the M icrobispora corallina rel A mutant compared with the wild‐type strain (WT). The strains were spotted on SMMS agar medium and incubated at 30°C for 10 days. The plate was then overlaid with a lawn of M icrococcus luteus and incubated overnight at 30°C before zones of inhibition were visualised. C. RT‐PCR analysis of gene expression in all six of the mib operons in the wild‐type and relA mutant strains of M . corallina. The individual genes chosen for analysis are given; mib Rp2 indicates that the amplified region corresponds to sequences present in the transcript initiating at mib Rp2 that are not present in the mib Rp1 transcript.
Deletion of relA in M . corallina abolishes microbisporicin production
To determine whether microbisporicin production was RelA‐dependent in the natural producer, we adopted the Meganuclease strategy (Fernández‐Martínez and Bibb, 2014) to construct a markerless relA deletion mutant of M. corallina (see Experimental Procedures). M. corallina is a difficult strain to work with; it grows slowly, taking 2–3 weeks to obtain workable colonies and it sporulates poorly, necessitating the use of mycelial fragments for most genetic manipulations. Consequently, the availability of the Meganuclease system that allows the selection of second crossover events during gene deletion by homologous recombination is extremely valuable. The resulting relA mutant, M1596, was grown adjacent to the wild‐type strain on nitrogen‐limited Supplemented liquid Minimal Medium Solid (SMMS) agar (Chakraburtty et al., 1996). After 10 days of incubation, the strains were overlaid with the sensitive indicator strain Micrococcus luteus (Fig. 3B). While wild‐type M. corallina showed a clear zone of inhibition, the relA mutant was devoid of antibiotic activity. To assess the effect of deletion of relA on transcription of the mib cluster, RT‐PCR experiments were carried out using RNA extracted from M1596 and from the wild‐type strain 48 h after inoculation in SMM liquid medium using a series of oligonucleotide primers (Foulston and Bibb, 2010) covering regions of each of the six mib cluster operons (Fig. 1A). Deletion of relA resulted in no detectable transcription from the mibRp2 promoter nor of any of the other mib operons except for mibXW, where transcription appeared to be reduced, but not abolished.
A model for the regulation of microbisporicin production in M . corallina
Based on the results presented so far, we propose an update to our previous model (Foulston and Bibb, 2011) for the regulation of microbisporicin production that explains its growth phase‐dependence (Fig. 4). During growth under conditions of nutrient sufficiency, the system is poised for activation with MibX, produced from a basal level of expression, sequestered at the membrane by its cognate anti‐sigma factor MibW (mibXW are co‐transcribed) (Foulston and Bibb, 2010; 2011). Under conditions of nitrogen limitation, uncharged tRNAs bind to the ribosomal A‐site, activating the ribosome‐bound RelA resulting in ppGpp synthesis. This then leads to a low level of expression of mibR from the relA‐dependent mibRp2 promoter, and consequently, a low level of transcription of the mibABCDTUV operon and the production of a small amount of the immature and less active form of the lantibiotic lacking the chlorination of tryptophan at position 4, and the di‐hydroxylation of proline at position 14 (Lazzarini et al., 2005) This less active form of the lantibiotic is exported out of the cell by the ABC transporter MibTU (Foulston and Bibb, 2010) where it could interact either with MibW or its target lipid II resulting in cell envelope stress potentially sensed by MibW. In either case, MibX is released from MibW leading to transcription of all of the MibX‐dependent genes and operons, which include mibR, resulting in high levels of mibR expression and consequently, high‐level expression of the mibABCDTUV operon and production of the mature antibiotic. Thus microbisporicin production, at least under conditions on nitrogen limitation, is triggered by nutrient limitation sensed by RelA; the likely subsequent ppGpp synthesis then leads to a feed‐forward regulatory mechanism that results in high levels of lantibiotic biosynthesis.
Figure 4.
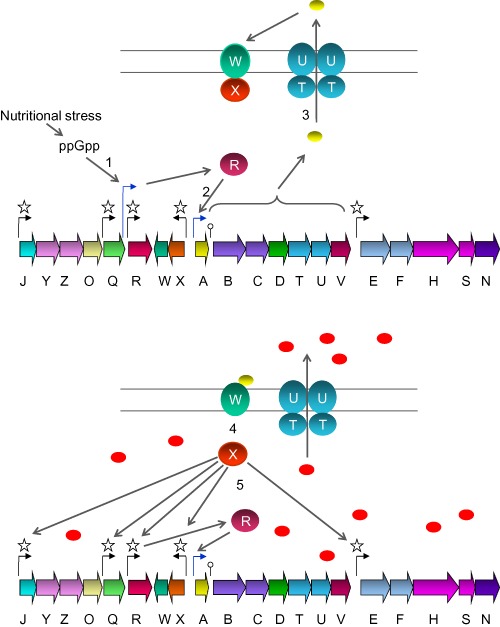
Model for the regulation of microbisporicin biosynthesis. Top: Prior to detectable microbisporicin production, MibW sequesters MibX at the membrane, preventing its interaction with target promoters. ppGpp, induced by nutrient limitation, activates transcription of mib Rp2 (1). MibR then activates transcription of the mib ABCDTUV operon (2) leading to production of the immature, less active form of microbisporicin (yellow ovals) which is the exported by MibTU (3). Bottom: Interaction of the peptide with MibW, or a low level of inhibition of peptidoglycan biosynthesis that may be perceived by the anti‐sigma factor, results in the release of MibX (4), and high‐level expression of the entire mib gene cluster (5) resulting in the formation of the fully modified and active form of microbisporicin (red ovals).
Constitutive expression of mibR or mibX triggers early and enhanced production of microbisporicin
The data and model presented thus far indicate that both MibR and MibX function as transcriptional activators to trigger microbisporicin biosynthesis. To assess the effect of constitutive expression of mibX and mibR on microbisporicin production, pIJ12572, containing mibR under the control of the thiostrepton‐inducible tipA promoter and pIJ12574, containing mibX under the control of the constitutive ermE* promoter, were integrated into the ΦC31 and ΦBT1 attB sites of M. corallina, respectively. Constitutive expression of mibR (the tipA promoter exhibits basal levels of expression in the absence of thiostrepton; Murakami et al., 1989; Ali et al., 2002) resulted in precocious and increased levels of microbisporicin production, which were further enhanced in the presence of the inducer (Fig. 5A; confirmed by Matrix‐Assisted Laser Desorption/Ionisation‐Time of Flight (MALDI‐ToF) analysis, data not shown). Constitutive expression of mibX also caused precocious microbisporicin production and at much higher levels than in the wild‐type strain (Fig. 5B). Simultaneous constitutive expression of both mibR and mibX resulted in even higher levels of microbisporicin biosynthesis (Fig. 5B). These results confirm that both MibR and MibX function as positively acting regulators of microbisporicin biosynthesis in M. corallina.
Figure 5.

A. Antimicrobial activity of M1592 (M icrobispora corallina containing mib R expressed from tip Ap) compared with the wild‐type strain (WT). The strains were grown in VSPA at 30°C and culture supernatants sampled at different time points. M1592 was also grown at two different concentrations of thiostrepton (inducer of tip Ap). Forty microlitre of samples of culture supernatants were applied to filter paper discs, which were dried and placed on a lawn of M icrococcus luteus. The plate was incubated overnight at 30°C before zones of inhibition were visualised. B. Antimicrobial activity of M . corallina constitutively expressing mibR (M1592), mibX (M1593) and both mibR and mibX simultaneously (M1597) compared with the wild‐type strain (WT). Strains were grown in VSPA liquid medium at 30°C and 40 μl of samples of culture supernatants were applied to filter paper discs which were dried and placed on a lawn of M icrococcus luteus. The plate was incubated overnight at 30°C before zones of inhibition were visualised.
Microbisporicin acts as a signalling molecule in M . corallina to induce its own production
The model presented earlier for the regulation of microbisporicin production is essentially an example of auto‐induction, where the production of a small amount of the immature form of the lantibiotic functions in a feed‐forward mechanism initiated by nutrient limitation and ppGpp synthesis to subsequently trigger high levels of production. But in principle, once transported out of the cell, both forms of the lantibiotic could interact with other members of the M. corallina community that are not nutrient limited to coordinate microbisporicin production, perhaps in an attempt to achieve ecologically relevant levels of antibiotic activity. To address this possibility, M. corallina M1592 (the mibR over‐expression strain), which produces microbisporicin precociously, was spotted on V0.1 agar (Marcone et al., 2010) plates in close proximity to streaks of the wild‐type M. corallina strain and the ΔmibA non‐producing mutant M1127 (Foulston and Bibb, 2010). After 5 days of incubation, when the wild‐type strain had not commenced microbisporicin biosynthesis (usually detected from 6 days onwards), the plates were overlaid with soft nutrient agar containing the indicator strain Micrococcus luteus (Fig. 6 left plate). The inverted pear‐shaped zone of inhibition revealed induction of microbisporicin biosynthesis in the wild‐type mycelia closest to M1592 while the inhibition zone next to the non‐producer control strain remained circular. To confirm that this induction of production was due to microbisporicin, 2.5 μg of the lantibiotic were spotted onto an antibiotic assay disc next to streaks of wild‐type M. corallina and the ΔmibA non‐producing mutant. Again, the inverted pear‐shaped inhibition zone indicated autoinduction of microbisporicin production (Fig. 6, right plate) in the wild‐type strain, but not in the mibA mutant. A range of other antibiotics was also tested for their ability to induce microbisporicin production, including the cell wall biosynthesis inhibitors vancomycin, planosporicin, fosfomycin, bacitracin, carbenicillin and tunicamycin and the protein synthesis inhibitor apramycin (data not shown). A range of concentrations were used, but none induced precocious microbisporicin production even at concentrations at which some of the antibiotics inhibited growth of M. corallina. These results demonstrate that microbisporicin can induce its own synthesis at subinhibitory concentrations and in a highly specific manner.
Figure 6.
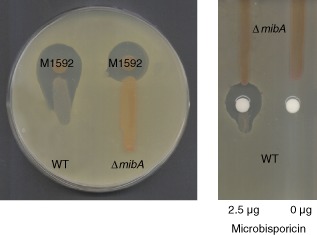
Microbisporicin induces its own production. Left plate: M icrobispora corallina M1592 constitutively expressing mibR and precociously producing the lantibiotic was spotted next to streaks of (left) the wild‐type (WT) strain and (right) a non‐producing Δmib A mutant. Right plate: Addition of purified microbisporicin to a filter paper disc triggers precocious microbisporicin biosynthesis in the M . corallina wild‐type strain, but not in the Δmib A mutant. The strains were grown on MV01 agar medium at 30°C for 5 days and then overlaid with a lawn of M icrococcus luteus in soft nutrient agar. The plates were incubated overnight at 30°C before zones of inhibition were visualised. The inverted pear‐shaped inhibition zones indicate precocious induction of microbisporicin production in the mycelium located proximal to the source of the compound.
MibEF are likely to be involved in immunity to microbisporicin in M . corallina
Previous bioinformatic analysis of the mib gene cluster identified three pairs of genes encoding ABC transporters: mibTU, mibEF and mibYZ. While deletion of mibTU had no apparent effect on microbisporicin production, the latter was essentially abolished in a mibEF mutant (Foulston and Bibb, 2010). To assess the possible role of MibEF in immunity, wild‐type M. corallina and the ΔmibEF mutant (M1131) were grown on V0.1 agar containing increasing concentrations of microbisporicin (Fig. 7A). The wild‐type strain grew well on 0.75 μg ml−1 of microbisporicin, whereas the ΔmibEF mutant failed to grow on 0.025 μg ml−1, exhibiting at least 30‐fold greater sensitivity towards the lantibiotic, suggesting a role for MibEF in immunity. Consistent with this, expression of mibEF from the constitutive ermE* promoter in pIJ12743 in S. coelicolor M145 led to a twofold increase in microbisporicin resistance (Fig. 7B) when compared with M145 containing the empty vector pIJ10257. Interestingly, homologues of MibZY, the transmembrane protein MibJ, and MibQ have also been implicated in immunity to microbisporicin in Microbispora sp. ATCC PTA5024 (Stegmann et al., 2014).
Figure 7.
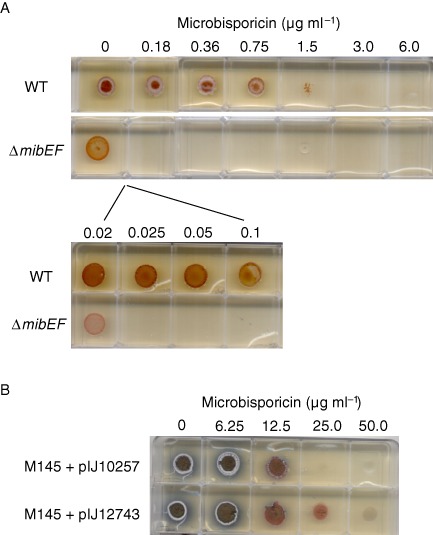
A. Wild‐type M icrobispora corallina (WT) and the mib EF mutant (Δmib EF) were grown on MV01 agar medium containing increasing concentrations of microbisporicin. The wild‐type strain was able to grow readily at 0.75 μg ml−1 microbisporicin while the Δmib EF mutant failed to grow at 0.025 μg ml−1 indicating an approximate 30‐fold increase in sensitivity to the compound. B. S treptomyces coelicolor M145 containing pIJ10257 (empty vector) or pIJ12743 (with mib EF expressed constitutively from erm E*p) were grown on R5 agar medium containing increasing concentrations of microbisporicin. Expression of mib EF resulted in an approximate twofold reduction in sensitivity to microbisporicin.
Discussion
In this study, we demonstrate that RelA, and presumably ppGpp synthesis, can activate the complex regulatory pathway that leads to the biosynthesis of microbisporicin, a potent lantibiotic currently undergoing preclinical trials. Our model suggests that an immature and less active form of microbisporicin serves as the initial extracellular signalling molecule to coordinate production throughout the M. corallina population. This may reflect a mechanism adopted by the organism to protect itself and its siblings from the highly potent mature lantibiotic, ensuring expression of mibTU before committing to the production of microbisporicin. A similar feed‐forward regulatory mechanism has been proposed for the lantibiotic planosporicin (Sherwood and Bibb, 2013). Interestingly, in this case, planosporicin, which is considerably less active than microbisporicin (Sherwood and Bibb, 2013), serves as the specific signalling molecule.
Auto‐induction of antibiotic biosynthesis has also been observed in low‐GC Gram‐positive lantibiotic producing bacteria; examples include nisin (Kleerebezem, 2004), subtilin (Stein et al., 2002) and mersacidin (Schmitz et al., 2006), where at least some of the compounds have been suggested to function as quorum sensors that monitor population size and density (Stein et al., 2002; Kleerebezem, 2004). In contrast, in M. corallina and potentially other actinomycetes, auto‐induction is triggered by nutrient limitation which we propose serves to coordinate production of the antibiotic throughout the mycelial population, not all of which may be experiencing starvation, with the aim of achieving ecologically effective levels of the antibiotic.
We have shown that deletion of mibEF in M. corallina results in increased sensitivity to microbisporicin. Furthermore, heterologous over‐expression of mibEF in S. coelicolor resulted in an increase in resistance to the lantibiotic, suggesting that MibEF play a role in conferring immunity to microbisporicin in the natural producer. In previous work, deletion of mibEF essentially abolished microbisporicin production (Foulston and Bibb, 2010). The closest homologues of MibEF (with the exception of transporters encoded by closely related actinobacterial lantibiotic gene clusters, such as that for planosporicin; Sherwood and Bibb, 2013) are the immunity ABC transporters found in low‐GC Gram‐positive lantibiotic producers. This raises the intriguing possibility of the existence, at least for microbisporicin, of a fail‐safe mechanism that ensures that the production of a potent antibiotic does not take place unless the corresponding immunity system is in place. How this potential mechanism influences the expression of mibR and/or mibX and hence, microbisporicin biosynthesis remains to be determined.
Preclinical trials suggest that microbisporicin is a promising candidate for clinical development, which will require the provision of significant amounts of the purified compound. In addition to deciphering the complex regulatory mechanism that triggers microbisporicin production, we have demonstrated, by constitutively expressing the genes encoding the two transcriptional activators present in the mib gene cluster, how that knowledge can be used to markedly increase the productivity of the wild‐type strain, hopefully contributing to the future clinical use of this compound.
The experiments reported here on M. corallina were also carried out on the commercial producer of microbisporicin (NAI‐107) Microbispora sp. ATCC PTA5024 with essentially the same results. For continuity with previous work (Foulston and Bibb, 2010; 2011), only the M. corallina results are reported here.
Experimental procedures
Strains and general methods
The strains used and generated in this study are listed in Table 2. E. coli strains were grown and manipulated following standard methods (Sambrook et al., 1989; Gust et al., 2003). For conjugation, M. corallina NRRL 30420 spores were germinated for 21 h in 10 ml Difco Nutrient Broth (DNB, Becton Dickinson, Sparks, Maryland, USA), resuspended in 500 μl DNB, mixed with E. coli S17 (Simon et al., 1983) carrying the relevant conjugative and integrative vector and plated on Soya Flour Mannitol (SFM) agar containing 10mM MgCl2. After growth at 30°C for 20 h, the plates were overlaid with 100 μl of fosfomycin (100 mg ml−1) and the appropriate concentration of the antibiotic used to select for the vector. Plates were grown at 30°C until putative exconjugants were visible (3–5 weeks). Microbisporicin was detected as described previously (Foulston and Bibb, 2010). S. coelicolor strains were grown and manipulated as described previously (Kieser et al., 2000). Plasmids and oligonucleotides are described in Tables 1 and S1, respectively.
Table 2.
Strains used and constructed in this study
| Strain | Genotype | Reference |
|---|---|---|
| Escherichia coli DH5α | F− φ80 lacZΔM15 Δ(lacZYA‐argF)U169 recA1 endA1 hsdR17 (rk −, mk +) phoA supE44 thi‐1 gyrA96 relA1 λ−) | Invitrogen™ |
| E. coli ET12567 | dam‐13:: Tn9 dcm‐6 hsdM CHLR, carrying helper plasmid pUZ8002 | MacNeil et al., 1992 |
| E. coli S17 | E. coli strain carrying an integrated RP4 derivative | Simon et al., 1983 |
| Microbispora corallina NRRL 30420 | M. corallina wild‐type strain | Nakajima et al., 1999 |
| M. corallina M1592 | M. corallina + pIJ12572 | This work |
| M. corallina M1593 | M. corallina + pIJ12574 | This work |
| M. corallina M1597 | M. corallina + pIJ12572 + pIJ12574 | This work |
| M. corallina M1596 | M. corallina ΔrelA | This work |
| M. corallina M1127 | M.corallina ΔmibA::aac(3)IV | Foulston and Bibb, 2010 |
| M. corallina M1131 | M.corallina ΔmibEF::aac(3)IV | Foulston and Bibb, 2010 |
| Micrococcus luteus ATCC 4698 | Bioassay indicator microorganism | ATCC |
| Streptomyces coelicolor M145 | S. coelicolor A3(2) plasmid‐free derivative | Kieser et al., 2000 |
| S. coelicolor M145 + pIJ12743 | M145 + pIJ12743 | This work |
| S. coelicolor M145 + pIJ12586 | M145 + pIJ12586 | This work |
| S. coelicolor M571 | S.coelicolor M145 ΔrelA | R. Chakraburtty and M. J. Bibb, unpublished |
| S. coelicolor M571 + pIJ12586 | M571 + pIJ12586 | This work |
| S. coelicolor M1152 | M145 derivative Δact Δred Δcpk Δcda rpoB(C1298T) | Gomez‐Escribano and Bibb, 2011 |
| S. coelicolor M1598 | M1152 + pIJ10257 | This work |
| S. coelicolor M1594 | M1152 + pIJ12576 | This work |
| S. coelicolor M1595 | M1152 + pIJ12574 | This work |
| S. coelicolor M1594 derivatives | ||
| M1594 + pIJ12579 | This work | |
| M1594 + pIJ12580 | This work | |
| M1594 + pIJ12581 | This work | |
| M1594 + pIJ12582 | This work | |
| M1594 + pIJ12583 | This work | |
| M1594 + pIJ12584 | This work | |
| S. coelicolor M1595 derivatives | ||
| M1595 + pIJ12579 | This work | |
| M1595 + pIJ12580 | This work | |
| M1595 + pIJ12581 | This work | |
| M1595 + pIJ12582 | This work | |
| M1595 + pIJ12583 | This work | |
| M1595 + pIJ12584 | This work | |
CHL, chloramphenicol.
Construction of a relA mutant of M . corallina
Chromosomal regions flanking the relA coding sequence (Sosio et al., 2014) were amplified by PCR and cloned into pIJ12738 (Fernández‐Martínez and Bibb, 2014). The 5′ flanking region was amplified to generate a 1902 bp fragment with terminal NcoI and EcoRI sites while the 3′ region was amplified to generate a 1945 bp fragment with terminal EcoRI and XbaI sites. These two fragments were cloned into pIJ12738 digested with NcoI and XbaI to generate pIJ12749 with the I‐SceI site adjacent to the introduced fragments. pIJ12749 was then introduced into E. coli S17 by transformation. Conjugation between the E. coli S17 derivative and M. corallina was carried out as described earlier. Chromosomal integration of pIJ12749, confirmed by PCR analysis (data not shown), generated M. corallina ΔrelA_int, which still contained relA. A 806 bp NdeI‐SacII fragment containing the I‐SceI Meganuclease gene codon‐optimised for expression in actinomycetes was excised from pIJ12739 (Fernández‐Martínez and Bibb, 2014) and cloned into pIJ10257 to generate pIJ12750, which was then conjugated into M. corallina ΔrelA_int to induce a double‐strand break at the introduced I‐SceI site. Three individual exconjugants were streaked on SFM agar containing hygromycin (10 μg ml−1) and grown at 30°C until sporulation. PCR analysis showed that one of these exconjugants, M1596, lacked the chromosomal relA sequence (data not shown).
gusA transcriptional fusions
Promoter regions from the mib gene cluster (PmibJ 299bp, PmibQ 166 bp, PmibR 384 bp, PmibX 254 bp, PmibA 244 bp and PmibE 440 bp) were amplified using oligonucleotides with 5′ XbaI and 3′ KpnI sites, confirmed by nucleotide sequencing and ligated into pGUS (Myronovskyi et al., 2011) digested with the same enzymes to generate the following plasmids containing gusA under the control of each of the promoters: pIJ12579 (PmibA‐gusA), pIJ12580 (PmibE‐gusA), pIJ12581 (PmibJ‐gusA), pIJ12582 (PmibQ‐gusA), pIJ12583 (PmibR‐gusA) and pIJ12584 (PmibX‐gusA). The plasmids were integrated at the ΦC31 attB site of S. coelicolor M1152 (Gomez‐Escribano and Bibb, 2011) after conjugation and at the same site in S. coelicolor M1594 and M1595, strains carrying constructs based on pIJ10257 (Hong et al., 2005) in which mibR or mibX, respectively, were expressed constitutively from ermE*p. Exconjugants were selected using apramycin (25 μg ml−1) and confirmed by PCR.
β‐Glucuronidase assays
Spectrophotometric β‐glucuronidase assays were carried out as described previously (Sherwood and Bibb, 2013). Enzymatic activity was plotted as Miller units calculated as 1000 × (OD420 of sample − OD420 of blank)/[time of reaction in minutes × volume of culture assayed (in ml)] and expressed per milligram of protein.
Constitutive expression of mibR and mibX in M . corallina and S . coelicolor M1152
PCR products containing mibR or mibX (extending from start to stop codons) were generated by high‐fidelity PCR using the primers listed in Table S1 and confirmed by nucleotide sequencing. The mibR fragment was cloned into the integrative vector pIJ8600 (Takano et al., 1995) to fuse mibR to the inducible tipA promoter generating pIJ12572, which was introduced into M. corallina by conjugation. For introduction into S. coelicolor M1152, the same mibR fragment was cloned into the integrative vector pIJ10257 to fuse mibR to the constitutive ermE* promoter generating pIJ12576. Similarly, the mibX fragment was also cloned into the integrative vector pIJ10257 to fuse mibX to the constitutive ermE* promoter generating pIJ12574. These constructs were transferred into M. corallina by conjugation from E. coli S17 and into S. coelicolor M1152 by conjugation from E. coli ET12567/pUZ8002 as described previously (Kieser et al., 2000).
Expression of mibEF in S . coelicolor M145
A PCR product containing mibEF (extending from the start codon of mibE to the stop codon of mibF) was generated by high‐fidelity PCR using the primers listed in Table S1 and confirmed by nucleotide sequencing (data not shown). This fragment was cloned into the integrative vector pIJ10257 to fuse mibEF to the constitutive ermE* promoter generating pIJ12743. This plasmid was transferred into S. coelicolor M145 by conjugation from E. coli ET12567/pUZ8002 as described previously (Kieser et al., 2000).
RT‐PCR analysis
The M. corallina wild‐type strain was grown in 10 ml VSP liquid medium (Marcone et al., 2010) until an OD600 of 0.4 was reached (after 3–4 days of growth) and then 1 ml of this culture was transferred to 50 ml VSP liquid medium (designated time 0). For nested RT‐PCR analysis of the mibR promoter (Fig. 2), RNA was extracted from the mycelia from 4 ml of culture sampled after 16, 24, 40 and 48 h of growth from time 0 (Tunca et al., 2007). For RT‐PCR analysis of the effect of deletion of relA on mib gene expression, M. corallina wild‐type and the relA mutant strain were grown in 10 ml SMM medium (Kieser et al., 2000) to an OD600 of 0.4 (reached after 6–7 days of growth) and then 1 ml of this culture was transferred to 50 ml SMM liquid medium (designated time 0). RNA was extracted from the mycelia from 4 ml of culture sampled after 48 h of growth from time 0 (Tunca et al., 2007). Mycelial pellets were resuspended in 1 ml RTL buffer with lysing matrix B (MP Biomedicals, Solon, Ohio, USA) and homogenised using a Camlab Omni Bead Ruptor 24 Drive Unit (Camlab, Cambridge, UK). Two pulses of 30 s of intensity 6.0 were applied with cooling down for 1 min on ice between pulses. Supernatants were centrifuged for 10 min at 13,000 r.p.m. and then treated according to the instructions given in the RNA Easy Kit (Qiagen, Crawley, UK). The RNA samples were treated with DNase I (Invitrogen) until they were free of DNA contamination (determined by PCR amplification). RNA was quantified and equal amounts from each sample were converted to cDNA following the manufacturer's instructions (SuperScript®, Invitrogen). Oligonucleotide pairs listed in Table S1 were used to amplify nested fragments of the mibR promoter region. The oligonucleotide pairs used to analyse expression of the mib cluster in the wild‐type and relA mutant strains were described previously (Foulston and Bibb, 2010). Amplification was also attempted using the same oligonucleotide pairs on RNA samples that had not been treated with reverse transcriptase to confirm lack of DNA contamination.
RACE
The 5′ end of the mibRp2 transcript was identified by using a 5′ RACE (rapid amplification of cDNA ends) kit (Invitrogen, Paisley, UK, version 2.0) following the manufacturer's instructions. Briefly, first‐strand cDNA synthesis was carried out using 5 μg of RNA, reverse transcriptase and the oligonucleotide primer RACE_mibR_R1 (Table S1). cDNA was purified using the SNAP columns provided in the kit, and poly(dC) tails were added to the 3′ ends using terminal deoxynucleotidyl transferase. PCR amplification of the tailed cDNA was carried out initially using the 5′ RACE abridged anchor primer with the first‐strand primer RACE_mibR_R1 (Table S1). A dilution of the PCR mixture was then subjected to a second amplification using the abridged anchor primer with the second nested primer RACE_mibR_R2 (Table S1). The PCR product was gel‐purified and a portion sequenced directly using oligonucleotide RACE_mibR_R2 as primer (RACE1 in Fig. S1B). Another portion was used for cloning into pGEM‐TEasy (Promega UK, Southampton, UK) and the cloned PCR fragment sequenced using RACE_mibR_R2 as primer (RACE2 in Fig. S1B).
Induction of microbisporicin production in M . corallina
To test for induction of microbisporicin production in wild‐type M. corallina, either M. corallina M1592 (mibR expression strain) or 2.5 μg of purified microbisporicin (in 10% DMSO) dried on a filter paper disc were placed on V0.1 (Marcone et al., 2010) agar plates adjacent to streaks of wild‐type M. corallina and a non‐producing mutant [ΔmibA, (Foulston and Bibb, 2010) ] as a control; the plates were overlaid with Micrococcus luteus after incubation at 30°C for 5 days.
Supporting information
Supporting information
Acknowledgements
This work was supported financially by grants from the European Commission (LAPTOP‐project, contract number 245066 for FP7‐KBBE‐2009‐3) and the UK Biotechnological and Biological Sciences Research Council (BBSRC) [Institute Strategic Programme Grant ‘Understanding and Exploiting Plant and Microbial Secondary Metabolism’ (BB/J004561/1) ]. We thank Margherita Sosio and Sonia Maffioli (NAICONS) for the gift of purified microbisporicin and members of the LAPTOP consortium for their interest in this work.
References
- Ali, N. , Herron, P.R. , Evans, M.C. , and Dyson, P.J. (2002) Osmotic regulation of the Streptomyces lividans thiostrepton‐inducible promoter, ptipA . Microbiology 148: 381–390. [DOI] [PubMed] [Google Scholar]
- Arnison, P.G. , Bibb, M.J. , Bierbaum, G. , Bowers, A.A. , Bugni, T.S. , Bulaj, G. , et al (2013) Ribosomally synthesized and post‐translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30: 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb, M.J. (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8: 208–215. [DOI] [PubMed] [Google Scholar]
- Bremer, H. , and Ehrenberg, M. (1995) Guanosine tetraphosphate as a global regulator of bacterial RNA synthesis: a model involving RNA polymerase pausing and queuing. Biochim Biophys Acta 1262: 15–36. [DOI] [PubMed] [Google Scholar]
- Brown, K.L. , Wood, S. , and Buttner, M.J. (1992) Isolation and characterization of the major vegetative RNA polymerase of Streptomyces coelicolor A3(2); renaturation of a sigma subunit using GroEL. Mol Microbiol 6: 1133–1139. [DOI] [PubMed] [Google Scholar]
- Cashel, M. , Gentry, D.R. , Hernandez, V.J. , and Vinella, D. (1996) The Stringent Response in Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd edn Washington D.C.: ASM Press, pp. 1458–1496. [Google Scholar]
- Castiglione, F. , Lazzarini, A. , Carrano, L. , Corti, E. , Ciciliato, I. , Gastaldo, L. , et al (2008) Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol 15: 22–31. [DOI] [PubMed] [Google Scholar]
- Chakraburtty, R. , and Bibb, M. (1997) The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol 179: 5854–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty, R. , White, J. , Takano, E. , and Bibb, M. (1996) Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2). Mol Microbiol 19: 357–368. [DOI] [PubMed] [Google Scholar]
- Chatterjee, C. , Paul, M. , Xie, L. , and van der Donk, W.A. (2005) Biosynthesis and mode of action of lantibiotics. Chem Rev 105: 633–684. [DOI] [PubMed] [Google Scholar]
- Donadio, S. , Sosio, M. , Serina, S. , and Mercorillo, D. (2009) Genes and proteins for the biosynthesis of the lantibiotic 107891. CA2695487 A1.
- Fernández‐Martínez, L.T. , and Bibb, M.J. (2014) Use of the Meganuclease I‐SceI of Saccharomyces cerevisiae to select for gene deletions in actinomycetes. Sci Rep 4: 7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulston, L. , and Bibb, M. (2011) Feed‐forward regulation of microbisporicin biosynthesis in Microbispora corallina . J Bacteriol 193: 3064–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulston, L.C. (2010) Cloning and analysis of the microbisporicin lantibiotic gene cluster from Microbispora corallina . PhD thesis.
- Foulston, L.C. , and Bibb, M.J. (2010) Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc Natl Acad Sci USA 107: 13461–13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca, A.O. , Colomer‐Winter, C. , and Lemos, J.A. (2015) Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197: 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Escribano, J.P. , and Bibb, M.J. (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust, B. , O'Rourke, S. , Bird, N. , Kieser, T. , and Chater, K.F. (2003) Recombineering in Streptomyces coelicolor. Norwich, UK: John Innes Foundation. [Google Scholar]
- Hesketh, A. , Sun, J. , and Bibb, M. (2001) Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII‐orf4 transcription and actinorhodin biosynthesis. 39: 136–144. [DOI] [PubMed] [Google Scholar]
- Hong, H.J. , Hutchings, M.I. , Hill, L.M. , and Buttner, M.J. (2005) The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor . J Biol Chem 280: 13055–13061. [DOI] [PubMed] [Google Scholar]
- Jabés, D. , Brunati, C. , Candiani, G. , Riva, S. , Romanó, G. , and Donadio, S. (2011) Efficacy of the new lantibiotic NAI‐107 in experimental infections induced by multidrug‐resistant Gram‐positive pathogens. Antimicrob Agents Chemother 55: 1671–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser, T. , Bibb, M.J. , Buttner, M.J. , Chater, K.F. , and Hopwood, D.A. (2000) Practical Streptomyces genetics. Norwich, UK, Norwich: John Innes Foundation. [Google Scholar]
- Kleerebezem, M. (2004) Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25: 1405–1414. [DOI] [PubMed] [Google Scholar]
- Krásný, L. , and Gourse, R.L. (2004) An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23: 4473–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarini, A. , Gastaldo, L. , Candiani, G. , Ciciliato, I. , Losi, D. , Marinelli, F. , et al (2005) Antibiotic 107891, its factors A1 and A2, 8 pharmaceutically acceptable salts and compositions, and use thereof. WO/2005/014628.
- Li, X. , and O'Sullivan, D.J. (2012) Contribution of the Actinobacteria to the growing diversity of lantibiotics. Biotechnol Lett 34: 2133–2145. [DOI] [PubMed] [Google Scholar]
- MacNeil, D.J. , Occi, J.L. , Gewain, K.M. , MacNeil, T. , Gibbons, P.H. , Ruby, C.L. , and Danis, S.J. (1992) Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115: 119–125. [DOI] [PubMed] [Google Scholar]
- Maffioli, S.I. , Iorio, M. , Sosio, M. , Monciardini, P. , Gaspari, E. , and Donadio, S. (2014) Characterization of the congeners in the lantibiotic NAI‐107 complex. J Nat Prod 77: 79–84. [DOI] [PubMed] [Google Scholar]
- Magnusson, L.U. , Farewell, A. , and Nyström, T. (2005) ppGpp: a global regulator in Escherichia coli . Trends Microbiol 13: 236–242. [DOI] [PubMed] [Google Scholar]
- Marcone, G.L. , Carrano, L. , Marinelli, F. , and Beltrametti, F. (2010) Protoplast preparation and reversion to the normal filamentous growth in antibiotic‐producing uncommon actinomycetes. J Antibiot (Tokyo) 63: 83–88. [DOI] [PubMed] [Google Scholar]
- Martín, J.F. , and Liras, P. (2012) Cascades and networks of regulatory genes that control antibiotic biosynthesis. Subcell Biochem 64: 115–138. [DOI] [PubMed] [Google Scholar]
- Martínez‐Costa, O.H. , Fernández‐Moreno, M.A. , and Malpartida, F. (1998) The relA/spoT‐homologous gene in Streptomyces coelicolor encodes both ribosome‐dependent (p)ppGpp‐synthesizing and ‐degrading activities. J Bacteriol 180: 4123–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, T. , Holt, T.G. , and Thompson, C.J. (1989) Thiostrepton‐induced gene expression in Streptomyces lividans . J Bacteriol 171: 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch, D. , Müller, A. , Schneider, T. , Kohl, B. , Wenzel, M. , Bandow, J.E. , et al (2014) The lantibiotic NAI‐107 binds to bactoprenol‐bound cell wall precursors and impairs membrane functions. J Biol Chem 289: 12063–12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myronovskyi, M. , Welle, E. , Fedorenko, V. , and Luzhetskyy, A. (2011) Beta‐glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol 77: 5370–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, Y. , Kitpreechavanich, V. , Suzuki, K. , and Kudo, T. (1999) Microbispora corallina sp. nov., a new species of the genus Microbispora isolated from Thai soil. Int J Syst Bacteriol 49 (Part 4): 1761–1767. [DOI] [PubMed] [Google Scholar]
- Ochi, K. (2007) From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem 71: 1373–1386. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn New York: Cold Spring Harbor. [Google Scholar]
- Schmitz, S. , Hoffmann, A. , Szekat, C. , Rudd, B. , and Bierbaum, G. (2006) The lantibiotic mersacidin is an autoinducing peptide. Appl Environ Microbiol 72: 7270–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood, E.J. , and Bibb, M.J. (2013) The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba . Proc Natl Acad Sci USA 110: E2500–E2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. , and Pühler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram‐negative bacteria. Bio/Technology 1: 784–791. [Google Scholar]
- Sosio, M. , Gallo, G. , Pozzi, R. , Serina, S. , Monciardini, P. , Bera, A. , et al (2014) Draft Genome Sequence of the Microbispora sp. Strain ATCC‐PTA‐5024, Producing the Lantibiotic NAI‐107. Genome Announc 2: e01198–13. doi: 10.1128/genomeA.01198‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann, E. , Frasch, H.J. , Kilian, R. , and Pozzi, R. (2014) Self‐resistance mechanisms of actinomycetes producing lipid II‐targeting antibiotics. Int J Med Microbiol 305: 190–195. [DOI] [PubMed] [Google Scholar]
- Stein, T. , Borchert, S. , Kiesau, P. , Heinzmann, S. , Klöss, S. , Klein, C. , et al (2002) Dual control of subtilin biosynthesis and immunity in Bacillus subtilis . Mol Microbiol 44: 403–416. [DOI] [PubMed] [Google Scholar]
- Takano, E. , and Bibb, M.J. (1994) The stringent response, ppGpp and antibiotic production in Streptomyces coelicolor A3(2). Actinomycetologica 8: 1–16. [Google Scholar]
- Takano, E. , White, J. , Thompson, C.J. , and Bibb, M.J. (1995) Construction of thiostrepton‐inducible, high‐copy‐number expression vectors for use in Streptomyces spp . Gene 166: 133–137. [DOI] [PubMed] [Google Scholar]
- Tunca, S. , Barreiro, C. , Sola‐Landa, A. , Coque, J.J.R. , and Martín, J.F. (2007) Transcriptional regulation of the desferrioxamine gene cluster of Streptomyces coelicolor is mediated by binding of DmdR1 to an iron box in the promoter of the desA gene. FEBS J 274: 1110–1122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


