Highlight
Arabidopsis glycerol-3-phosphate acyltransferase 9 (GPAT9) is an sn-1 specific acyl-CoA:GPAT that contributes to intracellular glycerolipid biosynthesis in seeds, developing leaves and pollen grains, but not to extracellular glycerolipid biosynthesis.
Key words: Acyl-CoA specificity, acyl lipid biosynthesis, cutin, GPAT, lipid droplet, pollen grain, regio-specificity, surface wax.
Abstract
GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE (GPAT) genes encode enzymes involved in glycerolipid biosynthesis in plants. Ten GPAT homologues have been identified in Arabidopsis. GPATs 4–8 have been shown to be involved in the production of extracellular lipid barrier polyesters. Recently, GPAT9 was reported to be essential for triacylglycerol (TAG) biosynthesis in developing Arabidopsis seeds. The enzymatic properties and possible functions of GPAT9 in surface lipid, polar lipid and TAG biosynthesis in non-seed organs, however, have not been investigated. Here we show that Arabidopsis GPAT9 exhibits sn-1 acyltransferase activity with high specificity for acyl-coenzyme A, thus providing further evidence that this GPAT is involved in storage lipid biosynthesis. We also confirm a role for GPAT9 in seed oil biosynthesis and further demonstrate that GPAT9 contributes to the biosynthesis of both polar lipids and TAG in developing leaves, as well as lipid droplet production in developing pollen grains. Conversely, alteration of constitutive GPAT9 expression had no obvious effects on surface lipid biosynthesis. Taken together, these studies expand our understanding of GPAT9 function to include modulation of several different intracellular glycerolipid pools in plant cells.
Introduction
Glycerolipids, including storage triacylglycerol (TAG), various polar membrane lipids, and numerous signaling molecules associated with plant growth, development, and resistance to biotic and abiotic stress, are produced in plants through three main biosynthetic pathways that are compartmentalized within plastids, mitochondria, and the endoplasmic reticulum (ER) (Roughan and Slack, 1982; Browse et al., 1986). Glycerolipids produced within chloroplasts are converted mainly to galactolipids, which serve primarily as functional and structural components of photosynthetic membranes (Dörmann and Benning, 2002). Conversely, glycerolipids synthesized within mitochondria may contribute to the production of membrane lipids and the relative fatty acid (FA) composition of TAG in a number of plant organs (Zheng et al., 2003). Glycerolipids produced on ER membranes generally constitute cellular membrane phospholipids, and in developing seeds are synthesized primarily as precursors in support of TAG production (Ohlrogge and Browse, 1995). Each of these three pathways consists of a distinct set of enzymes that essentially mediate the stereospecific esterification of FAs to a glycerol backbone, with an extensive coordinated exchange of glycerolipid molecules between the three compartments (Browse et al., 1986; Kunst et al., 1988).
Glycerol-3-phosphate acyltransferase (GPAT) provides a crucial function in these glycerolipid biosynthetic pathways, catalyzing the transfer of a fatty acyl moiety from acyl-CoA, dicarboxylic acid (DCA)-CoA or acyl-acyl carrier protein to sn-glycerol-3-phosphate (G3P) to form lysophosphatic acid (LPA). The resulting LPA is an important intermediate for the formation of numerous glycerolipids, including extracellular lipid polyesters, membrane lipids and storage TAG. The metabolic fate of LPA is determined in part by the subcellular localization of the GPAT enzyme, with a soluble form being present in the plastid stroma, and membrane-bound forms localized to the mitochondria and ER (Chen et al., 2011a ).
In Arabidopsis, ten GPAT genes, ATS1 and GPAT1-9, have been identified to date (Nishida et al., 1993; Zheng et al., 2003; Beisson et al., 2007; Gidda et al., 2009). While ATS1 appears to function in the production of major phospholipids that make up plastidial membranes (Kunst et al., 1988), GPAT1 has been found to play an important role in the development of pollen grains (Zheng et al., 2003). Interestingly, a recent study has shown that GPAT1 possesses sn-2 acyltransferase activity and can utilize DCA-CoA substrates, which indicates that it is likely involved in the biosynthesis of extracellular lipids (Yang et al., 2012). Similarly, GPAT4–8 have also been found to exhibit sn-2 regio-specificity with a preference for DCA- or omega-OH-CoA as substrate and have been shown to play important roles in the production of the extracellular lipid barrier polyesters, cutin and/or suberin (Suh et al., 2005; Beisson et al., 2007; Li et al., 2007a, b ; Yang et al., 2012).
In addition to the essential roles of GPATs in the production of extracellular lipid polyesters in plants, an ER-bound GPAT is believed to be crucial for the synthesis of storage TAG, catalyzing the first acylation reaction of the Kennedy pathway (reviewed by Bates et al., 2013). Intriguingly, in castor (Ricinus communis), a homolog of Arabidopsis GPAT9 has been identified on purified ER membranes by proteomics (Brown et al., 2012). Furthermore, GPAT9 exhibits the closest evolutionary relationship of all Arabidopsis GPATs to mammalian GPAT3 (Gidda et al., 2009), which is known to play a crucial role in storage lipid biosynthesis (Cao et al., 2006; Shan et al., 2010). Indeed, both human GPAT3 and Arabidopsis GPAT9 contain an additional conserved motif above the four motifs that are typically characteristic of other GPAT enzymes (Lewin et al., 1999). Unlike Arabidopsis GPAT1–8, homologs of GPAT9 appear to be present in various algal species that also produce an abundance of TAG (Khozin-Goldberg and Cohen, 2011; Iskandarov et al., 2015), and a recent study demonstrated that the GPAT9 homolog from the oleaginous green microalga Lobosphaera incisa increased TAG content by up to 50% when heterologously expressed in Chlamydomonas reinhardtii (Iskandarov et al., 2015). Additionally, it has recently been reported that GPAT9 plays a role in TAG biosynthesis in Arabidopsis, wherein down-regulation of GPAT9 led to decreased seed oil content, and a gpat9 mutant exhibited both male and female gametophytic lethality phenotypes (Shockey et al., 2016).
However, while GPAT9 appears to contribute to seed oil accumulation in Arabidopsis, there remains a lack of direct evidence that GPAT9, unlike other GPATs, is an sn-1 acyltransferase with a preference for acyl-CoAs as its substrate. Moreover, there is a paucity of information regarding its function in the biosynthesis of lipids in non-seed organs. In this study, we carried out in vitro enzyme assays using a yeast-based expression system and provide detailed functional investigations of Arabidopsis lines in which GPAT9 was either overexpressed or down-regulated. Our findings indicate that GPAT9 exhibits sn-1 acyltransferase activity with high specificity for acyl-CoA, thus confirming its role in seed TAG biosynthesis, and provide comprehensive evidence in support of its role in the production of both polar and non-polar lipids in leaves, as well as lipid droplets in pollen. Furthermore, since GPAT1–8 have been reported to play important roles in the production of extracellular lipid barrier polyesters, we also investigated the possible contribution of GPAT9 to this process. The results of our studies not only yield a deeper understanding of the function of GPAT9, but also bring us closer to a full elucidation of intracellular and extracellular lipid biosynthesis in plants, which will be important for the future improvement of lipid content in oilseed crops.
Materials and methods
Heterologous expression of GPAT9 in yeast and in vitro enzyme assays
The full-length Arabidopsis GPAT9 cDNA was inserted downstream of the GAL1 promoter in the pYES2.1/V5-His-TOPO®TA yeast expression vector (Invitrogen) according to the manufacturer’s instructions. Saccharomyces cerevisiae strain gat1Δ (Matα, his3C1, leu2C0, lys2C0, ura3C0, YKR067w::kanMX4; Zheng and Zou, 2001) was transformed with both the experimental construct as well as the empty vector control, respectively. Microsomal fractions containing recombinant Arabidopsis GPAT9 were used in the enzyme assays.
While unsubstituted acyl-CoA substrates (16:0-CoA, 18:0-CoA, 18:1Δ9cis-(hereafter 18:1)-CoA, 18:2 Δ9cis,12cis-(hereafter 18:2)-CoA, and 18:3Δ9cis,12cis,15cis (hereafter 18:3)-CoA were purchased directly (Avanti Polar Lipids, Inc.), 16:0-DCA-CoA was synthesized as described by Kawaguchi et al. (1981) and Yang et al. (2010). The in vitro GPAT enzyme assay was performed essentially as described previously (Yang et al., 2010; Chen et al., 2014).
Plant growth conditions
Arabidopsis seeds were cold-treated at 4 ºC in the dark for 3–5 d prior to their placement in a growth chamber at 22 ºC. In most cases, plants were grown with a photoperiod of 18h day/6h night and 250 µmol m−2 s−1 light intensity for the remainder of their development. Plants utilized for wax analyses were grown in a growth chamber with a 12-h-light/ 12-h-dark cycle and a light intensity of approximately 700 µmol m−2 s−1. In all instances, plants were regularly watered and fertilized.
RNA extraction, first-strand cDNA synthesis, and quantitative real-time RT-PCR
Various tissues were harvested from Arabidopsis plants (all within a Col-0 background) at a number of different developmental stages, flash frozen in liquid nitrogen, and stored at −80 ºC until further use. Total RNA was extracted using the Sigma Spectrum Plant Total RNA Kit (Sigma-Aldrich Canada Co., Oakville, ON) and contaminating DNA was removed using the TURBO DNA-free system (Ambion, Life Technologies Inc., Burlington, ON). First-strand cDNA synthesis was carried out using the Superscript III first-strand cDNA synthesis kit according to the manufacturer’s instructions (Invitrogen, Life Technologies Inc.).
Quantitative real-time RT-PCR assays were performed with SYBR green PCR master mix on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). The primers are AtGPAT9_QF2 (5ʹ–CGG TGA AAC AGG AAT TGA ATT TG–3ʹ) and AtGPAT9_QR2 (5ʹ–AGA CCC GCC CGA AGA GAT A–3ʹ). Primers AtPP2AAF1 (5ʹ–TCA ATC CGT GAA GCT GCT GCA AAC–3ʹ) and AtPP2AAR1 (5ʹ–ACT GCA CGA AGA ATC GTC ATC CGA–3ʹ) were utilized to amplify a 146-nt fragment of the constitutively expressed PROTEIN PHOSPHATASE 2A SUBUNIT 3 (PP2AA3) transcript (Czechowski et al., 2005), which was utilized as an internal control. Levels of gene expression were obtained using the standard curve method and SDS v2.4 software (Applied Biosystems). The Arabidopsis Genome Initiative numbers for the gene sequences utilized are AT5G60620 (GPAT9) and AT1G13320 (PP2AA3).
Generation of transgenic Arabidopsis
Five plant transformation constructs including one GPAT9-GUS (β-glucuronidase) translational fusion vector, two GPAT9 overexpression vectors [constitutive (GPAT9-OE) and seed-specific (GPAT9-SS-OE), respectively], and two GPAT9 RNAi vectors [constitutive (GPAT9-RNAi) and seed-specific (GPAT9-SS-RNAi), respectively], were generated.
The GPAT9-GUS vector was produced by first amplifying a 3345-bp fragment of the Arabidopsis GPAT9 gene, along with primers AtGPAT9F4SalI (5ʹ–ATA GTC GAC GAG AAG ACG ACG AGA AGA GC–3ʹ) and AtGPAT9R4BamHI (5ʹ–AGG GAT CCC TCT GCG AAA CTC TGT TGC–3ʹ), which contain SalI and BamHI restriction sites near their 5ʹ ends, respectively (indicated in bold). This fragment was inserted upstream of the GUSAint sequence (encoding β-glucuronidase) with its start codon removed and Nopaline synthase transcriptional terminator (nos-t) to create an in-frame GPAT9-GUS translational fusion within the pGreen 0029 background (Hellens et al., 2000).
To generate GPAT9 overexpression constructs, the GPAT9 coding sequence inserted between the constitutive tobacco tCUP3 promoter (Wu et al., 2003) and Pisum sativum Ribulose-1,5-bisphosphate carboxylase transcriptional terminator (rbcS-t) to generate the GPAT9-OE vector, or between the seed-specific Brassica napus Napin promoter (Josefsson et al., 1987) and rbcS-t transcriptional terminator to produce the GPAT9-SS-OE vector.
To produce the GPAT9 RNAi vectors, a 348-bp fragment near the 3ʹ end of the GPAT9 coding region was amplified from the GPAT9 cDNA clone with primers GPAT9RNAiF1SalI (5ʹ–GTC GAC GGG TGC TTT TGA ATT GGA CTG C–3ʹ) and GPAT9RNAiR1EcoRI (5ʹ–GAA TTC TCT TCC AAT CTA GCC AGG ATC G–3ʹ), as well as GPAT9RNAiF1XbaI (5ʹ–TCT AGA GGG TGC TTT TGA ATT GGA CTG C– 3ʹ) and GPAT9RNAiR1BamHI (5ʹ–GGA TCC TCT TCC AAT CTA GCC AGG ATC G–3ʹ) to generate sense and antisense copies. Although the GPAT9 coding sequence displays virtually no significant similarities to any other Arabidopsis GPAT sequence (Gidda et al., 2009), we further confirmed a lack of potential off-target effects by these fragments in Arabidopsis using the dsCheck software (http://dscheck.rnai.jp/). The GPAT9 RNAi fragments were inserted in opposite orientations between the constitutive tCUP3 promoter and intronic spacer from the pHannibal plasmid, and rbcS-t transcriptional terminator and intronic spacer, respectively. These same sense and antisense GPAT9 fragments were also inserted between the seed-specific Phaseolus vulgaris β-Phaseolin promoter (Frisch et al., 1995) and pHannibal intronic spacer, and the β-Phaseolin transcriptional terminator and intronic spacer, respectively, in a pPZP-RSC1 background (Hajdukiewicz et al., 1994) to yield the GPAT9-SS-RNAi vector.
Vectors were introduced into Agrobacterium tumefaciens strain GV3101 via electroporation, and in the case of pGreen-derived vectors, the helper plasmid pSoup (Hellens et al., 2000) was co-transformed simultaneously. The resulting recombinant bacteria were used for the transformation of Arabidopsis ecotype Col-0 using the floral dip method (Clough and Bent, 1998). The presence of target constructs in transgenic plants was confirmed by PCR and homozygous lines were identified using segregation analyses. For every experiment, transgenic experimental lines were grown in the same growth chamber at the same time as their corresponding negative control lines. In the case of all experiments involving T1 lines, plants transformed with empty vector were utilized as the wild-type control, while in experiments involving subsequent generations, wild-type controls comprised null-segregants.
Morphological analyses and histochemical staining
T3 seed weights from homozygous transgenic lines and wild-type plants were calculated by first weighing small batches of seeds, followed by particle counting using a FuorChem SP Imager and AlphaEase software (Alpha Innotech Corp., San Leandro, CA). Three technical replicates were used in each case. T3 seed areas from the same lines were determined using the particle analysis function of ImageJ software (http://imagej.nih.gov/ij).
Various tissues from at least 15 independent T1 transgenic lines bearing the GPAT9-GUS vector were stained for GUS activity essentially as described by Jefferson et al. (1987). Images were obtained using an Olympus SZ61 microscope with attached digital camera (Olympus Canada Inc., Richmond Hill, ON).
Toluidine blue staining for estimation of cuticle permeability was carried out by dipping rosette leaves [28 d after planting (DAP)] and open flowers from 10 GPAT9-RNAi T2 lines and wild-type plants, respectively, into 0.05% (w/v) toluidine blue for 2min at room temperature, followed by a rinse with distilled water. Pollen staining was conducted with anthers from 10 T2 GPAT9-RNAi lines, multiple plants from two T3 GPAT9-RNAi lines, and wild-type plants according to the method described by Peterson et al. (2010).
Lipid extraction and GC-MS analyses
Arabidopsis seed lipid extractions were carried out using mature seeds from GPAT9-OE, GPAT9-SS-OE, GPAT9-RNAi, GPAT9-SS-RNAi and wild-type plants as described in Pan et al. (2013). Total leaf lipids were extracted from developing GPAT9-OE, GPAT9-RNAi, and wild-type leaves (35 DAP). In each case, two leaves (leaves 5 and 6) from three individual T3 plants were utilized for lipid extractions using the protocol described by the Kansas Lipidopmics Research Center (https://www.k-state.edu/lipid/lipidomics/leaf-extraction.html). Internal standards of 0.0025mg 17:0 TAG and 0.05mg 19:0 phosphatidylcholine (PC) (Avanti Polar Lipids Inc., Alabaster, AL) were included in each reaction. Leaf lipids were separated into lipid classes by one-dimensional TLC on silica gel plates (SIL G25, 0.25mm, Macherey-Nagel, Düren, Germany) using hexane/diethyl ether/glacial acetic acid (70:30:1, v/v) as the solvent as described by Mietkiewska et al. (2011). Lipid classes were visualized under UV irradiation following treatment with 0.05% (w/v) primuline and spots corresponding to TAG and polar lipids (PLs) were scraped off and transmethylated with 3N methanolic HCl at 80 ºC for 1h. The resulting FA methyl esters were extracted twice with hexanes, evaporated under a stream of nitrogen gas and resuspended in 500 µl iso-octane with methyl heneicosanoin (21:0 methyl ester, 0.1mg/ml; Nu-Chek Prep Inc.) as a second internal standard.
All extracted FA methyl esters were analyzed using an Agilent 6890 Network GC system equipped with a DB-23 capillary column (30 m × 0.25mm × 0.25 µm) and a 5975 inert XL Mass Selective Detector (Agilent Technologies Canada Inc., Mississauga, ON). The following temperature program was utilized: 100 ºC, hold for 4min, 10 ºC min−1 to 180 ºC, hold for 5min, and 10 ºC min−1 to 230 ºC, hold for 5min.
Transmission electron microscopy
To visualize morphological changes in the pollen of GPAT9-OE and GPAT9-RNAi lines compared to wild-type plants, flower buds were fixed with 2.5% gluteraldehyde and 2% paraformaldehyde in 0.1M phosphate buffer (pH 7.2–7.4) at room temperature overnight and then stored at 4 ºC. Fixed samples were then post-fixed in 1% (w/v) osmium tetroxide in 0.1M phosphate buffer (pH 7.2–7.4) for 3h at room temperature. The samples were then dehydrated in a graded ethanol series, infiltrated with propylene oxide, and embedded using the Spurr’s Low Viscosity embedding kit (Electron Microscopy Sciences, Hatfield, PA). Ultrathin (80nm) sections were generated using an Ultracut E ultramicrotome (Reichert Inc., Depew, NY) and were stained with 4% uranyl acetate and lead citrate solution (Reynolds, 1963) for TEM microscopy using a Morgagni 268 TEM microscope (FEI Company, Hillsboro, OR) at an accelerating voltage of 80kV. Images were photographed using an attached Orius CCD camera (Gatan Inc., Pleasanton, CA).
Analysis of epicuticular waxes
Surface waxes were extracted and analyzed as described previously (Jenks et al., 1995) with slight modifications. In brief, each sample was submerged in 10ml hexanes and gently agitated for 45s. The samples were then removed from the solvent with forceps and placed on a transparency sheet for scanning. Three internal standards (10 µg of nonadecanoic acid, 10 µg of tetracosane and 20 µg of pentacosanol) were added to each sample vial. Wax extracts were heated at 60 °C then the solvent was reduced under N2 until the volume could be transferred into a 2ml glass vial. The scintillation vials were rinsed once with hexanes, the volume transferred again, and then evaporated to dryness.
For each wax sample, 100 µl of N, O-bis (trimethylsilyl) trifluoroacetamide (BSTFA) and 100 µl hexanes were added for a total volume of 200 µl. The samples were analyzed on an Agilent 7890A gas chromatograph equipped with a 5975C mass spectrometer and an onboard heater and shaker. Four leaves were harvested from five replicate plants 35 DAP, while stem samples were collected from the same plants 42 DAP. Surface areas were determined from sample scans using ImageJ. Each leaf value was multiplied by two to account for both surfaces and stem scans were multiplied by π. Quantified wax values are expressed as µg dm−2. Least squares means (P<0.05) were calculated for statistical analysis (P<0.05).
Isolation and analysis of cutin
Cutin monomers from Arabidopsis leaves harvested 28 DAP were isolated and analyzed as described by Bonaventure et al. (2004) and Chen et al. (2011b ). GC-MS analysis was performed using an Agilent 6890N gas chromatograph with an Agilent 5975 Inert Mass Selective Detector as described previously (Chen et al., 2011b ). Chromatographic separation was achieved using an HP-5MS capillary column (30 m, 0.25mm, 0.25mm; Agilent Technologies) with a temperature program of 140 ºC to 300 ºC at a rate of 3ºC min−1. The inlet was operated in split mode (10:1 split ratio, 1ml injection) at 310 ºC. For the mass spectra conditions, the solvent delay was 4min, ionization energy was 70eV, and data were acquired in scan mode with a range of 35 to 500 atomic mass units.
Results
Arabidopsis GPAT9 is an sn-1 acyltransferase with high specificity for acyl-CoA
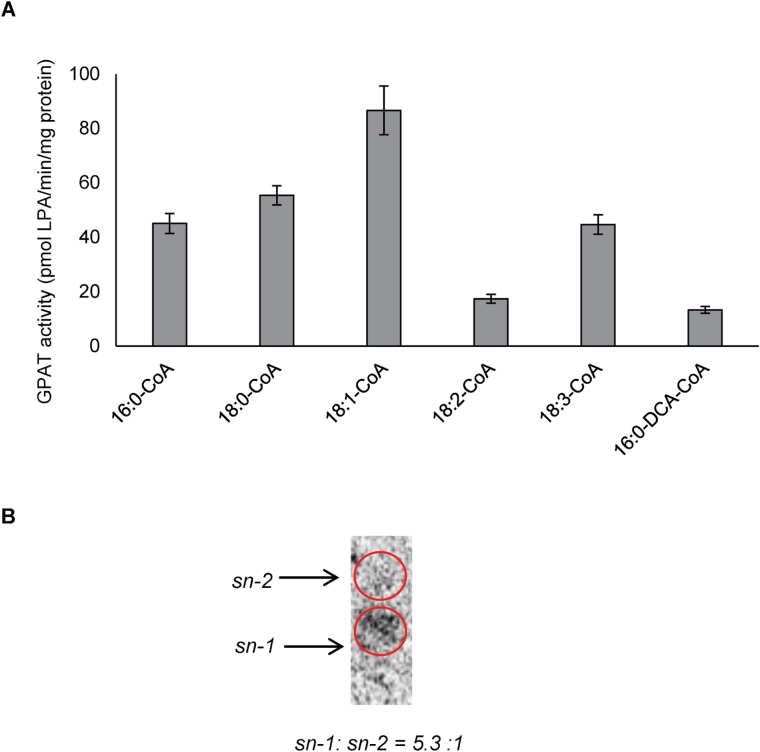
The in vitro enzyme assays of recombinant GPAT9 were initially carried out at 22 ºC using 16:0-CoA and G3P as substrate for 0, 5, 10, 20 and 60min yielding 0, 213.4±11.0, 389.2±20.5, 900.5±42.7 and 2622.7±65.0 pmol LPA/mg protein (corrected in each case for residual background activity in empty vector controls), respectively, indicating that GPAT9 is indeed a functional GPAT enzyme. Subsequent assays using various other substrates, including different molecular species of acyl-CoA and 16:0-DCA-CoA, further demonstrated that GPAT9 prefers acyl-CoAs over 16:0-DCA-CoA as its substrate (Fig. 1A), with 18:1-CoA resulting in the highest activity of all acyl-CoA substrates tested. GPAT9 was not found to possess any phosphatase activity, since monoacylglycerol was not produced in the reaction mixtures of these assays. Analysis of the regio-specificity of GPAT9 indicated that the majority of the acylation reactions catalyzed by this enzyme took place at the sn-1 position rather than the sn-2 position (5.3:1 ratio; Fig. 1B). Taken together, the preference of GPAT9 for acyl-CoA as its substrate, and its sn-1 regio-specificity, provide support that it plays an important role in the Kennedy pathway of glycerolipid biosynthesis.
Fig. 1.
Substrate specificity and regio-specificity of Arabidopsis GPAT9. (A) In vitro enzyme assays of GPAT9 indicate that it prefers acyl-CoA over dicarboxylic acid (DCA)-CoA as a substrate. Bars represent mean enzyme activities when the enzyme was assayed at 22 ºC for 20min. Standard errors of three independent enzyme preparations are also shown. Microsomes were prepared from yeast cells. Saccharomyces cerevisiae strain gat1Δ (Matα, his3C1, leu2C0, lys2C0, ura3C0, YKR067w::kanMX4; Zheng and Zou, 2001) was transformed with Arabidopsis GPAT9 and an empty vector control, respectively. (B) In vitro regio-specificity assays of GPAT9 reveal the enzyme is an sn-1 acyltransferase. Regio-specificity was determined by de-phosphorylating the lysophosphatidic acid (LPA) product, generated in the enzyme assays, using a commercial E. coli alkaline phosphatase and subsequently separating the resulting sn-1 and sn-2 monoacylglycerol on a borate-TLC plate.
GPAT9 is expressed throughout Arabidopsis development
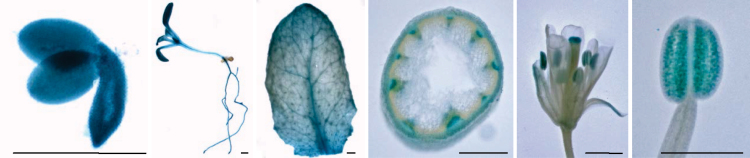
Both quantitative real-time RT-PCR analyses and GUS staining of GPAT9-GUS translational fusion lines indicated that the Arabidopsis GPAT9 gene is expressed in a relatively constitutive manner (Fig. 2, Supplementary Fig. S1at JXB online). Interestingly, the expression pattern of GPAT9 appears to be much broader than that of LYSOPHOSPHATIDIC ACID ACYLTRANSFERASES, DIACYLGLYCEROL ACYLTRANSFERASES (DGATs), PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFE RASE, and PHOSPHATIDIC ACID PHOSPHATASES when compared to previous microarray data (Supplementary Table S1). Indeed, comparable levels of GPAT9 expression were noted in the majority of tissues tested, including seedlings, rosette and cauline leaves (28 DAP), stems, roots, floral buds, open flowers, pollen, and siliques/seeds/embryos at various developmental stages. The highest levels of expression appeared to be evident in leaves and developing siliques [at ~9 d after flowering (DAF)]. Within siliques, developing embryos displayed strong GUS staining in the mid-stages of embryo development, during the time of abundant glycerolipid biosynthesis in this tissue. Within stem cross-sections, GUS staining was apparent in both the phloem and xylem, but appeared to be weak or absent in epidermal cells. In floral tissues, the majority of GPAT9 expression was restricted to anthers (and more specifically pollen), while little or no GUS staining was observed in sepals or petals. This expression profile of GPAT9 in floral tissues is not entirely consistent with the relatively constitutive expression pattern observed in previous microarray assays (Schmid et al., 2005; Winter et al., 2007), which may reflect differences in the timing/developmental stage used in the GUS staining experiments. It is also possible that the promoter fragment utilized for our GUS assays lacks certain cis-acting elements present in the native gene context.
Fig. 2.
GUS staining of transgenic GPAT9-GUS translational fusion lines. Pictures are representative of at least 15 independent T1 lines. From left to right, tissues include a developing embryo, young seedling, rosette leaf, stem cross-section, flower, and anther. Scale bars, 10 μm.
Validation of constitutive GPAT9 overexpression and RNAi Arabidopsis lines
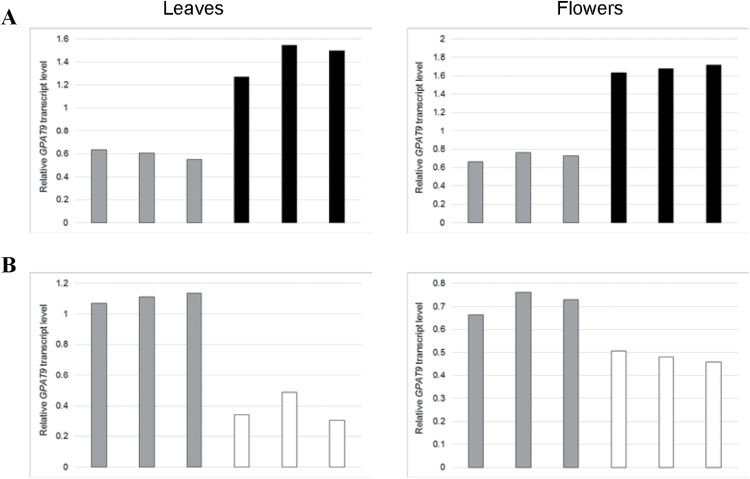
Transgenic lines constitutively overexpressing or down-regulating GPAT9 in Arabidopsis were generated (Supplementary Fig. S2) to elucidate the function of this gene at various stages of plant development in planta. Initially, leaf and floral tissues were harvested from a selection of independent T1 lines, as well as wild-type plants, and levels of GPAT9 transcripts were assayed using quantitative real-time RT-PCR. In the case of GPAT9-OE lines, GPAT9 transcripts were increased by between 113% and 159% (leaf tissue), and 128% and 139% (floral tissue), compared to wild type, while in GPAT9-RNAi lines, GPAT9 transcripts were reduced by between 56% and 72% (leaf tissue), and 30% and 36% (floral tissue), compared to wild type (Fig. 3). Furthermore, developing T2 siliques containing T3 seeds (14 DAF) were also harvested from two independent transgenic GPAT9-OE and GPAT9-RNAi lines (utilized throughout this study), respectively, and following the identification of homozygous lines in each case, appropriate alterations in GPAT9 expression were also confirmed in this tissue type (Supplementary Fig. S3). In GPAT9-OE siliques, GPAT9 transcripts were increased by 127% (GPAT9-OE-6) and 73% (GPAT9-OE-7) compared to wild-type levels, while in GPAT9-RNAi siliques, GPAT9 transcripts were diminished by 34% (GPAT9-RNAi-10) and 52% (GPAT9-RNAi-21) compared to wild-type lines (Supplementary Fig. S3). No obvious morphological changes were observed visually in vegetative or floral tissues from any independent line of any generation. Furthermore, neither germination (which occurred within 24h in all transgenic lines and wild-type plants) nor growth rates were altered in transgenic lines (Supplementary Fig. S9).
Fig. 3.
Confirmation of alterations in constitutive GPAT9 expression in leaves and flowers of GPAT9-OE and GPAT9-RNAi lines compared to wild type. Quantitative real-time RT-PCR analysis of GPAT9 expression in independent T1 GPAT9-OE (A), GPAT9-RNAi (B) and wild-type lines. Blocks denote mean GPAT9 transcript levels from three technical replicates relative to the internal control, PP2AA3. Gray blocks represent wild-type plants, black blocks represent independent GPAT9-OE lines, and white blocks represent independent GPAT9-RNAi lines.
Overexpression and down-regulation of GPAT9 in Arabidopsis results in changes in seed size, as well as seed oil content and composition
To gain insight into the possible role of GPAT9 in Arabidopsis seeds, we subjected homozygous T3 GPAT9-OE and GPAT9-RNAi seeds to an in-depth analysis of seed morphology. Interestingly, relative mean seed weights were increased by 13.4% (GPAT9-OE-6) and 9.4% (GPAT9-OE-7) in the GPAT9-OE lines, while GPAT9-RNAi lines exhibited an 18.7% (GPAT9-RNAi-10) and 11.0% (GPAT9-RNAi-21) reduction in seed weight compared to wild type (Supplementary Fig. S4A). In line with this, mean seed areas in GPAT9-OE lines were increased 8.4% (GPAT9-OE-6) and 2.2% (GPAT9-OE-7), while GPAT9-RNAi lines displayed mean seed areas that were decreased by 10.3% (GPAT9-RNAi-10) and 9.7% (GPAT9-RNAi-21) compared to wild-type seeds (Supplementary Fig. S4B). No significant differences were noted in seed yields between either T3 homozygous GPAT9-OE (255.3mg/plant ±28.7 SE, n=12) and wild-type (253.7mg/plant ±17.1 SE, n=14) lines, or GPAT9-RNAi (304.4mg/plant ±12.1 SE, n=12) and wild-type (330.8mg/plant ±18.0 SE, n=14) lines.
Similarly, while overexpression of GPAT9 in GPAT9-OE lines resulted in increased seed oil content, down-regulation of this gene in GPAT9-RNAi lines yielded significant decreases in seed oil content (Supplementary Figs S5–S8). To ascertain that any changes in lipid production in transgenic seeds were not a result of phenotypic alterations in vegetative or floral tissues, we also produced lines that overexpressed or down-regulated GPAT9 in a seed-specific manner (GPAT9-SS-OE and GPAT9-SS-RNAi; Supplementary Fig. S2) and confirmed these alterations in expression via qRT-PCR (Supplementary Fig. S3). As was the case for constitutive lines, both seed-specific overexpressing and RNAi lines exhibited small but significant increases and decreases in oil content, respectively, compared to wild-type seeds (Supplementary Figs S5–S8). Furthermore, alterations in seed oil composition were evident in both overexpression and RNAi lines, with overall increases in 18:1 and 22:0, and decreases in 20:1 in overexpression lines (Supplementary Figs S5, S6) and increases in C18:0, C18:1 and C18:3, and reductions in 16:0, 18:2, 20:1, 22:0 and 22:1 in RNAi lines (Supplementary Figs S7, S8). These results are consistent with a recent report of GPAT9 down-regulation via amiRNA (Shockey et al., 2016). Further details regarding these seed-based results can be found in Supplementary Data S1.
Overexpression of GPAT9 in Arabidopsis increases triacylglycerol and polar lipid levels in leaves
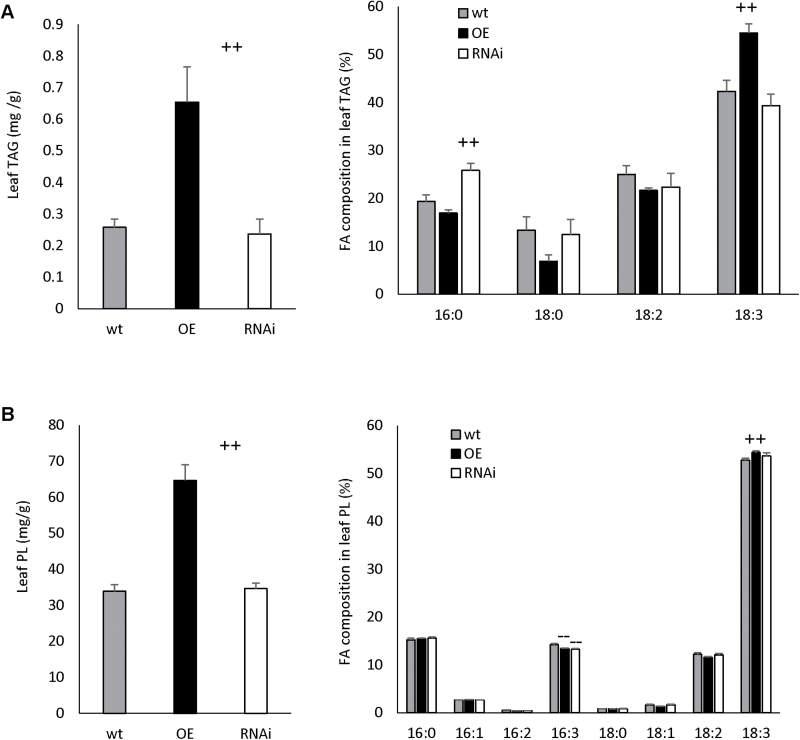
Leaf lipids were analyzed to gain insight to the possible role of GPAT9 in lipid biosynthesis in non-seed organs. No significant differences were observed in the levels of either TAG or PL in GPAT9-RNAi lines compared to wild-type plants. On the contrary, GPAT9-OE leaves exhibited a 153.3% relative increase in TAG and a 91.0% relative increase in PL compared to wild-type plants (Fig. 4).
Fig. 4.
Lipid content and composition in constitutive GPAT9-OE, GPAT9-RNAi and wild-type 35 DAP rosette leaves. (A) Triacylglycerol (TAG) content and fatty acid composition. (B) Polar lipid (PL) content and fatty acid composition. Blocks represent the mean values of three biological replicates from two independent homozygous T3 lines in each case. Bars denote standard errors. Significant increases and decreases compared to wild type (as measured by Student’s t-test) are indicated by ++ and - -, respectively (P≤0.01). DW, dry weight; FA, fatty acid; OE, GPAT9-OE lines; RNAi, GPAT9-RNAi lines; wt, wild type.
In addition to increases in the lipid content of GPAT9-OE leaves, alterations in the FA composition of both lipid classes were also observed in these lines. In the case of TAG, a significant 28.8% relative increase in 18:3 was observed in GPAT9-OE lines compared to wild type, with concomitant decreases in 16:0 (12.4%), 18:0 (48.6%) and 18:2 (13.3%; although these three latter changes were not significant). In leaf PL of GPAT9-OE lines, there was a significant decrease of 5.5% relative to wild type in 16:3Δ7cis,10cis,13cis and a significant increase of 3.1% in 18:3. GPAT9-RNAi lines also displayed changes in FA composition within leaf tissues, with a significant increase in 16:0 (33.6%) in leaf TAG and a significant decrease in 16:3 (6.7%) in PL relative to wild-type plants (Fig. 4).
Altering the constitutive expression of GPAT9 in Arabidopsis enhances production of lipid droplets in pollen grains
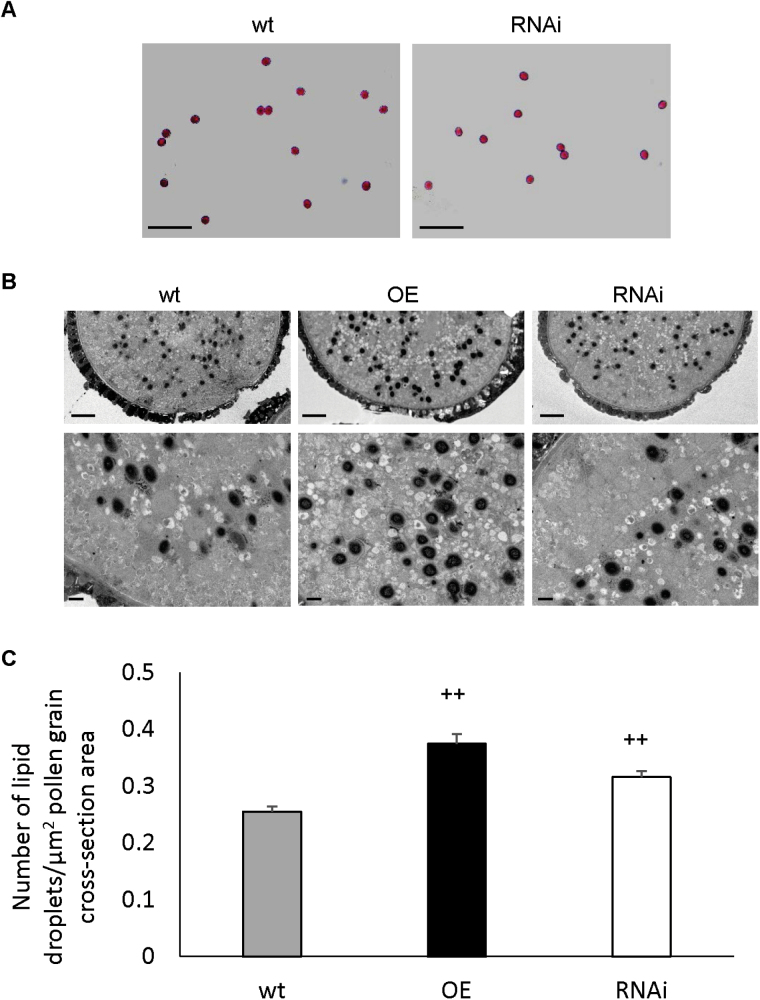
Since knockout of gpat9 has been shown to result in a male gametophytic lethality phenotype (Shockey et al., 2016) and GPAT9 has a high expression level in pollen (Fig. 2), we further hypothesized that GPAT9 may contribute to lipid biosynthesis in pollen grains. Since no differences were observed in the fertility of GPAT9-RNAi lines compared to wild type in this study, we further conducted Alexander staining (Alexander, 1969) of pollen grains from homozygous constitutive GPAT9-RNAi and wild-type plants to ensure pollen viability was not subtly affected by down-regulation of GPAT9 expression. As expected, pollen from both GPAT9-RNAi and wild-type plants appeared morphologically normal and exhibited pink staining, confirming viability of the pollen grains in both cases (Fig. 5A).
Fig. 5.
Effect of constitutive GPAT9 overexpression and down-regulation on pollen development in Arabidopsis. (A) Alexander staining of pollen derived from homozygous wild-type and GPAT9-RNAi lines indicates that pollen viability was not affected by down-regulation of GPAT9. Scale bars, 100 μm. (B) Representative TEM micrographs of sectioned pollen grains derived from homozygous GPAT9-OE, GPAT9-RNAi and wild-type lines. As osmium tetroxide was utilized as a staining agent, lipid droplets are apparent as dark spots. Scale bars, 2 μm (top panel) and 0.5 μm (bottom panel). (C) Number of lipid droplets per square unit area of pollen grain cross-section. Blocks represent mean values from GPAT9-OE (n=29), GPAT9-RNAi (n=36) and wild-type (n=21) pollen grains obtained from at least three separate plants in each case. Bars indicate standard errors. Significant increases compared to wild-type (as measured by Student’s t-test) are indicated by ++ (P≤0.01). OE, GPAT9-OE lines, RNAi, GPAT9-RNAi lines; wt, wild-type.
To gain a deeper understanding of a possible role for GPAT9 in lipid biosynthesis within pollen grains, which are known to accumulate relatively high levels of TAG (Murphy, 2005), we also carried out TEM of pollen sections from homozygous constitutive GPAT9-OE, GPAT9-RNAi and wild-type lines. Examination of the resulting micrographs revealed pollen that was undistinguishable in morphology between both transgenic and wild-type lines, with no obvious alterations in the production of the extracellular exine lipid layer (Fig. 5B). Interestingly, the number of internal lipid droplets observed per unit area of pollen grain cross-section was significantly increased in both GPAT9-OE (46.9% relative increase) and GPAT9-RNAi (24.0% relative increase) lines compared to wild type (Fig. 5C); however, the increase detected in the overexpression lines was significantly greater than that observed in the RNAi lines. In GPAT9-OE pollen, lipid droplets also appeared slightly larger on average than in wild-type or RNAi plants (Fig. 5B), although we found that accurate measurements of droplet areas were not possible due to the fact that their borders were not sufficiently well-defined. Furthermore, while lipid droplets in wild-type lines were generally absent in a band near the outer edges of the grain, those from overexpression lines tended to accumulate indiscriminately throughout the pollen grain (Fig. 5B). Taken together, these results show that perturbation of GPAT9 through either overexpression or knockdown disrupts normal lipid droplet accumulation in pollen grains.
Cuticular and epicuticular lipids are not affected in constitutive GPAT9-OE or GPAT9-RNAi lines
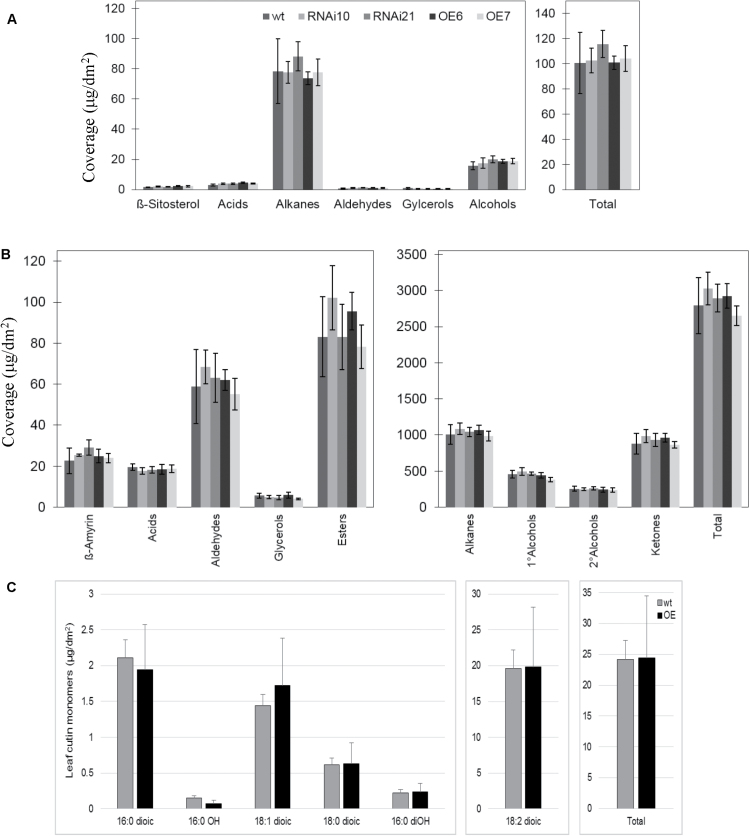
In order to determine whether there was any obvious reduction in cuticular lipids in constitutive GPAT9-RNAi lines, we carried out toluidine blue staining of both rosette leaves (28 DAP) and open flowers. This method is utilized extensively to examine cuticle defects in plant tissues (Tanaka et al., 2004). No staining was observed in either wild-type or GPAT9-RNAi lines, indicating that permeability of the cuticular layer was not reduced to any appreciable extent in the RNAi lines in either leaves or floral tissues (data not shown). In agreement with these observations, extraction and biochemical analysis of epicuticular wax components from leaves and stems showed no obvious changes in content or composition between wild-type and GPAT9-RNAi lines (Fig. 6A, B, respectively, P<0.05). Furthermore, no obvious changes in either surface waxes or cutin monomers were observed in plant lines constitutively overexpressing GPAT9 (Fig. 6C; P<0.05). Taken together, these data reveal that modulation of GPAT9 expression has no obvious effects on the production of lipids that are destined for the plant surface.
Fig. 6.
Epicuticular wax and cutin monomer content and composition of wild-type Arabidopsis and Arabidopsis with modulated constitutive GPAT9 expression. (A) Epicuticular wax profile of leaves of wild-type (wt), GPAT9-RNAi (RNAi), and GPAT9-OE (OE) Arabidopsis. Data are presented as average ±standard deviation of five replicates. (B) Epicuticular wax profile of stem of wt, RNAi, and OE Arabidopsis. Data are presented as average±standard deviation of five replicates. C, Leaf cutin monomers from WT and OE lines. Two technical replicates were carried out in each case. Blocks represent mean values from three biological replicates and bars denote standard errors.
Discussion
The ER-localized Kennedy pathway for the synthesis of glycerolipids is believed to play a key role in the production of TAG and membrane lipids in most organisms. The first acylation step of this pathway is believed to be catalyzed by an ER-bound sn-1 regio-specific GPAT with a preference for acyl-CoA as its substrate in various organisms; however, there has been much debate regarding the particular enzyme that catalyzes this reaction in plants (Chen et al., 2015). Very recently, knockdown of Arabidopsis GPAT9 has been found to result in reduced seed oil content (Shockey et al., 2016), which suggests that this gene is involved in seed TAG biosynthesis in plants. However, direct evidence for such a role in the form of enzymatic activity, as well as its function in non-seed tissues, is lacking. As such, we have carried out an in-depth characterization of this gene and the enzyme it encodes in an attempt to gain further insight into its function in plants.
Unlike all previously characterized Arabidopsis GPAT enzymes, which demonstrate sn-2 regio-specificity and a preference for omega-oxidized-acyl-CoAs as their substrates (Yang et al., 2012), our in vitro enzyme assays revealed that GPAT9 exhibits sn-1 regio-specificity and a preference for acyl-CoAs rather than DCA-CoA as a substrate (Fig. 1B). These results provide direct evidence that GPAT9 catalyzes the first step in the Kennedy pathway and regulates the in vivo biosynthesis of intracellular lipids in Arabidopsis. Furthermore, phosphatase activity has been confirmed previously in Arabidopsis GPAT4, GPAT6 (Yang et al., 2010) and GPAT8 (Yang et al., 2012), resulting in the conversion of a proportion of the LPA product to monoacylglycerol, which has been suggested to be important for polyester assembly (Yang et al., 2010). However, in silico analysis of the deduced GPAT9 amino acid sequence revealed that several amino acids known to be essential for this phosphatase activity in other plant GPATs, including DxD residues in motif I (DxD[T/V][L/V]) and K-D-D residues in motif III (K-[G/S][D/S]xxx[D/N] (Yang et al., 2010), were altered in GPAT9. Further confirmation of the lack of phosphatase activity was obtained via in vitro GPAT9 enzyme assays, and is in line with expectations for a GPAT involved in the Kennedy pathway.
It has been suggested previously that the plant GPAT responsible for the first acylation step of the Kennedy pathway might not exhibit a strong substrate preference, and instead acyl-CoA substrate availability might determine the acyl composition of the sn-1 position of TAG (Snyder et al., 2009). Conversely, in this study, our enzyme assay data indicate that of the acyl-CoA substrates tested, GPAT9 exhibited the highest activity with 18:1-CoA (Fig. 1A). This correlated well with our GPAT9-OE and GPAT9-SS-OE seed lipid data (Supplementary Figs S5, S6), which also showed consistent significant increases in the proportion of 18:1 compared to wild-type plants. Taken together, these findings imply that Arabidopsis GPAT9 enzymes may indeed display preferences for certain FA substrates. A comparative study of substrate specificity and selectivity of GPAT9s from diverse oilseed plants and oleaginous microalgae (such as the recently reported L. incisa GPAT9-like GPAT; Iskandarov et al., 2015) could further expand our understanding of the role of these enzymes in TAG biosynthesis and their potential application for the future improvement of oilseed crop quality.
As both our quantitative real-time RT-PCR and GUS fusion results indicated that GPAT9 is expressed at relatively high levels in developing leaves (Fig. 2, Supplementary Fig. S1), we sought to understand its possible function in this tissue type by analyzing the production of both extracellular and intracellular lipids in GPAT9-OE and GPAT9-RNAi lines. In line with our in vitro enzyme assay results, quantification of cutin monomer and epicuticular wax content and composition in transgenic lines indicated that alteration of GPAT9 transcript levels did not have any effect on the production of surface lipids (Fig. 6). This, along with the fact that we did not detect significant amounts of GPAT9 expression in epidermal cells (Fig. 2), implies that GPAT9 does indeed have a very different function in lipid biosynthesis from the previously characterized GPAT1 and GPAT4–8 enzymes.
In plant leaves, intracellular lipids are synthesized via two complementary pathways associated with the chloroplast (prokaryotic pathway) and ER (eukaryotic pathway), and mainly comprise thylakoid membrane galactolipids and other polar membrane lipids. A very small proportion of TAG is also generated in leaf tissues and is thought to participate in carbon storage and/or membrane lipid remodeling, but as of yet, its exact role remains elusive (Murphy and Parker, 1984; Murphy, 2001; Kaup et al., 2002; Lin and Oliver, 2008; Slocombe et al., 2009).
Previously, mutation or overexpression of the final ER-bound acyltransferase in the Kennedy pathway (DGAT) has been shown to affect the levels of TAG in leaves (Bouvier-Navé et al., 2000; Slocombe et al., 2009), which hinted at the possibility that this may also be the case for GPAT9. Indeed, in 35 DAP leaves from GPAT9-OE lines, we found a significant and substantial increase in both PL and TAG levels compared to wild-type lines (Fig. 4), suggesting that GPAT9 contributes to TAG and membrane lipid biosynthesis in Arabidopsis leaves. Since both PL and TAG would be expected to be derived, at least in part, from pathways that initiated with the sn-1 acylation of G3P by an ER-bound GPAT, it is not surprising that both lipid fractions were increased in transgenic overexpression lines. Since polar lipids, including galactolipids and phospholipids, have important roles in leaf development and function, GPAT9 activity may also affect the cellular and physiological performance of leaves. Therefore, further analysis of various subclasses of polar lipids, including galactolipids, in GPAT9-OE leaves would be an interesting next step in future investigations of GPAT9 function in leaf development.
We also observed a significant increase in 18:3 and concomitant decrease in 16:3 in the PL fraction from GPAT9-OE leaves compared to wild type, which strongly implied a shift towards the eukaryotic pathway (18:3) from the prokaryotic pathway (16:3), providing further confirmation that GPAT9 plays a role in the ER-localized eukaryotic glycerolipid biosynthesis pathway (Fig. 4B). Interestingly, a similar phenomenon was observed in a previous study when the plastidial Arabidopsis GPAT (ATS1), which is involved in the prokaryotic pathway of leaf lipid biosynthesis, was knocked out in Arabidopsis: the mutant plant was essentially converted from a 16:3 to a 18:3 plant (Kunst et al., 1988).
While no significant differences were noted in the levels of TAG or PL in GPAT9-RNAi leaves compared to wild type, FA compositional changes were evident in both lipid types (Fig. 4). Since GPAT9 is expressed at moderate levels in a relatively constitutive manner (Fig. 2), but knockout is known to be lethal (Shockey et al., 2016), it appears that down-regulation of GPAT9 transcripts by 34% and 52% yields enough GPAT9 activity to maintain normal plant morphology and lipid production. It is also possible that other enzymes, or perhaps an alternative lipid biosynthetic pathway, contribute in the compensation for down-regulation of GPAT9 in leaf tissues.
Interestingly, the changes in fatty acid composition in PL from both GPAT9-OE and GPAT9-RNAi leaves were very small compared to those observed in TAG. Since fatty acid composition in PL can significantly affect the function of cell membranes, and there is substantial interchange of polar lipids between the ER and chloroplasts in leaves, it is feasible that plants possess mechanisms to minimize fatty acid compositional changes in PL to maintain membrane function.
In addition to their presence in leaves and seeds, lipids are also deposited both externally on the pollen surface as a complex mixed polymer termed exine, as well as internally in the form of lipid droplets, which consist mainly of storage TAG (Stanley and Linskens, 1974; Piffanelli et al., 1998; Murphy, 2006). Our GUS fusion results (Fig. 2) demonstrated that within floral tissues, GPAT9 was expressed preferentially in pollen grains, suggesting a possible role in the production of these pollen lipids. While exine formation did not appear to be affected in GPAT9-OE lines, we found that the number of intracellular lipid droplets was significantly increased, and the droplets themselves seemed to be larger, compared to wild-type plants (Fig. 5B, C). Taken together, these results point to a role for GPAT9 in TAG biosynthesis within pollen grains as has been found previously to be the case for the Kennedy pathway DGAT1 in Arabidopsis (Zhang et al., 2009).
Unexpectedly, we also found an increase in lipid droplet production in our GPAT9-RNAi lines compared to wild type, although this enhancement was significantly less than that observed in the overexpression lines (Fig. 5C). These results were in line with the fact that we saw no abnormalities in pollen morphology or viability within these lines (Fig. 5A), but do not appear to corroborate the precise role of GPAT9 in lipid biosynthesis implied by our overexpression lines. Intriguingly, a similar phenomenon was noted previously following the knockout of GPAT1 in Arabidopsis (Zheng et al., 2003). GPAT1 was found to play a crucial role in pollen development and morphology, whereby the size of lipid droplets within pollen grains was found to be significantly increased compared to wild type (Zheng et al., 2003). These findings could possibly be attributed to an enhancement or activation of other GPAT genes, which in turn over-compensated for down-regulation of GPAT1 and GPAT9. Alternatively, other lipid biosynthetic pathways may be capable of taking over in such instances, as has been found to be the case for the acyl-CoA-dependent Kennedy pathway DGAT1 and acyl-CoA-independent phospholipid:diacylglycerol acyltransferase in pollen lipid biosynthesis, which requires both genes encoding these TAG-biosynthetic enzymes to be down-regulated/inactivated for pollen lipid biosynthesis to be affected (Zhang et al., 2009).
Significant increases and decreases in the content of seed oil, as well as seed size, were also noted in both constitutive and seed-specific GPAT9 overexpression and RNAi lines, respectively, compared to wild type (Supplementary Figs S4–S8). These findings confirm the recent Arabidopsis GPAT9 knockdown results obtained by Shockey et al. (2016) and demonstrate that this gene also plays a role in seed oil biosynthesis. Taken together, our results provide clear evidence that GPAT9 contributes to glycerolipid biosynthesis in various tissue types via a role in the Kennedy pathway.
Supplementary data
Supplementary data are available at JXB online.
Data S1. Description of supplementary results.
Table S1. Comparison of transcript levels from selected lipid biosynthetic genes.
Figure S1. Quantitative real-time RT-PCR analysis of Arabidopsis GPAT9 expression.
Figure S2. Schematic representations of experimental plant transformation constructs.
Figure S3. Confirmation of alterations in GPAT9 expression in siliques from GPAT9-OE and GPAT9-RNAi homozygous lines compared to wild type.
Figure S4. Seed weight and seed area in homozygous GPAT9-OE, GPAT9-RNAi and wild-type lines.
Figure S5. Oil content and fatty acid composition of T2 GPAT9-OE, GPAT9-SS-OE and wild-type seeds.
Figure S6. Seed oil content and composition in homozygous T3 GPAT9-OE and wild-type lines.
Figure S7. Oil content and fatty acid composition of T2 GPAT9-RNAi, GPAT9-SS-RNAi and wild-type seeds.
Figure S8. Seed oil content and composition in homozygous T3 GPAT9-RNAi and wild-type lines.
Figure S9. Growth rates of GPAT9-OE, GPAT9-RNAi and wild-type seedlings.
Figure S10. Confirmation of alterations in GPAT9 expression in GPAT9-SS-OE and GPAT9-SS-RNAi lines compared to wild type.
Acknowledgements
The authors would like to thank Arlene Oatway for her help with the microscopy work carried out in this study, as well as Annie Wong for her technical assistance throughout. RJW acknowledges the support provided by the Alberta Innovates Bio Solutions, Alberta Enterprise and Advance Education, the Canada Foundation for Innovation, the Canada Research Chairs Program and the Natural Sciences and Engineering Research Council of Canada.
Glossary
Abbreviations:
- DAF
days after flowering
- DAP
days after planting
- DCA
dicarboxylic acid
- DGAT
diacylglycerol acyltransferase
- ER
endoplasmic reticulum
- FA
fatty acid
- G3P
sn-glycerol-3-phosphate
- GPAT
glycerol-3-phosphate acyltransferase
- GUS
β-glucuronidase
- LPA
lysophosphatic acid
- PC
phosphatidylcholine
- PL
polar lipids
- PP2AA3
PROTEIN PHOSPHATASE 2A SUBUNIT 3
- TAG
triacylglycerol.
References
- Alexander MP. 1969. Differential staining of aborted and nonaborted pollen. Stain Technology 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Bates PD, Stymne S, Ohlrogge J. 2013. Biochemical pathways in seed oil synthesis. Current Opinion in Plant Biology 16, 358–364. [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. 2007. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19, 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M. 2004. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant Journal 40, 920–930. [DOI] [PubMed] [Google Scholar]
- Bouvier-Navé P, Benvenist P, Oelkers P, Sturley SL, Schaller H. 2000. Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferase. European Journal of Biochemistry 267, 85–96. [DOI] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR. 1986. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the 16:3 plant Arabidopsis thaliana . Biochemical Journal 235, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AP, Kroon JTM, Swarbreck D, Febrer M, Larson TR, Graham IA, Caccamo M, Slabas AR. 2012. Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. Plos One 7, e30100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Li JL, Li D, Tobin JF, Gimeno RE. 2006. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proceedings of the National Academy of Sciences, USA 103, 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Woodfield HK, Pan X, Harwood JL, Weselake RJ. 2015. Acyl-trafficking during plant oil accumulation. Lipids 50, 1057–1068. [DOI] [PubMed] [Google Scholar]
- Chen X, Chen G, Truksa M, Snyder CL, Shah S, Weselake RJ. 2014. Glycerol-3-phosphate acyltransferase 4 is essential for the normal development of reproductive organs and the embryo in Brassica napus . Journal of Experimental Botany 65, 4201–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Snyder CL, Truksa M, Shah S, Weselake RJ. 2011. a sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signal Behavior 6, 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Truksa M, Snyder CL, El-Mezawy A, Shah S, Weselake RJ. 2011. b Three homologous genes encoding sn-glycerol-3-phosphate acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus . Plant Physiology 155, 851–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Benning C. 2002. Galactolipids rule in seed plants. Trends in Plant Science 7, 112–118. [DOI] [PubMed] [Google Scholar]
- Frisch DA, van der Geest AHM, Dias K, Hall TC. 1995. Chromosomal integration is required for spatial regulation of expression from the β-phaseolin promoter. Plant Journal 7, 503–512. [Google Scholar]
- Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT. 2009. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiology and Biochemistry 47, 867–879. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molecular Biology 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Iskandarov U, Sitnik S, Shtaida N, Didi-Cohen S, Leu S, Khozin-Goldberg I, Cohen Z, Boussiba S. 2015. Cloning and characterization of a GPAT-like gene from the microalga Lobosphaera incisa (Trebouxiophyceae): overexpression in Chlamydomonas reinhardtii enhances TAG production. Journal of Applied Phycology 28, 907–919 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MA, Tuttle HA, Eigenbrode SD, Feldmann KA. 1995. Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiology 108, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson LG, Lenman M, Ericson ML, Rask L. 1987. Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus . The Journal of Biological Chemistry 262, 12196–12201. [PubMed] [Google Scholar]
- Kaup MT, Froese CD, Thompson JE. 2002. A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiology 129, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Yoshimura T, Okuda S. 1981. A new method for the preparation of acyl-CoA thioesters. Journal of Biochemistry 89, 337–339. [DOI] [PubMed] [Google Scholar]
- Khozin-Goldberg I, Cohen Z. 2011. Unraveling algal lipid metabolism: recent advances in gene identification. Biochimie 93, 91–100. [DOI] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C. 1988. Altered regulation of lipid biosynthesis in a mutant Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proceedings of the National Academy of Sciences, USA 85, 4143–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin TM, Wang P, Coleman RA. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38, 5764–5771. [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina L, Pollard M, Ohlrogge JB. 2007. a Identification of acyltransferases required for cutin synthesis and production of cutin with suberin-like monomers. Proceedings of the National Academy of Sciences, USA 104, 18339–18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Ohlrogge J, Pollard M. 2007. b Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiology 144, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Oliver DJ. 2008. Role of triacylglycerols in leaves. Plant Science 175, 233–237. [Google Scholar]
- Mietkiewska E, Siloto RMP, Dewald J, Shah S, Brindley DN, Weselake RJ. 2011. Lipins from plants are phosphatidate phosphatases that restore lipid synthesis in a pah1Δ mutant strain of Saccharomyces cerevisiae . FEBS Journal 278, 764–775. [DOI] [PubMed] [Google Scholar]
- Murphy DJ. 2006. The extracellular pollen coat in members of the Brassicaceae: Composition, biosynthesis, and functions in pollination. Protoplasma 228, 31–39. [DOI] [PubMed] [Google Scholar]
- Murphy DJ. 2005. Plant lipids. biology, utilisation and manipulation. Oxford, UK: Blackwell Publishing Ltd. [Google Scholar]
- Murphy DJ. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Progress in Lipid Research 40, 325–438. [DOI] [PubMed] [Google Scholar]
- Murphy GJP, Parker ML. 1984. Lipid composition and carbon turnover of wheat leaf oleosomes. Journal of Experimental Botany 35, 348–355. [Google Scholar]
- Nishida I, Tasaka Y, Shiraishi H, Murata N. 1993. The gene and the RNA for the precursor to the plastid-located glycerol-3-phosphate acyltransferase of Arabidopsis thaliana . Plant Molecular Biology 21, 267–277. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. 1995. Lipid biosynthesis. Plant Cell 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Siloto RMP, Wickramarathna AD, Mietkiewska E, Weselake RJ. 2013. Identification of a pair of phospholipid:diacylglycerol acyltransferases from developing flax. Linum usitatissimum L. seed catalyzing the selective production of trilinolenin. Journal of Biological Chemistry 288, 24173–24188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C. 2010. A simplified method for differential staining of aborted and non-aborted pollen grains. International Journal of Plant Biology 1, e13. [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ. 1998. Biogenesis and function of the lipidic structures of pollen grains. Sexual Plant Reproduction 11, 65–80. [Google Scholar]
- Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. Journal of Cell Biology 17, 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Slack CR. 1982. Cellular organization of glycerolipid metabolism. Annual Review of Plant Biology 33, 97–132. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Shan D, Li JL, Wu L, Li D, Hurov J, Tobin JF, Gimeno RE, Cao J. 2010. GPAT3 and GPAT4 are regulated by insulin-stimulated phosphorylation and play distinct roles in adipogenesis. Journal of Lipid Research 51, 1971–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J, Regmi A, Cotton K, Adhikari N, Browse J, Bates PD. 2016. Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiology 170, 163–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe SP, Cornah J, Pinfield-Wells H, Soady K, Zhang QY, Gilday A, Dyer JM, Graham IA. 2009. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnology Journal 7, 694–703. [DOI] [PubMed] [Google Scholar]
- Snyder CL, Yurchenko OP, Siloto RM, Chen X, Liu Q, Mietkiewska E, Weselake RJ. 2009. Acyltransferase action in modification of seed oil biosynthesis. New Biotechnology 26, 11–16. [DOI] [PubMed] [Google Scholar]
- Stanley RG, Linskens HF. 1974. Pollen. New York: Springer-Verlag. [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. 2005. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology 139, 1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. 2004. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant Journal 37, 139–146. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An electronic fluorescent pictograph browser for exploring and analyzing large-scale biological data sets. Plos One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Hu M, Martin T, Wang C, Li XQ, Tian L, Brown D, Miki B. 2003. The cryptic enhancer elements of the tCUP promoter. Plant Molecular Biology 51, 351–362. [DOI] [PubMed] [Google Scholar]
- Yang WL, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge J. 2010. A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proceedings of the National Academy of Sciences, USA 107, 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB. 2012. A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference and evolution. Plant Physiology 160, 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB. 2009. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21, 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J. 2003. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15, 1872–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Zou J. 2001. The initial step of the glycerolipid pathway: Identification of glycerol-3-phosphate/dihydroxyacetone phosphate and dual substrate acyltransferases in Saccharomyces cerevisiae. Journal of Biological Chemistry 276, 41710–41716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.