Highlight
PSE1 regulates Pb tolerance mainly through GSH-dependent phytochelatin synthesis by activating the expression of the genes involved in phytochelatin synthesis and at least partially through activating the expression of PDR12.
Key words: Arabidopsis, glutathione, lead tolerance, phytochelatin, PSE1, PDR12.
Abstract
Lead (Pb) is a dangerous heavy metal contaminant with high toxicity to plants. However, the regulatory mechanism of plant Pb tolerance is poorly understood. Here, we showed that the PSE1 gene confers Pb tolerance in Arabidopsis. A novel Pb-sensitive mutant pse1-1 (Pb-sensitive1) was isolated by screening T-DNA insertion mutants. PSE1 encodes an unknown protein with an NC domain and was localized in the cytoplasm. PSE1 was induced by Pb stress, and the pse1-1 loss-of-function mutant showed enhanced Pb sensitivity; overexpression of PSE1 resulted in increased Pb tolerance. PSE1-overexpressing plants showed increased Pb accumulation, which was accompanied by the activation of phytochelatin (PC) synthesis and related gene expression. In contrast, the pse1-1 mutant showed reduced Pb accumulation, which was associated with decreased PC synthesis and related gene expression. In addition, the expression of PDR12 was also increased in PSE1-overexpressing plants subjected to Pb stress. Our results suggest that PSE1 regulates Pb tolerance mainly through glutathione-dependent PC synthesis by activating the expression of the genes involved in PC synthesis and at least partially through activating the expression of the ABC transporter PDR12/ABCG40.
Introduction
Because of human activities and industrial processes, toxic heavy metals have been released or leached into the environment, which causes adverse effects on ecosystems (Ross, 1994; Steinnes, 2013). Among the heavy metals, lead (Pb) is a dangerous heavy metal contaminant that results in inhibition of metabolic processes, plant growth, and productivity (Meagher, 2000; Hall, 2002; Sharma and Dubey, 2005; Israr et al., 2011; Ali et al., 2014; Yang et al., 2015). At the cellular and molecular level, Pb toxicity leads to disturbed water balance, mineral nutrition, and membrane organization; it also inhibits photosynthetic and enzyme activity, and reduces cell division (Uveges et al., 2002; Israr et al., 2011; Pourrut et al., 2011). Furthermore, Pb causes many serious health problems in growing children and adults through food chain transfer (Kim et al., 1996; Dudka and Miller, 1999; Duruibe et al., 2007).
In order to protect against Pb stress, plants have developed a system of tolerance strategies, which include pumping out Pb at the plasma membrane; reducing the uptake of Pb; chelating or binding Pb to various thiol compounds in the cytosol, such as glutathione (GSH), phytochelatins (PCs), and metallothioneins (MTs), and then sequestering these into inactive organelles; and detoxifying Pb-induced reactive oxygen species (ROS) (Clemens, 2001; Hall, 2002; Verma and Dubey, 2003; Kim et al., 2006; Zafari et al., 2016).
One such strategy involves the induction of specific low molecular weight chelators, such as GSH and PCs, to bind and sequester Pb (Mehra et al., 1995). GSH, a γ-Glu-Cys-Gly tripeptide, can act as a metal chelator through its thiol groups, as a cellular antioxidant, and as a ROS signaling molecule (Jozefczak et al., 2012; Lin and Aaers, 2012; Seth et al., 2012). Therefore, GSH plays an important role in the detoxification of heavy metals and in the metal-induced oxidative stress response. Many studies have confirmed that reduced GSH is the precursor of PCs, which are enzymatically synthesized oligomers containing (γ-Glu-Cys)n-Gly (n=2–11) (Rauser, 1990, 1995, 1999; Zenk, 1996; Cobbett, 2000). PCs sequester Pb to form a complex that is then transported into the vacuole by ATP-binding cassette (ABC) transporters, such as ABCC1 and ABCC2 (Cobbett and Goldsbrough, 2002; Clemens, 2006; Song et al., 2010; Park et al., 2012). Some transporters (such as ABCC1 and ABCC2) sequester heavy metals into inactive organelles (such as the vacuole), and other transporters have also been shown to extrude them across the plasma membrane, such as PDR12 (an ABC transporter, also named ABCG40) (Lee et al., 2005; Verrier et al., 2008).
Phytoremediation is one of the important ways to decontaminate Pb pollution (Suresh and Ravishankar, 2004; Pilon-Smits, 2005; Aken, 2008; M. Chen et al., 2015). The identification of genes involved in Pb accumulation and tolerance is the first step towards their application in phytoremediation. It has been shown that some genes confer Pb tolerance, such as EIN2, ABCP1, ATM3, PDR12, and SbLRR2 (Lee et al., 2005; Kim et al., 2006; Xiao et al., 2008; Cao et al., 2009; Zhu et al., 2013). However, many novel genes involved in Pb tolerance require further identification. In this study, we identified a Pb-sensitive mutant, pse1-1 (Pb-sensitive1), from the Arabidopsis Biological Resource Center (ABRC) stocks. PSE1 encoded an unknown protein with an NC domain and was localized in the cytoplasm. We demonstrated that the transcription of the PSE1 gene was induced by Pb stress, and overexpression of the PSE1 gene led to increased Pb tolerance. The PSE1 gene triggered GSH-dependent PC synthesis by activating the expression of the genes involved in the PC synthesis pathway, which resulted in enhanced Pb accumulation and tolerance. In addition, the expression of PDR12 was increased in PSE1-overexpressing plants subjected to Pb stress, suggesting that a PDR12-dependent mechanism was partially involved in PSE1-mediated Pb tolerance.
Materials and methods
Plant materials, growth conditions, and treatments
Arabidopsis thaliana seeds of wild-type (WT; Col-0), pse1-1 (SALK_046412; Alonso et al., 2003), and transgenic plants were surface-sterilized and then germinated on half-strength Murashige and Skoog (1/2 MS) medium containing 1% (w/v) agar and 2% (w/v) sucrose at pH 5.8. The seeds were vernalized at 4 °C for 3 d in the dark. The plants were grown in a controlled culture room at 22 °C under long-day (16h of light/8h of dark) conditions with a light intensity of 100 µmol m−2 s−1.
For the Pb tolerance test, seeds of the WT, the pse1-1 mutant, and the transgenic plants were germinated and grown vertically on 1/2 MS medium in the absence or presence of the indicated concentrations of Pb(NO3)2 or the glutathione synthesis inhibitor, buthioninesulphoximine (BSO; Sigma) for 2 weeks, to check whether Pb tolerance in PSE1-overexpressing plants is glutathione dependent. Then, the plants were weighed and their root lengths were measured. There were triplicate replicates for the Pb tolerance test, and ~30 plants were used for each measurement. For Pb-inducible gene expression analysis, the plants were grown for 2 weeks on 1/2 MS medium and then exposed to a solution of 0.5mM Pb(NO3)2 or water (control) at the designated time points specified in the text.
Generation of PSE1-overexpressing plants and PSE1- complemented plants
To generate PSE1-overexpressing plants, the coding region of PSE1 (At5g06370) was amplified by PCR using the specific primers PSE1-FP-XhoI and PSE1-RP-HindIII (see Supplementary Table S1 at JXB online), followed by cloning into the pCAMBIA1301 vector at the XhoI and HindIII restriction sites. To generate PSE1-complemented plants, the promoter of PSE1 was amplified by the primers PSE1pro-FP-KpnI and PSE1pro-RP-XhoI (Supplementary Table S1) and then cloned into pXB93 (a pART27 derivative including MCS-GUS, kindly supplied by X.B. Peng and M.X. Sun) using the KpnI and XhoI restriction sites. Then, the amplified fragment of PSE1 cDNA was cloned into the above construct at the XhoI and HindIII restriction sites. The 35S::PSE1 and PSE1 pro ::PSE1 constructs were introduced into the Agrobacterium GV3101 strain, which was then used to transform the WT or the pse1-1 mutant using the floral dip method (Clough et al., 1998). All transgenic lines used in this study were T3 homozygous plants with single-copy insertions, from which two lines (OE-1 and OE-2; COM1 and COM2) were selected for further analysis.
RNA extraction, and quantitative RT-PCR (qRT-PCR) analysis
Total seedling RNA was extracted using Trizol Reagent (Invitrogen, CA) and used in a reverse transcription reaction using PrimeScript™ Reverse Transcriptase (TAKARA, Japan) and the oligo(dT)15 primer (TAKARA, Japan) following the manufacturer’s instructions. For qRT-PCR analysis, the reaction was performed according to the instructions provided for the Bio-Rad iCycleriQ system (Bio-Rad Laboratories, CA, USA) and platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, CA, USA). All experiments were performed at least in triplicate, and the Actin11 (At3g12110) gene was used as an internal control. The primers used are listed in Supplementary Table S1.
GUS (β-glucuronidase) staining and activity assay
In order to generate PSE1 pro ::GUS transgenic plants, a fragment of the PSE1 promoter was amplified by PCR using the primers PSE1pro-FP-KpnI and PSE1pro-RP-XhoI (Supplementary Table S1). The PCR product was cloned into pXB93 using the KpnI and XhoI restriction sites. The construct was transferred into the WT plants using the Agrobacterium strain GV3101 (Clough et al., 1998). Homozygous transgenic plants were germinated on 1/2 MS medium for 2 weeks or grown in a greenhouse for 4–6 weeks. All samples were collected and stained with 5-bromo4-chloro-3-indoyl-β-d-glucuronide (X-Gluc) as reported previously (Fan et al., 2013).
Subcellular localization of PSE1
To build up the construct of 35S::PSE1-GFP, the PSE1 coding region was obtained by PCR using specific primers and then cloned as a KpnI/XhoI fragment into pXB94 (a pART27 derivative including 35S:MCS-eGFP, kindly supplied by X.B. Peng and M.X. Sun). The construct was transferred into the Agrobacterium strain GV3101 by electrophoresis, and the WT plants were transformed. The transgenic lines were used for subcellular localization analysis. The subcellular localization of PSE1 was visualized using confocal microscopy (Leica TCS-SP8; Wetzlar, Hesse-Darmstadt, Germany).
Measurement of Pb content
The WT, PSE1-OE, and pse1-1 mutant plants were grown on 1/2 MS medium in the absence or presence of 0.5mM Pb(NO3)2 for 2 weeks and then sampled to determine the Pb content, which was analysed using the method described by Kim et al. (2006). Digested samples were analysed using an atomic absorption spectrometer (Solaar M6, Thermo Fisher, Waltham, MA, USA). A standard reference material was not used to assess the efficiency of the acid digestion procedure; therefore, while the relative concentrations of Pb in WT, PSE1-OE, and pse1-1 mutant plants are reliable, the actual concentrations reported may be less than the true values.
Determination of GSH and PC contents
The WT, PSE1-OE, and pse1-1 mutant plants were grown on 1/2 MS medium for 2 weeks and treated or not treated with 0.5mM Pb(NO3)2 for 12h, after which they were sampled to determine the GSH and PC contents. The method used to determine the GSH/PC contents followed J. Chen et al. (2015).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GeneBank/EMBL database under the following accession numbers: PSE1 (At5g06370), GSH1 (At4g23100), GSH2 (At5g27380), PCS1 (At5g44070), PCS2 (At1g03980), GR1 (At3g24170), GR2 (At3g54660), ABCC1 (At1g30400), ABCC2 (At2g34660), PDR8 (At1g59870), ATM3 (At5g58270), PDR12 (At1g15520), ACBP1 (At5g53470), and ACTIN11 (At3g12110).
Results
The pse1-1 mutant is Pb sensitive
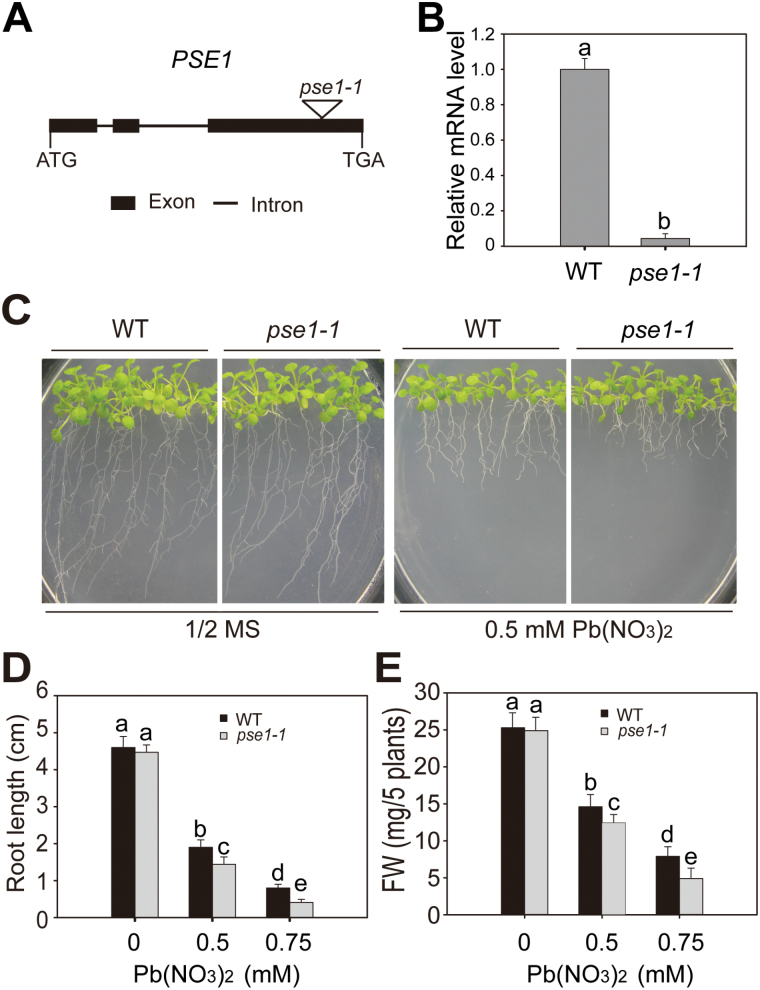
To identify novel genes involved in regulating the response to Pb stress in Arabidopsis, the mutants from the ABRC stock were screened. One T-DNA insertion mutant, designated pse1-1 (Pb-sensitive1; SALK_046412; Alonso et al., 2003), was more sensitive to Pb stress than the WT (Fig. 1). Using PCR analysis, we found that the T-DNA of the pse1-1 mutant was inserted in At5g06370. The At5g06370 gene is a single-copy gene possessing three exons and two introns, and is located on chromosome 5 (Fig. 1A). The transcript expression of PSE1 in pse1-1 was significantly reduced according to the qRT-PCR results (P<0.05; Fig. 1B). There were no significant differences in growth between the WT and pse1-1 plants grown on 1/2 MS medium. However, when 0.5mM Pb(NO3)2 was added, the pse1-1 mutant was more sensitive to Pb stress than the WT (Fig. 1C). The root length and fresh weight of the pse1-1 mutant were significantly inhibited compared with the WT plants in a Pb concentration-dependent manner (P<0.05; Fig. 1D, E). We further generated genetically complemented pse1-1 mutant plants using the full-length PSE1 under the PSE1 promoter. The transcript levels of PSE1 were similar or slightly higher in the transgenic than in the WT plants (P>0.05; Supplementary Fig. S1A). The complemented lines (COM1 and COM2) reversed the Pb-sensitive phenotype of the pse1-1 mutant to the level of the WT (Supplementary Fig. S1B, C).Together, these results indicate that loss-of-function of PSE1 is responsible for the Pb-sensitive phenotype.
Fig. 1.
Pb sensitivity of the pse1-1 mutant. (A) A schematic of the genomic region flanking the T-DNA insertion site in the pse1-1 mutant. Black boxes and black lines indicate exons and introns, respectively, in PSE1. The triangle indicates the site of T-DNA insertion. (B) qRT-PCR analysis of PSE1 transcription levels in 2-week-old seedlings of the pse1-1 mutant and wild-type (WT) plants. ACTIN11 was used as the internal control. Data are presented as the means SE of three replicate experiments. Statistical significance was determined by Student’s t-test; a significant difference (P<0.05) is indicated by different lower case letters. (C) Phenotypes of the pse1-1 mutant under Pb stress. Seedlings were grown on 1/2 MS medium with or without 0.5mM Pb(NO3)2 for 2 weeks. (D and E) Root length (D) and fresh weight (E) of the WT and pse1-1 grown on 1/2 MS medium with or without 0.5/0.75mM Pb(NO3)2 for 2 weeks. Three independent experiments were performed with similar results, each with three biological replicates. Five plants per genotype from one plate were measured for each replicate. Data are presented as the means ±SE, n=3. Statistical significance was determined using ANOVA in combination with post-hoc tests (Tukey); significant differences between the WT and pse1-1 (P<0.05) are indicated by different lower case letters. (This figure is available in colour at JXB online.)
To test whether pse1-1 was also tolerant to other stresses, the pse1-1 mutant was grown on 1/2 MS medium containing CdCl2, Na3AsO4, ZnSO4, and H2O2. Upon treatment with CdCl2, Na3AsO4, and ZnSO4, no significant difference was observed between the pse1-1 mutant and the WT plants (P>0.05; Supplementary Fig. S2A–C). However, the pse1-1 mutant was more sensitive to H2O2 than the WT (P<0.05; Supplementary Fig. S2D). In this study, we focused on the PSE1-mediated Pb tolerance mechanism.
Overexpression of PSE1 results in increased Pb tolerance
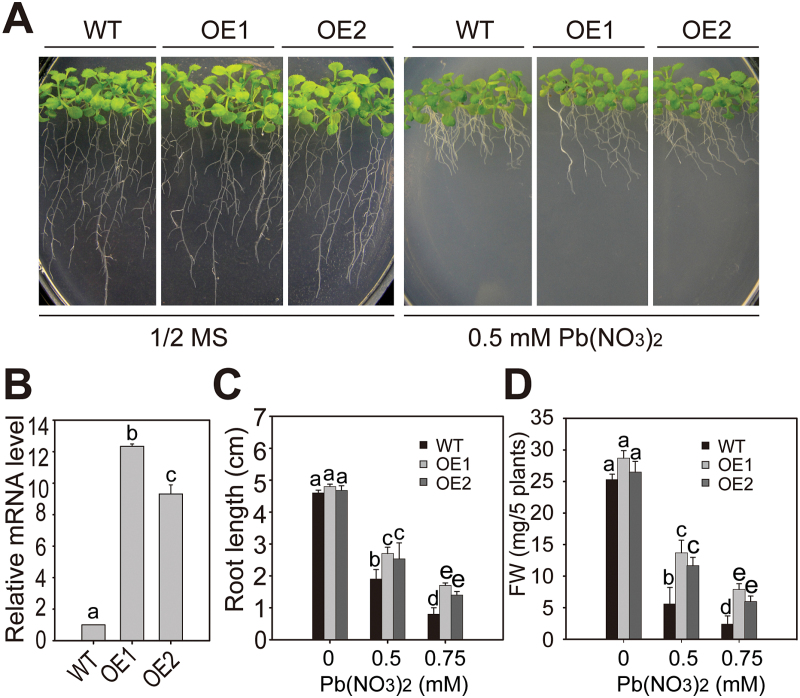
To determine further the role of PSE1 in Pb tolerance, we generated PSE1-overexpressing transgenic plants. The full-length cDNA was cloned under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. At least 10 transgenic lines were obtained, and two independent T3 homozygous lines (OE1 and OE2) were used for further analyses (Fig. 2). The transcript level of PSE1 was higher in PSE1-overexpressing transgenic plants than in the WT plants (P<0.05; Fig. 2A). In 1/2 MS medium, the growth of the WT and PSE1-overexpressing plants was not different; however, in 1/2 MS medium containing Pb(NO3)2, the PSE1-overexpressing plants were more tolerant than the WT plants (P<0.05; Fig. 2B–D). Under normal growth conditions, the pse1-1 mutant, PSE1-overexpressing plants, and the WT exhibited similar growth and development in both vegetative and reproductive phases (Supplementary Fig. S3). Together, these results suggest that PSE1 plays an important role in regulating Pb tolerance.
Fig. 2.
Pb tolerance in the PSE1-overexpressing lines. (A) Growth of wild-type (WT) and PSE1-overexpressing lines (OE1 and OE2) under Pb stress. Seedlings were grown on 1/2 MS medium with or without 0.5mM Pb(NO3)2 for 2 weeks. (B) qRT-PCR analysis of PSE1 transcription levels in 2-week-old seedlings of the WT and PSE1-overexpressing lines (OE1 and OE2). ACTIN11 was used as the internal control. Data are presented as the means ±SE of three replicate experiments. Statistical significance was determined by Student’s t-test; a significant difference (P<0.05) is indicated by different lower case letters. (C and D), Root length (C) and fresh weight (D) of the WT and PSE1-overexpressing lines (OE1 and OE2) grown on 1/2 MS medium with or without 0.5/0.75mM Pb(NO3)2 for 2 weeks. Three independent experiments were performed with similar results, each with three biological replicates. Five plants per genotype from one plate were measured for each replicate. Data are presented as the means ±SE, n=3. Statistical significance was determined using ANOVA in combination with post-hoc (Tukey) tests; significant differences between the WT and PSE1-overexpressing lines (P<0.05) are indicated by different lower case letters. (This figure is available in colour at JXB online.)
The PSE1 gene is induced by Pb stress
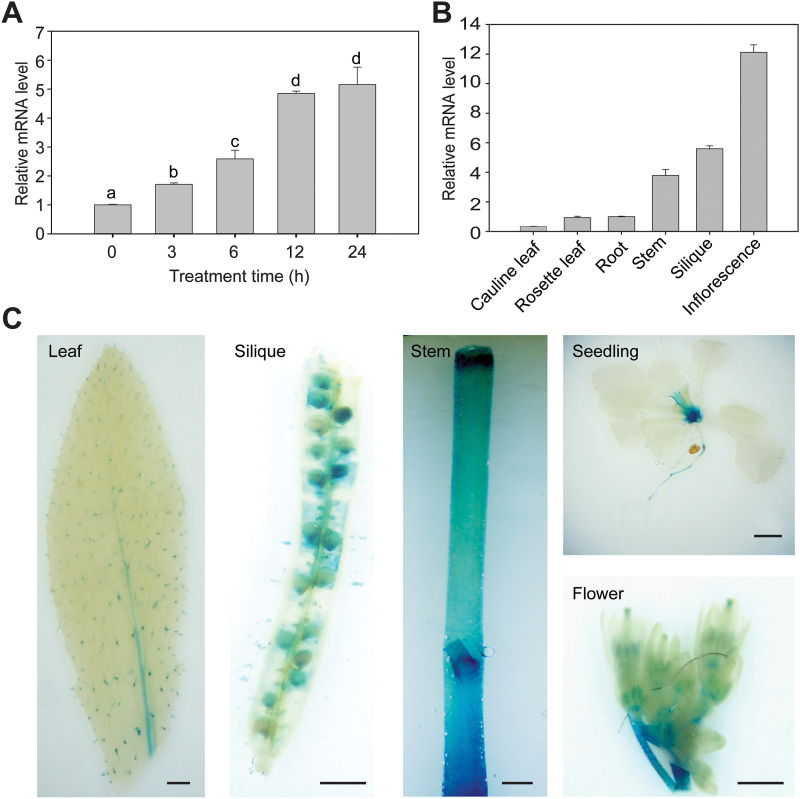
To examine whether the expression of PSE1 is induced by Pb stress, the WT seedlings were treated at different time points after Pb treatment. As shown in Fig. 3A, the expression of PSE1 was significantly induced by Pb stress (P<0.05). To test the expression profile of PSE1 in various tissues, the transcript levels of PSE1 were quantified using qRT-PCR. PSE1 was found to be expressed in most of the examined tissues and highly expressed in the inflorescence, silique, and stem (Fig. 3B). To explore further the spatial expression pattern, we generated transgenic Arabidopsis seedlings expressing a PSE1 pro ::GUS transgene. Histochemical analyses showed that GUS activity was detected in most organs, especially in the inflorescence, silique, and stem (Fig. 3C), which is consistent with the qRT-PCR results shown in Fig. 3B.
Fig. 3.
Expression pattern of the PSE1 gene. (A) Expression of PSE1 in response to Pb stress. Two-week-old wild-type (WT) seedlings grown on 1/2 MS medium were treated with Pb(NO3)2 (0.5mM) for 0, 3, 6, 12, and 24h, and then the tissues were harvested for qRT-PCR analysis. ACTIN11 was used as the internal control. Data are presented as the means ±SE of three replicate experiments. Statistical significance was determined using ANOVA in combination with post-hoc (Tukey) tests; significant differences between treated and control plants (P<0.05) are indicated by different lower case letters. (B) qRT-PCR analysis of PSE1 transcription in different tissues of WT plants. mRNAs were isolated from cauline leaves, rosette leaf, root, stem, silique, and inflorescences of 6-week-old WT plants. ACTIN11 was used as the internal control. (C) Histochemical analysis of PSE1 Pro ::GUS transgenic lines. Leaf (4-week-old), silique (6-week-old), stem (6-week-old), seedling (2-week-old), and flower (6-week-old) samples after 6h of GUS staining are shown in the panels from left to right. Scale bar=1mm. (This figure is available in colour at JXB online.)
PSE1 is localized in the cytoplasm
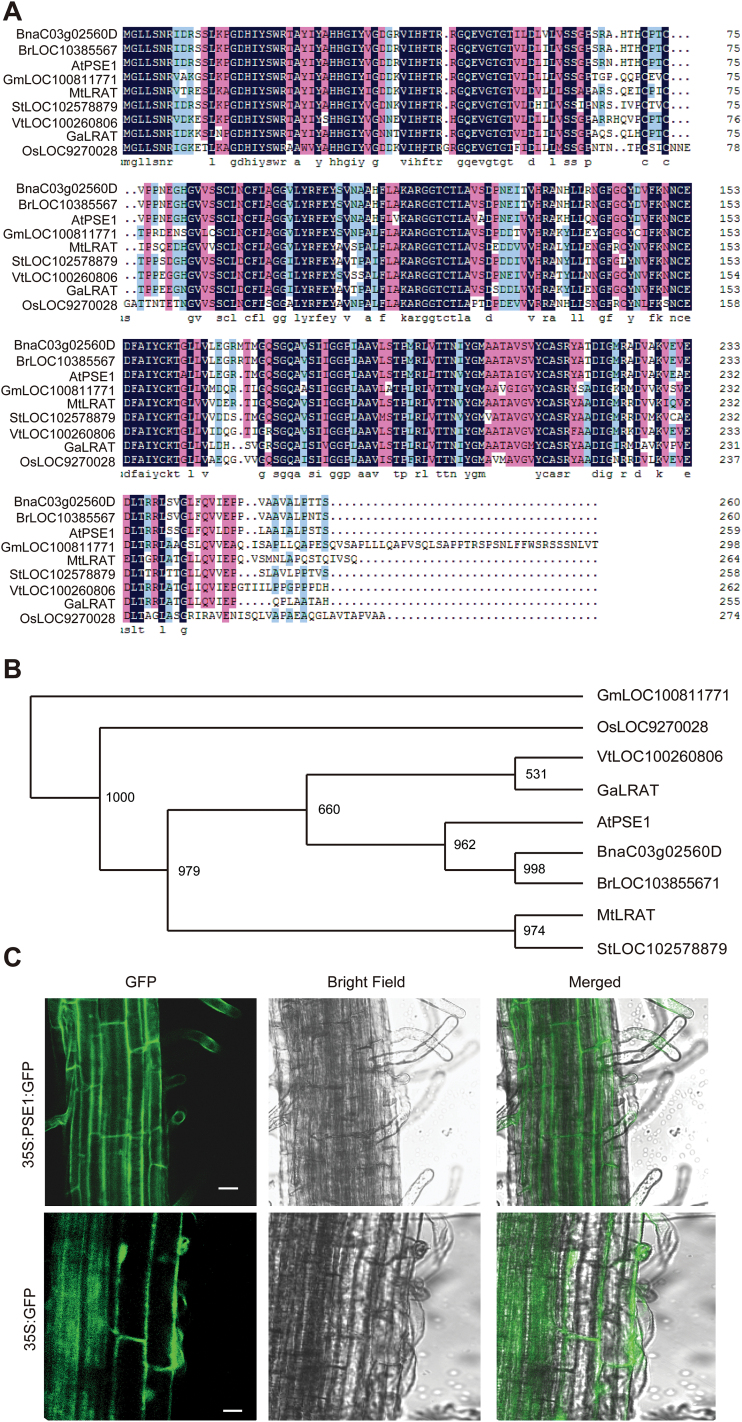
The PSE1 gene contains three exons and two introns, and encodes a 259 amino acid protein with one predicted NC domain (IPR0077053; Tsai et al., 2009). BLAST analysis of PSE1 in the non-redundant database of NCBI revealed that it shares >70% sequence similarity with other proteins such as BnaC03g02560D (ID:CDX81002, Brassica napus), BrLOC103855671 (ID:XP_009130938, Brassica rapa), VtLOC100260806 (ID:XP_002266364, Vitis vinifera), OsLOC9270028 (ID:XP_015640129, Oryza sativa Japonica Group), GmLOC100811771 (ID: XP_014623907, Glycine max), GaLRAT (ID:KHG06143, Gossypium arboreum), MtLRAT(ID:XP_013467890, Medicago truncatula), and StLOC102578879 (ID: XP_006348469, Solanum tuberosum) (Fig. 4A, B).
Fig. 4.
Subcellular localization of PSE1. (A) Multiple sequence alignment of PSE1 and several homologues from other species through DNAMAN software. (B) Phylogenic tree of PSE1 and homologues using Clustalx in combination with Phylip. (C) Subcellular localization of the 35S::PSE1–GFP fusion protein in transgenic lines. The expression of 35S::GFP was used as a control. Scale bar=20 μm. (This figure is available in colour at JXB online.)
PSE1 is predicted by the SubCellular Proteomic Database (SUBA3) (http://suba.plantenergy.uwa.edu.au/flatfile.php?id=AT5G06370.1) to be localized in the cytoplasm. To test this prediction, we fused the coding region of PSE1 at the N-terminus of a green fluorescent protein (GFP) fragment, and the fusion construct was transformed into WT A. thaliana. The fluorescent signal was clearly visible in the cytoplasm (Fig. 4C). Thus, our data demonstrate that PSE1 is indeed localized in the cytosplasm.
PSE1-mediated Pb tolerance is GSH dependent
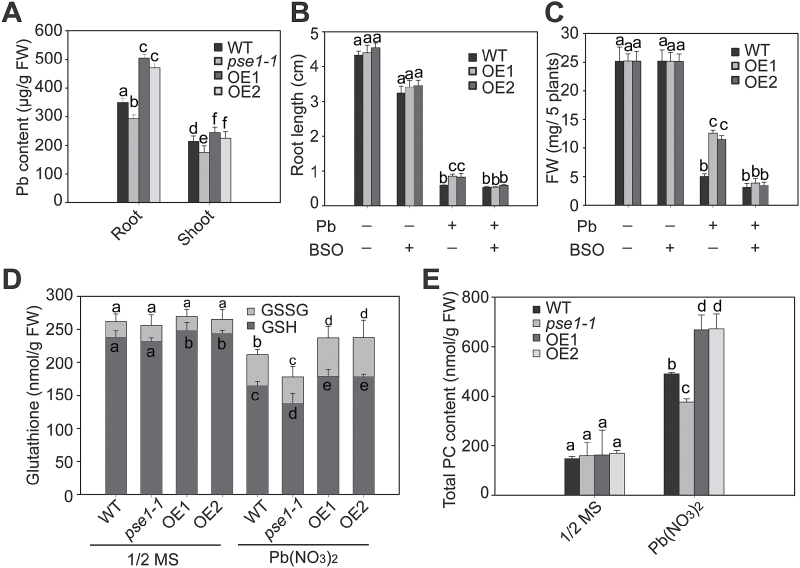
To investigate whether the PSE1-mediated Pb tolerance is associated with Pb accumulation, we analysed the Pb content in the WT, pse1-1 mutant, and PSE1-overexpressing plants that were treated with Pb(NO3)2. We found that the Pb contents were significantly increased in the PSE1-overexpressing plants and significantly decreased in the pse1-1 mutant in comparison with the WT control (P<0.05; Fig. 5A). Thus, a sequestration mechanism might be involved in PSE1-mediated Pb tolerance.
Fig. 5.
PSE1 regulates Pb tolerance through the GSH-dependent pathway. (A) Measurements of Pb contents in wild-type (WT), pse1-1 mutant, and PSE1-OE lines. These plants were grown on 1/2 MS medium with 0.5mM Pb(NO3)2 for 2 weeks, and roots and shoots of these samples were collected for Pb content measurements. (B and C) Effect of BSO on root length (B) and fresh weight (C) of WT and PSE1-OE plants. Seedlings were grown on 1/2 MS medium with (+) or without (–) 0.5mM Pb(NO3)2 or 0.1mM BSO for 2 weeks, and then their root lengths and fresh weights were measured. Three independent experiments were performed with similar results, each with three biological replicates. Five plants per genotype from one plate were measured for each replicate. Data are presented as the means ±SE, n=3. (D and E) Glutathione (D) and PC (E) contents in the pse1-1 mutant and the PSE1-OE line. Seedlings were grown on 1/2 MS medium for 2 weeks and then treated or not with 0.5mM Pb(NO3)2 for 12h, and their GSH/GSSG (D) and PC (E) contents were quantified. Data are presented as the means ±SE of three replicate experiments. Statistical significance was determined using ANOVA in combination with post-hoc (Tukey) tests; significant differences (P<0.05) between parameters are indicated by different lower case letters.
GSH is important for heavy metal detoxification via vacuolar sequestration (Lin et al., 2012; J. Chen et al., 2015), and it can bind Pb(II) ions as a precursor of phytochelation (Grill et al., 1989; Cobbett and Goldsbrough, 2002; Noctor et al., 2012). To test whether PSE1-mediated Pb tolerance is related to GSH, a GSH synthesis inhibitor, BSO, was used to treat the plants. WT and PSE1-overexpressing plants did not show significant differences in growth on medium containing BSO alone; however, the root length and fresh weight were higher in PSE1-overexpressing plants than in the WT plants in the medium containing Pb(NO3)2 (Fig. 5B, C). When BSO was added together with Pb(NO3)2, the phenotype of elongated root and increased fresh weight in the PSE1-overexpressing plants disappeared (Fig. 5B, C). These results suggest that the mechanism of PSE1-mediated Pb tolerance is GSH dependent.
To confirm further the role of GSH in PSE1-mediated Pb tolerance, we measured the concentrations of GSH in the WT, pse1-1 mutant, and PSE1-overexpressing plants in response to Pb stress. Without Pb treatment, no significant difference was detected in total glutathione (P>0.05, GSH plus 2GSSG) among the WT, PSE1-overexpressing plants, and the pse1-1 mutant; however, the PSE1-overexpressing plants contained a more reduced form of GSH whereas the pse1-1 mutant possessed less GSH in comparison with the WT (Fig. 5D). When challenged by Pb stress, the GSH concentrations decreased significantly in the PSE1-overexpressing plants, the pse1-1 mutant, and the WT; however, compared with the WT plants, a slight decrease was observed in the PSE1-overexpressing plants while a higher decrease existed in the pse1-1 mutant (P<0.05; Fig. 5D).
The total PC content was also analysed in the PSE1-overexpressing plants, pse1-1 mutant, and WT after treatment with Pb stress. Compared with the WT, the total PC content was significantly increased in the PSE1-overexpressing plants but decreased in the pse1-1 mutant in the presence of Pb (P<0.05; Fig. 5E). These results suggest that PSE1-mediated Pb accumulation and tolerance is associated with increased PC synthesis.
PSE1 positively regulates Pb tolerance by activating the expression of PC synthesis-related genes
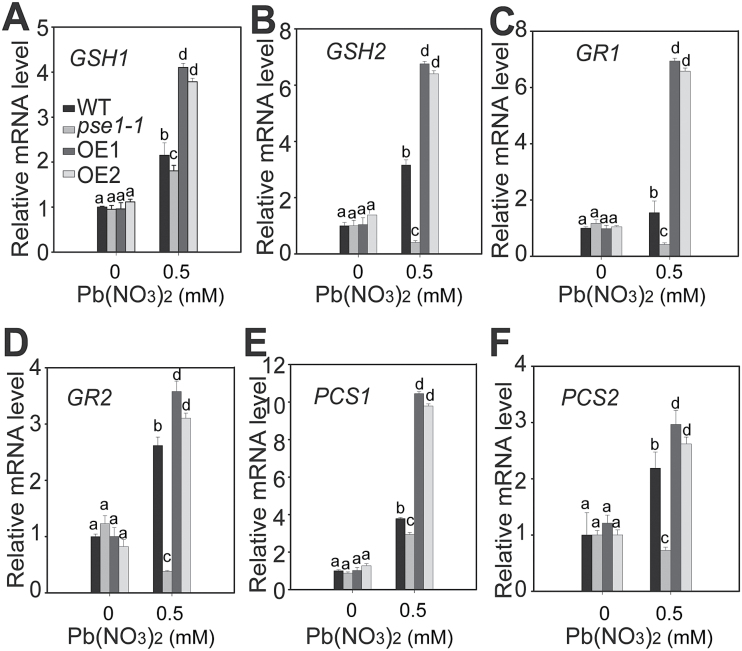
The above results suggest that PSE1 regulates Pb tolerance through the GSH-dependent PC synthesis pathway (Xiang and Oliver, 1998; Zhu et al., 1999a, b ; Kim et al., 2006; Verbruggen et al., 2009a, b ; Noctor et al., 2012; Seth et al., 2012). Therefore, we analysed the genes involved in this pathway. Under Pb stress, higher transcription levels of a few key genes were detected, including GSH1, GSH2, GR1, GR2, PCS1, and PCS2, in the PSE1-overexpressing plants than in the WT plants; in contrast, their transcription levels were significantly decreased in the pse1-1 mutant in comparison with the WT control (P<0.05; Fig. 6). In addition, the transcription levels of other genes involved in regulating Pb tolerance, including PDR12, ATM3, ACBP1, ABCC1, ABCC2, and PDR8 (Lee et al., 2005; Kim et al., 2006, 2007; Xiao et al., 2008; Park et al., 2012), were analysed. There were no significant differences in the genes ATM3, ACBP1, ABCC1, ABCC2, and PDR8 among the WT, PSE1-overexpressing plants, and the pse1-1 mutant (Supplementary Fig. S4). However, the induction of PDR12 was significantly higher in the PSE1-overexpressing plants but lower in the pse1-1 mutant (Supplementary Fig. S4). These results suggest that the GSH-dependent PC synthesis pathway is a major mechanism contributing to PSE1-mediated Pb tolerance, and PDR12-dependent Pb tolerance might be another complementary mechanism. Moreover, PDR8 was also induced but had no significant difference among the WT, PSE1-overexpressing plants, and the pse1-1 mutant. This may explain why there was no significant difference in growth between the pse1-1 mutant and the WT plants in the presence of Cd.
Fig. 6.
qRT-PCR analysis of the genes involved in PC synthesis. Quantitative analysis of the transcription of genes involved in GSH/PC synthesis in the pse1-1 mutant and PSE1-OE lines, including GSH1 (A), GSH2 (B), GR1 (C), GR2 (D), PCS1 (E), and PCS2 (F). The wild type (WT), the pse1-1 mutant and PSE1-OE lines were grown on 1/2 MS medium for 2 weeks and treated or not with 0.5mM Pb(NO3)2 for 12h, after which their mRNAs were isolated for qRT-PCR analysis. ACTIN11 was used as the internal control. Data are presented as the means ±SE of three replicate experiments.. Statistical significance was determined using ANOVA in combination with post-hoc (Tukey) tests; significant differences (P<0.05) between the pse1-1/PSE1-OE line and the WT are indicated by different lower case letters.
Discussion
In this study, we explored the role of PSE1 in Pb tolerance in Arabidopsis by investigating the effects of gain and loss of function of this gene on plant responses to Pb stress. Our results demonstrated that PSE1 contributed to Pb tolerance: Pb stress rapidly induced PSE1, which then triggered GSH-dependent PC synthesis and GSH-independent PDR12 transport through the co-ordinated regulation of the expression of PC synthesis-related genes and PDR12. This cascade of events resulted in enhanced Pb accumulation and tolerance (Fig. 7).
Fig. 7.
A working model for the role of PSE1 in regulating Pb accumulation and tolerance. Pb stress rapidly induced the expression of PSE1, which triggered GSH-dependent PC synthesis by activating the expression of the genes involved in the PC synthesis pathway. Moreover, the induced expression of PSE1 also led to increased expression of the Pb pump PDR12. Together, these factors triggered Pb-activated PC synthesis and the pumping out of Pb, which resulted in enhanced Pb accumulation and tolerance. Solid lines indicate direct regulation between upstream and downstream factors, while dashed lines indicate indirect regulation between these factors.
The molecular mechanisms for Pb tolerance are still not well understood in plants, and only a few genes have been identified and characterized (Clemens, 2001; Hall, 2002; Lee et al., 2005; Kim et al., 2006; Xiao et al., 2008; Cao et al., 2009). Here, we showed that PSE1 encodes a member of the NC protein family that plays an important role in Pb tolerance. First, the root length and fresh weight of the pse1-1 mutant grew less than those of the WT in the presence of Pb, while the complemented plants had a reversed Pb sensitivity phenotype (Fig. 1; Supplementary Fig. S1). Secondly, overexpression of PSE1 led to improved Pb tolerance (Fig. 2). Thirdly, the expression of PSE1 was induced by Pb (Fig. 3). Thus, PSE1 is required for Pb tolerance in Arabidopsis.
A GSH-dependent PC pathway is one of the most important mechanisms contributing to Pb resistance (Mehra et al., 1995). Many studies have shown that PCs play an important role in heavy metal detoxification in living organisms (Verbruggen et al., 2009a, b ; Lin et al., 2012). PCs are induced by a wide range of heavy metals (Grill et al., 1987; Maitani et al., 1996). In addition, they bind to Pb to form stable metal–PC complexes and are sequestered in the vacuole to inhibit Pb toxicity in the cell (Cobbett and Goldsbrough, 2002; Clemens, 2006; Song et al., 2010; Park et al., 2012). GSH, as the metabolic precursor of PCs, is involved in heavy metal detoxification in plants (Cobbett, 2002; Ball et al., 2004). Overexpression of GSH1, GSH2, and PCS1 involved in PC biosynthesis has been shown to increase Cd accumulation and tolerance (Zhu et al., 1999a, b ; Brunetti et al., 2011). Our results demonstrated that PSE1 enhanced Pb accumulation and tolerance though the induction of PC synthesis and the activation of the expression of genes involved in PC synthesis, including GSH1, GSH2, PCS1, PCS2, GR1, and GR2 (Figs 5, 6). These results suggest that PSE1 is an important regulator of the GSH-dependent PC synthesis pathway that contributes to Pb accumulation and tolerance. Many metal transporters have been identified as important components involved in heavy metal detoxification in diverse organisms; these transporters remove heavy metals from the cytoplasm to avoid toxicity in cells. Some transporters extrude them across the plasma membrane and others sequester them into inactive organelles, such as the vacuole (Lee et al., 2005). In Arabidopsis, AtPDR12, AtATM3, ABCC1, ABCC2, and AtPDR8 were reported to be associated with Pb tolerance (Cobbett and Goldsbrough, 2002; Lee et al., 2005; Clemens, 2006; Kim et al., 2006, 2007; Song et al., 2010; Park et al., 2012). AtPDR12 was identified as the first transporter gene that served specifically as an efflux pump for Pb exclusion, and the AtPDR12-dependent mechanism is a GSH-independent mechanism (Lee et al., 2005). AtATM3 responded to both Pb and Cd stresses and may mediate GSH-conjugated Cd transport across the mitochondrial membrane (Kim et al., 2006). AtPDR8 is induced by both Pb and Cd, and is involved as an efflux pump of Cd or Cd conjugates at the plasma membrane (Kim et al., 2007). ABCC1 and ABCC2 transported a PC–Pb complex into the vacuole (Cobbett and Goldsbrough, 2002; Clemens, 2006; Song et al., 2010; Park et al., 2012). It is noteworthy that the expression of PDR12 was induced in the PSE1-overexpressing plants (see Supplementary Fig. S4). However, as shown in Fig. 5A–C, the concentration of Pb in the PSE1-overexpressing plants was much higher than that in the WT, and the Pb tolerance phenotype of the PSE1-overexpressing plants almost disappeared in the presence of BSO. These results suggest that PSE1-mediated Pb tolerance may be mainly related to a GSH-dependent mechanism and, at least in part, depends on a PDR12-dependent detoxification mechanism. Based on the facts that the expression of genes involved in PC synthesis and AtPDR12 was activated by PSE1 and that PSE1 was localized in the cytoplasm, we speculated that PSE1 might interact with some special transcription factors or other signalling components to regulate the expression of these genes, thereby leading to the activation of the GSH-dependent PC synthesis and PDR12-dependent detoxification pathways. However, these interacting partners require further identification.
The multiple sequence alignment and phylogenic tree results showed that PSE1 belongs to the NC protein family along with LRAT, an expanding family of homologous proteins (Fig. 4). However, the biochemical function of PSE1 requires further study in future research. Retinoid-inducible gene 1 (RIG1) has recently been classified into this family, and Tsai et al. (2009) reported that the NC domain, especially the NC motif, plays the major role in RIG1-mediated pro-apoptotic activity. The cysteine residues in the amino acid sequence of PSE1 suggest that PSE1 might be redox regulated, and the glutathione/glutaredoxin system has been reported to regulate the protein redox state (Cheng et al., 2006; Zaffagnini et al., 2012; Krasensky et al., 2014). This indicates that PSE1 may play a role in the oxidative stress response. Interestingly, we found that the growth of the pse1-1 mutant was not different from that of the WT plants under Cd, Zn, or As stress, but pse1-1 was more sensitive to H2O2 than the WT plants. The possible mechanism of PSE1-mediated H2O2 tolerance may be related to ROS production or anti-oxidative capacity. However, further research is needed to ascertain a possible link between ROS and Pb tolerance conferred by PSE1.
In summary, our results suggest that the PSE1 gene plays an important role in regulating Pb tolerance in Arabidopsis through the GSH-dependent PC synthesis and PDR12-dependent detoxification pathways.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The PSE1-complemented plants exhibited a Pb-insensitive phenotype different from that of the pse1-1 mutant.
Figure S2. Phenotype of wild-type (WT) and pse1-1 mutant seedlings in response to Cd, As, Zn, or H2O2 stress.
Figure S3. Phenotypes of wild-type (WT) and pse1-1 plants in soil-filled pots.
Figure S4. Transcript levels of Pb stress-related genes in WT, pse1-1 mutant, and PSE1-overexpressing plants subjected to Pb stress.
Table S1. Primer sequences used for cloning and qRT-PCR.
Acknowledgements
We would like to thank Wangchun Dong, Na Li, Lingxia Guan, and Guanglang Chen for their technical assistance. This work was supported by the National Natural Science Foundation of China (grant nos 31571250, 31500213, and 31300989), the Fundamental Research Fund for the Central Universities (2014HGCH0001), Anhui Provincial Natural Science Foundation (1508085QC50), and Scientific and Technological Project of Anhui Province (grant nos 1201a0301009, 1401032006, and 15czz03120).
References
- Aken BV. 2008. Transgenic plants for phytoremediation: helping nature to clean up environmental pollution. Trends in Biotechnology 26, 225–227. [DOI] [PubMed] [Google Scholar]
- Ali B, Xu X, Gill RA, Yang S, Ali S, Tahir M, Zhou W. 2014. Promotive role of 5-aminolevulinic acid on mineral nutrients and antioxidative defense system under lead toxicity in Brassica napus. Industrial Crops and Products 52, 617–626. [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechtold U, et al. 2004. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. The Plant Cell 16, 2448–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti P, Zanella L, Proia A, De Paolis A, Falasca G, Altamura MM, Sanita di Toppi L, Costantino P, Cardarelli M. 2011. Cadmium tolerance and phytochelatin content of Arabidopsis seedlings overexpressing the phytochelatin synthase gene AtPCS1. Journal of Experimental Botany 62, 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SQ, Chen ZY, Liu GQ, Jiang L, Yuan HB, Ren G, Bian XH, Jian HY, Ma XL. 2009. The Arabidopsis Ethylene-Insensitive 2 gene is required for lead resistance. Plant Physiology and biochemistry 47, 308–312. [DOI] [PubMed] [Google Scholar]
- Chen J, Yang LB, Gu J, et al. 2015. MAN3 gene regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis thaliana . New Phytologist 205, 570–582. [DOI] [PubMed] [Google Scholar]
- Chen M, Zhang LL, Li J, He XJ, Cai JC. 2015. Bioaccumulation and tolerance characteristics of a submerged plant Ceratophyllumdemersum L. exposed to toxic metal lead. Ecotoxicology and Environmental Safety 122, 313–321. [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Brock A, Nelson RS, Hirschi KD. 2006. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. Journal of Biological Chemistry 281, 26280–26288. [DOI] [PubMed] [Google Scholar]
- Clemens S. 2001. Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212, 475–486. [DOI] [PubMed] [Google Scholar]
- Clemens S. 2006. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88, 1707–1719. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cobbett CS. 2000. Phytochelatins and their roles in heavy metal detoxification. Plant Physiology 123, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P. 2002. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology 53, 159–182. [DOI] [PubMed] [Google Scholar]
- Dudka S, Miller W. 1999. Accumulation of potentially toxic elements in plants and their transfer to human food chain. Journal of Environmental Science and Health 34, 681–708. [DOI] [PubMed] [Google Scholar]
- Duruibe J, Ogwuegbu M, Egwurugwu J. 2007. Heavy metal pollution and human biotoxic effects. International Journal of Physical Science 2, 112–118. [Google Scholar]
- Fan TT, Zhai HH, Shi WW, Wang J, Jia HL, Xiang Y, An LZ. 2013. Overexpression of profilin 3 affects cell elongation and F-actin organization in Arabidopsis thaliana . Plant Cell Reports 32, 149–160. [DOI] [PubMed] [Google Scholar]
- Grill E, Loffler S, Winnacker EL, Zenk MH. 1989. Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific gamma-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proceedings of the National Academy of Sciences, USA 86, 6838–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Winnacker EL, Zenk MH. 1987. Phytochelatins, a class of heavy-metal-binding peptides from plants, are functionally analogous to metallothioneins. Proceedings of the National Academy of Sciences, USA 84, 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JL. 2002. Cellular mechanisms for heavy metal detoxification and tolerance. Journal of Experimental Botany 53, 1–11. [PubMed] [Google Scholar]
- Israr M, Jewell A, Kumar D, Sahi SV. 2011. Interactive effects of lead, copper, nickel and zinc on growth, metal uptake and antioxidative metabolism of Sesbania drummondii . Journal of Hazardous Materials 186, 1520–1526. [DOI] [PubMed] [Google Scholar]
- Jozefczak M, Remans T, Vangronsveld J, Cuypers A. 2012. Glutathione is a key player in metal-induced oxidative stress defenses. International Journal of Molecular Sciences 13, 3145–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Rotnitzky A, Sparrow D. 1996. A longitudinal study of low level lead exposure and impairment of renal function: a normative aging study. Journal of the American Heart Association 275, 1177–1181. [PubMed] [Google Scholar]
- Kim DY, Bovet L, Kushnir S, Noh EW, Martinoia E, Lee Y. 2006. AtATM3 is involved in heavy metal resistance in Arabidopsis . Plant Physiology 140, 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y. 2007. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. The Plant Journal 50, 207–218. [DOI] [PubMed] [Google Scholar]
- Krasensky J, Broyart C, Rabanal FA, Jonak C. 2014. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxidants and Redox Signaling 21, 1289–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Lee K, Lee J, Noh EW, Lee Y. 2005. AtPDR12 contributes to lead resistance in Arabidopsis . Plant Physiology 138, 827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Aaers M. 2012. The molecular mechanism of zinc and cadmium stress response in plants. Cellular and Molecular Life Sciences 69, 3187–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitani T, Kubota H, Sato K, Yamada T. 1996. The composition of metals bound to class III metallothionein (phytochelatin and its desglycyl peptide) induced by various metals in root cultures of Rubia tinctorum . Plant Physiology 110, 1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R. 2000. Phytoremediation of toxic elemental and organic pollutants. Current Opinion in Plant Biology 3, 153–162. [DOI] [PubMed] [Google Scholar]
- Mehra RK, Kodati VR, Abdullah R. 1995. Chain length-dependent Pb(II)-coordination in phytochelatins. Biochemical and Biophysical Research Communations 215, 730–736. [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH. 2012. Glutathione in plants: an integrated overview. Plant, Cell and Environment 35, 454–484. [DOI] [PubMed] [Google Scholar]
- Park J, Song W-Y, Ko D, Eom Y, Hansen TH, Schiller M, Lee TG, Martinoia E, Lee Y. 2012. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. The Plant Journal 69, 278–288. [DOI] [PubMed] [Google Scholar]
- Pilon-Smits E. 2005. Phytoremediation. Annual Review of Plant Biology 56, 15–39. [DOI] [PubMed] [Google Scholar]
- Pourrut B, Shahid M, Dumat C, Winterton P, Pinelli E. 2011. Lead uptake, toxicity, and detoxification in plants. Reviews of Environmental Contamination and Toxicology 213, 113–136. [DOI] [PubMed] [Google Scholar]
- Rauser WE. 1990. Phytochelatins. Annual Review of Biochemistry 59, 61–86. [DOI] [PubMed] [Google Scholar]
- Rauser WE. 1995. Phytochelatins and related peptides: structure, biosynthesis, and function. Plant Physiology 109, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauser WE. 1999. Structure and function of metal chelators produced by plants: the case for organic acids, amino acids, phytin and metallothioneins. Cell Biochemistry and Biophysics 31, 19–48. [DOI] [PubMed] [Google Scholar]
- Ross SM. 1994. Sources and forms of potentially toxic metal in soil–plant systems. In Ross SM, ed, Toxic metals in soil–plant systems. New York: John Wiley & Sons, 3–25. [Google Scholar]
- Seth CS, Remans T, Keunen E, Jozefczak M, Gielen H, Opdenakker K, Weyens N, Vangronsveld J, Cuypers A. 2012. Phytoextraction of toxic metals: a central role for glutathione. Plant, Cell and Environment 35, 334–346. [DOI] [PubMed] [Google Scholar]
- Sharma P, Dubey R. 2005. Lead toxicity in plants. Brazilian Journal of Plant Physiology 17, 35–52. [Google Scholar]
- Song WY, Park J, Mendoza-Cózatl DG, Suter-Grotemeyer M, Shim D, Hörtensteiner S, Geisler M, Weder B, Rea PA, Rentsch D. 2010. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proceedings of the National Academy of Sciences, USA 107, 21187–21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinnes E. 2013. Lead. In: Alloway BJ, ed. Heavy metals in soils: trace metals and metalloids in soils and their bioavailability. Dordrecht: Springer, 395–409. [Google Scholar]
- Suresh B, Ravishankar GA. 2004Phytoremediation—a novel and promising approach for environmental clean-up. Critical Reviews in Biotechnology 24, 97–124. [DOI] [PubMed] [Google Scholar]
- Tsai FM, Shyu RY, Lin SC, Wu CC, Jiang SY. 2009. Induction of apoptosis by the retinoid inducible growth regulator RIG1 depends on the NC motif in HtTA cervical cancer cells. BMC Cell Biology 10, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uveges JL, Corbett AL, Mal TK. 2002. Effects of Pb contamination on the growth of Lythrumsalicaria . Environmental Pollution 120, 319–323. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. 2009. a Molecular mechanisms of metal hyperaccumulation in plants. New Phytologist 181, 759–776. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. 2009. b Mechanisms to cope with arsenic or cadmium excess in plants. Current Opinion in Plant Biology 12, 364–372. [DOI] [PubMed] [Google Scholar]
- Verma S, Dubey RS. 2003. Lead toxicity induces lipid peroxidation and alters the activities of antioxidant enzymes in growing rice plants. Plant Science 164, 645–655. [Google Scholar]
- Verrier PJ, Bird D, Burla B, et al. 2008. Plant ABC proteins—a unified nomenclature and updated inventory. Trends in Plant Science 13, 151–159. [DOI] [PubMed] [Google Scholar]
- Xiang CB, Oliver DJ. 1998. Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. The Plant Cell 10, 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML. 2008. Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis . The Plant Journal 54, 141–151. [DOI] [PubMed] [Google Scholar]
- Yang YR, Han XZ, Liang Y, Ghosh A, Chen J, Tang M. 2015. The combined effects of arbuscular mycorrhizal fungi (AMF) and lead (Pb) stress on Pb accumulation, plant growth parameters, photosynthesis, and antioxidant enzymes in Robinia pseudoacacia L. PLoS One 10, e0145726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafari S, Sharifi M, Chashmi NA, Mur LAJ. 2016. Modulation of Pb-induced stress in Prosopis shoots through an interconnected network of signaling molecules, phenolic compounds and amino acids. Plant Physiology and Biochemistry 99, 11–20. [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Marchand CH, Couturier JR, Gao XH, Rouhier N, Trost P, Lemaire SP. 2012Glutaredoxins: unique properties for redox signaling. Antioxidants and Redox Signaling 16, 17–32. [DOI] [PubMed] [Google Scholar]
- Zenk MH. 1996. Heavy metal detoxification in higher plants: a review. Gene 179, 21–30. [DOI] [PubMed] [Google Scholar]
- Zhu YL, Pilon-Smits EA, Jouanin L, Terry N. 1999. a Overexpression of glutathione synthetase in Indian mustard enhances cadmium accumulation and tolerance. Plant Physiology 119, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Pilon-Smits EA, Tarun AS, Weber SU, Jouanin L, Terry N. 1999. b Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing γ-glutamylcysteine synthetase. Plant Physiology 121, 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FY, Li L, Lam PY, Chen ML, Lo C. 2013. Sorghum extracellular leucine-rich repeat protein SbLRR2 mediates lead tolerance in transgenic Arabidopsis. Plant and Cell Physiology 54, 1549–1559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.