ABSTRACT
Phytohormones act in concert to coordinate plant growth and the response to environmental cues. Gibberellins (GAs) are growth-promoting hormones that recently emerged as modulators of plant immune signaling. By regulating the stability of DELLA proteins, GAs intersect with the signaling pathways of the classical primary defense hormones, salicylic acid (SA) and jasmonic acid (JA), thereby altering the final outcome of the immune response. DELLA proteins confer resistance to necrotrophic pathogens by potentiating JA signaling and raise the susceptibility to biotrophic pathogens by attenuating the SA pathway. Here, we show that JUB1, a core element of the GA - brassinosteroid (BR) - DELLA regulatory module, functions as a negative regulator of defense responses against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and mediates the crosstalk between growth and immunity.
KEYWORDS: Arabidopsis, defense, DELLA proteins, gibberellin, jasmonic acid, pathogens, salicylic acid, transcription factor
Introduction
The final outcome of a plant-pathogen interaction depends on the competence of the host to recognize the pathogen and trigger an appropriate, rapid defense response. Crosstalk between the different hormone signaling pathways plays an essential role in fine-tuning the plant's response against pathogens. Several studies demonstrated the mutually antagonistic interaction of jasmonic acid (JA) and salicylic acid (SA) in the response to pathogens.1-3 JA-mediated signaling elevates the resistance to necrotrophic pathogens such as Botrytis cinerea or Alternaria brassicicola, whereas the SA pathway is associated with enhanced resistance to (hemi)biotrophs such as Pseudomonas syringae.4-7 Thus, elevated resistance to necrotrophs is often correlated with an increased susceptibility to (hemi)biotrophs, and vice versa.8 Emerging evidence indicates that gibberellic acids (GAs), which are essential growth regulators, also play a central role in the plant´s response to pathogen attack. The role of GAs in the regulation of growth and disease response is attributed to the degradation of DELLA proteins, the major suppressors of GA signaling.9 It has been shown that DELLA proteins can influence disease outcome by modulating the balance between SA and JA signaling.10,11 In Arabidopsis thaliana, DELLA proteins amplify JA-dependent defense signaling, while they antagonize SA-mediated defense, thereby promoting resistance to necrotrophic pathogens but increasing susceptibility to biotrophic pathogens.10,11
It has been demonstrated that DELLA proteins interact with jasmonate ZIM domain (JAZ) proteins known as repressors of JA signaling.12,13 In the absence of GA, interaction with DELLA proteins releases the inhibitory effect of JAZ on JA-responsive transcription factors (TFs) such as MYC2, a basic helix-loop-helix (bHLH) TF, thus resulting in the activation of JA-responsive gene expression.14-16 DELLA-enhanced expression of JA-responsive defense genes, but suppression of SA signaling genes contributes to an enhanced resistance to necrotrophic pathogens and increased susceptibility to biotrophic pathogens.10
We have recently discovered a novel function of the NAC transcription factor JUNGBRUNNEN1 (JUB1) in balancing growth and stress responses in Arabidopsis.17 We have shown that JUB1 exerts its cellular function partly in a DELLA-dependent manner via the synergistic regulation of brassinosteroid (BR) and GA biosynthesis/signaling coordinated through direct repression of the key GA and BR biosynthesis genes GA3ox1 and DWF4, respectively, and activation of DELLAs.17 Repression of GA and BR biosynthesis by JUB1 results in the accumulation and activation of DELLA proteins in JUB1 overexpressor (JUB1-OX) plants. We showed that the repressive effect of high JUB1 expression can be fully relieved by concomitant overexpression of GA3ox1 (GA3ox1-OX) and DWF4 (DWF4-OX) in the JUB1-OX background (Triple-OX lines): the GA and BR deficiency as well as the enhanced abiotic stress tolerance phenotype of JUB1-OX plants was reversed to normal in the Triple-OX plants, showing the critical contribution of the GA and BR biosynthesis genes to the phenotypes observed in JUB1-OX plants.
In the current study, we identify JUB1 as a negative regulator of the defense response against the bacterial (hemi)biotrophic pathogen Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). We demonstrate that suppression of the Pst DC3000-induced defense response in JUB1-OX plants is mainly attributed to the accumulation of DELLA proteins, resulting in potentiation of JA signaling and therefore promotion of the susceptibility to Pst DC3000. Our data thus identify JUB1 as a transcriptional regulator acting at the interface of growth regulation (via affecting GA and BR synthesis/signaling) and the response to pathogens.
Results
Activation of JUB1 expression by biotrophic pathogens
Publicly available gene expression data (Arabidopsis eFP browser; www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) revealed that JUB1 expression is induced in response to infection with the biotrophic pathogens Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and P. syringae strain avrRPM1. Saga et al. (2012) also showed that expression of JUB1 is induced upon infection with Pst DC3000, and P. syringae strain avrRPT2, suggesting a role of JUB1 in regulating plant immune responses.18
Here, we confirmed induction of JUB1 by Pst DC3000 in Arabidopsis wild-type Col-0 (WT) plants sprayed with Pst DC3000 (108 cfu/mL), by quantitative RT-PCR (qRT-PCR). JUB1 expression is highly induced 6 and 24 hours post inoculation (hpi) (Fig. 1A). To analyze the spatial expression of JUB1 in response to Pst DC3000 inoculation, we used transgenic plants expressing GUS under control of the JUB1 promoter (ProJUB1:GUS).19 Analysis of GUS expression was carried out in leaves infiltrated with Pst DC3000 (106 cfu/mL) or MgCl2 (mock) 6 and 24 hpi. GUS activity slightly increased at 6 hpi, but strongly enhanced at 24 hpi in leaves infiltrated with Pst DC3000 compared to mock-treated leaves (Fig. 1B), confirming activation of the JUB1 promoter by Pst DC3000 treatment.
Figure 1.
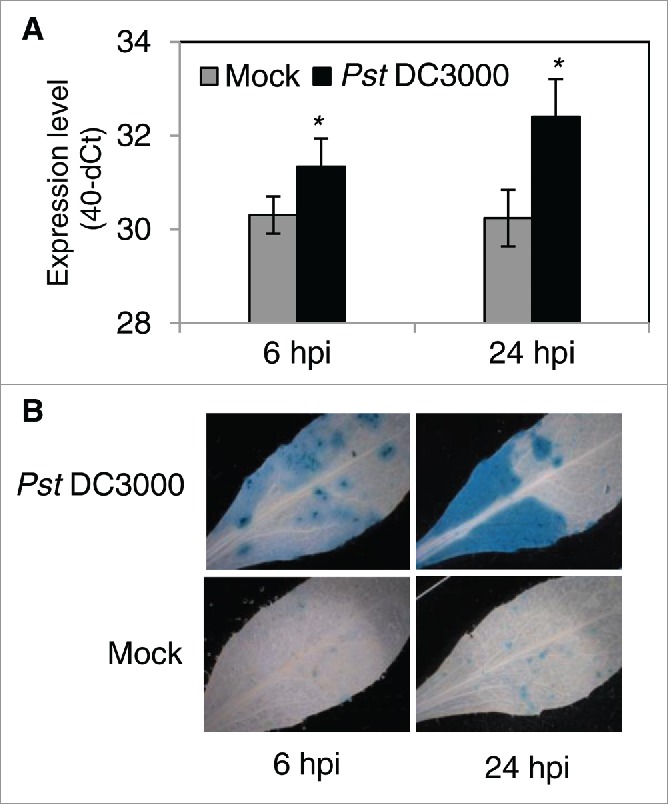
Induction of JUB1 upon infection with Pst DC3000. (A) Expression level of JUB1 measured by qRT-PCR in 5-week-old WT plants sprayed with Pst DC3000 compared to mock (10 mM MgCl2) treated plants, 6 and 24 hpi. Means ± SD (n = 3). Asterisks indicate a significant difference from Mock (*p < 0.05; Student´s t-test). (B) Histochemical analysis of GUS expression in rosette leaves of 5-week-old ProJUB1:GUS plants, 6 and 24 h after pressure infiltration with Pst DC3000 or MgCl2 (mock).
JUB1 functions as a negative regulator of the defense against Pst DC3000
Our previous study reported that transgenic Arabidopsis plants overexpressing the JUB1 gene (JUB1-OX) showed reduced growth but improved tolerance to different abiotic stresses,17,19,20 suggesting its involvement in the crosstalk between plant growth and stress response. We found that the stress- and growth-related phenotypes observed in JUB1-OX plants are mainly associated with the negative regulatory impact of JUB1 on the expression of key enzymes of GA and BR biosynthesis (GA3ox1 and DWF4, respectively) which ultimately results in accumulation and activation of DELLA proteins. To determine whether altered expression of JUB1 can also affect disease responses to Pst DC3000 and whether this is linked to the JUB1 regulation of growth via DELLA proteins we inoculated JUB1 transgenic plants (JUB1-OX, jub1-1 knockdown mutant,19 Triple-OX) as well as WT plants with Pst DC3000 by both, pressure infiltration and surface inoculation, and monitored the progress of infection over time (Fig. 2). Pressure infiltration of leaves with Pst DC3000 (1×106 cfu/mL) revealed differential defense responses in JUB1 transgenic plants. While an increased susceptibility was observed in JUB1-OX plants, jub1-1 and Triple-OX plants developed less disease symptoms compared to WT (Fig. 2A). Consistent with these results more bacterial growth was observed in JUB1-OX plants at 3 d post inoculation (dpi) compared to WT, but jub1-1 exhibited a bacterial titer that was lower than that of both, WT and Triple-OX plants. The number of bacteria present in Triple-OX plants was similar to that of WT (Fig. 2B). Next, we assessed the phenotype of JUB1 transgenics upon surface inoculation with Pst DC3000, which manifests the efficiency of bacteria to enter the plant tissues through the stomatal pores and to cause infection. To this end, plants were spray-inoculated with a bacterial suspension (1×108 cfu/mL). Development of disease symptoms was assessed at 5 dpi as disease severity index. While JUB1-OX plants developed severe necrotic lesions, jub1-1 plants exhibited less disease symptoms than both, Triple-OX and wild-type plants that show a similar response toward the treatment (Fig 2C, D). Recovery of the JUB1-OX stress response in Triple-OX plants confirms that the negative regulation of GA and BR pathways by JUB1 contributes to the biotic stress response against Pst DC3000. Taken together, our data indicate that JUB1 negatively regulates the defense response against Pst DC3000 in conjunction with hormonal networks.
Figure 2.
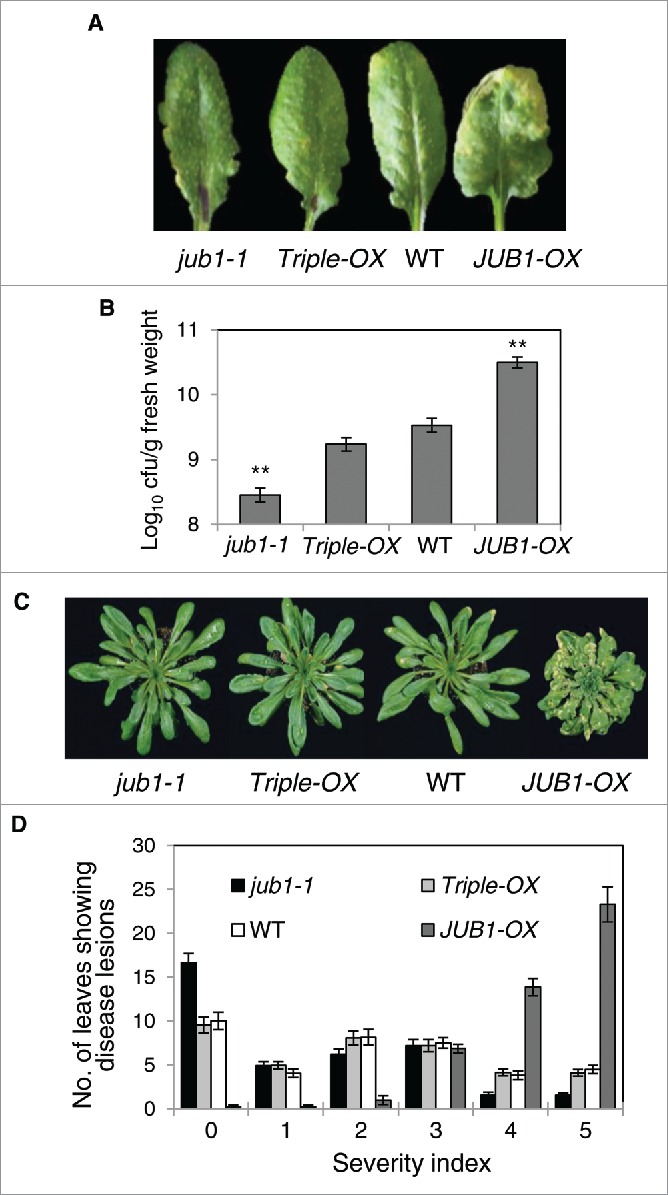
Defense responses observed in JUB1-OX are fully restored in Triple-OX plants. (A) Rosette leaves of 5-week-old plants pressure infiltrated with Pst DC3000 at 2 dpi. The experiments were repeated 3 times and similar results were obtained. (B) In planta growth of Pst DC3000 bacteria in JUB1 transgenics and WT, assayed 3 dpi. Data are from 2 experiments with at least 8 plants per genotype in each. Mean growth ± SD. Asterisks indicate a significant difference from WT (**p < 0.01; Student´s t-test). (C) Defense response of plants sprayed with Pst DC3000 at 5 dpi. The experiment was repeated 4 times with similar results. (D) Disease severity index (according to lesion diameters; 0, small chlorotic lesions; 5, large lesions) assessed in plants shown in (C). Data are from 3 experiments with at least 8 plants per genotype in each. Means are shown ± SD.
JUB1 regulates defense responses to Pst DC3000 mainly through the accumulation of DELLA proteins
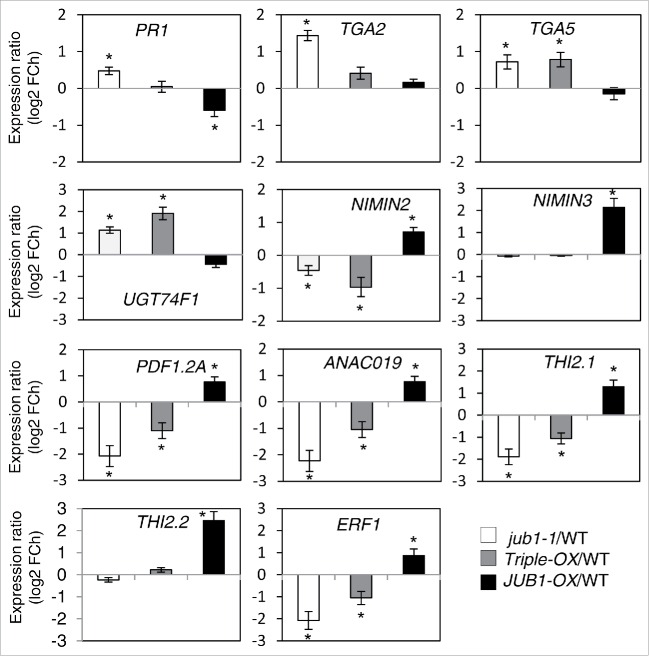
To evaluate the role of JUB1 for the regulation of defense response genes, we profiled the expression of several defense responsive genes in 5-week-old JUB1 transgenic and WT plants sprayed with Pst DC3000 at 6 hpi using qRT-PCR. We identified 11 genes differentially expressed between JUB1 transgenics and WT, including 5 JA- and 6 SA-responsive genes. Expression of the JA-responsive genes PDF1.2A, THI2.1, THI2.2, ANAC019, and ERF1 was enhanced in JUB1-OX plants, while expression of these genes was highly or moderately decreased in jub1-1 and Triple-OX plants, except for THI2.2, which was not altered (Fig. 3). Elevated expression of JA-responsive genes in JUB1-OX plants upon Pst DC3000 treatment may indicate the activation of JA-mediated defense responses resulting in the promotion of susceptibility toward Pst DC3000. Expression of the SA-defense marker gene PR1 was repressed in JUB1-OX plants, but induced in jub1-1 plants, whereas Triple-OX plants displayed a WT level of PR1 expression. Expression of TGA2 and TGA5, which encode TFs that interact with NPR1 and activate PR1,21 was induced in both jub1-1 and Triple-OX plants, whereas the expression of both genes was close to WT levels in JUB1-OX plants. Moreover, expression of NIMIN2 and NIMIN3, negative regulators of the SA-induced defense response that act through their differential interaction with NPR1,22 is induced in JUB1-OX plants but repressed or unchanged in jub1-1 and Triple-OX plants. Furthermore, UGT74F1 transcript level was higher in jub1-1 and Triple-OX plants, while it was slightly decreased in JUB1-OX plants. UGT74F1 encodes an UDP-glucosyl transferase involved in the glycosylation of SA to SA 2-O-β-D-glucose (SAG); the ugt47f1 mutant shows an increased susceptibility to Pst DC3000.23 The expression profile of these genes in JUB1 transgenic plants clearly reflects an attenuated SA defense pathway, which is either due to direct regulation by JUB1 or an indirect effect exerted by DELLAs (through direct repression or by activation of JA signaling). To understand this regulation, we analyzed the promoters of the genes differentially expressed between JUB1 transgenic and WT plants. The absence of JUB1 binding sites in the promoters of differentially expressed genes suggests their indirect regulation by JUB1. Previously we showed that the level of DELLA proteins is enhanced in JUB1-OX plants compared to jub1-1, Triple-OX and WT plants.17 DELLA accumulation by JUB1 occurs via suppression of GA biosynthesis and direct transcriptional activation of DELLA genes. Indirect regulation of defense responsive genes by JUB1 further highlights a role for DELLA proteins in the regulation of JUB1 stress responses against pathogens and the modulation of JA-SA antagonistic crosstalk.
Figure 3.
SA- and JA-responsive defense genes differentially expressed in JUB1 transgenic plants compared to WT. Gene expression was quantified by qRT-PCR in rosettes of 5-week-old JUB1 transgenics compared to WT, 6 h after spraying with Pst DC3000. FCh, fold change. Means ± SD (n = 3). Asterisks indicate a significant difference from WT (*p < 0.05; Student´s t-test).
Discussion
Here, we report that the transcription factor JUB1 regulates disease responses to Pst DC3000 likely through modulation of DELLA proteins. JUB1 expression is induced by the biotrophic pathogen Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) (Fig. 1; eFP browser). Overexpression of JUB1 results in attenuated resistance to Pst DC3000, while the jub1-1 knockdown mutant and Triple-OX plants (which concomitantly overexpress JUB1, GA3ox1 and DWF417) exhibit reduced disease symptoms and support less bacterial proliferation when surface-inoculated or pressure-infiltrated with Pst DC3000 (Fig. 2). Thus, JUB1 represents a negative regulator of plant defense against Pst DC3000. Expression profiling of several SA- and JA-responsive defense genes in JUB1 transgenics and WT plants upon Pst DC3000 treatment displayed differential regulation of 11 genes by JUB1 (Fig. 3).
Transcript abundance of JA-related defense genes such as PDF1.2, THI2.1, ANAC019 and ERF1 was enhanced in JUB1-OX plants, but attenuated in both, jub1-1 and Triple-OX plants. In contrast, expression of SA-marker gene PR1 was reduced in JUB1-OX plants, while it was increased in jub1-1 plants. Accordingly, transcript levels of TGA2 and TGA5, the positive transcriptional regulators of PR1, were enhanced in jub1-1 and Triple OX plants. Moreover, transcript levels of NIMIN2 and NIMIN3, which encode the negative regulators of PR1, were enhanced in JUB1-OX plants, but repressed in jub1-1 and Triple OX plants. Such patterns of gene expression indicate that JUB1 acts as a negative regulator of the SA pathway in response to Pst DC3000 infection. The SA-mediated pathway is crucial for the establishment of an efficient defense response against Pst DC3000. SA exists in free or conjugated forms, which involves glycosylation or methylation.24 Induced expression of UGT74F1, a UDP-glucosyl transferase that is involved in the glycosylation of SA, was observed in both, jub1-1 and Triple-OX plants, while its transcript level was decreased in JUB1-OX plants. Recently, Boachon et al. (2014)23 showed that the ugt74f1 mutant displays an increased susceptibility to Pst DC3000.
Although Pst DC3000 elevates both, JA and SA levels, the balance between these hormonal pathways is critical for establishing the output of the plant-pathogen interaction.22 Transcript changes observed in JUB1 transgenics clearly demonstrate that the JA pathway is favored over SA signaling leading to increased Pst DC3000 susceptibility in JUB1-OX plants. The JA-mediated defense pathway is crucial for the plant's response to necrotrophic pathogens. A previous study by Saga et al. (2012)18 showed that jub1 mutant lines were highly susceptible to infection by the necrotrophic fungus Alternaria brassicicola in part due to a defect in camalexin biosynthesis. Activation of JA-related defense genes may be a further mechanism through which JUB1 exerts the resistance against necrotrophs.
Recently, an antagonistic interaction of JA and GA was observed in the regulation of hypocotyl elongation, flowering, root growth as well as the defense response against hembiotrophic and necrotrophic pathogens.8,11-13,25 These reports suggest a role for both, GA and JA in the regulation of the tradeoff between plant growth and stress response where the interaction between DELLAs and JAZ proteins determines the output. Activation of JA signaling requires the release of JA-activated TFs such as MYC2 from the JAZ-mediated repression.14 Interaction of DELLAs with JAZ proteins leads to release of repression on JA-activated TFs, resulting in an activated JA response.12 Recently, we reported JUB1 as a core component of a regulatory module that triggers the accumulation of DELLA proteins.17 Activated JA signaling in JUB1 overexpressors upon Pst DC3000 infection can be attributed to increased levels of DELLA proteins in these plants,17 while reduced levels of DELLA proteins in jub1-1 and Triple-OX plants may result in attenuated JA signaling upon Pst DC3000 infection. Interestingly, Wild et al. (2012)8 reported that the rgl3-5 mutation enhances susceptibility to Botrytis cinerea but resistance to Pseudomonas syringae. Like in jub1-1 plants, ERF1 and PDF1.2 transcript levels were lower in infected rgl3-5 plants than in wild-type plants. JAZ proteins interact with various transcription factors, including EIN3, a positive regulator of ERF1 and PDF1.2.26 The competitive binding of DELLA proteins to JAZ proteins releases repression on EIN3 (or other JAZ-interacting factors). Consistently, overexpression of RGL3 enhances expression of EIN3-regulated genes, including ERF1 and PDF1.2, in transgenic Arabidopsis plants.27-29 In addition, JA-mediated induction of PDF1.2 was reduced in the rgl3-5 mutant in comparison to wild-type plants,27-29 whereas strong and earlier induction of PR1 and PR2 expression was reported in the quadruple della mutant compared to WT plants.11 These findings indicate that DELLAs play a critical role in modulating the defense response to Pst DC3000 by activating JA responses through their differential interaction with JAZ proteins and repressing the SA defense pathway.
Collectively, our study indicates that JUB1 participates in the SA-JA antagonism by favoring JA signaling over the SA pathway, mainly due to the accumulation of DELLA proteins (Fig. 4), leading to attenuated resistance against P. syringae pv. tomato DC3000, but enhanced resistance to necrotrophic fungi such as Alternaria brassicicola.
Figure 4.
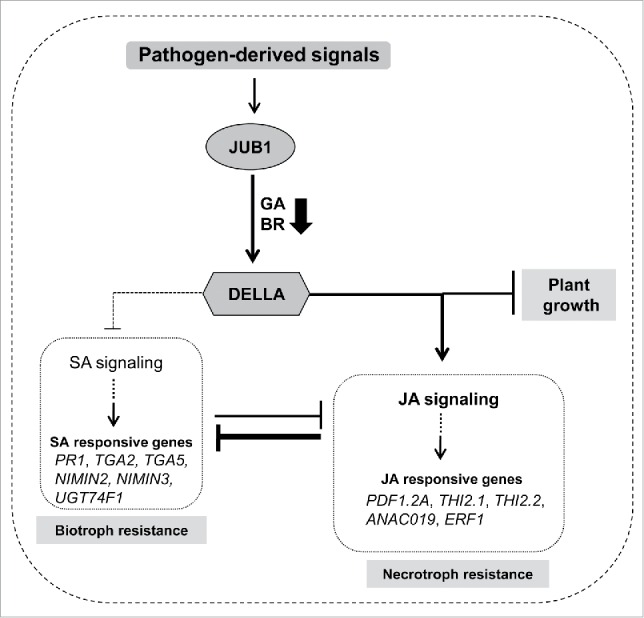
Model showing the function of JUB1 during pathogen attack. A working model showing the JUB1-mediated defense response mechanism; JUB1 expression is induced by various pathogen-derived signals, and it mediates the plant's response to pathogen attack through accumulation of DELLA proteins resulting in activated JA defense signaling, but an attenuated SA defense response leading to enhanced susceptibility to Pst DC3000. This study also emphasizes the role of JUB1 for the regulation of plant growth and defense crosstalk.
Material and methods
Plant materials and growth conditions
The ProJUB1:GUS, jub1-1 (SALK ID 036474), JUB1-OX and Triple-OX lines were reported previously.17,19 Seeds of Arabidopsis thaliana were germinated on half-strength Murashige and Skoog (MS) agar medium containing 1% (w/v) sucrose. Ten-day-old seedlings grown on sterile media were transferred to pots containing soil and were grown at 22˚C under an 8-h light/16-h dark cycle.
Treatments
For pathogen treatment, Pst DC3000 was grown on antibiotics-supplemented King's B medium at 28°C for 2 days.30 Pst DC3000 infection was carried out according to Perchepied et al. (2006).31 Briefly, bacterial cells were collected by centrifugation (2,500 g), and resuspended in 10 mM MgCl2. Pressure infiltration of 5-week-old plants was performed using a needleless syringe with Pst DC3000 (OD600nm = 0.002; 106 cfu/mL). Bacterial growth (cfu/g fresh weight−1) was measured at 3 dpi as described previously.31 For surface inoculation, 5-week-old plants were sprayed with a suspension of Pst DC3000 (OD600 nm = 0.2; 108 cfu/mL) containing 0.02% (v/v) Silwet L-77 and kept under a plastic cover. The disease severity index was scored according to Kover et al. (2005).32
Gene expression analysis
Procedures for total RNA extraction, synthesis of cDNA, and qRT-PCR using SYBR Green (Life Technologies) as well as sequences of gene-specific primers were reported previously.33 Real-time qPCR was performed using an ABI PRISM 7900 HT sequence detection system (Applied Biosystems Applera). Expression levels were normalized against the expression level of ACTIN2. The AGI codes and primer sequences of defense responsive genes used in this study are provided in Table S1.
Supplementary Material
Acknowledgments
We thank Barbara N. Kunkel (Department of Biology, Washington University, USA) for providing the Pst DC3000 strain, and Karin Koehl and colleagues from the Max Planck Institute of Molecular Plant Physiology (Potsdam, Germany) for plant care.
Funding
S.B. thanks the Deutsche Forschungsgemeinschaft (BA 4769/2-1) for funding.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 2011; 49:317-43; PMID:21663438; http://dx.doi.org/ 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- 2.Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 2012; 28:489-521; PMID:22559264; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- 3.Gimenez-Ibanez S, Solano R. Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front Plant Sci 2013; 4:72; PMID:23577014; http://dx.doi.org/ 10.3389/fpls.2013.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomma BP, Penninckx IA, Broekaert WF, Cammue BP. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 2001; 13:63-8; PMID:11154919; http://dx.doi.org/ 10.1016/S0952-7915(00)00183-7 [DOI] [PubMed] [Google Scholar]
- 5.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 2005; 43:205-27; PMID:16078883; http://dx.doi.org/ 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- 6.Koornneef A, Pieterse CMJ. Cross talk in defense signaling. Plant Physiol 2008; 146:839-44; PMID:18316638; http://dx.doi.org/ 10.1104/pp.107.112029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. Networking by small-molecule hormones in plant immunity. Nat Chem Biol 2009; 5:308-16; PMID:19377457; http://dx.doi.org/ 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- 8.Wild M, Davière J-M, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 2012; 24:3307-19; PMID:22892320; http://dx.doi.org/ 10.1105/tpc.112.101428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Bruyne L, Höfte M, De Vleesschauwer D. Connecting growth and defense: The emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol Plant 2014; 7:943-59; PMID:24777987; http://dx.doi.org/ 10.1093/mp/ssu050 [DOI] [PubMed] [Google Scholar]
- 10.Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 2008; 18:656-60; PMID:18450450; http://dx.doi.org/ 10.1016/j.cub.2008.04.034 [DOI] [PubMed] [Google Scholar]
- 11.Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 2008; 18:650-5; PMID:18450451; http://dx.doi.org/ 10.1016/j.cub.2008.03.060 [DOI] [PubMed] [Google Scholar]
- 12.Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 2010; 19:884-94; PMID:21145503; http://dx.doi.org/ 10.1016/j.devcel.2010.10.024 [DOI] [PubMed] [Google Scholar]
- 13.Yang D-L, Yao J, Mei C-S, Tong X-H, Zeng L-J, Li Q, Xiao L-T, Sun T, Li J, Deng X-W, et al.. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 2012; 109:E1192-200; PMID:22529386; http://dx.doi.org/ 10.1073/pnas.1201616109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al.. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666-71; PMID:17637675; http://dx.doi.org/ 10.1038/nature06006 [DOI] [PubMed] [Google Scholar]
- 15.Gfeller A, Dubugnon L, Liechti R, Farmer EE. Jasmonate biochemical pathway. Sci Signal 2010; 3:cm3; PMID:20159849 [DOI] [PubMed] [Google Scholar]
- 16.Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al.. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011; 23:701-15; http://dx.doi.org/ 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shahnejat-Bushehri S, Tarkowska D, Sakuraba Y, Balazadeh S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat Plants 2016; 2:16013; http://dx.doi.org/ 10.1038/nplants.2016.13 [DOI] [PubMed] [Google Scholar]
- 18.Saga H, Ogawa T, Kai K, Suzuki H, Ogata Y, Sakurai N, Shibata D, Ohta D. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol Plant Microbe Interact 2012; 25:684-96; PMID:22295908; http://dx.doi.org/ 10.1094/MPMI-09-11-0244 [DOI] [PubMed] [Google Scholar]
- 19.Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor M-I, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, et al.. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012; 24:482-506; PMID:22345491; http://dx.doi.org/ 10.1105/tpc.111.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahnejat-Bushehri S, Mueller-Roeber B, Balazadeh S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal Behav 2012; 7:1518-21; PMID:23073024; http://dx.doi.org/ 10.4161/psb.22092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 2000; 13:191-202; PMID:10659709; http://dx.doi.org/ 10.1094/MPMI.2000.13.2.191 [DOI] [PubMed] [Google Scholar]
- 22.Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 2008; 3:348-51; PMID:18541211; http://dx.doi.org/ 10.1016/j.chom.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 23.Boachon B, Gamir J, Pastor V, Erb M, Dean JV, Flors V, Mauch-Mani B. Role of two UDP-glycosyltransferases from the L group of Arabidopsis in resistance against Pseudomonas syringae. Eur J Plant Pathol 2014; 139:707-20; http://dx.doi.org/ 10.1007/s10658-014-0424-7 [DOI] [Google Scholar]
- 24.Lee HI, León J, Raskin I. Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci USA 1995; 92:4076-9; PMID:11607533; http://dx.doi.org/ 10.1073/pnas.92.10.4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou X, Ding L, Yu H. Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep 2013; 32:1067-74; PMID:23525761; http://dx.doi.org/ 10.1007/s00299-013-1423-4 [DOI] [PubMed] [Google Scholar]
- 26.Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim J-M, To TK, Li W, et al.. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 2011; 108:12539-44; PMID:21737749; http://dx.doi.org/ 10.1073/pnas.1103959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 1998; 10:2103-13; PMID:9836748; http://dx.doi.org/ 10.1105/tpc.10.12.2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 1998; 12:3703-14; PMID:9851977; http://dx.doi.org/ 10.1101/gad.12.23.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wild M, Achard P. The DELLA protein RGL3 positively contributes to jasmonate/ethylene defense responses. Plant Signal Behav 2013; 8:e23891; PMID:23425858; http://dx.doi.org/ 10.4161/psb.23891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA 2005; 102:1791-6; PMID:15657122; http://dx.doi.org/ 10.1073/pnas.0409450102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perchepied L, Kroj T, Tronchet M, Loudet O, Roby D. Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS One 2006; 1:e123; PMID:17205127; http://dx.doi.org/ 10.1371/journal.pone.0000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kover PX, Wolf JB, Kunkel BN, Cheverud JM. Genetic architecture of Arabidopsis thaliana response to infection by Pseudomonas syringae. Heredity (Edinb) 2005; 94:507-17; PMID:15770233; http://dx.doi.org/ 10.1038/sj.hdy.6800651 [DOI] [PubMed] [Google Scholar]
- 33.Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) 2008; 10 Suppl 1:63-75; PMID:18721312; http://dx.doi.org/ 10.1111/j.1438-8677.2008.00088.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.