ABSTRACT
VIP1 (VIRE2-INTERACTING PROTEIN 1) is a bZIP transcription factor in Arabidopsis thaliana. VIP1 and its close homologs (i.e., Arabidopsis group I bZIP proteins) are present in the cytoplasm under steady conditions, but are transiently localized to the nucleus when cells are exposed to hypo-osmotic conditions, which mimic mechanical stimuli such as touch. Recently we have reported that overexpression of a repression domain-fused form of VIP1 represses the expression of some touch-responsive genes, changes structures and/or local auxin responses of the root cap cells, and enhances the touch-induced root waving. This raises the possibility that VIP1 suppresses touch-induced responses. VIP1 should be useful to further characterize touch responses of plants. Here we discuss 2 seemingly interesting perspectives about VIP1: (1) What factors are involved in regulating the nuclear localization of VIP1?; (2) What can be done to further characterize the physiological functions of VIP1 and other Arabidopsis group I bZIP proteins?
KEYWORDS: Arabidopsis thaliana, bZIP transcription factor, calcium signaling, cell death, cell wall, hypo-osmotic stress, mechanical stimuli, protein phosphorylation, root cap, root waving
VIP1 (VIRE2-INTERACTING PROTEIN 1) was originally identified as an Arabidopsis thaliana bZIP protein interacting with the Agrobacterium tumefaciens protein VirE2,1 and has been suggested to play pleiotropic roles.1-12 Previously we reported that VIP1 exists mainly in the cytosol under steady conditions but transiently accumulates in the nucleus when cells are exposed to hypo-osmotic conditions,9 that close homologs of VIP1 (i.e., Arabidopsis group I bZIP proteins13) also exhibit such a pattern of subcellular localization,11 and that overexpression of a repression domain-fused form of VIP1 (VIP1-SRDX) enhances touch-induced root waving.14 Touch as well as the hypo-osmotic conditions seems to induce the nuclear localization of VIP1 (Fig. 1). VIP1 is a novel regulator of touch responses of Arabidopsis, and should be useful to further characterize them. Here we discuss perspectives for further studies on VIP1.
Figure 1.
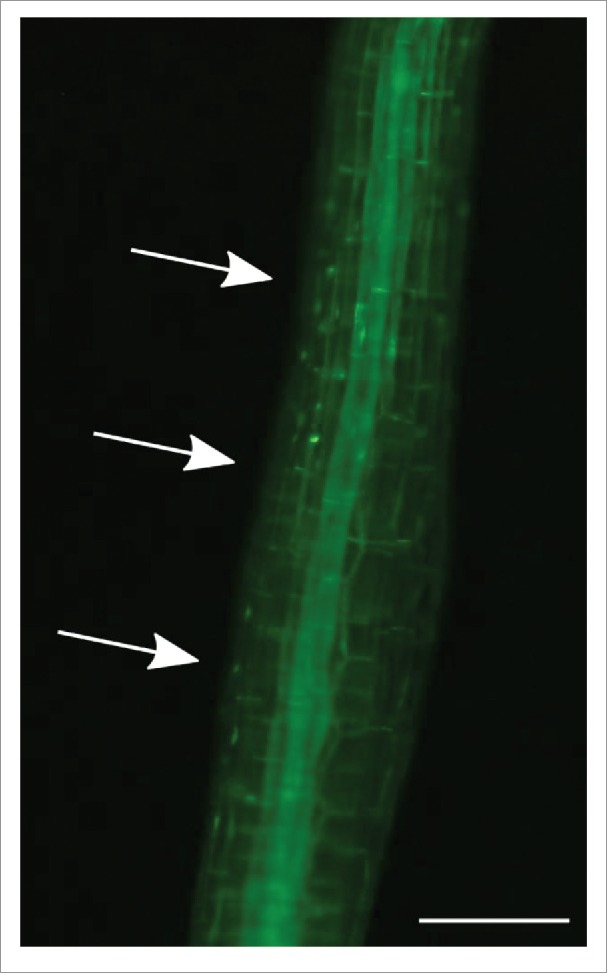
Touch-induced nuclear localization of VIP1. Transgenic Arabidopsis plants expressing the VIP1-GFP fusion protein were grown for 3 d in darkness. VIP1-GFP signals were observed approximately 3 minutes after their hypocotyls were touched by a needle tip. More than 5 plants were used, and a representative result is presented. Arrows indicate the approximate position where the hypocotyl was touched. Scale bar = 200 μm.
What factors are involved in regulating the nuclear localization of VIP1?
Previously a treatment with a microbe associated molecular pattern, flg22, caused VIP1 to be localized to the nucleus,6 and a treatment with a gibberellin biosynthesis inhibitor, uniconazole-P, caused the putative tobacco VIP1 ortholog RSG (REPRESSION OF SHOOT GROWTH) to be localized to the nucleus.15 However, in our experiments, water itself, which was used to dilute flg22 and uniconazole-P stocks in those experiments, can induce the nuclear localization of VIP1 (ref. 10 and unpublished data), thus the effects of these chemicals may have to be re-evaluated.
VIP1 is thought to accumulate in the nucleus when its 79th serine is phosphorylated by MPK3 (MITOGEN-ACTIVATED PROTEIN KINASE 3),6 although this idea has been questioned.11 On the other hand, RSG is thought to accumulate in the nucleus when its phosphorylated 114th serine is either dephosphorylated or replaced by alanine.15 Although replacing the VIP1 115th serine, which corresponds to the RSG 114th serine, with alanine does not affect the subcellular localization of VIP1,11 our unpublished data suggest that VIP1 has multiple putative phosphorylation sites, and that these sites are dephosphorylated when VIP1 is localized to the nucleus. The RSG 114th serine is phosphorylated by the calcium-dependent protein kinase CDPK1 in tobacco,16 thus a CDPK1 homolog may phosphorylate VIP1 in Arabidopsis. Our unpublished data also suggest that calcium signaling regulates both the nuclear import and the nuclear export of VIP1. It should be interesting to identify protein kinases and protein phosphatases regulating VIP1 phosphorylation states, and to identify the phosphorylation sites in VIP1.
The mechanosensitive calcium channels MCA1 and MCA2 (MID-COMPLEMENTING ACTIVITY 1 and 2, respectively), the 7-transmembrane domain proteins MLO4 and MLO11 (MILDEW RESISTANCE LOCUS O 4 and 11, respectively), and the receptor kinase FERONIA have been identified as candidate mechanical stress sensors regulating calcium signaling and root tropisms.17-20 TOUCH2 and TOUCH4, which are up-regulated by the FERONIA-mediated touch-responsive signaling,20 are unlikely to be VIP1 target genes.14 However, it would be worth examining whether VIP1 interacts with these proteins and/or acts under the control of them. To identify novel regulators for VIP1, it would also be helpful and practical to screen for chemicals and genetic mutations that affect the subcellular localization of VIP1.
What can be done to further characterize the physiological functions of VIP1 and other Arabidopsis group I bZIP proteins?
VIP1 and its close homologs in tomato, tobacco, and rice, have been identified in different studies as a regulator of certain physiological responses.15,16,21-26 This would support the idea that such VIP1 homologs have important, pleiotropic roles. Thus far only 2 Arabidopsis group I bZIP protein genes, VIP1 and PosF21, have been associated with physiological roles.1-12,14,27 However, at least 5 of the other group I bZIP protein genes are expressed as highly as VIP1 and PosF21,11 and they could function redundantly. In our recent study, expressing the VIP1-GFP fusion protein suppressed the VIP1-SRDX-induced enhancement of root waving.14 It would be interesting to examine whether other group I bZIP proteins can also suppress the VIP1-SRDX-induced enhancement of root waving. In our preliminary experiments, the triple mutant that has T-DNA in VIP1, PosF21, and bZIP29 (another group I bZIP protein gene) was similar to the wild type in phenotypes including root waving, but knocking out various combinations of the group I bZIP protein genes should also help to elucidate the physiological roles of them.
The local auxin responses in the root tip are different between wild-type plants and VIP1-SRDX-overexpressing (VIP1-SRDXox) plants, and this may be attributed to the difference in adhesion and/or removal of the root cap cells. Expression levels of some mechanical stimulus-induced genes that should regulate cell wall properties are lower in VIP1-SRDXox plants than in the wild-type plants, and this may cause the abnormal root cap cell adhesion/removal in VIP1-SRDXox plants.14 On the other hand, cell death mediated by the NAC (NO APICAL MERISTEM)-family transcription factor SOMBRERO and the S1-P1 nuclease-family protein BFN1 (BIFUNCTIONAL NUCLEASE 1) is necessary for the removal of the lateral root cap cells.28,29 It would be interesting to characterize the cell wall properties and cell death in the root cap cells of VIP1-SRDXox plants, and to examine genetic and physical interactions between the group I bZIP proteins and the above regulators of cell wall properties and cell death. It would also be important to further evaluate how mechanical stimuli affect cell wall properties and cell death in root cap cells.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Grant-in-Aid for JSPS Fellows (grant number 25-7247) to D.T.
References
- 1.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 2001; 20:3596-607; PMID:11432846; http://dx.doi.org/ 10.1093/emboj/20.13.3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc Natl Acad Sci USA 2002; 99:10435-40; PMID:12124400; http://dx.doi.org/ 10.1073/pnas.162304099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 2004; 431:87-92; PMID:15343337; http://dx.doi.org/ 10.1038/nature02857 [DOI] [PubMed] [Google Scholar]
- 4.Lacroix B, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J 2005; 24:428-37; PMID:15616576; http://dx.doi.org/ 10.1038/sj.emboj.7600524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA 2005; 102:5733-8; PMID:15824315; http://dx.doi.org/ 10.1073/pnas.0404118102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 2007; 318:453-6; PMID:17947581; http://dx.doi.org/ 10.1126/science.1148110 [DOI] [PubMed] [Google Scholar]
- 7.Pitzschke A, Djamei A, Teige M, Hirt H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci USA 2009; 106:18414-9; PMID:19820165; http://dx.doi.org/ 10.1073/pnas.0905599106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Zhao Q, Gao L, Yu XM, Fang P, Oliver DJ, Xiang CB. Isolation and characterization of low-sulphur-tolerant mutants of Arabidopsis. J Exp Bot 2010; 61:3407-22; PMID:20547563; http://dx.doi.org/ 10.1093/jxb/erq161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsugama D, Liu S, Takano T. A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiol 2012; 159:144-55; PMID:22452852; http://dx.doi.org/ 10.1104/pp.112.197020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacroix B, Citovsky V. Characterization of VIP1 activity as a transcriptional regulator in vitro and in planta. Sci Rep 2013; 3:2440; PMID:23942522; http://dx.doi.org/ 10.1038/srep02440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsugama D, Liu S, Takano T. Analysis of functions of VIP1 and its close homologs in osmosensory responses of Arabidopsis thaliana. PLoS One 2014; 9:e103930; PMID:25093810; http://dx.doi.org/ 10.1371/journal.pone.0103930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Lee LY, Gelvin SB. Is VIP1 important for Agrobacterium-mediated transformation? Plant J 2014; 79:848-60; PMID:24953893; http://dx.doi.org/ 10.1111/tpj.12596 [DOI] [PubMed] [Google Scholar]
- 13.Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, bZIP Research Group . bZIP transcription factors in Arabidopsis. Trends Plant Sci 2002; 7:106-11; PMID:11906833; http://dx.doi.org/ 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- 14.Tsugama D, Liu S, Takano T. The bZIP protein VIP1 is involved in touch responses in Arabidopsis roots. Plant Physiol 2016. Apr 6; PMID:27208231; http://dx.doi.org/15377759 10.1104/pp.16.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishida S, Fukazawa J, Yuasa T, Takahashi Y. Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 2004; 16:2641-51; PMID:15377759; http://dx.doi.org/ 10.1105/tpc.104.024604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida S, Yuasa T, Nakata M, Takahashi Y. A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 2008; 20:3273-88; PMID:19106376; http://dx.doi.org/ 10.1105/tpc.107.057489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa Y, Katagiri T, Shinozaki K, Qi Z, Tatsumi H, Furuichi T, Kishigami A, Sokabe M, Kojima I, Sato S, et al.. Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc Natl Acad Sci USA 2007; 104:3639-44; PMID:17360695; http://dx.doi.org/ 10.1073/pnas.0607703104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Noir S, Kwaaitaal M, Hartmann HA, Wu MJ, Mudgil Y, Sukumar P, Muday G, Panstruga R, Jones AM. Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell 2009; 21:1972-91; PMID:19602625; http://dx.doi.org/ 10.1105/tpc.108.062653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamanaka T, Nakagawa Y, Mori K, Nakano M, Imamura T, Kataoka H, Terashima A, Iida K, Kojima I, Katagiri T, et al.. MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol 2010; 152:1284-96; PMID:20097794; http://dx.doi.org/25127214 10.1104/pp.109.147371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol 2014; 24:1887-92; PMID:25127214; http://dx.doi.org/ 10.1016/j.cub.2014.06.064 [DOI] [PubMed] [Google Scholar]
- 21.Torres-Schumann S, Ringli C, Heierli D, Amrhein N, Keller B. In vitro binding of the tomato bZIP transcriptional activator VSF-1 to a regulatory element that controls xylem-specific gene expression. Plant J 1996; 9:283-96; PMID:8919907; http://dx.doi.org/ 10.1046/j.1365-313X.1996.09030283.x [DOI] [PubMed] [Google Scholar]
- 22.Yin Y, Zhu Q, Dai S, Lamb C, Beachy RN. RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J 1997; 16:5247-59; PMID:9311985; http://dx.doi.org/ 10.1093/emboj/16.17.5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringli C, Keller B. Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol Biol 1998; 37:977-88; PMID:9700070; http://dx.doi.org/ 10.1023/A:1006030007333 [DOI] [PubMed] [Google Scholar]
- 24.Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 2000; 12:901-15; PMID:10852936; http://dx.doi.org/ 10.1105/tpc.12.6.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai S, Zhang Z, Chen S, Beachy RN. RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc Natl Acad Sci USA 2004; 101:687-92; PMID:14704272; http://dx.doi.org/ 10.1073/pnas.0307687100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai S, Wei X, Alfonso AA, Pei L, Duque UG, Zhang Z, Babb GM, Beachy RN. Transgenic rice plants that overexpress transcription factors RF2a and RF2b are tolerant to rice tungro virus replication and disease. Proc Natl Acad Sci USA 2008; 105:21012-6; PMID:19104064; http://dx.doi.org/ 10.1073/pnas.0810303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Oosten MJ, Sharkhuu A, Batelli G, Bressan RA, Maggio A. The Arabidopsis thaliana mutant air1 implicates SOS3 in the regulation of anthocyanins under salt stress. Plant Mol Biol 2013; 83:405-15; PMID: 23925404; http://dx.doi.org/18603613 10.1007/s11103-013-0099-z [DOI] [PubMed] [Google Scholar]
- 28.Farage-Barhom S, Burd S, Sonego L, Perl-Treves R, Lers A. Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J Exp Bot 2008; 59:3247-58; PMID:18603613; http://dx.doi.org/ 10.1093/jxb/ern176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, et al.. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 2014; 24:931-40; PMID:24726156; http://dx.doi.org/ 10.1016/j.cub.2014.03.025 [DOI] [PubMed] [Google Scholar]


