ABSTRACT
In recent years, many small RNAs and long non-protein-coding RNAs (lncRNAs) have been identified and characterized. They have been proved to play essential regulatory roles in gene expression in both primary and secondary metabolisms. In nature, many plants produce alkaloids. However, there are only few reports on the involvement of non-coding RNAs in alkaloid biosynthesis. Nicotine is major alkaloid in tobacco plants. Its biosynthesis and regulation in tobacco (Nicotiana tabacum) have been well studied; and major structural genes involved in the nicotine biosynthesis and transcriptional regulators related to its biosynthesis have been identified and characterized. In our recent studies, we identified a microRNA (nta-miRX27) and also a lncRNA (nta-eTMX27) as an endogenous target mimicry (eTM) in tobacco targeting the nicotine biosynthesis key gene QPT2 encoding quinolinate phosphoribosyltransferase (QPT) and thereby regulating the nicotine content. Their regulatory pattern leads us to conclude that nicotine biosynthesis is regulated by 2 more layers besides previously known mechanisms. Future study on the relationship between the non-coding RNAs and transcription factors in nicotine biosynthesis was discussed in this article.
KEYWORDS: Alkaloid, endogenous target mimicry, long non-protein-coding RNA, microRNA, nicotine biosynthesis, tobacco
Abbreviations
- AP2
APETALA2
- ERF
ethylene response factor
- eTM
endogenous target mimicry
- lncRNAs
long non-protein-coding RNAs
- MYC2/bHLH
MYC2-like basic helix-Loop-helix
- QPT
quinolinate phosphoribosyltransferase
In nature, about 20% of plant species produce more than 12,000 alkaloids,1 which are important compounds for human health (For a review see refs. 2,3) as well as for plant defense.4 A more comprehensive understanding of their biosynthesis and regulation at molecular level is essential to facilitate genetic engineering to modify alkaloid production. Among alkaloid producing plants, tobacco (Nicotiana tabacum) is a good model plant to study alkaloid biosynthesis, transportation, accumulation and degradation. Tobacco plants produce very high levels of alkaloid about 2–4% of dried leaves and nicotine accounts for 90% of the total alkaloids.5 Major structural genes involved in the nicotine biosynthesis have been identified and characterized (For a review see ref. 6). In recent years, transcriptional regulators of nicotine biosynthesis have been widely studied resulting in identification of 2 distinct families of transcription factors - the APETALA2 (AP2)/ethylene response factor (ERF) and MYC2-like basic helix-Loop-helix (bHLH) families (For a review see refs. 6,7). These studies provided a wealth of inofmration at molecular level to understand observations made from classic genetic and biochemical studies regarding the regulation of nicotine biosynthesis by 2 distinct nicotine loci NICOTINE1 and NICOTINE2 (originally called the A and B loci) and wounding induced jasmonic acid elevation. In addition, it is not surprising to find that nicotine permeation, transportation and accumulation8-12 could also affect the expression of the nicotine biosynthesis pathway genes via the cellular nicotine levels.
Benefiting from advancement in sequencing techniques and with the available genome data for tobacco and its 2 progenitors, we recently not only identified diverse microRNAs (miRNAs) responsive to wounding or topping treatments13 but also a miRNA (nta-miRX27) and its endogenous target mimicry (eTM) nta-eTMX27 targeting key nicotine biosynthesis pathway gene QPT2 encoding quinolinate phosphoribosyltransferase (QPT).14 These studies demonstrated that the nicotine biosynthesis is regulated by 2 more layers distinct from previously known mechanisms. One layer is through a miRNA-mediated gene silencing mechanism to degrade the transcripts of QPT2 by miRNA nta-miRX27 (Fig. 1). MiRNAs have been proved to play regulatory roles in plant development, signal transduction and responses to biotic and abiotic stresses15,16 as well as secondary metabolism.17-19 In the nicotine biosynthesis pathway, QPT is a key enzyme catalyzing the rate limited step by converting quinolinic acid to nicotinic acid mononucleotide and serving as the entry point into the pyridine nucleotide cycle.6 There are 2 members in QPT family: QPT1 and QPT2. QPT2 has been shown to be mainly responsible for nicotine biosynthesis20,21 while QPT1 is not because it is unresponsive to the genetic, biotic and abiotic factors that regulate nicotine production.22 By knocking down QPT2, we showed that the nicotine levels could be reduced about 95%,23 further confirming that QPT2 is a key gene in nicotine biosynthesis pathway. Therefore, any interference of its expression would affect nicotine biosynthesis.
Figure 1.
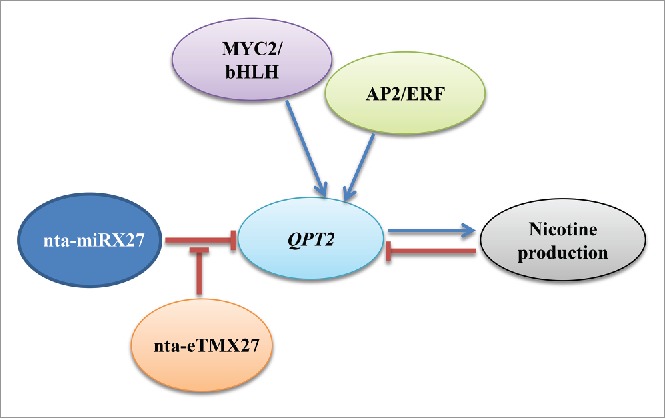
Schematic representation of regulatory network of QPT2 and nicotine biosynthesis.
The mature nta-miRX27 that we identified in our study14 is derived from a 131 bp precursor having 21 nucleotides, which fits a typical miRNA length of 20–22 nucleotides.15 By prediction, the nta-miRX27 could bind QPT2 at 2 sites located at exon 2 and intron 8 with targeting scores of 4.5 and 2.5, respectively. Topping experiments indicated that the expressions of QPT2 and nta-miRX27 were negatively correlated (Fig. 1) with the correlation coefficiency of −0.949 when their transcription levels in tobacco roots were measured at 3, 6, 12, 24 and 48 h after topping. Our degradome data supported the idea that the predicted binding site at exon 2 is very likely a target for the cleavage by nta-miRX27. However, binding site at intron 8 cannot be ruled out because RLM-RACE experiments using different primers were not successful. Nevertheless, our results related to overexpression or silencing of nta-miRX27 demonstrated that nta-miRX27 indeed negatively regulated QPT2 transcript levels to cause corresponding nicotine level changes in tobacco leaves.
A more surprising discovery was identification of an eTM for nta-miRX27 designated as nta-eTMX27, which acted as a decoy for nta-miRX27.14 We discovered that the cleavage activity of nta-miRX27 was negatively regulated by nta-eTMX27 leading us to propose that another layer of control is exerted over QPT2 expression and nicotine biosynthesis via an eTM. An eTM acting as a decoy for the microRNA to buffer its target gene is a newly emerging regulation mechanism on gene expression. Since the first eTM from a long non-protein-coding mRNA (lncRNA) gene INDUCED BY PHOSPHATE STARVTION1 was reported by Franco-Zorrilla et al.24 in Arabidopsis to sequester miR399 and to disrupt its cleavage activity, non-coding eTMs for various miRNAs have been reported in several plant species.25-27 Tobacco nta-eTMX27 is a 1,213 bp long lncRNA having a binding site for nta-miRX27 with a 3-nucleotide bulge between the ninth and 10th position. The topping experiment showed that the expression levels of nta-eTMX27 and nta-miRX27 were negatively correlated with correlation coefficiency of −0.956 while those of nta-eTMX27 and QPT2 were correlated positively with correlation coefficiency of 0.986 when their transcriptional levels were quantified at 3, 6, 12, 24 and 48 h after topping. Moreover, nta-eTMX27 overexpressing lines had higher expression levels of QPT2 along with higher nicotine contents via the sequestration of nta-miRX27 away from QPT2 whereas nta-eTMX27 RNAi lines had increased expressions of nta-miRX27 and lower levels of QPT2, and subsequently lower nicotine contents comparing to vector control lines. These results clearly demonstrated that both nta-miRX27 and nta-eTMX27 regulate QPT2 and nicotine biosynthesis, but with opposite effects.
Previous studies have shown that QPT2 expression is up-regulated by topping and wounding, but inhibited by auxin.20,21 Topping by removal of the plant/flower head has both transient wounding effect as well as lasting low auxin effect on QPT2 expression and nicotine production. So far, the expressions of nta-miRX2713,14,28 and nta-eTMX2714 respond to the topping treatment. Whether or not wounding and auxin affect nta-miRX27 and nta-eTMX27 expressions remains to be determined. Further studies with created transgenic lines with overexpressing or suppressing nta-miRX27 and nta-eTMX27 are warranted to fully understand how they respond to wounding and low auxin levels. This will allow us to know how nta-miRX27 and nta-eTMX27 regulate QPT2 expression and nicotine biosynthesis spatiotemporally. Additionally, structural genes of the nicotine biosynthesis pathway including QPT2 are regulated by 2 families of transcription factors AP2/ERF and MYC2/bHLH.6,7 It is worth to investigate whether the expression of nta-miRX27 and nta-eTMX27 is also regulated by these transcription factors or their regulatory roles on nicotine biosynthesis run parallel to these transcription factors.
Recent advances in metabolic engineering have shown great potential to more effectively maximize the capacity of alkaloid biosynthesis and to produce specific alkaloids in plants.29 Currently, metabolic engineering of alkaloid production is focused primarily on manipulation of genes in secondary metabolic pathways29,30 and their related transcription factors.12,31,32 Genes involved in rate-limiting steps or at metabolic branch points are the most common targets for genetic modifications.23,29,31,33 To reduce the content of certain alkaloid by interrupting the rate-limiting gene is relatively easy.23,33 However, increasing levels of a certain alkaloid by overexpressing one or several key genes remains a real challenge2 because of emerging new rate-limiting steps in the metabolic pathways and some unknown factors. Fully understanding how alkaloid metabolic pathways are regulated, and exploring other ways to manipulate them are essential for improving alkaloid production by metabolic engineering.
There is a growing body of literature describing miRNAs involved in secondary metabolism, but the total number of identified miRNAs for secondary metabolic pathway genes is still very small with only a few of them functionally characterized. This is mainly because of lack of genome information for alkaloid-producing plants. As more and more plant genomes sequences become available in the future, more miRNAs for alkaloid biosynthesis are expected to be discovered. Moreover, although eTMs have been suggested to exist widely in plant species, nta-eTMX27, to the best of our knowledge, is the first eTM identified and confirmed to involve in alkaloid metabolism. With further discovery and functional characterization of new non-coding RNAs from secondary metabolism, it may be possible to precisely manipulate alkaloid biosynthesis pathways in plants to produce specific alkaloids as well as to maximize alkaloid production.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Farooqahmed S. Kittur for critical reading of the manuscript.
References
- 1.Faccini PJ. Alkaloid biosynthesis in plants: Biochemistry, cell biology, molecular regulation and metabolic engineering applications. Annu Rev Plant Physiol Plant Mol Biol 2001; 52:29-66; PMID:11337391; http://dx.doi.org/ 10.1146/annurev.arplant.52.1.29 [DOI] [PubMed] [Google Scholar]
- 2.Verpoorte R, Memelink J. Engineering secondary metabolite production in plants. Curr Opin Biotechnol 2002; 13:181-87; PMID:11950573; http://dx.doi.org/ 10.1016/S0958-1669(02)00308-7 [DOI] [PubMed] [Google Scholar]
- 3.Ziegler J, Facchini PJ. Alkaloid biosynthesis: metabolism and trafficking. Annu Rev Plant Biol 2008; 6:212-9; PMID:18251710; http://dx.doi.org/11418346 10.1146/annurev.arplant.59.032607.092730 [DOI] [PubMed] [Google Scholar]
- 4.Baldwin IT, Halitschke R, Kessler A, Schittko U. Merging molecular and ecological approaches in plant-insect interactions. Curr Opin Plant Biol 2001; 4:351-58; PMID:11418346; http://dx.doi.org/ 10.1016/S1369-5266(00)00184-9 [DOI] [PubMed] [Google Scholar]
- 5.Saitoh F, Nona M, Kawashima N. The alkaloid contents of sixty Nicotiana species. Phytochemistry 1985; 24:477-80; http://dx.doi.org/ 10.1016/S0031-9422(00)80751-7 [DOI] [Google Scholar]
- 6.Dewey RE, Xie J. Molecular genetics of alkaloid biosynthesis in Nicotiana tabacum. Phytochemistry 2013; 94:10-27; PMID:23953973; http://dx.doi.org/ 10.1016/j.phytochem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 7.Shoji T, Hashimoto T. Smoking out the masters: transcriptional regulators for nicotine biosynthesis in tobacco. Plant Biotechnol 2013; 30:217-24; http://dx.doi.org/ 10.5511/plantbiotechnology.13.0221a [DOI] [Google Scholar]
- 8.Morita M, Shitan N, Sawada K, Van Montagu MCE, Inze D, Rischer H, Goossens A, Oksman-Caldentey KM, Moriyama Y, Yazaki K. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc Natl Acad Sci USA 2009; 106:2447-52; PMID:19168636; http://dx.doi.org/19098091 10.1073/pnas.0812512106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoji T, Inai K, Yazaki Y, Sato Y, Takase H, Shitan N, Yazaki K, Goto Y, Toyooka K, Matsuoka K, et al.. Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol 2009; 149:708-18; PMID:19098091; http://dx.doi.org/ 10.1104/pp.108.132811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildreth SB, Gehman EA, Yang H, Lu RH, Ritesh KC, Harich KC, Yu S, Lin J, Sandoe JL, Okumoto S, et al.. Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc Natl Acad Sci USA 2011; 108:18179-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato K, Shoji T, Hashimoto T. Tobacco nicotine uptake permease regulates the expression of a key transcription factor gene in the nicotine biosynthesis pathway. Plant Physiol 2014; 166:2195-204; PMID:25344505; http://dx.doi.org/ 10.1104/pp.114.251645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang BW, Lewis RS, Shi J, Song Z, Gao Y, Li W, Chen H, Qu R. Genetic factors for enhancement of nicotine levels in cultivated tobacco. Sci Rep 2015; 5:17360; PMID:26626731; http://dx.doi.org/ 10.1038/srep17360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang She, Yu Wang, Zefeng Li, Yijie Gui, Bingguang Xiao, Jiahua Xie, Qian-Hao Zhu, Longjiang Fan Identification of wounding and topping responsive small RNAs in tobacco (Nicotiana tabacum). BMC Plant Biol 2012; 12: 28; PMID:22353177; http://dx.doi.org/26246450 10.1186/1471-2229-12-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F, Wang W, Zhao N, Xiao B, Cao P, Wu X, Ye C, Shen E, Qiu J, Zhu QH, Xie J, Zhou X, Fan L. Regulation of nicotine biosynthesis by an endogenous target mimicry of miRNA in tobacco. Plant Physiol 2015; 169:1062-71; PMID:26246450; http://dx.doi.org/ 10.1104/pp.15.00649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips JR, Dalmay T, Bartels D. The role of small RNAs in abiotic stress. FEBS Lett 2007; 581:3592-7; PMID:17451688; http://dx.doi.org/ 10.1016/j.febslet.2007.04.007 [DOI] [PubMed] [Google Scholar]
- 16.Khraiwesh B, Zhu J-K, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochem Biophys Acta 2012; 1819:137-48; PMID:21605713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng DW, Zhang C, Miller M, Palmer G, Whiteley M, Tholl D, Chen ZJ. Cis-and trans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 2011; 23:1729-40; PMID:21602291; http://dx.doi.org/ 10.1105/tpc.111.083915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robert-Seilaniantz A, MacLean D, Jikumaru Y, Hill L, Yamaguchi S, Kamiya Y, Jones JD. The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J 2011; 67:218-31; PMID:21457368; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04591.x [DOI] [PubMed] [Google Scholar]
- 19.Boke H, Ozhuner E, Turktas M, Parmaksiz I, Ozcan S, Unver T. Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnol J 2015; 13:409-20; PMID:25735537 [DOI] [PubMed] [Google Scholar]
- 20.Song W. Molecular characterizations of two tobacco root-specific genes: TobRB7 and NtQPT1. Ph.D. thesis, North Carolina State University, USA, 1997 [Google Scholar]
- 21.Sinclair SJ, Murphy KJ, Birch CD, Hamill JD. Molecular characterization of quinolinate phosphoribosyltransferase (QPRTase) in Nicotiana. Plant Mol Biol 2000; 44:603-17; PMID:11198422 [DOI] [PubMed] [Google Scholar]
- 22.Shoji T, Hashimoto T. Recruitment of a duplicated primary metabolism gene into the nicotine biosynthesis regulon in tobacco. Plant J 2011; 67:949-59; PMID:21605206 [DOI] [PubMed] [Google Scholar]
- 23.Xie J, Song W, Maksymowicz W, Jin W, Cheah K, Chen W, Carnes C, Ke J, Conkling M. Biotechnology: a tool for reduced risk tobacco products-the nicotine experience from test tube to cigarette pack. Rev Adv Tob Sci 2004; 30:17-37 [Google Scholar]
- 24.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 2007; 39:1033-7; PMID:17643101 [DOI] [PubMed] [Google Scholar]
- 25.Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 2010; 6:e1001031; PMID:20661442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivashuta S, Banks IR, Wiggins BE, Zhang Y, Ziegler TE, Roberts JK, Heck GR. Regulation of gene expression in plants through miRNA inactivation. PLoS One 2011; 6:e21330; PMID:21731706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H-J, Wang Z-M, Wang M, Wang X-J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 2013; 161:1875-84; PMID:23429259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo H, Kan Y, Liu W Differential expression of miRNAs in response to topping in flue-cured tobacco (Nicotiana tabacum) roots. PLoS One 2011; 6:e28565; http://dx.doi.org/ 10.1371/journal.pone.0028565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu SQ, Chappell J Metabolic engineering of natural products in plants; tools of the trade and challenges for the future. Curr Opin Biotechnol 2008; 19:145-52; http://dx.doi.org/ 10.1016/j.copbio.2008.02.007 [DOI] [PubMed] [Google Scholar]
- 30.Ogita S, Uefuji H, Yamaguchi Y, Koizumi N, Sano H Producing decaffeinated coffee plants. Nature 2003; 423:823; http://dx.doi.org/ 10.1038/423823a [DOI] [PubMed] [Google Scholar]
- 31.van der Fits L, Memelink J ORCA3, a Jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism Science 2000; 889:295-7; http://dx.doi.org/ 10.1126/science.289.5477.295 [DOI] [PubMed] [Google Scholar]
- 32.Grotewold E Transcription factors for predictive plant metabolic engineering: are we there yet?. Curr Opin Biotechnol 2008; 19:138-44 [DOI] [PubMed] [Google Scholar]
- 33.Hibi N, Higashiguchi S, Hashimoto T, Yamada Y Gene expression in tobacco low-nicotine mutants. Plant Cell 1994; 6:723–35; http://dx.doi.org/ 10.1105/tpc.6.5.723 [DOI] [PMC free article] [PubMed] [Google Scholar]


