Summary
In Primula vulgaris outcrossing is promoted through reciprocal herkogamy with insect‐mediated cross‐pollination between pin and thrum form flowers. Development of heteromorphic flowers is coordinated by genes at the S locus. To underpin construction of a genetic map facilitating isolation of these S locus genes, we have characterised Oakleaf, a novel S locus‐linked mutant phenotype.
We combine phenotypic observation of flower and leaf development, with classical genetic analysis and next‐generation sequencing to address the molecular basis of Oakleaf.
Oakleaf is a dominant mutation that affects both leaf and flower development; plants produce distinctive lobed leaves, with occasional ectopic meristems on the veins. This phenotype is reminiscent of overexpression of Class I KNOX‐homeodomain transcription factors. We describe the structure and expression of all eight P. vulgaris PvKNOX genes in both wild‐type and Oakleaf plants, and present comparative transcriptome analysis of leaves and flowers from Oakleaf and wild‐type plants.
Oakleaf provides a new phenotypic marker for genetic analysis of the Primula S locus. We show that none of the Class I PvKNOX genes are strongly upregulated in Oakleaf leaves and flowers, and identify cohorts of 507 upregulated and 314 downregulated genes in the Oakleaf mutant.
Keywords: heterostyly, KNOX genes, Oakleaf, Primula vulgaris, S locus
Introduction
Observations on different forms of Primula flowers date back nearly 400 yr (van Dijk, 1943; P.M. Gilmartin, unpublished). The development of two distinct floral forms, known as pin and thrum, attracted the attention of Darwin, who recognised and described their relevance and significance in his detailed studies of P. vulgaris and P. veris (Darwin, 1862). Primula produce either pin or thrum flowers, which exhibit reciprocal herkogamy and show different degrees of self‐incompatibility (Darwin, 1862, 1877). Pin flowers have a long style with the stigma at the corolla mouth and anthers attached midway down the corolla tube; thrum flowers have anthers which are positioned at the corolla mouth and a short style which presents the stigma midway up the corolla tube (Darwin, 1862; Webster & Gilmartin, 2006). Elevation of the anthers in thrum flowers is caused by increased cell division in the corolla tube below their point of attachment, whilst in pin flowers the style is extended by increased cell elongation (Heslop‐Harrison et al., 1981; Webster & Gilmartin, 2006). Differential floral architecture is orchestrated by different cellular mechanisms affecting anther elevation and style elongation (Webster & Gilmartin, 2006). Other morph‐specific differences include pollen size, corolla opening diameter, stigma shape, stigmatic papillae length and style cross‐section (Darwin, 1877; Haldane, 1933; Dowrick, 1956; Dulberger, 1975; Heslop‐Harrison et al., 1981; Richards, 1997; Webster & Gilmartin, 2006). Darwin observed that within‐morph pin–pin or thrum–thrum crosses were less fertile than intermorph pin–thrum or thrum–pin crosses (Darwin, 1877). This observation is underpinned by the presence of a sporophytic incompatibility system that in combination with the structural differences between the two forms of flower inhibits self‐pollination and promotes outcrossing (Shivanna et al., 1981; Richards, 1997).
Floral heteromorphy in Primula is controlled by the S locus; pins are homozygous recessive (s/s), thrums heterozygous (S/s) (Bateson & Gregory, 1905; Haldane, 1933; Dowrick, 1956). Studies by Ernst (Ernst, 1928, 1933) and others (Pellow, 1928; Haldane, 1933; Dowrick, 1956; Lewis & Jones, 1993) suggested that the S locus comprises three dominant genetic functions: G, which suppresses style elongation; P, responsible for enlarged pollen; and A, which controls anther elevation. These genes represent a co‐adapted linkage group. Other genes responsible for male and female sporophytic self‐incompatibility functions are also linked (Lewis, 1949; Lewis & Jones, 1993; de Nettancourt, 1997; Richards, 1997). Tight linkage of the GPA gene cluster maintains coupling and cosegregation of the dominant alleles. The classical model is that thrum plants have genotype GPA/gpa and pin plants gpa/gpa (Dowrick, 1956; Lewis & Jones, 1993; Richards, 1997).
Several genes linked to the S locus in P. sinensis and P. vulgaris, but not directly involved in floral heteromorphy, have been identified through analysis of mutants and phenotypic variation, including flower pigment genes (Gregory et al., 1923; De Winton & Haldane, 1933, 1935; Kurian, 1996), Hose in Hose (Ernst, 1942; Webster & Grant, 1990; Webster & Gilmartin, 2003; Webster, 2005) and sepaloid (Webster & Gilmartin, 2003; Webster, 2005; Li et al., 2008). Related studies on differential gene expression also identified genes that are differentially regulated in response to the S locus (McCubbin et al., 2006), and genes and polymorphisms located at, or close to, the S locus (Manfield et al., 2005; Li et al., 2007). However, the key S locus genes that orchestrate floral heteromorphy in Primula remain to be identified.
Heterostyly is not restricted to the Primulaceae but found in over 28 families (Ganders, 1979; Barrett & Shore, 2008) including Primula (Darwin, 1862), Turnera (Barrett, 1978) and Fagopyrum (Garber & Quisenberry, 1927). Progress has been made towards characterisation of the genes responsible for heterostyly in F. esculentum and T. subulata which both produce dimorphic flowers showing reciprocal herkogamy, as well as in Linum grandiflorum which exhibits stigma height dimorphism without anther height variation (Darwin, 1863; Lewis, 1943; Barrett, 2010). Studies in T. subulata based on a genetic map, chromosome deletion mutants and a BAC contig spanning the S locus (Woo et al., 1999; Labonne et al., 2008, 2009, 2010) enabled positional closing of the s haplotype (Labonne & Shore, 2011). In F. esculentum similar mapping approaches (Matsui et al., 2004; Yasui et al., 2004, 2008; Konishi et al., 2006), together with transcriptome sequencing, identified a candidate gene, S‐ELF3, for the short‐styled buckwheat phenotype (Yasui et al., 2012). Molecular analysis of protein and transcript profiles in long‐styled and short‐styled L. grandiflorum flowers also identified candidates for the control of dimorphic style development (Ushijima et al., 2012). The polyphyletic origin of heterostyly and the different floral architectures in different species suggest different molecular mechanisms underpinning heterostylous flower development. Parallel analyses of different heterostylous species are therefore important to facilitate comparative analyses of mechanisms that evolved to promote outbreeding.
A key step towards defining the key S locus genes in Primula is the identification of genetic markers for the S locus. Here we describe a new S locus‐linked mutant phenotype, which we call Oakleaf. We explore the molecular basis of Oakleaf through a candidate gene approach, and via transcriptomic and genomic analyses to profile the molecular phenotype as a prelude to construction of a genetic map of the Primula S locus. Oakleaf provides an important marker that will facilitate identification of key genes orchestrating distyly in Primula.
Materials and Methods
Plant material and linkage analysis
Plants used in this study are wild‐type Primula vulgaris Huds. and derived commercial cultivars. Primula vulgaris Oakleaf plants were originally obtained from Richards Brumpton (Woodborough Nurseries, Nottingham, UK) in 1999 and maintained by Margaret Webster as part of the National Collection of Primula, British Floral Variants. Plants were grown as described previously (Webster & Gilmartin, 2006). Hose in Hose, Jack in the Green and Jackanapes (Webster & Grant, 1990; Webster & Gilmartin, 2003) were crossed with Oakleaf, and controlled crosses between Oakleaf and wild‐type were performed, in insect‐free environments following emasculation of pollen recipients by removal of corolla and anthers. Seed was harvested from ripe seed capsules and stored at c. 4°C in air‐tight containers.
Scanning electron microscopy (SEM)
Floral apical meristems and developing buds were dissected using scalpels and razor blades with a ×20 hand lens. Samples were prepared for cryo‐SEM, analysed and images recorded as described previously (Webster & Gilmartin, 2003).
Draft genome sequence acquisition
Paired‐end and mate‐pair genomic DNA sequence reads were generated by Illumina HiSeq2000 at The Genome Analysis Centre, Norwich Research Park, Norwich, UK. DNA was isolated from leaves of inbred self‐fertile long homostyle P. vulgaris originating from Wyke Champflower, Somerset, UK (Crosby, 1940) for paired‐end read sequencing. This genotype was chosen due to homozygosity compared with outbreeding pin and thrum plants. The assembly was scaffolded with mate‐pair reads from a 9 kb thrum genomic DNA library. The paired‐end reads provided ×60 genome coverage, and the mate‐pair reads provided ×26 read coverage after filtering. A draft assembly was generated using ABySS v1.3.4 (Simpson et al., 2009) (kmer length = 81) to assemble paired‐end reads, then SOAPdenovo v2.0.4 (Luo et al., 2012) to scaffold contigs using mate‐pair reads (kmer length = 71). This process generated an assembly of 424 Mb comprising 102 442 sequences and a scaffold N50 of 47.8 kb. This draft assembly was used to identify the full complement of PvKNOX‐like sequences and gene model assemblies for differential transcript analysis. Full details of the fully assembled and annotated P. vulgaris genome will be published elsewhere.
Gene model predictions for P. vulgaris KNOX (PvKNOX) genes
Arabidopsis thaliana KNOX proteins, KNAT1, KNAT2 (Lincoln et al., 1994), KNAT3, KNAT4, KNAT5 (Serikawa et al., 1996), KNAT6 (Belles‐Boix et al., 2006), KNAT7 (Li et al., 2011) and STM1 (Long et al., 1996), were aligned to the draft P. vulgaris genome with Exonerate v2.2.0 (Slater & Birney, 2005) (http://ccb.jhu.edu/software/tophat/index.shtml). Primula vulgaris KNOX loci were identified and gene models confirmed by transcript evidence from TopHat v2.0.8 and Cufflinks v2.1.1 (http://ccb.jhu.edu/software/tophat/index.shtml; http://cole-trapnell-lab.github.io/cufflinks/) (Trapnell et al., 2013) and by homology of the predicted proteins to KNOX proteins from the TAIR10 protein database (https://www.arabidopsis.org/). Parameters for protein sequence comparisons were ≥ 50% identity with ≥ 30% coverage of the KNOX query sequence. Gene models were curated manually where necessary with GenomeView (http://genomeview.org/). Sequences corresponding to PvKNL1 were initially identified on two genomic contigs. The gene model was resolved as one locus by alignment to a Trinity (http://trinityrnaseq.github.io/) (Grabherr et al., 2011) assembly of the same Illumina RNA‐Seq paired‐end read data from pin and thrum flower RNA, as used for the Cufflinks analysis (Supporting Information Table S1). [Correction added after online publication 9 April 2015; in this section, URLs to TopHat and Trinity have been updated.]
Generation of the PvKNOX phylogenetic tree
Multiple sequence alignment of Zea mays KNOTTED1, A. thaliana KNOX proteins, and predicted PvKNOX protein sequences was carried out in MEGA6 using MUSCLE (Edgar, 2004; Tamura et al., 2013). To obtain phylogeny support, Bayesian analyses were performed using MrBayes v3.2.2 (Ronquist et al., 2012) and output files visualised in FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). The mixed amino acid substitution model was used, and the first 25% of samples were discarded as burn‐in. The consensus tree was obtained after 1000 000 generations, with the average standard deviation of split frequencies below 0.01 to ensure convergence. In addition, a acid sequence alignments of predicted protein sequences were generated with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) (Fig. S2).
Analysis of differential gene expression between Oakleaf and wild‐type plants
RNA was isolated from leaves and open flowers of Oakleaf and wild‐type pin plants, and from mixed stage pin and thrum flowers for RNA‐Seq using Illumina HiSeq2000 (Table S1). RNA‐Seq reads were aligned to draft P. vulgaris genome contigs using TopHat v2.0.8 (http://ccb.jhu.edu/software/tophat/index.shtml) (Trapnell et al., 2012), followed by construction and merging of the transcriptome using Cufflinks v2.1.1 (Trapnell et al., 2013) (http://cole-trapnell-lab.github.io/cufflinks/). [Correction added after online publication 9 April 2015; in this section, URLs to TopHat and Cufflinks have been updated.] RNA‐Seq reads from mixed stage pin and thrum flowers were used for transcriptome assembly but not subsequent expression analysis. HTSeq (Anders et al., 2014) was used to count raw read numbers per gene using RNA‐Seq data from Oakleaf and wild‐type leaf and flower samples. These read counts were normalised by estimating the effective library size with DESeq v1.16.0 (Anders & Huber, 2010) which was used to carry out differential expression analysis. Genes upregulated by a ×2 log2 fold‐change in both Oakleaf leaves and Oakleaf flowers were characterised by BlastX analysis (e‐value 1 × 10−4) (Camacho et al., 2009) to identify related sequences in the TAIR10 (https://arabidopsis.org/) and NCBI nonredundant (nr) protein databases, the latter being used as an input for Blast2GO (Conesa et al., 2005) All sequences have been deposited in NCBI under Bioproject number PRJNA260472. [Correction added after online publication 9 April 2015; the Bioproject number has been corrected.]
Results
The Oakleaf mutant phenotype
The Oakleaf phenotype was identified in 1999 amongst commercial ornamental Primula plants. The pedigree and cultivar of Oakleaf are unknown. A division of the original mutant plant was obtained by Margaret Webster and an Oakleaf population established which was used in this study, alongside development of Oakleaf in polyanthus form as a commercial variety.
The Oakleaf phenotype is first visible in seedlings which sometimes produce normal and sometimes lobed cotyledons (Fig. 1a). However, the first true leaves consistently show the lobed appearance characteristic of Quercus species (Fig. 1a). The phenotype is variable but distinctive and easily recognisable. Mature leaves have an angular lobed appearance and contain wider and thicker leaf veins than wild‐type (Fig. 1b). The lamina of the leaf is thicker and firmer than wild‐type and the abaxial surface is pubescent. The effects of the mutation are not limited to the leaves; Oakleaf plants typically produce a distinctive floral phenotype. Oakleaf flowers are smaller than wild‐type, typically 2 cm in diameter, and calyces are frequently split (Fig. 1c) with occasional yellow petaloid material in the sepals. The severity of the floral phenotype varies as seen in the F1 siblings from an Oakleaf × wild‐type cross (Fig. 1d–f). The most extreme floral phenotype presents five narrow straight‐edged separate petals that look like the spokes of a wheel (Fig. 1d). Some plants produce an intermediate phenotype with attenuated rounded and separated petals (Fig. 1e), and in the least severe form, petals are similar to wild‐type but sometimes with splits in the corolla to give partially separated petals (Fig. 1f). Oakleaf plants are fully fertile as both male and female parents.
Figure 1.

Developmental phenotypes of Primula vulgaris Oakleaf. (a) Seedlings from a wild‐type × Oakleaf cross showing wild‐type and mutant phenotypes, arrows indicate Oakleaf seedlings. (b) Leaf from Oakleaf plant. (c) Primula vulgaris Oakleaf mutant showing leaves and flowers. (d) Example of F1 plant from wild‐type × Oak Leaf cross showing extreme attenuated petals phenotype. (e) Example of F1 plant from wild‐type × Oakleaf cross showing partially attenuated petals. (f) Example of F1 plant from wild‐type × Oakleaf cross showing near normal petals. (g) Leaves emerging from an ectopic meristem (indicated by arrow) on the main vein of an Oakleaf leaf. (h) Flower bud (arrow) emerging from an ectopic meristem on the tip of an Oakleaf leaf. (i) Seed capsule (arrow) arising from ectopic flower shown in (h) following pollination. (j) Flower on Hose in Hose – Oakleaf double mutant plant. (k) Flower on Jack in the Green – Oakleaf double mutant plant. (l) Flower on Hose in Hose – Jack in the Green – Oakleaf triple mutant plant. Bars, 1 cm.
We previously documented wild‐type Primula flower development by cryo‐SEM (Webster & Gilmartin, 2003). To investigate the timing of Oakleaf action and any impact on early flower development, we observed Oakleaf flowers from late stage 3 to late stage 4 (Fig. 2a). In both wild‐type (Webster & Gilmartin, 2003) and Oakleaf flowers, sepals and anthers initiate at late stage 3 (Fig. 2a). Carpel development initiates at early stage 4 (Fig. 2a,d) and is accompanied by petal primordia bulges on the abaxial side of stamen primordia by mid stage 4 (Fig. 2a). Oakleaf does not therefore interfere with organ initiation or timing of development in early flower buds. However, at flower stage 6, the impact of Oakleaf on reduced petal and sepal development is visible. In Oakleaf, stage 6 petals are attenuated and the sepals have not expanded to engulf the developing stamens and carpels (Fig. 2b,c) as seen in wild‐type flowers at this stage (Webster & Gilmartin, 2003). Comparison to wild‐type flowers at mid stage 5 (Fig. 2d) reveals that by this earlier stage in wild‐type, the sepals have already enclosed the flower. Standardisation of developmental stage comparisons between Oakleaf and wild‐type were defined by equivalence of carpel development in Oakleaf and wild‐type flowers; Oakleaf does not affect carpel development. There is no difference in the Oakleaf phenotype between pin and thrum plants.
Figure 2.
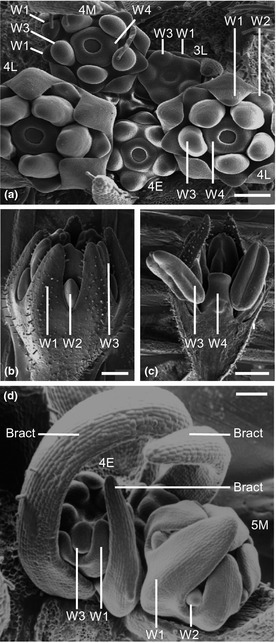
Early development of Primula vulgaris Oakleaf flowers. (a) Scanning electron micrograph (SEM) of P. vulgaris Oakleaf flowers at late stage 3 (3L), early stage 4 (4E), middle stage 4 (4M) and late stage 4 (4L). Sepal (W1), petal (W2), stamen (W3) and carpel (W4) primorida are indicated. (b) Lateral view of stage 6 Oakleaf flower, showing attenuation of sepals (W1) and petals (W2). Developing anthers (W3) are also visible. (c) Bisected stage 6 Oakleaf flower showing normal development of anthers (W3) and carpels (W4). (d) Wild‐type flowers at early stage (4E) and mid stage 5 (5M). Bracts are indicated; these were removed before cryo‐SEM from the Oakleaf samples shown in (a–c). Bars, 200 μm.
Oakleaf plants occasionally produce ectopic meristems on the veins of leaves. These ectopic meristems can be vegetative, giving rise to leaves (Fig. 1g), or floral (Fig. 1h), leading to seed pods (Fig. 1i) but without viable seeds. Some aspects of the Oakleaf phenotype are reminiscent of the effects of ectopic expression of Class I KNOX homeodomain genes in Arabidopsis (Lincoln et al., 1994; Chuck et al., 1996; Hay & Tsiantis, 2010), and their role during normal development of lobed leaves in tomato and Cardamine hirsuta (Hareven et al., 1996; Bharathan et al., 2002; Hay & Tsiantis, 2006; Shani et al., 2009).
In order to explore the influence of Oakleaf on leaf and petal development and to examine whether the effects are organ‐specific or whorl‐specific, we combined Oakleaf with the following mutant phenotypes: Hose in Hose (Webster & Grant, 1990; Li et al., 2010), a dominant mutant phenotype in which sepals are converted to petals; Jack in the Green (Webster & Gilmartin, 2003), a dominant mutant phenotype in which sepals undergo a homeotic transformation to leaves; and Jackanapes (Webster & Gilmartin, 2003), a double mutant carrying both Jack in the Green and Hose in Hose dominant alleles, which produces hybrid petal/leaf structures in the first floral whorl.
Progeny from crosses of Oakleaf and Hose in Hose produce flowers with two whorls of Oakleaf type petals (Fig. 1j); progeny from crosses between Oakleaf and Jack in the Green produce flowers with characteristic Oakleaf petals surrounded by a calyx of miniature Oakleaf leaves (Fig. 1k); progeny from crosses between Oakleaf and Jackanapes produce flowers with a corolla of Oakleaf petals surrounded by a calyx comprising hybrid Oakleaf leaves and yellow petaloid tissue (Fig. 1l). Appearance of the Oakleaf phenotype in combination with other mutant phenotypes in F1 progeny indicates that the Oakleaf allele is dominant to wild‐type and that its effect on petal and leaf development is determined by organ identity and not organ position.
Inheritance of Oakleaf
Preliminary analyses in horticultural crosses suggested that Oakleaf was dominant to wild‐type (R. Brumpton, pers. comm.). Our crosses between Oakleaf and floral mutants reinforce this observation. To fully explore the inheritance of Oakleaf we undertook a series of controlled crosses. The first crosses (Fig. 3) used an Oakleaf thrum as both pollen recipient (Cross 1) and pollen donor (Cross 2) with a wild‐type P. vulgaris pin plant. Seed from the Oakleaf thrum parent (Cross 1) yielded 44 progeny: 23 Oakleaf and 21 wild‐type based on seedling phenotype (Fig. 3a); chi‐squared analysis supports a 1 : 1 ratio (P > 0.70). Four Oakleaf and 14 wild‐type plants were subsequently lost between seedling stage and flowering. The excess of pin Oakleaf and thrum wild‐type progeny indicate linkage of Oakleaf to the S locus with coupling to the recessive s allele; three thrum Oakleaf progeny reveal recombination of Oakleaf from the recessive s allele to the dominant S allele. These small numbers suggest a map distance for Oakleaf to S of 11.5 cM.
Figure 3.
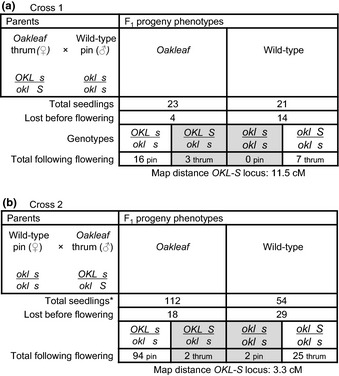
Genetic analysis of reciprocal crosses between Primula vulgaris Oakleaf and wild‐type plants. The results of reciprocal crosses between a P. vulgaris Oakleaf thrum and a wild‐type pin plant are shown. (a) Cross 1, Oakleaf as female parent. (b) Cross 2, Oakleaf as male parent. The phenotypes and genotypes, with respect to leaf shape (wild‐type or Oakleaf), and the S locus (pin or thrum) of parent plants are indicated. The phenotypes, and predicted genotypes, of F1 progeny are shown, along with numbers of progeny classified initially only with respect to leaf shape. The number of each class of progeny lost before flowering is shown, as well as the number of pin‐ and thrum‐type flowers found on Oakleaf and wild‐type plants. Oakleaf (OKL) is shown in coupling to the recessive s allele of the S locus in the original plant based on the assumption that minor progeny classes represent recombinants; genotypes of recombinant chromosomes in progeny and numbers of recombinant progeny are shaded grey. *Does not include 45 seedlings that died before forming secondary leaves.
The reciprocal cross (Cross 2) with Oakleaf as pollen donor, confirmed linkage of Oakleaf to S and coupling to the recessive s allele (Fig. 3b). Of the 258 seeds planted, 45 germinated but died before producing secondary leaves and could not be scored. Of the remaining 112 Oakleaf and 54 wild‐type plants, a further 18 Oakleaf and 29 wild‐type plants died before flowering. These data (Fig. 3b) suggest a significant deviation from the anticipated 1 : 1 ratio (P < 0.001) of Oakleaf to wild‐type. Linkage of Oakleaf to the S locus is supported by the excess of Oakleaf pin and wild‐type thrum plants. Four progeny, two Oakleaf thrums and two wild‐type pins (Fig. 3b), are recombinants; these larger progeny numbers give a map distance between Oakleaf and the S locus of 3.3 cM.
In order to further confirm linkage to the S locus we backcrossed an Oakleaf thrum progeny plant to a wild‐type pin plant. With the Oakleaf thrum as pollen acceptor (Cross 3), 28 progeny were obtained: 12 Oakleaf and 16 wild‐type yielding the anticipated 1 : 1 ratio (P > 0.3) (Fig. 4a). Three Oakleaf plants died before flowering and one Oakleaf plant produced pin flowers, revealing recombination between S and Oakleaf bringing Oakleaf back in coupling with s (Fig. 4a). The reciprocal cross (Cross 4) yielded 20 Oakleaf and 39 wild‐type plants; eight plants were lost before flowering and no recombinants were found in the remainder (Fig. 4b). These data indicate distortion of the anticipated 1 : 1 ratio (P > 0.01) of Oakleaf to wild‐type, with Oakleaf progeny underrepresented. In this cross we did not observe any losses of plants at the seedling stage.
Figure 4.
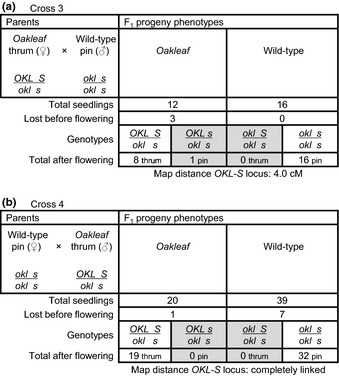
Confirmation of linkage between Primula vulgaris Oakleaf and the S locus. A recombinant P. vulgaris Oakleaf thrum plant was used in a reciprocal back cross with a wild‐type pin plant. (a) Cross 3, Oakleaf as female parent. (b) Cross 4, Oakleaf as male parent. The phenotypes and genotypes, with respect to leaf shape (wild‐type or Oakleaf), and the S locus (pin or thrum) of parent plants are indicated. The phenotypes, and predicted genotypes, of F1 progeny are shown along with numbers of progeny classified initially only with respect to leaf shape. The number of each class of progeny lost before flowering is shown, as well as the number of pin‐ and thrum‐type flowers found on Oakleaf and wild‐type plants. Based on data from Fig. 4, the Oakleaf parent used in this cross carries the OKL locus in coupling to the dominant S allele of the S locus; genotypes of recombinant chromosomes in progeny and numbers of recombinant progeny are shaded grey. The map distance in cM between OKL and the S locus are indicated.
In order to investigate deviation from the anticipated 1 : 1 ratio of Oakleaf to wild‐type plants in progeny from Cross 2 and Cross 4, we undertook further analyses. Reciprocal crosses between an Oakleaf pin and an Oakleaf thrum, with Oakleaf in coupling to S, were established with the thrum as pollen donor (Cross 5) and pollen recipient (Cross 6) (Fig. 5). These crosses were predicted to yield a 3 : 1 ratio of Oakleaf to wild‐type plants which would be characteristic of a cross between two heterozygotes each carrying a dominant allele. From Cross 5 we obtained 15 Oakleaf and five wild‐type plants and from Cross 6 we obtained 21 Oakleaf and five wild‐type plants, both results being consistent with the expected 3 : 1 ratio (P > 0.95 and P > 0.50, respectively) (Fig. 5a,b).
Figure 5.
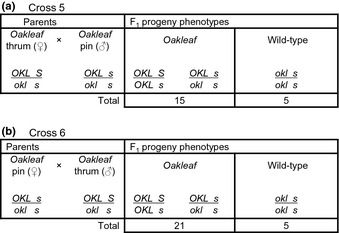
Reciprocal crosses between Primula vulgaris Oakleaf pin and Oakleaf thrum plants to assess viability of Oakleaf homozygotes. The results of reciprocal crosses between a P. vulgaris Oakleaf thrum, with OKL in coupling with the dominant S allele, and an Oakleaf pin, with OKL in coupling to the recessive s allele, are presented. (a) Cross 5, Oakleaf thrum as female parent. (b) Cross 6, Oakleaf pin as female parent. The phenotypes and genotypes, with respect to leaf shape (wild‐type or Oakleaf), and the S locus (pin or thrum) of parent plants are indicated. The number of F1 progeny of each phenotype, classified with respect to leaf phenotype, and their predicted genotypes are shown; progeny were not scored with respect to flower morph.
Characterisation of the PvKNOX gene family
Aspects of the Oakleaf phenotype – namely lobed leaves, ectopic meristems and dominance – are reminiscent of the consequences of ectopic overexpression of Class I KNOX genes in A. thaliana (Lincoln et al., 1994; Chuck et al., 1996; Hay & Tsiantis, 2010). We therefore set out to explore whether Oakleaf results from a constitutive overexpression mutation of a KNOX homeodomain gene. We considered and explored three possibilities: that the phenotype is caused by upregulation of a PvKNOX gene in mature leaves and flowers of Oakleaf plants; that the phenotype arises from a mutation in a PvKNOX gene that does not affect expression but confers a dominant gain of function in protein activity; that the dominant mutation is caused by upregulation of a gene unrelated to the PvKNOX gene family.
The KNOX homeodomain gene family in Maize (Vollbrecht et al., 1991), Arabidopsis (Lincoln et al., 1994; Long et al., 1996; Serikawa et al., 1996; Belles‐Boix et al., 2006; Li et al., 2011) and other species (Bharathan et al., 1999; Hay & Tsiantis, 2010) have been characterised and classified as Class I or Class II based on phylogenetic relationships and expression dynamics (Kerstetter et al., 1994; Bharathan et al., 1999). We used this framework to define the full complement of Class I and Class II PvKNOX genes. Illumina RNA‐Seq analysis of wild‐type P. vulgaris leaf and flower transcriptomes, together with transcriptome analysis of Oakleaf mutant leaves and flowers, was used to generate a transcriptome dataset. We also included RNA‐Seq datasets obtained from pin and thrum mixed stage flower samples to maximise the opportunity for PvKNOX related gene identification; these mixed pin and thrum flower RNA‐Seq samples were not included in subsequent comparative expression analyses. A summary of read number, base coverage and transcript assemblies from these six RNA samples is presented in Table S1.
In parallel, we used Illumina sequencing to generate a draft P. vulgaris genome sequence. The full assembly and annotation of the genome will form the basis of a subsequent publication. We screened this draft genome assembly with A. thaliana KNOX protein sequences using Exonerate c2.2.0 (Slater & Birney, 2005) and identified nine genomic contig assemblies with KNOX gene homology. Within these contigs we defined gene models using the RNA‐Seq dataset with Tophat v2.0.8 (Trapnell et al., 2012) and Cufflinks v2.1.1 (Trapnell et al., 2013). Seven of the genomic contigs were predicted to contain full‐length PvKNOX gene models. Of the two remaining contigs, one contained three exons representing the 5′‐end of a PvKNOX gene, the other contained two exons corresponding to the 3′‐homeodomain region. It was not initially clear whether these models represented two partial loci or one locus split between two contigs due to an incomplete genome assembly. Both partial models were supported by RNA‐Seq data. We therefore screened a de novo Trinity (Grabherr et al., 2011) transcript assembly generated from RNA‐Seq data of the P. vulgaris pin and thrum mixed stage flower bud RNA samples, and identified a single Trinity transcript assembly derived from a single locus (PvKNKL1) bridging the unjoined genomic contigs. This finding resolved that the P. vulgaris genome encodes eight PvKNOX genes; the predicted gene structures are shown in Fig. 6(a). Figure S1 presents the predicted amino acid sequence from each gene; a Clustal Omega sequence alignment of the eight proteins with conserved protein domains indicated is shown in Fig. S2.
Figure 6.
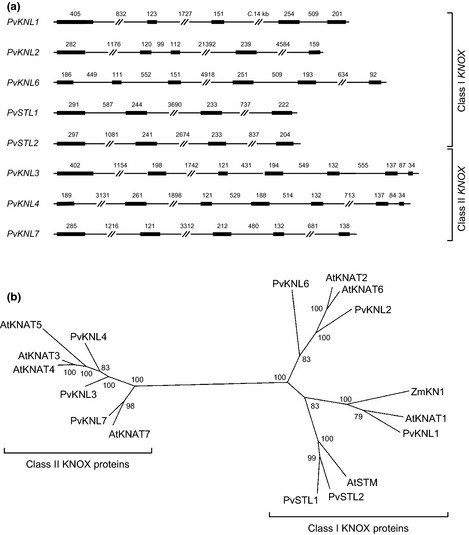
Classification of the PvKNOX gene family. (a) Predicted gene structures of eight Primula vulgaris gene models that encode proteins with amino acid similarity to the Arabidopsis thaliana knotted‐homeodomain (KNOX) gene family members KNAT and STM; Primula genes were named Knotted‐Like (PvKNL) and Shootmeristem‐Like (PvSTL). Gene structures are represented by thick lines (exons) and thin lines (introns); the number of bases in each intron and exon is shown. Genes are shown grouped as Class I and Class II KNOX genes. Accession numbers: PvKNL1, KM586811; PvKNL2, KM586816; PvKNL3, KM586814; PvKNL4, KM586817; PvKNL6, KM586812; PvKNL7, KM586810; PvSTL1, KM586815; PvSTL2, KM586813. (b) Unrooted phylogenetic tree based on amino acid sequence of the eight P. vulgaris PvKNL and PvSTL homeodomain proteins in comparison to A. thaliana KNAT and STM proteins, with Zea Mays KNOTTED1. Class I and Class II KNOX protein clades are identified. Posterior probabilities for clades are shown as percentages.
Figure 6(b) shows a phylogenetic analysis of the eight predicted PvKNOX proteins (Fig. S1) in comparison to the A. thaliana KNOX protein family, comprising seven KNAT proteins (Lincoln et al., 1994; Serikawa et al., 1996; Belles‐Boix et al., 2006; Li et al., 2011) and STM1 (Long et al., 1996), together with KNOTTED‐1 from Zea mays (Vollbrecht et al., 1991). Following this analysis we named the PvKNOX genes and their encoded proteins Knotted‐like (PvKNL) and Shootmeristem‐like (PvSTL) based on encoded protein sequence similarity. Primula vulgaris does not have a homologue of AtKNAT5, but contains two STM‐like genes; it therefore has five Class I and three Class II PvKNOX genes.
Expression analysis and sequence comparison of PvKNOX genes in wild‐type and Oakleaf
Identification of the full complement of Class I and Class II PvKNOX genes enabled us to compare the expression of each gene in leaves and flowers of wild‐type and Oakleaf to determine whether constitutive upregulation of a PvKNOX gene was associated with the Oakleaf phenotype. Based on previous studies of overexpression of Class I KNOX genes in other species (Smith et al., 1992; Lincoln et al., 1994; Chuck et al., 1996; Hareven et al., 1996; Bharathan et al., 2002; Hay & Tsiantis, 2006; Shani et al., 2009) we explored whether the Oakleaf phenotype also resulted from constitutive upregulation of a PvKNOX‐like gene. Our gene expression analyses, described earlier, used HTSeq to create a data file of RNA‐Seq reads aligned to each locus. We then used DESeq to compare RNA‐Seq read counts for each locus in Oakleaf leaves, Oakleaf flowers, wild‐type leaves and wild‐type flowers (Anders & Huber, 2010). Graphical representation of the data is shown in Fig. 7. Normalised read counts for each gene in each tissue, and the log2 fold‐change between Oakleaf and wild‐type leaves, and Oakleaf and wild‐type flowers, are shown in Table S2.
Figure 7.
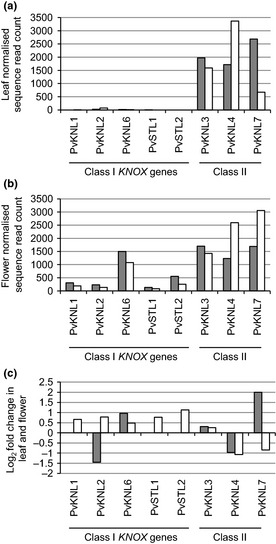
Differential expression of the PvKNOX gene family in Primula vulgaris Oakleaf and wild‐type plants. Expression of the eight genes represented by normalized Illumina RNA‐Seq read count from (a) RNA isolated from P. vulgaris Oakleaf leaves (closed bars) and wild‐type leaves (open bars); (b) RNA isolated from Oakleaf flowers (closed bars) and wild‐type flowers (open bars). (c) the Log2 fold increase or decrease in expression levels between Oakleaf leaf and wild‐type leaves (closed bars) and Oakleaf and wild‐type flowers (open bars). The wild‐type was a pin plant. Class I and Class II PvKNOX genes are indicated.
The five Class I PvKNOX genes are expressed at very low levels in leaves of both wild‐type and Oakleaf plants (Fig. 7a). Only PvKNL2 and PvKNL6 produce measurable read counts from leaves (Table S2). Higher expression levels are observed for the Class II PvKNOX genes in wild‐type and Oakleaf flowers (Fig. 7b). When relative expression levels are compared between Oakleaf and wild‐type, all Class I PvKNOX genes show higher expression levels in Oakleaf flowers than wild‐type; only PvKNL6 shows higher read counts in Oakleaf leaves (Fig. 7c; Table S2), but the normalised read counts of only 15 and 7 reads, respectively, are only just above background. None of the Class I PvKNOX genes are strongly upregulated in Oakleaf leaves (Fig. 7c; Table S2).
In contrast to the Class I PvKNOX genes, the three Class II PvKNOX genes – PvKNL3, PvKNL4 and PvKNL7 – show strong expression in both leaves and flowers of Oakleaf and wild‐type plants (Fig. 7a,b). Only one gene, PvKNL3, is upregulated in both leaves and flowers of Oakleaf (Fig. 7; Table S2). PvKNL3 expression in Oakleaf and wild‐type leaves is represented by normalised read counts of 1971 and 1591 reads, respectively. Normalised read counts for Oakleaf and wild‐type flowers are 1704 and 1424, respectively (Table S2). These values give Log2 fold upregulation in Oakleaf of 0.31 for leaves and 0.26 in flower (Fig. 7c; Table S2).
It is possible that a dominant phenotype could arise through a splicing mutation that results in a protein lacking a critical regulatory domain. We therefore compared RNA‐Seq read abundance profiles across all predicted exons of all PvKNOX loci and saw no difference between Oakleaf and wild‐type that would indicate alternate splicing profiles. We did, however, identify 18 polymorphisms between seven PvKNOX genes in Oakleaf and the wild‐type PvKNOX sequences from the genome assembly that would cause amino acid substitutions (Table S3). The Oakleaf plant used was heterozygous for the Oakleaf locus in a pin genetic background. We therefore then compared the Oakleaf single nucleotide polymorphisms (SNPs) with PvKNOX genes expressed in the flowers and leaves of a wild‐type pin plant to determine whether the SNP was Oakleaf‐specific. Three SNPs in PVKNL2 and PvSTL1 were predicted to result in truncated proteins (Table S3). For the seven remaining SNPs in PvSTL1, PvKNL3, PvKNL4 and PvKNL7, the potential impact of amino acid substitution was analysed using the SIFT prediction tool (Ng & Henikoff, 2003). Five SNPs were predicted to result in tolerated amino acid substitutions which would represent conservative changes (Table S3) and the remaining two, Leu335‐Ser in PvKNL3 and Gly6‐Glu in PvKNL7, are predicted to result in nontolerated amino acid substitutions (Table S3) and could therefore affect protein function (Ng & Henikoff, 2003).
Differential gene expression between Oakleaf and wild‐type plants
KNOX proteins are transcriptional regulators and we would therefore anticipate wider changes in patterns of gene expression of both direct and indirect target genes in response to any aberrant expression of a PvKNOX gene in Oakleaf. It is also possible that Oakleaf is caused by mutation of an unrelated gene that results in a similar phenotype to that predicted from overexpression of a PvKNOX gene. Either possibility would result in transcript profile changes between Oakleaf and wild‐type plants. We therefore used Oakleaf and wild‐type flower and leaf RNA‐Seq data to explore global transcriptome changes between Oakleaf and wild‐type plants.
Assembly of the RNA‐Seq datasets through alignment to the draft P. vulgaris genome identified a total of 39 193 transcript models and created a data file of all RNA‐Seq reads aligned to each of the corresponding loci. HTSeq and DESeq (Anders & Huber, 2010; Anders et al., 2014) were then used to generate normalised counts of RNA‐Seq reads corresponding to each locus for each of the four RNA‐Seq samples from leaves and flowers of wild‐type and Oakleaf plants. Analysis using a log2 fold‐change threshold > 2 identified 1313 genes upregulated in Oakleaf leaves and 2854 genes upregulated in Oakleaf flowers. Of these genes, 507 were common to both tissues. Parallel analyses using the same threshold identified 2099 genes downregulated in Oakleaf leaves and 1285 downregulated in Oakleaf flowers, of which 314 were represented in both tissues. These data are summarised in Fig. 8. None of the P. vulgaris KNOX genes are included in these samples as the fold‐change in expression for these genes is below the two‐fold cut‐off used. Summaries of genes which are upregulated, or downregulated, in both leaves and flowers of Oakleaf, including BlastX analysis of nonredundant protein and Arabidopsis TAIR databases, as well as Gene Ontology assignments, are presented in Table S4 and S5, respectively.
Figure 8.
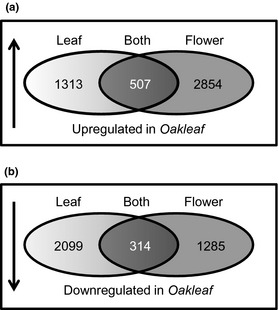
Identification of up‐ and downregulated genes in Primula vulgaris Oakleaf compared with wild‐type. Venn diagrams showing: (a) numbers of genes upregulated in Oakleaf leaves (light grey) and Oakleaf flowers (mid‐grey) compared with pin wild‐type leaves and flowers. The numbers of genes upregulated in both organs (dark grey) are shown. (b) Numbers of genes downregulated in Oakleaf leaves (light grey) and Oakleaf flowers (mid grey) compared with pin wild‐type leaves and flowers. The numbers of genes downregulated in both organs (dark grey) are shown.
Discussion
Records of mutant phenotypes in Primula date back over 400 yr (Gerard, 1597; van de Passe, 1614; Parkinson, 1629) and predominantly affect floral phenotype. More recently identified mutants in P. sinensis include flower and leaf phenotypes (De Winton & Haldane, 1933, 1935), some of which are linked to the S locus. Contemporary studies in P. vulgaris (Webster, 2005) include two phenotypes linked to the S locus, Hose in Hose (Ernst, 1942; Webster & Grant, 1990; Li et al., 2010) and sepaloid (Webster, 2005; Li et al., 2008); others such as double are not linked to the S locus (Webster, 2005). Oakleaf is the third S locus‐linked developmental phenotype in P. vulgaris. Oakleaf was identified as a spontaneous mutation; it is dominant and affects both flower and leaf morphology. A P. sinensis mutation described in 1911, and designated o caused oak‐shaped leaves and affected flower morphology, but was recessive and not linked to the S locus (Gregory, 1911). The shape and character of the lobed leaves in Oakleaf are variable but their presence is characteristic of the mutation. The attenuated petal phenotype is also variable as seen in the F1 siblings from an Oakleaf × wild‐type cross (Fig. 1d–f). This observation may reflect differences in expressivity of the mutant locus in different organs in response to genetic background.
The mutation sometimes increases separation and size of sepals, but does not cause lobed sepals. Crosses of Oakleaf to other floral mutants reveal the organ‐specificity of Oakleaf action (Fig. 1). In combination with Hose in Hose, both first and second whorls of petals show attenuation characteristic of Oakleaf petals (Fig. 1j). In combination with Jack in the Green, the leaves that replace sepals have the lobed appearance of Oakleaf leaves (Fig. 1k). These two examples, and that of Oakleaf combined with Jackanapes (Fig. 1l), reveal that Oakleaf action is organ‐ and not whorl‐specific. Oakleaf does not always affect cotyledons but is consistently presented in the primary leaves. The developmental profile of Oakleaf suggests either organ‐specific expression of the dominant locus, or restricted expression, or action, of downstream network components.
Genetic analyses with Oakleaf as the female parent, where Oakleaf is either in repulsion (Fig. 3a) or coupling (Fig. 4b) to the S locus, pollinated from a wild‐type pin, show that Oakleaf is inherited as a single dominant locus (Figs 1, 2, 3). However, in the reciprocal crosses, with Oakleaf as the male parent (Figs 3b, 4b) we observed progeny numbers that deviated from the anticipated 1 : 1 ratio. In both cases, the missing progeny were consistent with reduced transmission of the dominant thrum S allele. Such distorted segregation ratios were not observed in all crosses (Fig. 5): we are unaware of other examples where the pin : thrum ratio distorts from the anticipated equal transmission of dominant and recessive S alleles (Darwin, 1862; Bateson & Gregory, 1905). It is therefore unlikely that the distorted ratios are due to poor transmission of the dominant S allele.
The data presented in Fig. 3(b) show a significant deviation from the anticipated 1 : 1 ratio (P < 0.001) of Oakleaf to wild‐type progeny. The reason for this is unclear, but in this cross 45 seedlings were lost before secondary leaf development. Intriguingly, chi‐squared analysis of progeny numbers, including the 45 lost seedlings as wild‐type, support a 1 : 1 ratio (P > 0.30). Primula seedlings are susceptible to ‘damping off’ due to bacterial or fungal infection before secondary leaves emerge. Leaves of Oakleaf plants are thicker and firmer than wild‐type. In three of four crosses (Figs 3, 4), progeny losses before flowering were higher for wild‐type than Oakleaf. We speculate that if the Oakleaf mutation gives greater resilience to seedling loss under unfavourable conditions, or in response to pathogen exposure, this could account for the ratio distortion. Indeed, previous studies of asymmetric leaves 1 (as1) mutants in Arabidopsis, Antirrhinum and tobacco showed enhanced resistance to necrotrophic fungi (Nurmberg et al., 2007). AS1 is involved in repression of KNOX gene expression, and as1 mutants have similar phenotypes to KNAT1 overexpression lines (Hay et al., 2002). This hypothesis for seedling resilience in Oakleaf needs to be tested. The reason for underrepresentation of Oakleaf progeny in Cross 4 (Fig. 4b) is unclear, and could reflect a statistical consequence of the small progeny numbers.
Based on data obtained from backcrosses (Figs 3a, 4a), and the reciprocal crosses between heterozygous Oakleaf plants (Fig. 5) which produce the predicted 1 : 1 and 3 : 1 progeny ratios, respectively, we conclude that Oakleaf is caused by a single dominant locus. Linkage of Oakleaf to the S locus is demonstrated by predominant cosegregation of Oakleaf with pin or thrum phenotypes in specific crosses, together with small numbers of recombinants. These crosses suggest a range of potential map distances, but the cross with the largest number of progeny (Fig. 3b) gives a map distance of 3.3 cM. This map distance is possibly an underestimate as the total progeny numbers do not include the 92 plants lost as seedling or before flowering.
By analogy to Hose in Hose, where upregulated expression of a transcription factor is responsible for the phenotype (Li et al., 2010), and based on similarities to the phenotype of Class I KNOX homeodomain gene overexpression in A. thaliana (Lincoln et al., 1994; Chuck et al., 1996; Hay & Tsiantis, 2010), we considered three possibilities as the basis for Oakleaf : dominant upregulation of a PvKNOX homeodomain gene; mutation in a PvKNOX gene that confers a dominant gain of function on the encoded protein, such as a point mutation that introduces an amino acid change, or through a splice site mutation that yields a truncated protein with dominant function; and dominant mutation of a gene unrelated to the PvKNOX homeodomain gene family.
We used a combination of de novo genome assembly and RNA‐Seq to identify the full complement of eight PvKNOX genes (Figs 6b, S1, S2). A fully assembled and annotated P. vulgaris genome will form the basis of a future publication. Phylogenetic analysis (Fig. 6b) shows that P. vulgaris has five Class I and three Class II PvKNOX genes (Kerstetter et al., 1994; Bharathan et al., 1999). Alignment of RNA‐Seq datasets from Oakleaf and wild‐type leaves and flowers enabled us to investigate expression of each gene in Oakleaf and wild‐type leaves and flowers (Fig. 7; Table S2). We also explored whether any of the PvKNOX genes showed constitutive upregulation in mature leaves and flowers of Oakleaf. In line with previous observations on the localised expression of Class I KNOX genes in A. thaliana (Bharathan et al., 1999; Hay & Tsiantis, 2010), we observed low expression of Class I PvKNOX genes in wild‐type Primula leaves (Fig. 7; Table S2); none is strongly upregulated in Oakleaf leaves. Only PvKNL6 has higher sequence read counts in both Oakleaf leaves and flowers (Table S2) but expression in leaves was low with only 15 and 7 reads in Oakleaf and wild‐type, respectively. None of the Class I PvKNOX genes show strong upregulation in both flowers and leaves of Oakleaf.
Analysis of Class II PvKNOX gene expression (Fig. 7; Table S2) shows comparable expression levels in leaf and flower tissue and this is consistent with observations of broader expression profiles for Class II PvKNOX genes compared with Class I genes (Serikawa et al., 1997; Bharathan et al., 1999; Truernit et al., 2006). In A. thaliana, Class II KNOX genes have distinct functions from the Class I genes; KNAT3, KNAT4 and KNAT5 are implicated in root development (Truernit et al., 2006) and KNAT7 in secondary cell wall formation (Li et al., 2011, 2012). Of the three Class II PvKNOX genes, only PvKNL3 is potentially upregulated in both leaves and flowers of Oakleaf; however, because A. thaliana Class II KNOX genes do not have roles in apical meristem identity, we do not consider PvKNL3 as a strong candidate for Oakleaf. None of the PvKNOX genes is strongly upregulated in Oakleaf leaves and we conclude that dominant constitutive overexpression of a PvKNOX gene is not a basis of the Oakleaf phenotype.
In order to establish whether mutation within a PvKNOX gene is responsible for Oakleaf, we analysed RNA‐Seq read profiles against PvKNOX gene models. We speculated that a change in amino acid sequence or expression of a truncated polypeptide might lead to a dominant gain‐of‐function. Analysis of Oakleaf RNA‐Seq read profiles for the eight PvKNOX genes did not reveal differential splicing between Oakleaf and wild‐type that might cause expression of a variant protein. However, several SNPs were identified between Oakleaf and the corresponding wild‐type genome sequence. Those SNPs that were homozygous in Oakleaf, that were also found in RNA‐Seq data from wild‐type pin flowers, or that were predicted to lead to conservative amino acid substitutions, were discounted as the possible basis for Oakleaf (Table S3). Three SNPs in PvKNL2 and PvSTL1, all heterozygous in Oakleaf, would cause truncation of the encoded polypeptide, and two further heterozygous SNPs in PvKNL3 and PvKNL7 cause nonconservative amino acid substitutions. Although these five SNPs might affect KNOX protein function, those in PvKNL2 and PvSTL1 were observed only in Oakleaf flower but not leaf transcripts, and those in PvKNL3 and PvKNL7 were only observed in Oakleaf leaf but not flower transcripts; for PvKNL7 there were no RNA‐Seq reads over this SNP in flower. It seems unlikely given the absence of the SNP in both flower and leaf samples that these are responsible for the dominant Oakleaf phenotype. However, the availability of a P. vulgaris genome sequence, and availability of SNPs for each gene will enable future segregation analyses to determine whether any of the PvKNOX genes are linked to the S locus.
Transcriptome analysis of Oakleaf and wild‐type identified cohorts of genes that are differentially up‐ and downregulated. These studies provide not only candidates for genes controlled by Oakleaf, but also potential candidates for Oakleaf if it proves not to be a PvKNOX gene. The 507 genes which are upregulated and 314 genes downregulated (Log2 fold cut‐off > 2) (Tables S4, S5) represent a broad spectrum of predicted function and we can only speculate which genes are the likely players in the regulatory networks operating downstream of Oakleaf. It has been shown that networks operating downstream of Class I KNOX genes in A. thaliana involve upregulation of GA2 oxidase and downregulation of GA20 oxidase, alongside upregulation of IPT7, which alter gibberellin and cytokinin concentrations, respectively; genes involved in lignin synthesis such as COMT1, CCoAOMT and AtP12 are also downregulated by Class I KNOX genes (Hay & Tsiantis, 2010). Analysis of the differentially expressed genes in Oakleaf (Tables S4, S5) does not reveal the P. vulgaris homologues for these A. thaliana genes. It is possible that Oakleaf is not caused by overexpression of a Class I PvKNOX gene, but is instead a phenocopy caused by a different pathway, as in the case of Wavy auricle in blade 1, a dominant mutant phenotype (Hay & Hake, 2004).
Here we have identified Oakleaf as a new S locus‐linked phenotype that has enabled us to develop a genetic map of the S locus (Li et al., 2015). We have explored three possible explanations for the Oakleaf phenotype based on analysis of the complete PvKNOX gene family and have identified other potential candidates for Oakleaf, as well as candidate Oakleaf‐regulated genes using RNA‐Seq analysis. Future studies, facilitated by a Primula genome assembly and SNP analysis of candidate genes, will reveal potential candidates for Oakleaf on the basis of their linkage to the S locus.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Predicted amino acid sequences of PvKNOX proteins.
Fig. S2 Multiple sequence alignment of PvKNOX proteins.
Table S1 RNA‐Seq read data from six paired‐end read libraries
Table S2 Differential expression of PvKNOX genes
Table S3 Analysis of single nucleotide polymorphisms in PvKNL genes
Table S4 Genes upregulated in Primula vulgaris Oakleaf as compared with wild‐type
Table S5 Genes downregulated in Primula vulgaris Oakleaf as compared with wild‐type
Acknowledgements
We are grateful to Dr Richard Brumpton for the original Oakleaf plants and thank Martin Lappage, Mike Hughes and Pam Wells for horticultural support, and colleagues at The Genome Analysis Centre (Norwich, UK) for genome sequencing. This work was supported by BBSRC grant BB/H019278/2. We thank the University of Leeds, Durham University and Gatsby Foundation for support during early stages of this work. We thank the University of East Anglia for support and the John Innes Centre for hosting P.M.G.'s laboratory under the UEA‐JIC Norwich Research Park collaboration.
References
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2014. HTSeq – a Python framework to work with high‐throughput sequencing data. bioRxiv 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH. 1978. Heterostyly in a tropical weed: the reproductive biology of the Turnera ulmifolia complex (Turneraceae). Canadian Journal of Botany 56: 1713–1725. [Google Scholar]
- Barrett SCH. 2010. Darwin's legacy: the forms, functrion and sexual divesity of flowers. Philosophical Transactions of the Royal Society B 365: 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH, Shore JS. 2008. Self‐incompatibility in flowering plants – evolution, diversity and mechanisms. Berlin, Germany: Springer. [Google Scholar]
- Bateson W, Gregory RP. 1905. On the inheritance of heterostylism in Primula . Proceedings of the Royal Society of London B Series 76: 581–586. [Google Scholar]
- Belles‐Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V. 2006. KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. 2002. Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Bharathan G, Janssen BJ, Kellogg EA, Sinha N. 1999. Phylogenetic relationships and evolution of the KNOTTED class of plant homeodomain proteins. Molecular Biology and Evolution 16: 553–563. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST plus: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. 1996. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis . Plant Cell 8: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia‐Gomez JM, Terol J, Talon M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Crosby JL. 1940. High proportions of homostyle plants in populations of Primula vulgaris . Nature 145: 672–673. [Google Scholar]
- Darwin CR. 1862. On the two forms or dimorphic condition in the species of Primula, and on their remarkable sexual relations. Journal of the Proceedings of the Linnean Society, Botany 6: 77–96. [Google Scholar]
- Darwin CR. 1863. On the existence of two forms, and on their reciprocal sexual relation, in several species of the genus Linum . Journal of the Proceedings of the Linnean Society, Botany 7: 69–83. [Google Scholar]
- Darwin CR. 1877. The different forms of flowers on plants of the same species. London, UK: John Murray. [Google Scholar]
- De Winton D, Haldane JBS. 1933. The genetics of Primula sinensis. II. Segregation and interaction of factors in the diploid. Journal of Genetics 27: 1–44. [Google Scholar]
- De Winton D, Haldane JBS. 1935. The genetics of Primula sinensis. III. Linkage in the diploid. Journal of Genetics 31: 67–100. [Google Scholar]
- van Dijk W. 1943. La decóuverte de l'hétérostylie chez Primula par Ch. de l'Écluse et P. Reneaulme. Nedelandsch Kruidkundig Archief 53: 81–85. [Google Scholar]
- Dowrick VPJ. 1956. Heterostyly and homostyly in Primula obconica . Heredity 10: 219–236. [Google Scholar]
- Dulberger R. 1975. S‐gene action and the significance of characters in the heterostylous syndrome. Heredity 35: 407–415. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A. 1928. Zur Vererbung der morphologischen Heterostylie merkmale. Berichte der Deutschen Botanischen Gesellschaft 46: 573–588. [Google Scholar]
- Ernst A. 1933. Weitere Untersuchungen zur phananalyse zum fertilitatsproblem und zur genetik heterostyler Primeln. I. Primula viscose . Archive der Julius Klaus Stiftung für Vererbungsforschung Sozialanthropologie und Rassenhygiene 8: 1–215. [Google Scholar]
- Ernst A. 1942. Vererbung durch labile gene. Archive der Julius Klaus Stiftung für Vererbungsforschung Sozialanthropologie und Rassenhygiene 17: 1–567. [PubMed] [Google Scholar]
- Ganders FR. 1979. The biology of heterostyly. New Zealand Journal of Botany 17: 607–635. [Google Scholar]
- Garber R, Quisenberry KS. 1927. Self‐fertilization in buckwheat. Journal of Agricultural Research 34: 185–190. [Google Scholar]
- Gerard J. 1597. The herball or generall historie of plantes. London, UK: John Norton. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al 2011. Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RP. 1911. Experiments with Primula sinensis . Journal of Genetics 1: 73–132. [Google Scholar]
- Gregory RP, De Winton D, Bateson MA. 1923. Genetics of Primula sinensis . Journal of Genetics 13: 219–253. [Google Scholar]
- Haldane JBS. 1933. Two new allelomorphs for heterostylism in primula. American Naturalist 67: 559–560. [Google Scholar]
- Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. 1996. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84: 735–744. [DOI] [PubMed] [Google Scholar]
- Hay A, Hake S. 2004. The dominant mutant Wavy auricle in blade1 disrupts patterning in a lateral domain of the maize leaf. Plant Physiology 135: 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. 2002. The gibberellin pathway mediates KNOTTED1‐type homeobox function in plants with different body plans. Current Biology 12: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2006. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta . Nature Genetics 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165. [DOI] [PubMed] [Google Scholar]
- Heslop‐Harrison Y, Heslop‐Harrison J, Shivanna KR. 1981. Heterostyly in Primula. 1. Fine‐structural and cytochemical features of the stigma and style in Primula vulgaris Huds. Protoplasma 107: 171–187. [Google Scholar]
- Kerstetter R, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S. 1994. Sequence‐analysis and expression patterns divide the maize KNOTTED1‐like homeobox genes into 2 classes. Plant Cell 6: 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Iwata H, Yashiro K, Tsumura Y, Ohsawa R, Yasui Y, Ohnishi O. 2006. Development and characterization of microsatellite markers for common buckwheat. Breeding Science 56: 277–285. [Google Scholar]
- Kurian V. 1996. Investigations into the breeding system supergene in Primula. PhD thesis, University of Newcastle upon Tyne, Newcastle upon Tyne, UK. [Google Scholar]
- Labonne JDJ, Shore JS. 2011. Positional cloning of the s haplotype determining the floral and incompatibility phenotype of the long‐styled morph of distylous Turnera subulata . Molecular Genetics and Genomics 285: 101–111. [DOI] [PubMed] [Google Scholar]
- Labonne JDJ, Tamari F, Shore JS. 2010. Characterization of X‐ray‐generated floral mutants carrying deletions at the S locus of distylous Turnera subulata . Heredity 105: 235–243. [DOI] [PubMed] [Google Scholar]
- Labonne JDJ, Vaisman A, Shore JS. 2008. Construction of a first genetic map of distylous Turnera and a fine‐scale map of the S locus region. Genome 51: 471–478. [DOI] [PubMed] [Google Scholar]
- Labonne JJD, Goultiaeva A, Shore JS. 2009. High‐resolution mapping of the S locus in Turnera leads to the discovery of three genes tightly associated with the S alleles. Molecular Genetics and Genomics 281: 673–685. [DOI] [PubMed] [Google Scholar]
- Lewis D. 1943. The physiology of incompatibility in plants II. Linum grandiflorum . Annals of Botany 7: 115–122. [Google Scholar]
- Lewis D. 1949. Incompatibility in flowering plants. Biological Reviews 24: 471–496. [DOI] [PubMed] [Google Scholar]
- Lewis D, Jones DA. 1993. The genetics of heterostyly In: Barrett SCH, ed. Evolution and function of heterostyly. Berlin, Germany: Springer, 129–150. [Google Scholar]
- Li E, Bhargava A, Qiang W, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ. 2012. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus . New Phytologist 194: 102–115. [DOI] [PubMed] [Google Scholar]
- Li E, Wang S, Liu Y, Chen J‐G, Douglas CJ. 2011. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana . Plant Journal 67: 328–341. [DOI] [PubMed] [Google Scholar]
- Li J, Dudas B, Webster MA, Cook HE, Davies BH, Gilmartin PM. 2010. Hose in Hose, an S locus‐linked mutant of Primula vulgaris is caused by an unstable mutation at the Globosa locus. Proceedings of the National Academy of Sciences, USA 107: 5664–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Webster M, Dudas B, Cook H, Manfield I, Davies B, Gilmartin PM. 2008. The S locus‐linked Primula homeotic mutant sepaloid shows characteristics of a B‐function mutant but does not result from mutation in a B‐function gene. Plant Journal 56: 1–12. [DOI] [PubMed] [Google Scholar]
- Li J, Webster MA, Furuya M, Gilmartin PM. 2007. Identification and characterization of pin and thrum alleles of two genes that co‐segregate with the Primula S locus. Plant Journal 51: 18–31. [DOI] [PubMed] [Google Scholar]
- Li J, Webster MA, Wright J, Cocker JM, Smith MC, Badakshi F, Heslop‐Harrison P, Gilmartin PM. 2015. Integration of genetic and physical maps of the Primula vulgaris S locus and localization by chromosome in situ hybridisation. New Phytologist 208: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. 1994. A KNOTTED1‐like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when over‐expressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis . Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y et al 2012. SOAPdenovo2: an empirically improved memory‐efficient short‐read de novo assembler. GigaScience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Pavlov VK, Li JH, Cook HE, Hummel F, Gilmartin PM. 2005. Molecular characterization of DNA sequences from the Primula vulgaris S locus. Journal of Experimental Botany 56: 1177–1188. [DOI] [PubMed] [Google Scholar]
- Matsui K, Nishio T, Tetsuka T. 2004. Genes outside the S supergene suppress S functions in buckwheat (Fagopyrum esculentum). Annals of Botany 94: 805–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin AG, Lee C, Hetrick A. 2006. Identification of genes showing differential expression between morphs in developing flowers of Primula vulgaris . Sexual Plant Reproduction 19: 63–72. [Google Scholar]
- de Nettancourt D. 1997. Incompatibility in angiosperms. Sexual Plant Reproduction 10: 185–199. [Google Scholar]
- Ng PC, Henikoff S. 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research 31: 2812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmberg PL, Knox KA, Yun B‐W, Morris PC, Shafiei R, Hudson A, Loake GJ. 2007. The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proceedings of the National Academy of Sciences, USA 104: 18 795–18 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. 1629. Paradisus in sole paradisus terrestris. London, UK: Humfrey Lownes & Robert Young. [Google Scholar]
- van de Passe C. 1614. Hortus floridus. Utrecht, the Netherlands: University Library Utrecht. [Google Scholar]
- Pellow C. 1928. Report for the year 1928. Merton, UK: The John Innes Horticultural Institution. [Google Scholar]
- Richards AJ. 1997. Plant breeding systems, 2nd edn London, UK: Chapman & Hall. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikawa KA, MartinezLaborda A, Kim HS, Zambryski PC. 1997. Localization of expression of KNAT3, a class 2 Knotted1‐like gene. Plant Journal 11: 853–861. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, MartinezLaborda A, Zambryski P. 1996. Three knotted1‐like homeobox genes in Arabidopsis . Plant Molecular Biology 32: 673–683. [DOI] [PubMed] [Google Scholar]
- Shani E, Burko Y, Ben‐Yaakov L, Berger Y, Amsellem Z, Goldshmidt A, Sharon E, Ori N. 2009. Stage‐specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1‐like homeobox proteins. Plant Cell 21: 3078–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivanna KR, Heslop‐Harrison J, Heslop‐Harrison Y. 1981. Heterostyly in Primula. 2. Sites of pollen inhibition, and effects of pistil constituents on compatible and incompatible pollen tube growth. Protoplasma 107: 319–337. [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Research 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. 1992. A dominant mutation in the maize homeobox gene KNOTTED‐1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pacher L. 2013. Differential analysis of gene regulation at transcript resolution with RNA‐seq. Nature Biotechnology 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nature Protocols 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Siemering KR, Hodge S, Grbic V, Haseloff J. 2006. A map of KNAT gene expression in the Arabidopsis root. Plant Molecular Biology 60: 1–20. [DOI] [PubMed] [Google Scholar]
- Ushijima K, Nakano R, Bando M, Shigezane Y, Ikeda K, Namba Y, Kume S, Kitabata T, Mori H, Kubo Y. 2012. Isolation of the floral morph‐related genes in heterostylous flax (Linum grandiflorum): the genetic polymorphism and the transcriptional and post‐transcriptional regulations of the S locus. Plant Journal 69: 317–331. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. 1991. The developmental gene KNOTTED‐1 is a member of a maize homeobox gene family. Nature 350: 241–243. [DOI] [PubMed] [Google Scholar]
- Webster MA. 2005. Floral morphogenesis in Primula: inheritance of mutant phenotypes, heteromorphy, and linkage analysis. PhD thesis, University of Leeds, Leeds, UK. [Google Scholar]
- Webster MA, Gilmartin PM. 2003. A comparison of early floral ontogeny in wild‐type and floral homeotic mutant phenotypes of Primula . Planta 216: 903–917. [DOI] [PubMed] [Google Scholar]
- Webster MA, Gilmartin PM. 2006. Analysis of late stage flower development in Primula vulgaris reveals novel differences in cell morphology and temporal aspects of floral heteromorphy. New Phytologist 171: 591–603. [DOI] [PubMed] [Google Scholar]
- Webster MA, Grant CJ. 1990. The inheritance of calyx morph variants in Primula vulgaris (Huds). Heredity 64: 121–124. [Google Scholar]
- Woo SH, Adachi T, Jong SK, Campbell CG. 1999. Inheritance of self‐compatibility and flower morphology in an inter‐specific buckwheat hybrid. Canadian Journal of Plant Science 79: 483–490. [Google Scholar]
- Yasui Y, Mori M, Aii J, Abe T, Matsumoto D, Sato S, Hayashi Y, Ohnishi O, Ota T. 2012. S‐LOCUS EARLY FLOWERING 3 is exclusively present in the genomes of short‐styled buckwheat plants that exhibit heteromorphic self‐incompatibility. PLoS ONE 7: e31264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y, Mori M, Matsumoto D, Ohnishi O, Campbell CG, Ota T. 2008. Construction of a BAC library for buckwheat genome research – an application to positional cloning of agriculturally valuable traits. Genes & Genetic Systems 83: 393–401. [DOI] [PubMed] [Google Scholar]
- Yasui Y, Wang YJ, Ohnishi O, Campbell CG. 2004. Amplified fragment length polymorphism linkage analysis of common buckwheat (Fagopyrum esculentum) and its wild self‐pollinated relative Fagopyrum homotropicum . Genome 47: 345–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Predicted amino acid sequences of PvKNOX proteins.
Fig. S2 Multiple sequence alignment of PvKNOX proteins.
Table S1 RNA‐Seq read data from six paired‐end read libraries
Table S2 Differential expression of PvKNOX genes
Table S3 Analysis of single nucleotide polymorphisms in PvKNL genes
Table S4 Genes upregulated in Primula vulgaris Oakleaf as compared with wild‐type
Table S5 Genes downregulated in Primula vulgaris Oakleaf as compared with wild‐type


