Summary
Quorum sensing was once considered a way in which a species was able to sense its cell density and regulate gene expression accordingly. However, it is now becoming apparent that multiple microbes can sense particular quorum‐sensing molecules, enabling them to sense and respond to other microbes in their neighbourhood. Such interactions are significant within the context of polymicrobial disease, in which the competition or cooperation of microbes can alter disease progression. Fungi comprise a small but important component of the human microbiome and are in constant contact with bacteria and viruses. The discovery of quorum‐sensing pathways in fungi has led to the characterization of a number of interkingdom quorum‐sensing interactions. Here, we review the recent developments in quorum sensing in medically important fungi, and the implications these interactions have on the host's innate immune response.
Introduction
Fungi and bacteria often occupy the same niche, whether in the environment, or in plant or animal hosts. The evolution of eukaryotes, both fungi and mammalian hosts, has therefore been heavily influenced by the close proximity of bacteria. Interactions between bacteria and fungi can be chemical, for example, quorum sensing (QS), a cell–cell communication mechanism, or physical, including coaggregation within a biofilm. Polymicrobial interactions are of great importance in a variety of fields. For example, in the food industry, interactions between lactic acid‐producing bacteria and yeasts are important in the production of baked goods (Gobbetti, 1998). In the dairy industry, interactions between yeasts and bacteria are important factors in fermented products and in the ripening of specific cheeses (Viljoen, 2001). In agriculture, polymicrobial interactions play an important role in the complex mycorrhizal network of economically important crops and plants (Deslandes et al., 2003; Bonfante and Anca, 2009; Newton et al., 2010). Finally, polymicrobial interactions have important consequences in veterinary and human medicine (Peleg et al., 2010).
Clinically, polymicrobial infections are harder to treat because of increased resistance to antimicrobial therapy, and as such, polymicrobial diseases can have increased mortality compared with their monomicrobial counterparts (McKenzie, 2006; Harriott and Noverr, 2011). For example, a recent review of polymicrobial bloodstream infections (BSIs) within an intensive care unit found that polymicrobial BSIs had a mortality rate of 47% compared with 19.6% of monomicrobial BSIs (Pammi et al., 2014). Despite this, we currently have limited understanding of the roles of these interactions in disease progression. Therefore, characterizing the complex interactions that occur in these mixed species communities is essential to provide alternative therapies for the treatment of individuals with polymicrobial disease.
Fungal–bacterial interactions vary in dynamics depending on species, strain and environment, but they can be endosymbiotic, synergistic or antagonistic. For example, the plant fungal pathogen Rhizopus microsporus has an endosymbiotic relationship with the Gram‐negative bacterium Burkholderia rhizoxinica and Burkholderia endofungorum, using the bacteria to produce rhizoxin, the cause of rice seedling blight (Partida‐Martinez and Hertweck, 2005; Partida‐Martinez et al., 2007). Interactions during dental plaque formation tend to be synergistic, promoting biofilm formation (Diaz et al., 2012; Nobbs and Jenkinson, 2015). Penicillium species are known to produce quorum‐sensing inhibitors to prevent bacterial communication, reducing their competitors virulence (Rasmussen et al., 2005).
While we acknowledge that the gut is a major site for interkingdom interactions, which are essential in maintaining homeostasis (Hooper and Gordon, 2001; Fujiya et al., 2007) and regulating immunity (Kau et al., 2011), this review will focus on the role of fungal QS in medically important polymicrobial infections and discuss the impact of these microbial signalling molecules on the host's immune system.
Polymicrobial infections involving fungi
Fungal–polymicrobial interactions are important in a variety of disease states and niches including, but not limited to, infections of the respiratory system [i.e. cystic fibrosis (CF) and ventilated‐associated pneumonia], formation of dental plaque, invasive disease, skin and mucosal infections, and bloodstream infections (Fig. 1) (reviewed in Peleg et al., 2010; Frey‐Klett et al., 2011). For example, the CF lung is a major site for polymicrobial infections. Although Pseudomonas aeruginosa is the major colonizer of the CF lung, Burkholderia cepacia complex and Staphylococcus aureus also predominate in these infections, with colonization of B. cepacia indicating chronic infection (Jones et al., 2004). Fungi also colonize the CF lung with Candida albicans, Aspergillus fumigatus and Scedosporium species being the most frequently observed (Bakare et al., 2003; Chmiel et al., 2014). Therefore, like the gut, the CF lung is a major site for interkingdom interactions. However, the occurrence of polymicrobial infections in other niches is often under‐reported, because of difficulties in diagnosing multiple pathogens via traditional culture techniques (McKenzie, 2006; Rolston et al., 2007; Chotirmall et al., 2010). To further compound this issue, fungal disease is also under‐reported, especially when the comparative burden on society is taken into account (Head et al., 2013). Advances in next generation sequencing have enabled efficient diagnosis of polymicrobial disease (Harris et al., 2007; Sibley et al., 2008; Mohammadi et al., 2015). Still, many studies focus on 16S ribosomal sequencing, therefore, missing out any fungal or other eukaryotic species that may be present. A combination of sequencing techniques must be used to better understand the full range of species present in a given disease.
Figure 1.
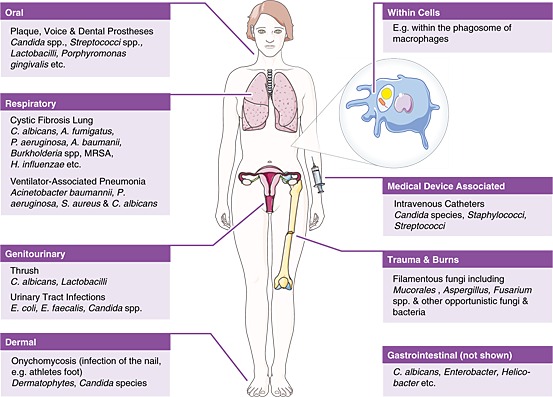
Common niches in which fungal quorum‐sensing interactions occur. Diagrammatic representation of the most common niches where polymicrobial interactions occur. Only key species are highlighted.
Quorum sensing
Microbes were once thought to act selfishly, but it is now accepted that populations can function cooperatively via QS. Through the density‐dependent accumulation of small diffusible molecules, microbes sense the size of the local population, and once the surrounding level of quorum‐sensing molecules (QSMs) reach a threshold concentration, a concerted change in gene expression occurs (Waters and Bassler, 2005). This can lead to a switch in the mode of growth, for example, a morphological switch, or biofilm formation, and the expression of virulence factors (De Sordi and Mühlschlegel, 2009). QS has the potential to be pathogenic by two means: firstly, through controlling the population‐wide expression of virulence factors, or secondly, in some instances, through QSMs themselves being directly toxic to host cells (Albuquerque and Casadevall, 2012). Because of their essential involvement in virulence, QS mechanisms are now being targeted for non‐lethal antimicrobial therapies (Raina et al., 2009).
Quorum sensing was originally thought to be specific to bacteria, but the investigation of the cell density‐dependent morphological switch in C. albicans led to the discovery that farnesol acts as QSM in this eukaryote (Hornby et al., 2001). Since the discovery of farnesol, QSMs have been described in a number of other fungal species (Hogan, 2006; Tseng and Fink, 2008; Albuquerque and Casadevall, 2012), which are involved in regulating growth, stress resistance, morphogenesis and biofilm formation (Table 1). So far, identified fungal QSMs include peptides, for example, quorum‐sensing‐like peptide 1 of Cryptococcus neoformans (Lee et al., 2007), oxylipins in Aspergillus nidulans (Affeldt et al., 2012), and alcohols and alcohol derivatives, such as tyrosol, a phenolic compound that induces filamentation of C. albicans (Chen et al., 2004). Although fungi have not been shown to produce analogues of the bacterial autoinducers (De Sordi and Mühlschlegel, 2009), research into fungal QS is still in its infancy, and it is likely that there are many QS systems still to be discovered.
Table 1.
Known effects of quorum‐sensing molecules on fungi.
| Class | Quorum‐sensing molecule | Known effects | Therapeutic potential | References |
|---|---|---|---|---|
| Alcohol derivatives | Farnesol | Inhibits morphogenesis and growth | Preventing biofilm formation of bacteria with fungi and augmenting antibiotics | (Hornby et al., 2001; Brehm‐Stecher and Johnson, 2003; Derengowski et al., 2009; Liu et al., 2009; Gomes et al., 2011; Brilhante et al., 2012; Cordeiro et al., 2012; Brasch et al., 2013) |
| Induces apoptosis | Anti‐tumorgenesis | (Semighini et al., 2006; Shirtliff et al., 2009; Joo and Jetten, 2010) | ||
| Role in oxidative stress resistance | (Westwater et al., 2005) | |||
| Tyrosol | Induces morphogenesis | (Chen et al., 2004) | ||
| Dodecanol | Inhibits morphogenesis | (Hogan et al., 2004) | ||
| Induces resistance to oxidative stress | (Hall et al., 2011) | |||
| Acyl‐homoserine lactones | 3‐Oxo‐C12 HSL | Inhibits morphogenesis and biofilm formation | (Hogan et al., 2004; Mowat et al., 2010) | |
| Unsaturated fatty acids | Burkholderia DSF | Inhibits morphogenesis | Inhibiting biofilm formation on abiotic surfaces | (Boon et al., 2008; Tian et al., 2013) |
| Stenotrophomonas DSF | Inhibits growth | (Kerr, 1996) | ||
| Peptides | Aggregatibacter actinomycetemcomitans AI‐2 | Inhibits morphogenesis | (Bachtiar et al., 2014) | |
| Streptococcus gordinii AI‐2 | Induces morphogenesis | (Bamford et al., 2009) | ||
| Cryptococcus neoformans QSP1 | Promotes growth and production of virulence factors (e.g. glucuronoxylomannan and melanin) | (Lee et al., 2007; Albuquerque et al., 2013) |
A summary of the current known effects of quorum‐sensing molecules on fungi, including key references.
HSLs, homoserine lactones; DSF, diffusible signal factor; AI, autoinducer; QSP, quorum‐sensing‐like peptide.
Farnesol
One of the major functions of farnesol is to regulate the morphogenic switch of C. albicans through modulation of the cAMP‐dependent PKA signalling pathway (Davis‐Hanna et al., 2008). Biochemical approaches confirmed that farnesol directly targets the active site of the soluble adenylyl cyclase, inhibiting cAMP production (Hall et al., 2011). The significance of farnesol in C. albicans pathogenicity is still under speculation. One plausible explanation is that farnesol enables yeast cell dissemination from biofilms (Ramage and Saville, 2002). However, the effect of farnesol is not limited to C. albicans. In fact, farnesol exerts effects on many other fungal species including perturbing the growth of C. neoformans (Cordeiro et al., 2012) and Penicillium expansum (Liu et al., 2009), inhibiting morphogenesis of Paracoccidioides brasiliensis (Derengowski et al., 2009) and inducing apoptosis in A. nidulans (Semighini et al., 2006). Furthermore, there is evidence to suggest that farnesol can affect cell wall and cytoskeletal integrity in A. fumigatus (Dichtl et al., 2010). Farnesol and other related alcohols produced by C. albicans can also inhibit the growth of dermatophytes (Brasch et al., 2013), which cause superficial skin and nail infections, including ringworm and athlete's foot (Soll, 2002). Importantly, at high concentrations, farnesol induces apoptosis in Candida species (Shirtliff et al., 2009), suggesting that farnesol can not only be used to gain a competitive advantage but also as a measure to restrict growth.
In addition to affecting fungal species, farnesol has been shown to mediate effects in bacterial species. For example, farnesol inhibits the production of the P. aeruginosa quinolone signal (PQS), through inhibition of PqsA (Cugini et al., 2007). However, farnesol can also restore PQS production in the absence of LasR, through reactive oxygen species (ROS)‐dependent activation of RhlR signalling (Cugini et al., 2010), further complicating the interaction between these two species. The activation of alternative signalling networks in the absence of LasR has been proposed to result from altered bacterial respiration (Cugini et al., 2010). Considering that LasR mutants are associated with chronic lung infection in CF patients, the role of these alternative pathways in mediating pathogenicity clearly warrants further investigation.
Because of the wide implications farnesol has on fungal and bacterial growth, it is now being investigated as a potential antimicrobial, including use as an adjuvant alongside antibiotics. For example, farnesol enhances the susceptibility of S. aureus to various antibiotics (Brehm‐Stecher and Johnson, 2003). In addition, farnesol exhibits synergy with nafcillin and vancomycin to inhibit biofilm formation of Staphylococcus epidermidis (Gomes et al., 2011; Gomes et al., 2011; Pammi et al., 2011). Furthermore, farnesol has been shown to augment the efficacy of B‐lactams against Burkholderia pseudomallei (Brilhante et al., 2012), highlighting not only the potential this QSM has in antimicrobial therapy but also the importance of knowing how these interactions impact on therapeutic treatment.
N‐Acyl homoserine lactones
N‐Acyl homoserine lactones (AHLs, also commonly referred to as homoserine lactones, HSLs) are a QSM produced by Gram‐negative bacteria such as P. aeruginosa. There are two main proteins involved in AHL‐based QS, LuxI and LuxR; homologues of which also exist in other species (Waters and Bassler, 2005). The AHLs regulate a number of virulence factors within Gram‐negative bacteria, including the expression of competitive antimicrobials, such as phenazines, and the maturation of biofilms (Williams and Cámara, 2009). In addition, they can also significantly alter signalling in eukaryotic cells, as discussed later.
The P. aeruginosa QSM, 3‐oxo‐C12 homoserine lactone (3‐oxo‐C12 HSL), can inhibit morphogenesis of C. albicans (Hogan et al., 2004) and the conidiation and biofilm formation of A. fumigatus (Mowat et al., 2010). In C. albicans, similar to farnesol, 3‐oxo‐C12 HSL mediates its effects through modulation of the fungal cAMP‐dependent PKA signalling pathway (Davis‐Hanna et al., 2008). This inhibition of cAMP signalling is due to 3‐oxo‐C12 HSL directly targeting the active site of the soluble adenylyl cyclase, thereby reducing cytoplasmic cAMP concentrations (Hall et al., 2011). This interaction is intriguing considering that during direct cell–cell interactions, P. aeruginosa can bind and kill only C. albicans hyphae (Hogan and Kolter, 2002), suggesting that the interaction between C. albicans and P. aeruginosa is more complex than first thought. One possibility is that C. albicans evolved this response to avoid being killed. On the other hand, it is possible that the type of interaction that occurs between these microbes is dependent on additional interactions within the environment.
Intriguingly, Peleg et al. (2008) observed that Acinetobacter baumanii, an emerging multi‐drug‐resistant pathogen found frequently in a nosocomial setting and in the CF lung, inhibited filamentation of C. albicans within a Caenorhabditis elegans infection model. However, deletion of LuxI failed to restore yeast morphogenesis, indicating that AHLs produced by A. baumanii do not modulate hyphal formation in this model. Therefore, it is possible that A. baumanii produces an alternative and currently unidentified QSM with activity against C. albicans morphogenesis, or that other factors within the C. elegans gut interfere with anticipated fungal–bacterial interactions.
In the evolutionary arms race, it appears that some fungi have evolved the ability to inhibit QS. A variety of plant‐mycorrhizal‐associated fungi, from the Ascomycota and Basidiomycota lineages, can directly interfere with bacterial QS through lactonase‐dependent degradation of QSMs (Uroz and Heinonsalo, 2008). Trichosporon loubieri can degrade bacterial AHLs through the production of lactonase (Wong et al., 2013). The discovery of lactonase genes in other medically related fungi suggests that AHL degradation may be another dynamic in fungal–bacterial interactions and may have important consequences in colonization and disease, although to our knowledge this area has not been investigated.
Diffusible signal factor
In addition to the AHLs, some Gram‐negative bacteria communicate using a group of QSMs named diffusible signal factors (DSFs). DSFs are cis‐unsaturated fatty acids (Ryan and Dow, 2011). This novel QS system was first described in plant pathogen Xanthomonas campestris pv. campestris (Xcc) (Barber et al., 1997) but has since been identified in human pathogens.
Cis‐2‐Dodecenoic acid (or Burkholderia diffusible signal factor, BDSF), a QSM produced by the B. cepacia complex, inhibits the filamentation of C. albicans (Boon et al., 2008; Deng et al., 2010). The precise mechanism through which this is achieved has not yet been elucidated. However, Hall et al. (2011) showed that BDSF did not act via the same pathway as farnesol or 3‐oxo‐C12 HSL, but worked via the transcriptional repressor Sfl1. Clinical applications based upon this interaction are already being investigated. A recent study indicated that the addition of BDSF could greatly reduce the binding and subsequent biofilm formation of C. albicans upon abiotic surfaces, including catheters (Tian et al., 2013). Stenotrophomonas maltophilia, another Gram‐negative bacteria associated within the CF lung also produces a DSF (Valenza et al., 2008; Ryan and Dow, 2011; Waters et al., 2011). While the interactions of this QSM with fungi has not yet been specifically characterized, it has been found to inhibit the growth of a number of Candida species (Kerr, 1996).
The QS interactions within the CF lung are complex and dynamic (Fig. 2). As well as the fungal–bacterial interactions already discussed, bacterial QSMs can also interact with each other. For example, the S. maltophilia DSF can alter the biofilm structure and increase the stress tolerance of P. aeruginosa (Ryan et al., 2008). Furthermore, BDSF can reduce the expression of P. aeruginosa QS systems and virulence factors, including the type 3 secretion system (Deng et al., 2013).
Figure 2.
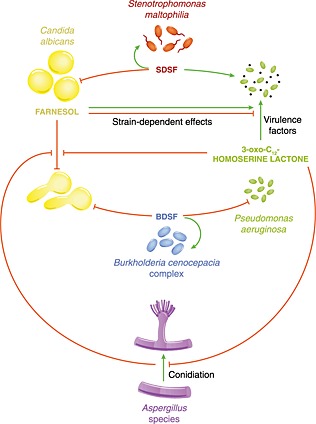
Key interkingdom quorum‐sensing interactions that occur in the cystic fibrosis lung. Diagrammatic representation of quorum‐sensing interactions occurring between fungal and bacterial colonizers of the cystic fibrosis lung. Green lines indicate where a quorum‐sensing molecule exerts a stimulatory effect (i.e. enhanced expression of virulence factors), while red lines indicate inhibition. SDSF, Stenotrophomonas diffusible signal factor; BDSF, Burkholderia diffusible signal factor, cis‐2‐dodecenoic acid.
Autoinducer‐2
Autoinducer‐2 (AI‐2), a family of cyclic oligopeptides, are widely conserved bacterial QSMs with a proposed role in inter‐bacterial communication (Sun et al., 2004; Waters and Bassler, 2005). Currently, the role of AI‐2 in fungal–bacterial interactions is confounding. AI‐2 produced by Aggregatibacter actinomycetemcomitans inhibits morphogenesis of C. albicans, similar to BDSF and 3‐oxo‐C12 HSL (Bachtiar et al., 2014). However, AI‐2 from Streptococcus gordinii promotes morphogenesis of C. albicans through modulating the effects of farnesol (Bamford et al., 2009). These results are intriguing considering that AI‐2 is thought to be structurally conserved among bacteria and is classed as a universal signal (Elias and Banin, 2012). The different responses could be due to study variation (i.e. different media and C. albicans strain), but a more interesting explanation would be that C. albicans could distinguish between AI‐2 molecules produced by different bacterial species and amount different responses. The ability of fungi to discriminate between closely related bacterial QSMs has not been investigated. However, given that C. albicans is a commensal of the gut, where it encounters thousands of bacterial species, the evolution of a system to discriminate between the bacteria would be beneficial to the fungus.
Enterococcus autoinducers
Enterococcus faecalis is a commensal, opportunistic Gram‐positive bacterium that is often found in the same niches as C. albicans, including the oral cavity and gastrointestinal tract (Cruz et al., 2013). Its primary QSM is the gelatinase biosynthesis‐activating cluster peptide, produce via the fsr QS system, which is homologous to the well‐characterized Staphylococcal agr QS system (Nakayama et al., 2006). In a C. elegans model, the fsr system was partially responsible for the inhibition of C. albicans filamentation, with a number of metabolic genes also playing a role (Cruz et al., 2013). Intriguingly, Enterococcus faecium, a closely related species, does not inhibit C. albicans filamentation within the C. elegans gut (Peleg et al., 2008), again suggesting that C. albicans may have the ability to distinguish between bacterial species.
Phenazines
Phenazines are secreted toxins. Although not technically QSMs, phenazines are regulated by QS systems and play important roles in fungal–bacterial interactions. For instance, phenazine‐1‐carboxamide produced by Pseudomonas chlororaphis has antifungal properties against Fusarium oxysporum (Chin‐A‐Woeng et al., 1998). In the clinical setting, the four phenazines produced by P. aeruginosa inhibit the growth of A. fumigatus through the production of ROS (Briard et al., 2015). However, at sub‐inhibitory concentrations, phenazines promote growth of A. fumigatus via enhanced iron uptake (Briard et al., 2015). Phenazine derivatives from P. aeruginosa are also fungicidal to C. albicans at high concentrations, but at lower concentrations inhibit fungal morphogenesis and reduce mitochondrial respiration (Morales et al., 2013). In fact, phenazine‐1‐carboxamide has been shown to be antifungal against a range of human pathogenic fungi including C. neoformans, Candida glabrata and A. nidulans (Tupe et al., 2015).
Impact of quorum sensing on the immune system
One important consideration is that the accumulation of these fungal and bacterial QSMs occurs inside the host, and as a result, these QSMs will also affect host cells. For example, farnesol stimulates the NF‐κB pathway via MEK1/2‐ERK1/2‐MSK1‐dependent phosphorylation of p65, leading to production of cytokines including interleukin (IL)‐6 and IL‐1α (Joo and Jetten, 2008). In the murine macrophage cell line RAW264.7, farnesol acts synergistically with yeast cell wall components (zymosan) to enhance the expression of proinflammatory cytokines (Ghosh et al., 2010). Furthermore, farnesol can alter the maturation of monocytes to dendritic cells (Leonhardt et al., 2015). When compared with control treatments, immature dendritic cells cultured in the presence of farnesol were shown to have altered cell surface markers, including increased CD86 and reduced CD1α, significantly reduced expression of multiple genes involved in cell adhesion and migration, including AMICA1 and MMP2, and reduced migrational behaviour (Leonhardt et al., 2015). These dendritic cells therefore had a reduced capability to recruit and activate T cells, dampening the adaptive immune response. This work highlights the need for a full understanding of the effects of QSMs upon both microbes and their host cells before they could be proposed for therapeutic uses.
Some QSMs can increase stress resistance in fungi, including protecting the fungus from ROS. For example, farnesol has been shown to enhance resistance of C. albicans to ROS (Westwater et al., 2005). This resistance was found to be due to the increased expression of protective catalase Cat1, primarily because of inhibition of the Ras1‐cAMP pathway and cross‐talk with Hog1 regulators (Deveau et al., 2010). ROS production is a common mechanism employed by innate immune cells to kill pathogens once inside the phagosome (Flannagan et al., 2009). Therefore, it is possible that exposure to QSMs during the course of infection promotes survival of the pathogen inside phagocytes. Farnesol is also able to induce apoptosis in mammalian cells via activation of ROS production (Abe et al., 2009). The apoptosis‐inducing effect of farnesol, including the ability to halt cell cycle progression, is being investigated for its anti‐tumorgenesis potential (Joo and Jetten, 2010). Therefore, QSMs may have therapeutic benefits besides controlling microbial growth, and these off‐target effects should be carefully considered before developing QSMs as antimicrobials.
Quorum sensing plays a major role in the immune response during CF (Winstanley and Fothergill, 2009). Similar to farnesol, Pseudomonas produced 3‐oxo‐C12 HSL can stimulate the production of cytokines in eukaryotic cells, including IL‐8 in lung fibroblasts and epithelial cells (DiMango et al., 1995; Smith et al., 2001). IL‐8 is a key cytokine involved in the migration of neutrophils (Huber et al., 1991), exacerbating destructive pulmonary inflammation that is a hallmark of CF (LiPuma, 2010).
Within the respiratory tract, mucus and trapped particles are directed away from the lungs by the continued beating on cilia on the surface of epithelial cells, which is dramatically reduced in CF patients. The beating of these cilia is regulated via cAMP (Schmid et al., 2007). Considering that both fungal and bacterial QSMs, that are found at high concentrations in CF sputum, have been shown to directly target the activity of adenylyl cyclase (Hall et al., 2011), it is tempting to speculate that these QSMs may also influence cilia dynamics, prolonging infection. However, other reports have shown that AHLs can stimulate calcium‐dependent nitric oxide production, increasing mucociliary clearance (Lee et al., 2013). These conflicting reports highlight the complexity of the role of QS in host–pathogen interactions.
Another interesting observation is the fact that fungal and bacterial QSMs induce acrosome loss and decrease sperm motility (Rennemeier et al., 2009). This suggests that the microbiota of the female reproductive tract may play a role in infertility. However, further studies into the role of the microbiome and QS in infertility are required to confirm these observations.
Conclusion
In nature, microbes seldom occur in isolation. Instead, microbes grow in communities that may be diverse in species and genera. This interplay has resulted in the evolution of interkingdom polymicrobial interactions. As we delve deeper into the interactions that occur between clinically relevant microorganisms, it becomes clear that these are complex interactions; the effects of which may be dependent on the environment and combination of species present within the niche. So far, research has generally been limited to studying the interactions between dual species (i.e. C. albicans and P. aeruginosa). However, in reality, each species is continuously interacting with multiple species at any given time, and currently, we have limited understanding of these dynamics. Because of the already identified cross‐talk between QS systems, it is likely that the outcome of these interactions surpasses the sum of the individual interactions. For example, many of the QSMs that have activity against C. albicans mediate their effects through modulation of the cAMP‐PKA pathway. Although other signalling cascades have been indicated in fungal QS (Sato et al., 2004; Kruppa et al., 2004; Kebaara et al., 2008), the Ras‐PKA signalling pathway is emerging as a general quorum‐sensing mechanism in fungi. The fungal soluble adenylyl cyclase is responsive to a number of environmental parameters including carbon dioxide (Klengel et al., 2005) and bacterial peptidoglycan (Xu et al., 2008), which mediate their effects through interactions with different domains of the enzyme. Because of the importance of this signalling pathway in both fungal pathogenesis and interkingdom communication, the fungal soluble adenylyl cyclase has been proposed as a coincidence detector of environmental signals (Hogan and Muhlschlegel, 2011). Further investigation into this area of biology will undoubtedly identify additional QS systems and interactions that play key roles in modulating colonization and disease progression.
It is becoming clear that these microbial communication molecules also exert effects on host cells. The discovery that farnesol enhances proinflammatory responses in macrophages, but prevents activation of cellular immunity, suggests that farnesol is also an immune modulatory signalling molecule. In addition, many pathogens have the ability to replicate within the macrophage phagosome, including B. cenocepacia (Saini et al., 1999), C. glabrata (Seider et al., 2010) and C. neoformans (Feldmesser et al., 2000). Therefore, the phagosome may serve as a micro‐niche enabling QS to occur within mammalian cells. However, our understanding of the role(s) of these QSMs in regulating host–pathogen interactions is still in its infancy. Understanding these interactions, both in terms of their effects on the microbes themselves and on the host, and the role(s) they play in disease is paramount for the development of novel antimicrobial therapies for the treatment of individuals with polymicrobial disease.
Acknowledgements
The authors would like to acknowledge all the contributions in the field that could not be included in the review because of space limitations. Elements of Fig. 1 were adapted from resources provided by Servier Medical Art, under a Creative Commons Attribution 3.0 Unported License (http://creativecommons.org/licenses/by/3.0/). Work in the Hall laboratory is supported through an MRC Career Development Award MR/L00903X/1 to R. A. H., and E. F. D. is supported by an MRC funded PhD studentship.
Dixon, E. F. , and Hall, R. A. (2015) Noisy neighbourhoods: quorum sensing in fungal–polymicrobial infections. Cellular Microbiology, 17: 1431–1441. doi: 10.1111/cmi.12490.
References
- Abe, S. , Tsunashima, R. , Iijima, R. , Yamada, T. , Maruyama, N. , Hisajima, T. , et al (2009) Suppression of anti‐Candida activity of macrophages by a quorum‐sensing molecule, farnesol, through induction of oxidative stress. Microbiol Immunol 53: 323–330. [DOI] [PubMed] [Google Scholar]
- Affeldt, K.J. , Brodhagen, M. , and Keller, N.P. (2012) Aspergillus oxylipin signaling in Cryptococcus neoformans and quorum sensing pathways depend on G protein‐coupled receptors. Toxins (Basel) 4: 695–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque, P. , and Casadevall, A. (2012) Quorum sensing in fungi – a review. Med Mycol 50: 337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque, P. , Nicola, A.M. , Nieves, E. , Costa Paes, H. , Williamson, P.R. , Silva‐Pereira, I. , and Casadevall, A. (2013) Quorum sensing‐mediated, cell density‐dependent regulation of growth and virulence in Cryptococcus neoformans . MBio 5: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtiar, E.W. , Bachtiar, B.M. , Jarosz, L.M. , Amir, L.R. , Sunarto, H. , Ganin, H. , et al (2014) AI‐2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front Cell Infect Microbiol 4: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakare, N. , Rickerts, V. , and Bargon, J. (2003) Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses 46: 19–23. [DOI] [PubMed] [Google Scholar]
- Bamford, C.V. , D'Mello, A. , Nobbs, A.H. , Dutton, L.C. , Vickerman, M.M. , and Jenkinson, H.F. (2009) Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77: 3696–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, C.E. , Tang, J.L. , Feng, J.X. , Pan, M.Q. , Wilson, T.J. , Slater, H. , et al (1997) A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol 24: 555–566. [DOI] [PubMed] [Google Scholar]
- Bonfante, P. , and Anca, I.‐A. (2009) Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol 63: 363–383. [DOI] [PubMed] [Google Scholar]
- Boon, C. , Deng, Y. , Wang, L.‐H. , He, Y. , Xu, J.‐L. , Fan, Y. , et al (2008) A novel DSF‐like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2: 27–36. [DOI] [PubMed] [Google Scholar]
- Brasch, J. , Horter, F. , Fritsch, D. , Beck‐Jendroschek, V. , Tröger, A. , and Francke, W. (2013) Acyclic sesquiterpenes released by Candida albicans inhibit growth of dermatophytes. Med Mycol 52: 1–10. [DOI] [PubMed] [Google Scholar]
- Brehm‐Stecher, B.F. , and Johnson, E.A. (2003) Sensitization of Staphylococcus aureus and Escherichia coli to antibiotics by the sesquiterpenoids nerolidol, farnesol, bisabolol, and apritone. Antimicrob Agents Chemother 47: 3357–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briard, B. , Bomme, P. , Lechner, B.E. , Mislin, G.L. , Lair, V. , Prévost, M.C. , et al (2015) Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci Rep 5: 8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhante, R.S.N. , Valente, L.G.A. , Rocha, M.F.G. , Bandeira, T.J.P.G. , Cordeiro, R.A. , Lima, R.A.C. , et al (2012) Sesquiterpene farnesol contributes to increased susceptibility to β‐lactams in strains of Burkholderia pseudomallei . Antimicrob Agents Chemother 56: 2198–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Fujita, M. , Feng, Q. , Clardy, J. , and Fink, G.R. (2004) Tyrosol is a quorum‐sensing molecule in Candida albicans . Proc Natl Acad Sci U S A 101: 5048–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin‐A‐Woeng, T.F.C. , Bloemberg, G.V. , van der Bij, A.J. , van der Drift, K.M.G.F. , Schripsema, J. , Kroon, B. , et al (1998) Biocontrol by phenazine‐1‐carboxamide‐producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. Radicis‐lycopersici 11: 1069–1077. [Google Scholar]
- Chmiel, J.F. , Aksamit, T.R. , Chotirmall, S.H. , Dasenbrook, E.C. , Elborn, J.S. , LiPuma, J.J. , et al (2014) Antibiotic management of lung infections in cystic fibrosis: part I. The microbiome, MRSA, Gram‐negative bacteria, and multiple infections. Ann Am Thorac Soc 11: 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotirmall, S. , Greene, C.M. , and McElvaney, N.G. (2010) Candida species in cystic fibrosis: a road less travelled. Med Mycol 48: 114–24. [DOI] [PubMed] [Google Scholar]
- Cordeiro, R.D.A. , Nogueira, G.C. , Brilhante, R.S.N. , Teixeira, C.E.C. , Mourão, C.I. , Castelo‐Branco, D.D.S.C.M. , et al (2012) Farnesol inhibits in vitro growth of the Cryptococcus neoformans species complex with no significant changes in virulence‐related exoenzymes. Vet Microbiol 159: 375–380. [DOI] [PubMed] [Google Scholar]
- Cruz, M.R. , Graham, C.E. , Gagliano, B.C. , Lorenz, M.C. , and Garsin, D.A. (2013) Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans . Infect Immun 81: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugini, C. , Calfee, M.W. , Farrow, J.M. , Morales, D.K. , Pesci, E.C. , and Hogan, D.A. (2007) Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa . Mol Microbiol 65: 896–906. [DOI] [PubMed] [Google Scholar]
- Cugini, C. , Morales, D.K. , and Hogan, D.A. (2010) Candida albicans‐produced farnesol stimulates Pseudomonas quinolone signal production in LasR‐defective Pseudomonas aeruginosa strains. Microbiology 156: 3096–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis‐Hanna, A. , Piispanen, A.E. , Stateva, L.I. , and Hogan, D.A. (2008) Farnesol and dodecanol effects on the Candida albicans Ras1‐cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 67: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Boon, C. , Chen, S. , Lim, A. , and Zhang, L.‐H. (2013) Cis‐2‐Dodecenoic acid signal modulates virulence of Pseudomonas aeruginosa through interference with quorum sensing systems and T3SS. BMC Microbiol 13: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Wu, J. , Eberl, L. , and Zhang, L.H. (2010) Structural and functional characterization of diffusible signal factor family quorum‐sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol 76: 4675–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derengowski, L.S. , De‐Souza‐Silva, C. , Braz, S.V. , Mello‐De‐Sousa, T.M. , Báo, S.N. , Kyaw, C.M. , and Silva‐Pereira, I. (2009) Antimicrobial effect of farnesol, a Candida albicans quorum sensing molecule, on Paracoccidioides brasiliensis growth and morphogenesis. Ann Clin Microbiol Antimicrob 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , et al (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A 100: 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau, A. , Piispanen, A.E. , Jackson, A.A. , and Hogan, D.A. (2010) Farnesol induces hydrogen peroxide resistance in Candida albicans yeast by inhibiting the Ras‐cyclic AMP signaling pathway. Eukaryot Cell 9: 569–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, P.I. , Xie, Z. , Sobue, T. , Thompson, A. , Biyikoglu, B. , Ricker, A. , et al (2012) Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun 80: 620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl, K. , Ebel, F. , Dirr, F. , Routier, F.H. , Heesemann, J. , and Wagener, J. (2010) Farnesol misplaces tip‐localized Rho proteins and inhibits cell wall integrity signalling in Aspergillus fumigatus . Mol Microbiol 76: 1191–1204. [DOI] [PubMed] [Google Scholar]
- DiMango, E. , Zar, H.J. , Bryan, R. , and Prince, A. (1995) Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin‐8. J Clin Invest 96: 2204–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias, S. , and Banin, E. (2012) Multi‐species biofilms: living with friendly neighbors. FEMS Microbiol Rev 36: 990–1004. [DOI] [PubMed] [Google Scholar]
- Feldmesser, M. , Kress, Y. , and Novikoff, P. (2000) Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68: 4225–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan, R.S. , Cosío, G. , and Grinstein, S. (2009) Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7: 355–66. [DOI] [PubMed] [Google Scholar]
- Frey‐Klett, P. , Burlinson, P. , Deveau, A. , Barret, M. , Tarkka, M. , and Sarniguet, A. (2011) Bacterial‐fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev 75: 583–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiya, M. , Musch, M.W. , Nakagawa, Y. , Hu, S. , Alverdy, J. , Kohgo, Y. , et al (2007) The Bacillus subtilis quorum‐sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1: 299–308. [DOI] [PubMed] [Google Scholar]
- Ghosh, S. , Howe, N. , Volk, K. , Tati, S. , Nickerson, K.W. , and Petro, T.M. (2010) Candida albicans cell wall components and farnesol stimulate the expression of both inflammatory and regulatory cytokines in the murine RAW264.7 macrophage cell line. FEMS Immunol Med Microbiol 60: 63–73. [DOI] [PubMed] [Google Scholar]
- Gobbetti, M. (1998) The sourdough microflora : interactions of lactic acid bacteria and yeasts. Trends Food Sci Technol 9: 267–274. [Google Scholar]
- Gomes, F. , Leite, B. , Teixeira, P. , Cerca, N. , Azeredo, J. , and Oliveira, R. (2011) Farnesol as antibiotics adjuvant in Staphylococcus epidermidis control in vitro . Am J Med Sci 341: 191–195. [DOI] [PubMed] [Google Scholar]
- Gomes, F. , Teixeira, P. , Cerca, N. , Azeredo, J. , and Oliveira, R. (2011) Effect of farnesol on structure and composition of Staphylococcus epidermidis biofilm matrix. Curr Microbiol 63: 354–359. [DOI] [PubMed] [Google Scholar]
- Hall, R.A. , Turner, K.J. , Chaloupka, J. , Cottier, F. , Sordi, L.D. , Sanglard, D. , et al (2011) The quorum‐sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans . Eukaryot Cell 10: 1034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott, M.M. , and Noverr, M.C. (2011) Importance of Candida‐bacterial polymicrobial biofilms in disease. Trends Microbiol 19: 557–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, J.K. , Groote, M.A.D. , Sagel, S.D. , Zemanick, E.T. , Kapsner, R. , Penvari, C. , et al (2007) Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci U S A 104: 20529–20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head, M.G. , Fitchett, J.R. , Atun, R. , and May, R.C. (2013) Systematic analysis of funding awarded for mycology research to institutions in the UK, 1997–2010. BMJ Open 4: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, D.A. (2006) Talking to themselves: autoregulation and quorum sensing in fungi. Eukaryot Cell 5: 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, D.A. , and Kolter, R. (2002) Pseudomonas–Candida interactions: an ecological role for virulence factors. Science 296: 2229–2232. [DOI] [PubMed] [Google Scholar]
- Hogan, D.A. , and Muhlschlegel, F.A. . (2011) Candida albicans developmental regulation: adenylyl cyclase as a coincidence detector of parallel signals. Curr Opin Microbiol 14: 682–686. [DOI] [PubMed] [Google Scholar]
- Hogan, D.A. , Vik, A. , and Kolter, R. (2004) A Pseudomonas aeruginosa quorum‐sensing molecule influences Candida albicans morphology. Mol Microbiol 54: 1212–23. [DOI] [PubMed] [Google Scholar]
- Hooper, L.V. , and Gordon, J.I. (2001) Commensal host‐bacterial relationships in the gut. Science 292: 1115–1118. [DOI] [PubMed] [Google Scholar]
- Hornby, J. , Jensen, E. , and Lisec, A. (2001) Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 67: 2982–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, A.R. , Kunkel, S.L. , Todd, R.F. , and Weiss, S.J. (1991) Regulation of transendothelial neutrophil migration by endogenous interleukin‐8. Science 254: 99–102. [DOI] [PubMed] [Google Scholar]
- Jones, A.M. , Dodd, M.E. , Govan, J.R.W. , Barcus, V. , Doherty, C.J. , Morris, J. , and Webb, A.K. (2004) Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59: 948–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, J.H. , and Jetten, A.M. (2008) NF‐κB‐dependent transcriptional activation in lung carcinoma cells by farnesol involves p65/RelA(Ser276) phosphorylation via the MEK‐MSK1 signaling pathway. J Biol Chem 283: 16391–16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, J.H. , and Jetten, A.M. (2010) Molecular mechanisms involved in farnesol‐induced apoptosis. Cancer Lett 287: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau, A.L. , Ahern, P.P. , Griffin, N.W. , Goodman, A.L. , and Gordon, J.I. (2011) Human nutrition, the gut microbiome and the immune system. Nature 474: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebaara, B.W. , Langford, M.L. , Navarathna, D.H.M.L.P. , Dumitru, R. , Nickerson, K.W. , and Atkin, A.L. (2008) Candida albicans Tup1 is involved in farnesol‐mediated inhibition of filamentous‐growth induction. Eukaryot Cell 7: 980–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, J.R. (1996) Inhibition of growth of fungi pathogenic to man by Stenotrophomonas maltophilia . J Med Microbiol 45: 380–382. [DOI] [PubMed] [Google Scholar]
- Klengel, T. , Liang, W.‐J. , Chaloupka, J. , Ruoff, C. , Schröppel, K. , Naglik, J.R. , et al (2005) Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 15: 2021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa, M. , Krom, B.P. , Chauhan, N. , Bambach, A. V, Cihlar, R.L. , and Calderone, R.A. (2004) The two‐component signal transduction protein Chk1p regulates quorum sensing in Candida albicans . Eukaryot Cell 3: 1062–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. , Chang, Y.C. , Nardone, G. , and Kwon‐Chung, K.J. (2007) TUP1 disruption in Cryptococcus neoformans uncovers a peptide‐mediated density‐dependent growth phenomenon that mimics quorum sensing. Mol Microbiol 64: 591–601. [DOI] [PubMed] [Google Scholar]
- Lee, R.J. , Chen, B. , Redding, K.M. , Margolskee, R.F. , and Cohen, N.A. (2013) Mouse nasal epithelial innate immune responses to Pseudomonas aeruginosa quorum‐sensing molecules require taste signaling components. Innate Immun 20: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt, I. , Spielberg, S. , Weber, M. , Albrecht‐eckardt, D. , Bläss, M. , Claus, R. , et al (2015) The fungal quorum‐sensing molecule farnesol activates innate immune cells but suppresses cellular adaptive immunity. MBio 6: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiPuma, J.J. (2010) The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23: 299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P. , Deng, B. , Long, C.A. , and Min, X. (2009) Effect of farnesol on morphogenesis in the fungal pathogen Penicillium expansum . Ann Microbiol 59: 33–38. [Google Scholar]
- McKenzie, F.E. (2006) Case mortality in polymicrobial bloodstream infections. J Clin Epidemiol 59: 760–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, R. , Badiee, P. , Badali, H. , Abastabar, M. , Safa, A.H. , Hadipour, M. , et al (2015) Use of restriction fragment length polymorphism to identify Candida species, related to onychomycosis. Adv Biomed Res 4: 95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, D.K. , Grahl, N. , Okegbe, C. , Dietrich, L.E. , Jacobs, N.J. , Hogan, D.A. , et al (2013) Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 4: e00526–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat, E. , Rajendran, R. , Williams, C. , McCulloch, E. , Jones, B. , Lang, S. , and Ramage, G. (2010) Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol Lett 313: 96–102. [DOI] [PubMed] [Google Scholar]
- Nakayama, J. , Chen, S. , Oyama, N. , Nishiguchi, K. , Azab, E.A. , Tanaka, E. , et al (2006) Revised model for Enterococcus faecalis fsr quorum‐sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis‐activating pheromone propeptide corresponding to staphylococcal AgrD. J Bacteriol 188: 8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, A.C. , Fitt, B.D.L. , Atkins, S.D. , Walters, D.R. , and Daniell, T.J. (2010) Pathogenesis, parasitism and mutualism in the trophic space of microbe‐plant interactions. Trends Microbiol 18: 365–373. [DOI] [PubMed] [Google Scholar]
- Nobbs, A.H. , and Jenkinson, H.F. (2015) Interkingdom networking within the oral microbiome. Microbes Infect 17: 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammi, M. , Liang, R. , Hicks, J.M. , Barrish, J. , and Versalovic, J. (2011) Farnesol decreases biofilms of Staphylococcus epidermidis and exhibits synergy with nafcillin and vancomycin. Pediatr Res 70: 578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammi, M. , Zhong, D. , Johnson, Y. , Revell, P. , and Versalovic, J. (2014) Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case‐control study. BMC Infect Dis 14: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida‐Martinez, L.P. , Groth, I. , Schmitt, I. , Richter, W. , Roth, M. , and Hertweck, C. (2007) Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant‐pathogenic fungus Rhizopus microsporous . Int J Syst Evol Microbiol 57: 2583–2590. [DOI] [PubMed] [Google Scholar]
- Partida‐Martinez, L.P. , and Hertweck, C. (2005) Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437: 884–888. [DOI] [PubMed] [Google Scholar]
- Peleg, A.Y. , Hogan, D.A. , and Mylonakis, E. (2010) Medically important bacterial–fungal interactions. Nat Rev Microbiol 8: 340–9. [DOI] [PubMed] [Google Scholar]
- Peleg, A.Y. , Tampakakis, E. , Fuchs, B.B. , Eliopoulos, G.M. , Moellering, R.C. , and Mylonakis, E. (2008) Prokaryote–eukaryote interactions identified by using Caenorhabditis elegans . Proc Natl Acad Sci U S A 105: 14585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina, S. , Vizio, D.D. , Odell, M. , Clements, M. , Vanhulle, S. , and Keshavarz, T. (2009) Microbial quorum sensing: a tool or a target for antimicrobial therapy? Biotechnol Appl Biochem 54: 65–84. [DOI] [PubMed] [Google Scholar]
- Ramage, G. , and Saville, S. (2002) Inhibition of Candida albicans biofilm formation by farnesol, a quorum‐sensing molecule. Appl Environ Microbiol 68: 5459–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, T.B. , Skindersoe, M.E. , Bjarnsholt, T. , Phipps, R.K. , Christensen, K.B. , Jensen, P.O. , et al (2005) Identity and effects of quorum‐sensing inhibitors produced by Penicillium species. Microbiology 151: 1325–1340. [DOI] [PubMed] [Google Scholar]
- Rennemeier, C. , Frambach, T. , Hennicke, F. , Dietl, J. , and Staib, P. (2009) Microbial quorum‐sensing molecules induce acrosome loss and cell death in human spermatozoa. Infect Immun 77: 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolston, K.V.I. , Bodey, G.P. , and Safdar, A. (2007) Polymicrobial infection in patients with cancer: an underappreciated and underreported entity. Clin Infect Dis 45: 228–233. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , and Dow, J.M. (2011) Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol 19: 145–152. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Fouhy, Y. , Garcia, B.F. , Watt, S.A. , Niehaus, K. , Yang, L. , et al (2008) Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa . Mol Microbiol 68: 75–86. [DOI] [PubMed] [Google Scholar]
- Saini, L.S. , Galsworthy, S.B. , John, M.A. , and Valvano, M.A. (1999) Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145: 3465–3475. [DOI] [PubMed] [Google Scholar]
- Sato, T. , Watanabe, T. , Mikami, T. , and Matsumoto, T. (2004) Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen‐activated protein kinase cascade. Biol Pharm Bull 27: 751–2. [DOI] [PubMed] [Google Scholar]
- Schmid, A. , Sutto, Z. , Nlend, M.‐C. , Horvath, G. , Schmid, N. , Buck, J. , et al (2007) Soluble adenylyl cyclase is localized to cilia and contributes to ciliary beat frequency regulation via production of cAMP. J Gen Physiol 130: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seider, K. , Heyken, A. , Lüttich, A. , Miramón, P. , and Hube, B. (2010) Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr Opin Microbiol 13: 392–400. [DOI] [PubMed] [Google Scholar]
- Semighini, C.P. , Hornby, J.M. , Dumitru, R. , Nickerson, K.W. , and Harris, S.D. (2006) Farnesol‐induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol Microbiol 59: 753–764. [DOI] [PubMed] [Google Scholar]
- Shirtliff, M.E. , Krom, B.P. , Meijering, R.A.M. , Peters, B.M. , Zhu, J. , Scheper, M.A. , et al (2009) Farnesol‐induced apoptosis in Candida albicans . Antimicrob Agents Chemother 53: 2392–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley, C.D. , Parkins, M.D. , Rabin, H.R. , Duan, K. , Norgaard, J.C. , and Surette, M.G. (2008) A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci U S A 105: 15070–15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R.S. , Fedyk, E.R. , Springer, T.A. , Mukaida, N. , Iglewski, B.H. , and Phipps, R.P. (2001) IL‐8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N‐3‐oxododecanoyl homoserine lactone is transcriptionally regulated by NF‐kappa B and activator protein‐2. J Immunol 167: 366–374. [DOI] [PubMed] [Google Scholar]
- Soll, D.R. (2002) Mixed Mycotic Infections In Polymicrobial Diseases. ASM Press, Washington, DC, USA: pp. 1–27. [Google Scholar]
- Sordi, L.D. , and Mühlschlegel, F.A. (2009) Quorum sensing and fungal‐bacterial interactions in Candida albicans: a communicative network regulating microbial coexistence and virulence. FEMS Yeast Res 9: 990–999. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Daniel, R. , Wagner‐Döbler, I. , and Zeng, A.‐P. (2004) Is autoinducer‐2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol Biol 4 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, J. , Weng, L.X. , Zhang, Y.Q. , and Wang, L.H. (2013) BDSF inhibits Candida albicans adherence to urinary catheters. Microb Pathog 64: 33–38. [DOI] [PubMed] [Google Scholar]
- Tseng, C.C. , and Fink, G.R. (2008) Quorum Sensing in Fungi In Chemical Communication Among Bacteria. Winans S.C., and Bassler B.L. (eds). Washington, DC: ASM Press. [Google Scholar]
- Tupe, S.G. , Kulkarni, R.R. , Shirazi, F. , Sant, D.G. , Joshi, S.P. , Deshpande, M.V. , et al (2015) Possible mechanism of antifungal phenazine‐1‐carboxamide from Pseudomonas sp. against dimorphic fungi Benjaminiella poitrasii and human pathogen Candida albicans. J Appl Microbiol 118: 39–48. [DOI] [PubMed] [Google Scholar]
- Uroz, S. , and Heinonsalo, J. (2008) Degradation of N‐acyl homoserine lactone quorum sensing signal molecules by forest root‐associated fungi. FEMS Microbiol Ecol 65: 271–278. [DOI] [PubMed] [Google Scholar]
- Valenza, G. , Tappe, D. , Turnwald, D. , Frosch, M. , König, C. , Hebestreit, H. , and Abele‐Horn, M. (2008) Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros 7: 123–127. [DOI] [PubMed] [Google Scholar]
- Viljoen, B.C. (2001) The interaction between yeasts and bacteria in dairy environments. Int J Food Microbiol 69: 37–44. [DOI] [PubMed] [Google Scholar]
- Waters, C.M. , and Bassler, B.L. (2005) Quorum sensing: cell‐to‐cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346. [DOI] [PubMed] [Google Scholar]
- Waters, V. , Yau, Y. , Prasad, S. , Lu, A. , Atenafu, E. , Crandall, I. , et al (2011) Stenotrophomonas maltophilia in cystic fibrosis: serologic response and effect on lung disease. Am J Respir Crit Care Med 183: 635–640. [DOI] [PubMed] [Google Scholar]
- Westwater, C. , Balish, E. , and Schofield, D.A. (2005) Candida albicans‐conditioned medium protects yeast cells from oxidative stress: a possible link between quorum sensing and oxidative stress resistance. Eukaryot Cell 4: 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P. , and Cámara, M. (2009) Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12: 182–91. [DOI] [PubMed] [Google Scholar]
- Winstanley, C. , and Fothergill, J.L. (2009) The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol Lett 290: 1–9. [DOI] [PubMed] [Google Scholar]
- Wong, C.S. , Koh, C.L. , Sam, C.K. , Chen, J.W. , Chong, Y.M. , Yin, W.F. , and Chan, K.G. (2013) Degradation of bacterial quorum sensing signaling molecules by the microscopic yeast Trichosporon loubieri isolated from tropical wetland waters. Sensors (Basel) 13: 12943–12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.‐L.L. , Lee, R.T.H. , Fang, H.‐M.M. , Wang, Y.Y.‐M.M. , Li, R. , Zou, H. , et al (2008) Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 4: 28–39. [DOI] [PubMed] [Google Scholar]


