Abstract
Lower birthweight is consistently associated with a higher risk of type 2 diabetes in observational studies, but the mechanisms underlying this association are not fully understood. Animal models and studies of famine-exposed populations have provided support for the developmental origins hypothesis, under which exposure to poor intrauterine nutrition results in reduced fetal growth and also contributes to the developmental programming of later type 2 diabetes risk. However, testing this hypothesis is difficult in human studies, which are mostly observational and have limited scope for causal inference due to the presence of confounding factors. In this issue of Diabetologia, Wang et al. [6] have used genetic variation associated with birth weight in a Mendelian randomization analysis to assess evidence of a causal link between fetal growth and type 2 diabetes. Mendelian randomization offers the potential to examine associations between exposures and outcomes in the absence of factors that would normally confound observational studies. This Commentary discusses the results of the Mendelian randomization study in relation to the study design and its limitations. Challenges and opportunities for future studies are also outlined.
Keywords: Birthweight, Fetal growth, Genetics, Genome-wide association study, Instrumental variable, Mendelian randomization, Pleiotropy, Type 2 diabetes
Type 2 diabetes is a complex disease with multiple contributory factors. The hypothesis that it might have origins in the developmental period is one possible explanation for an inverse observational association between birthweight and type 2 diabetes [1], which has been widely replicated [2]. Studies of populations exposed to famine support the idea that poor nutrition in utero, with its adverse effects on growth and development, result in permanent adaptations by a fetus, thereby programming the individual to poorer glucose tolerance in later life [3]. Additional support for a role of developmental programming in type 2 diabetes has come from studying monozygotic twin pairs discordant for type 2 diabetes: despite being genetically identical, the affected twins had lower birthweights than their unaffected partners [4]. Proof-of-concept support comes from studies using animal models, which have shown that undernutrition in pregnancy causes defects in glucose tolerance in the offspring [5]. However, the extent to which these findings can be applied to human pregnancy is unclear. We therefore rely heavily on observational studies, which are limited in their ability to control for confounding factors and in their potential for causal inference.
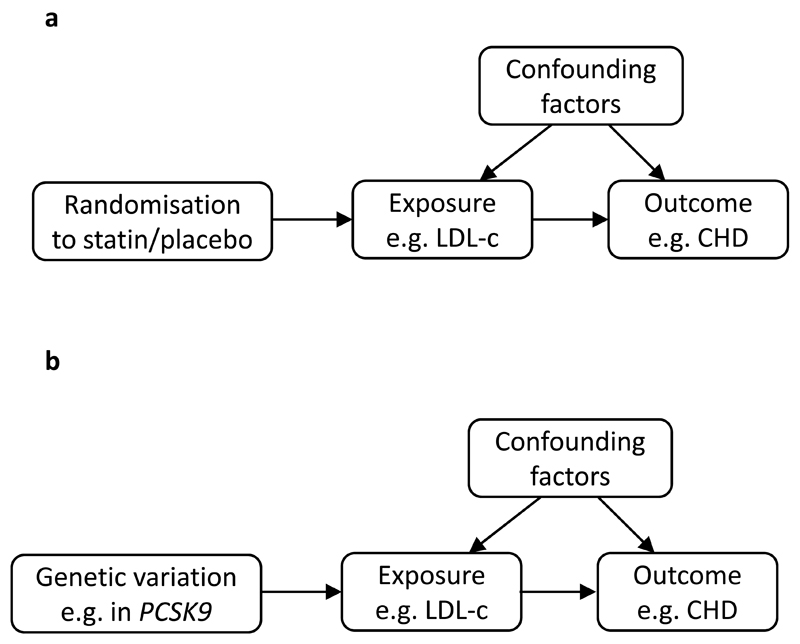
A study published in this issue of Diabetologia by Wang et al [6] has used an alternative approach, known as Mendelian randomisation [7], to assess evidence of a causal link between birthweight and type 2 diabetes. Mendelian randomisation may be likened to a randomised trial in which participants are randomly assigned to groups either exposed or unexposed to a therapy. Random allocation ensures that any potential confounders (measured or unmeasured) are not associated with the therapy. Any difference in the outcome in the exposed group relative to the unexposed group is, therefore, assumed to have been caused by the therapy. In a similar way, a Mendelian randomisation analysis tests for differences in outcome between groups of individuals who differ by genotype (see Fig. 1). Here, the genotype is used as a proxy for measuring the exposure of interest directly and is termed the ‘instrumental variable’ (IV). The IV could be the genotype at a single variant in the DNA sequence, or a number of variants grouped together (a ’genetic risk score’, or GRS) and it is analogous to the intervention in a randomised trial. For example, to test the effect of LDL cholesterol (LDL-c) levels on coronary heart disease, genetic variation in PCSK9, which is known to influence LDL-cholesterol levels, has been used in Mendelian randomisation analysis in a similar capacity to the way in which statin therapy may be used in a randomised trial [8]. Genotypes in a population should be randomised to any potentially confounding factors because the segregation of alleles into gametes at meiosis guards against confounding by environmental factors, while the independent assortment of genes guards against confounding from other traits. It follows that an association between genotype and outcome supports a causal association between the exposure and the outcome.
Fig. 1.
The principle of Mendelian randomisation and its similarity to a randomised trial. The diagrams, (a) and (b), are directed acyclic graphs (DAGs). Each arrow indicates a plausible causal relationship between the variables. The absence of an arrow between any two variables indicates that a causal relationship is not considered plausible. DAG (a) represents a randomised trial in which the effect of LDL cholesterol (LDL-c) levels on CHD risk is tested using randomisation to statin therapy or placebo. DAG (b) represents a Mendelian randomisation analysis in which the effect of LDL-c on CHD risk is tested using the PCSK9 genotype as a proxy for LDL-c levels [8]. In both examples, association between the IV (i.e. the statin in [a] and the genotype in [b]) and the outcome indicates a causal association between LDL-c and CHD if the following assumptions are upheld: (1) the IV is causally related to the exposure (LDL-c); (2) the IV is not associated with factors that confound the exposure outcome association; (3) the IV is only associated with the outcome via its effect on the exposure. In the Mendelian randomisation analysis, the size of the causal association between LDL-c and CHD may be calculated as the ratio of the genotype-CHD association over the genotype-LDL-c association
To assess evidence of a causal relationship between lower birthweight and type 2 diabetes, Wang and colleagues [6] selected five genetic variants known to be robustly associated with birthweight [9] and constructed a GRS in 3267 cases and 12,974 controls of European ancestry, from two prospective studies. They showed that this GRS was associated with a higher risk of type 2 diabetes in the pooled sample (OR 1.06 [95% CI 1.03, 1.09]), but not with potential confounders, such as smoking. In a formal IV analysis, a 1 SD lower birthweight was estimated to be causally associated with a 2.94 (95% CI 1.70, 5.16) times higher risk of type 2 diabetes. To complement this, the authors combined summary statistics from the same five variants in publicly available data from genome-wide association studies (GWAS) of birthweight [9] and type 2 diabetes [10] (26,488 cases and 83,964 controls) and used them to perform a two-sample IV analysis [11]. The effect size estimates were somewhat smaller, but consistent in direction with the authors’ own study, supporting a causal association (OR 1.70 per 1 SD lower birthweight [95% CI 1.32, 2.19]). The authors concluded that their findings provide ‘novel evidence to support the role of intrauterine exposures in the pathogenesis of type 2 diabetes’.
This study is noteworthy in being the first to apply a Mendelian randomisation approach to investigate the developmental origins of type 2 diabetes. Another recent study took a similar approach with publicly available data to evaluate the evidence for a causal effect of birthweight on adult fasting glucose level [12]. With the increasing sizes and availability of GWAS datasets and the continued identification of suitable genetic variants to use as instruments, there is great potential to apply this approach to other outcomes hypothesised to have developmental origins, such as hypertension. However, there are certain aspects of the relationship between birthweight and cardiometabolic disease in adulthood that make this area conceptually different from a more traditional application of Mendelian randomisation, such as the relationship between LDL-c and coronary heart disease (see Fig. 2). It is important to consider these differences, together with the potential limitations of the data reported by Wang et al [6], to understand what inferences we can reasonably make and to inform future study design.
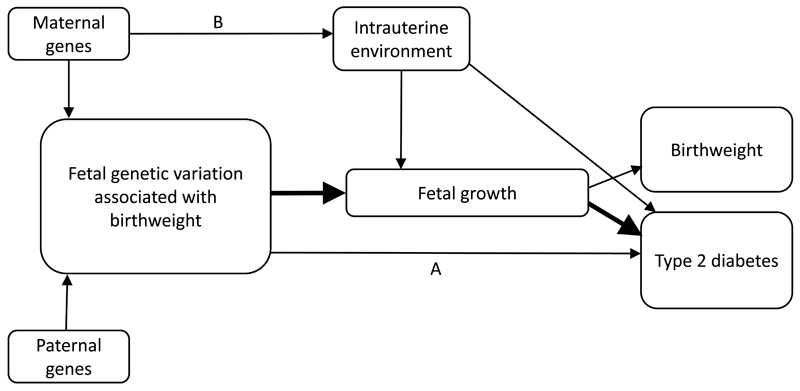
Fig. 2.
Schematic diagram showing relationships between genetic variation, birthweight and type 2 diabetes that are relevant to Mendelian randomisation analysis. The thick, black arrows show the genotype-exposure and exposure-outcome associations, while arrow A illustrates pathways between genotype and outcome that are independent of the exposure and therefore violate a key assumption of IV analysis (e.g. variants at CDKAL1 and ADCY5, which are associated with both type 2 diabetes and birthweight). Arrow B illustrates that maternal genes may influence fetal growth and birthweight indirectly via the intrauterine environment, and could also be associated with the outcome via pathways independent of fetal growth. The maternal genotype is therefore important to account for when interpreting the results of a Mendelian randomisation analysis
As noted by the authors, birthweight is not itself an exposure but an outcome measure reflecting a large number of intrauterine exposures that collectively influence fetal growth. What we are really asking when we use birthweight-associated variants for a Mendelian randomisation analysis is whether reduced intrauterine growth contributes causally to the later life risk of type 2 diabetes (denoted by thick arrows in Fig. 2). Evidence of causality would then suggest that improved fetal growth would reduce the risk of type 2 diabetes. However, birthweight is an end result of the developmental process, during which critical periods of growth and development may make the fetus particularly sensitive to environmental influences at certain times but not at others. Even if we conclude from the analysis that the association is causal, it does not demonstrate that raising birthweight by any possible intervention, pre or during pregnancy, would necessarily bring about reduced rates of diabetes. The question of whether a specific intrauterine exposure influencing fetal development has long term effects requires an instrumental variable for that exposure, not for birthweight. More readily interpretable results are therefore likely to come from analyses using maternal genotypes as instruments for intrauterine exposures.
A key assumption of Mendelian randomisation analysis (see Fig. 1) is that the IV (genotype) is associated with the outcome (type 2 diabetes) solely through its effect on the exposure (here, fetal growth). To uphold this, the authors excluded the known type 2 diabetes risk variants at ADCY5 and CDKAL1 from the GRS, since they do not influence type 2 diabetes risk via a primary effect on fetal growth. Rather, associations at these loci with birthweight and type 2 diabetes are likely to be a result of pleiotropic effects mediated by pancreatic beta cell function (see arrow A in Fig. 2) [9]. The genetic associations at ADCY5 and CDKAL1 indicate that a proportion of the overall covariance between birthweight and type 2 diabetes is genetically mediated [13].
To accept that the Mendelian randomisation analysis carried out by Wang et al [6] demonstrated a causal association, we must assume that the five genetic variants that were selected were not associated with type 2 diabetes through any pathway other than fetal growth. Understanding of these variants is not comprehensive, but four out of five of them do exhibit strong associations with other traits (e.g. blood pressure [ADRB1] [14], height [HMGA2 and LCORL] [15], leptin [CCNL1] [16]). It is therefore quite possible that they could influence type 2 diabetes risk via pathways other than intrauterine growth. If this were the case, the results of the study would not support a causal effect of fetal growth. If the genetic variants with larger birthweight effects had correspondingly larger associations with type 2 diabetes, this would support a causal effect of fetal growth and argue against pleiotropy. However, such a dose-response relationship was not observed in the current study. It is also worth noting that while exclusion of ADCY5 and CDKAL1 was justified in this study, the selection of genetic variants as instruments based on incomplete information could introduce bias and would in principle be less preferable to analyses incorporating all variants and using unbiased selection methods [11]. Recently developed statistical methods such as MR-Egger [17], which enables both detection of, and correction for, pleiotropic effects would be highly recommended for future Mendelian randomisation analyses, particularly when larger numbers of genetic variants are available.
Despite the use of thousands of cases and controls, statistical power was limited in Wang et al’s [6] Mendelian randomisation analysis, even when both groups were combined, because the genetic variants captured only a small amount of variation in the exposure. The authors performed a separate Mendelian randomisation analysis for each variant and then meta-analysed the causal estimates. Low power and weak instrument bias (where the Mendelian randomisation estimate is biased towards the confounded observational association [11]) are key issues and could be countered somewhat by combining single nucleotide polymorphisms (SNPs) into a score before performing the Mendelian randomisation analysis. It is encouraging that the larger two-sample IV analysis using public data gave a consistent result, though for the latter result to be robust, the birthweight and type 2 diabetes GWAS samples should not be overlapping [11].
In the study by Wang et al [6], no data were available on the parental genotypes of participants. This is not surprising as very few large family studies exist with birthweight and type 2 diabetes data in addition to genetic data. However, it is important to recognise that the fetal genetic variation that was studied by the authors is correlated with maternal genetic variation, which may influence fetal growth indirectly through the intrauterine environment (see arrow B in Fig. 2) and, as discussed above, may provide a more appropriate instrument for environmental exposures in a Mendelian randomization analysis. Whether using the maternal or the fetal genotype as the instrumental variable, it is preferable to take account of both of them in the analysis. A genetic variant which influences diabetes risk might be associated with higher birthweight when present in the mother (through raised fetal insulin secretion in response to maternal hyperglycaemia), but with lower birthweight when present in the fetus (due to reduced insulin secreting ability). This phenomenon is observed in mothers and babies with rare heterozygous GCK mutations [18]. In future Mendelian randomisation analyses the effects of both the maternal and fetal genotypes should be accounted for in large numbers so as to enable meaningful interpretation of the results.
Some observational studies suggest a non-linear association of birthweight with diabetes (particularly in populations with high type 2 diabetes prevalence [2]), which might be expected given the causal effect of maternal gestational diabetes or high glucose [19] on birthweight and the genetic and lifestyle risk factors for diabetes that mothers and their offspring will share. The analysis by Wang et al [6] assumed a linear relationship between birthweight and type 2 diabetes. Formal methods for causal analyses exist for non-linear relationships, but require very large sample sizes [20].
Overall, the study by Wang et al [6] demonstrates that genetics may prove to be a useful tool for causal inference in the relationship between birthweight and type 2 diabetes. This is much needed due to the nature of observational epidemiological data. Their results are suggestive of a causal association, but are limited in terms of the strength of the genetic instruments used and the possibility of pleiotropic effects. Future studies will benefit from using larger sample sizes and the identification of further genetic variants which will collectively capture more variation in birthweight. In addition, the use of statistical methods to account for pleiotropy will be vital, as will large studies with maternal genotype data. Finally, since it is unclear precisely which exposures are being tested when birthweight-associated genetic variation is used as an instrumental variable, the interpretation of such analyses will be challenging. The use of maternal genetic instruments for specific intrauterine exposures is likely to shed additional light on this area.
Acknowledgements
I am grateful to the following colleagues for helpful discussions and for their comments on the draft manuscript: T.M. Frayling, R.N. Beaumont, J. Tyrrell, A.T. Hattersley, M.N. Weedon (University of Exeter, Exeter, UK); D.A. Lawlor, N.J. Timpson, G. Davey Smith (University of Bristol, Bristol UK); D.M. Evans (University of Queensland, Brisbane, Australia); M.I. McCarthy, M. Horikoshi (University of Oxford, Oxford UK).
Funding:
RMF is a Sir Henry Dale Fellow (Wellcome Trust and Royal Society grant: 104150/Z/14/Z).
Abbreviations
- GRS
Genetic risk score
- GWAS
Genome-wide association studies
- LDL-c
LDL cholesterol
- IV
Instrumental variable
Footnotes
Duality of interest:
The author declares that there is no duality of interest associated with this manuscript.
Contribution statement:
The author was the sole contributor to this paper.
Published article: The final publication is available at Springer via http://dx.doi.org/10.1007/s00125-016-4057-6
References
- [1].Hales CN, Barker DJ, Clark PM, et al. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 1991;303:1019–1022. doi: 10.1136/bmj.303.6809.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harder T, Rodekamp E, Schellong K, Dudenhausen JW, Plagemann A. Birth weight and subsequent risk of type 2 diabetes: a meta-analysis. Am J Epidemiol. 2007;165:849–857. doi: 10.1093/aje/kwk071. [DOI] [PubMed] [Google Scholar]
- [3].de Rooij SR, Painter RC, Roseboom TJ, et al. Glucose tolerance at age 58 and the decline of glucose tolerance in comparison with age 50 in people prenatally exposed to the Dutch famine. Diabetologia. 2006;49:637–643. doi: 10.1007/s00125-005-0136-9. [DOI] [PubMed] [Google Scholar]
- [4].Poulsen P, Vaag AA, Kyvik KO, Moller Jensen D, Beck-Nielsen H. Low birth weight is associated with NIDDM in discordant monozygotic and dizygotic twin pairs. Diabetologia. 1997;40:439–446. doi: 10.1007/s001250050698. [DOI] [PubMed] [Google Scholar]
- [5].Warner MJ, Ozanne SE. Mechanisms involved in the developmental programming of adulthood disease. Biochem J. 2010;427:333–347. doi: 10.1042/BJ20091861. [DOI] [PubMed] [Google Scholar]
- [6].Wang T, Huang T, Li Y, et al. Low birth weight and risk of type 2 diabetes: a Mendelian randomization study. Diabetologia. 2016 doi: 10.1007/s00125-016-4019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Davey Smith G, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- [8].Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- [9].Horikoshi M, Yaghootkar H, Mook-Kanamori DO, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Replication DIG, Meta-analysis C, Asian Genetic Epidemiology Network Type 2 Diabetes C et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016 doi: 10.3945/ajcn.115.118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Thompson JR, Minelli C, Del Greco MF. Mendelian Randomization using Public Data from Genetic Consortia. Int J Biostat. 2016 doi: 10.1515/ijb-2015-0074. [DOI] [PubMed] [Google Scholar]
- [13].Hattersley AT, Tooke JE. The fetal insulin hypothesis: an alternative explanation of the association of low birthweight with diabetes and vascular disease. Lancet. 1999;353:1789–1792. doi: 10.1016/S0140-6736(98)07546-1. [DOI] [PubMed] [Google Scholar]
- [14].Johnson AD, Newton-Cheh C, Chasman DI, et al. Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension. 2011;57:903–910. doi: 10.1161/HYPERTENSIONAHA.110.158667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kilpelainen TO, Carli JF, Skowronski AA, et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat Commun. 2016;7:10494. doi: 10.1038/ncomms10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hattersley AT, Beards F, Ballantyne E, Appleton M, Harvey R, Ellard S. Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat Genet. 1998;19:268–270. doi: 10.1038/953. [DOI] [PubMed] [Google Scholar]
- [19].Tyrrell J, Richmond RC, Palmer TM, et al. Genetic Evidence for Causal Relationships Between Maternal Obesity-Related Traits and Birth weight. JAMA. 2016;315:1129–1140. doi: 10.1001/jama.2016.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burgess S, Davies NM, Thompson SG, Consortium EP-I Instrumental variable analysis with a nonlinear exposure-outcome relationship. Epidemiology. 2014;25:877–885. doi: 10.1097/EDE.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]