Abstract
Conformational phosphorylation and cleavage events drive the tau protein from a soluble, monomeric state to a relatively insoluble, polymeric state that precipitates the formation of neurofibrillary tangles (NFTs) in projection neurons in Alzheimer’s disease (AD), including the magnocellular perikarya located in the nucleus basal of Meynert (NBM) complex of the basal forebrain. Whether these structural changes in the tau protein are associated with pathogenic changes at the molecular and cellular level remains undetermined during the onset of AD. Here we examined alterations in gene expression within individual NBM neurons immunostained for pS422, an early tau phosphorylation event, or dual labeled for pS422 and TauC3, a later stage tau neoepitope, from tissue obtained postmortem from subjects who died with an antemortem clinical diagnosis of no cognitive impairment (NCI), mild cognitive impairment (MCI), or mild/moderate AD. Specifically, pS422-positive pretangles displayed an upregulation of select gene transcripts subserving protein quality control. On the other hand, late stage TauC3-positive NFTs exhibited upregulation of mRNAs involved in protein degradation but also cell survival. Taken together, these results suggest that molecular pathways regulating protein homeostasis are altered during the evolution of NFT pathology in the NBM. These changes likely contribute to the disruption of protein turnover and neuronal survival of these vulnerable NBM neurons during the progression of AD.
Introduction
Cholinergic neurons located within the region of the substantia innominata termed the nucleus basalis of Meynert (NBM) degenerate early in Alzheimer’s disease (AD) [1–5]. These neurons provide the primary source of acetylcholine to the entire cortical mantle [6,7] and their degeneration correlates with deficits of memory and attention in AD patients [8,9]. Several lines of evidence suggest that cholinergic NBM cytopathology begins prior to the onset of mild cognitive impairment (MCI), a putative prodromal stage of AD. For example, stereologic analysis revealed that NBM neurons display a phenotypic downregulation of the trkA cognate nerve growth factor receptor [4,10] and the p75NTR pan-neurotrophin receptor [5] in the MCI brain [11–13].
NBM neurons are exquisitely prone to neurofibrillary tangle (NFT) cytopathology [14], which correlates with tau pathology within their cortical projection fields in AD [1–5,15]. NFTs are primarily composed of abnormally phosphorylated aggregates of the microtubule-associated protein tau [6,7,16–18]. Biochemical and neuropathological studies indicate that conformational changes in the tau protein drive this molecule from the soluble, microtubule-bound state to the relatively insoluble polymeric form that precipitates NFT formation within NBM neurons in MCI and AD [3,8,9,19–21]. Antibodies that label the early stages of NFT formation such as AT8, Alz-50, and pS422, can be detected in NBM neurons in the MCI brain [3,4,10,22,23]. Interestingly, the appearance of pS422-immunoreactivity in NBM neurons is a stronger correlate of cognitive decline than later stage tau epitopes [5,19], suggesting that the appearance of discrete tau epitopes related to NFT evolution paces the cascade of pathophysiologic events leading to NBM neuronal and cortical axonal disconnection. However, whether progressive stages of NFT formation are associated with differential gene expression and cellular pathogenic alterations within NBM neurons during the progression of AD remains unknown.
In the present study, we quantified gene expression patterns of individual NBM neurons singly immunostained for the pS422 tau epitope, an early phosphorylation event preceding C-terminal truncation of tau at D421, or dual labeled for pS422 and TauC3, a later stage tau neoepitope revealed by tau truncation at D421 [11–13,24]. Individual NBM neurons were microdissected from tissue sections obtained postmortem from subjects who died with an antemortem clinical diagnosis of no cognitive impairment (NCI), MCI, or AD followed by custom-designed microarray analysis. Our data show that the dysregulation of select genes regulating protein modification, quality control, and turnover are differentially associated with the appearance of the pS422 and TauC3 epitopes, suggesting that these molecular pathways may be involved in early pathogenic mechanisms underlying selective neuronal vulnerability during the progression of AD.
Methods
Subjects
Custom-designed microarray analysis of single NBM neurons was performed using tissue obtained postmortem from 28 participates in the Rush Religious Orders Study (RROS) [14,25]. Demographic, clinical, and neuropathological characteristics of the subjects are summarized in Table 1. Details of cognitive evaluations and diagnostic criteria have been extensively published [25–28]. Briefly, a team of investigators performed annual neuropsychological performance testing including the Mini-Mental State Exam (MMSE) and 19 additional neuropsychological tests referable to five cognitive domains: orientation, attention, memory, language, and perception. A Global Cognitive Score (GCS), consisting of a composite z-score calculated from this test battery, was determined for each participant. Participants were clinically diagnosed within a year of death with NCI (n=10), MCI (n=10), or mild/moderate AD (n=8). A board-certified neurologist with expertise in the evaluation of the elderly made the clinical diagnosis based on impairments in each of the five cognitive domains and a clinical examination. The diagnosis of dementia or AD met recommendations by the joint working group of the National Institute of Neurologic and Communicative Disorders and Stroke/AD and Related Disorders Association (NINCDS/ADRDA) [29]. The MCI population was defined as subjects who exhibited cognitive impairment on neuropsychological testing but did not meet the criteria for AD or dementia. These criteria for MCI are consistent with those used by others in the field [30].
Table 1.
Clinical, demographic, and neuropathological characteristics by diagnosis category
| Clinical Diagnosis
|
P-value | Pair-wise comparison | |||
|---|---|---|---|---|---|
| NCI (N=10) | aMCI (N=10) | AD (N=8) | |||
| Age (years) at death: | |||||
| Mean ± SD | 84.6 ± 4.3 | 85.4 ± 3.9 | 84.7 ± 5.0 | 0.6a | -- |
| (Range) | (78–92) | (79–91) | (76–88) | ||
| Number (%) of males: | 5 (50%) | 5 (50%) | 3 (38%) | 0.5b | -- |
| Years of education: | |||||
| Mean ± SD | 18.7 ± 1.6 | 17.5 ± 4.3 | 19.1 ± 3.5 | 0.2a | -- |
| (Range) | (16–21) | (15–25) | (16–24) | ||
| Number (%) with ApoE ε4 allele: | 1 (10%) | 3 (33%) | 4 (50%) | 0.007b | NCI < AD |
| MMSE: | |||||
| Mean ± SD | 27.8 ± 1.6 | 27.4 ± 2.7 | 22.1 ± 5.3 | <0.001a | (NCI, MCI) > AD |
| (Range) | (26–30) | (22–30) | (15–26) | ||
| Global Cognitive Score: | |||||
| Mean ± SD | 0.04 ± 0.3 | −0.06 ± 0.3 | −1.3 ± 0.4 | <0.0001a | NCI > MCI > AD |
| (Range) | (−0.4–0.4) | (−1.2–−0.2) | (−2.0–−0.9) | ||
| Post-mortem interval (hours): | |||||
| Mean ± SD | 5.9 ± 2.5 | 6.0 ± 2.6 | 5.5 ± 4.0 | 0.5a | -- |
| (Range) | (3.3–9.0) | (2.0–10.0) | (2.5–12.0) | ||
| Distribution of Braak scores: | |||||
| 0 | 0 | 0 | 0 | 0.004a | NCI < (MCI, AD) |
| I/II | 4 | 2 | 1 | ||
| III/IV | 6 | 5 | 2 | ||
| V/VI | 0 | 3 | 5 | ||
| NIA Reagan diagnosis (likelihood of AD): | |||||
| No AD | 0 | 0 | 0 | 0.002a | NCI < (MCI, AD) |
| Low | 6 | 3 | 1 | ||
| Intermediate | 3 | 5 | 2 | ||
| High | 1 | 2 | 5 | ||
| CERAD diagnosis: | |||||
| No AD | 3 | 3 | 0 | 0.01a | (NCI, MCI) < AD |
| Possible | 4 | 3 | 1 | ||
| Probable | 3 | 3 | 3 | ||
| Definite | 1 | 1 | 4 | ||
Kruskal-Wallis test, with Bonferroni correction for multiple comparisons.
Fisher’s exact test, with Bonferroni correction for multiple comparisons.
Brains were processed at autopsy as previously published [26–28]. Tissue blocks containing the NBM were immersion-fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2 for 24–72 h at 4 °C, cryoprotected, and cut frozen into 40 μm thick sections. A board-certified neuropathologist blinded to the clinical diagnosis performed the neuropathological evaluation using paraffin-embedded sections prepared for visualization and quantification of amyloid plaques and NFTs. Cases were classified based on the NIA–Reagan, CERAD and Braak staging criteria [31–33].
Dual-label immunohistochemistry for microarray analysis
RNase-free precautions were used throughout the experimental procedures. Acridine orange histofluorescence [34] and bioanalysis (Agilent, Santa Clara, CA) were performed to confirm the presence of high-quality, intact RNA. Tissue sections containing the anteromedial NBM subfields were processed for pS422 and TauC3 dual-labeling immunohistochemistry. Sections were incubated in 0.1M Tris-buffered saline (TBS, pH 7.4) containing 0.1 M sodium periodate to eliminate endogenous peroxidase activity, blocked with 0.1 M TBS containing 0.25% Triton X-100 and 10% normal goat serum (NGS; Vector Labs, Burlingame, CA) for 1 h, and then incubated with a mouse monoclonal antibody raised against the tau D421 cleavage neoepitope, TauC3 (1:5,000) [35] in a 0.1 M TBS/1% NGS solution overnight at 4 °C. Sections were then incubated with biotinylated goat anti-mouse IgG (1:500; Vector Labs), and processed with avidin-biotin complex reagent (ABC; Vector Labs) and Vector SG peroxidase substrate (Vector Labs) to yield a dark blue reaction product labeling TauC3+ neurons. Subsequently, tissue sections were incubated with a rabbit polyclonal antibody directed against the phospho-tau epitope pS422 (1:15,000; Biosource/Invitrogen, Carlsbad, CA) in a 0.1 M TBS/1% NGS solution overnight at 4 °C. Sections were then incubated with biotinylated goat anti-rabbit IgG (1:500; Vector Labs), ABC kit, and visualized with 0.05% diaminobenzidine (DAB, Sigma, St. Louis, MO) containing 0.03% hydrogen peroxide to yield a reddish brown reaction product labeling pS422+ profiles. Tissue sections were slide mounted, Nissl-counterstained to reveal immunonegative NBM neurons and to aid in cytoarchitectonic analysis, and stored at 4 °C without cover-slipping prior to microaspiration.
For confocal microscopic analysis of singly- and dual-labeled neurons, tissue was blocked with 0.1 M TBS containing 0.05% Triton X-100, 10% normal goat serum, and 2% bovine serum albumin for 1 h, and then exposed to both primary antibodies diluted in 0.1 M TBS/0.05% Triton X-100/2% normal goat serum overnight (pS422, 1:2500; TauC3, 1:10,000). Tissue was rinsed with 0.1 M TBS/0.05% Triton X-100 and then incubated with secondary antibodies for 2 h. pS422 was labeled with Alexa Fluor 488 goat anti-rabbit, and TauC3 was developed with Alexa Fluor 594 goat anti-mouse (Invitrogen). Tissue was rinsed with the nuclear counterstain DAPI (1:10,000), mounted, dried overnight, dehydrated in 70% ethanol for 1 min, saturated in 2% Sudan Black/70% ethanol for 3 min, and dedifferentiated in 70% ethanol for 2 min before coverslipping with Vectashield HardSet mounting media (Vector Labs).
Single cell microaspiration, TC RNA amplification, and array hybridization
Individual Nissl-positive but immunonegative, pS422-reactive, dual-labeled pS422/TauC3, and singly TauC3-labeled NBM neurons were accessed using either a micromanipulator and micro-controlled vacuum source (Eppendorf, Westbury, NY) attached to a Nikon TE2000 inverted microscope (Fryer, Huntley, IL) [36–38] or an Arcturus XT laser capture microdissection (LCM) instrument (Applied Biosystems, Grand Island, NY). Approximately 50–60 neurons per phenotype were individually analyzed by the custom-designed microarrays [39–41].
RNA amplification from NBM neurons was performed using terminal continuation (TC) RNA amplification methodology [39,42,43]. Briefly, microaspirated NBM neurons were homogenized in 500 μL Trizol reagent (Invitrogen, Carlsbad, CA). RNAs were reverse transcribed in the presence of the poly d(T) primer (100 ng/μl) and TC primer (100 ng/μl) in 1× first strand buffer (Life Technologies), 2 μg of linear acrylamide (Applied Biosystems), 10 mM dNTPs, 100 μM DTT, 20 U of SuperRNase Inhibitor (Life Technologies), and 200 U of reverse transcriptase (Superscript III, Life Technologies). Single-stranded cDNAs were digested with RNase H and re-annealed with the primers in a thermal cycler: RNase H digestion step at 37 °C, 30 min; denaturation step 95 °C, 3 min; primer re-annealing step 60 °C, 5 min. This step generated cDNAs with double-stranded regions at the primer interface. Samples were then purified by column filtration (Montage PCR filters; Millipore, Billerica, MA). Hybridization probes were synthesized by in vitro transcription using 33P incorporation in 40 mM Tris (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine, 10 mM DTT, 2.5 mM ATP, GTP and CTP, 100 μM of cold UTP, 20 U of SuperRNase Inhibitor, 2 KU of T7 RNA polymerase (Epicentre, Madison, WI), and 120 μCi of 33P-UTP (Perkin-Elmer, Boston, MA) [36,37,39]. The reaction was performed at 37 °C for 4 h. Radiolabeled TC RNA probes were hybridized to custom-designed microarrays without further purification. Arrays were hybridized overnight at 42 °C in a rotisserie oven and washed sequentially in 2X SSC/0.1% SDS, 1X SSC/0.1% SDS, and 0.5X SSC/0.1% SDS for 20 min each at 42 °C. Arrays were placed in a phosphor screen for 24 h and developed on a Storm phosphor imager (GE Healthcare, Piscataway, NJ).
Custom-designed microarray platforms and data collection
Array platforms consisted of 1 μg linearized cDNA purified from plasmid preparations adhered to high-density nitrocellulose (Hybond WL, GE Healthcare). Approximately 576 cDNAs of interest to neurobiology were utilized on the array platform (See Supplementary Table 1). Hybridization signal intensity was determined using Image Quant-software (GE Healthcare) and quantified by subtracting background using an empty vector (pBluescript). Expression of TC amplified RNA bound to each linearized cDNA minus background was expressed as a ratio of the total hybridization signal intensity of the array (i.e., global normalization) [36,43]. The data analysis generated expression profiles of relative changes in mRNA levels among the phenotypically distinct NBM neurons dissected from each case within the clinical diagnostic groups. Each neuron was analyzed in triplicate via three independent probe amplifications and array hybridizations using the original neuronal cDNA pool as template.
Data analysis and statistics
Demographic variables (Table 1) were compared among clinical diagnostic groups by Kruskal-Wallis or Fisher’s Exact tests with Bonferroni correction for pairwise comparisons. Relative changes in total hybridization signal intensity and in individual mRNAs were analyzed by one-way ANOVA with post-hoc Newman-Keuls analysis for multiple comparisons. The level of statistical significance was set at p < 0.05. A false discovery rate controlling procedure was used to reduce type I errors due to the large number of genes analyzed simultaneously [40,42,44,45]. Expression levels of select mRNAs were clustered and displayed using a bioinformatics and graphics software package (GeneLinker Gold, Improved Outcomes, Kingston, ON). For each transcript of interest, we report both the F-test statistic for overall between-group differences as well as individual p values from post hoc comparisons.
Results
Subject Demographics
Demographic, clinical, and neuropathological characteristics of the 28 cases (10 NCI, 10 MCI, and 8 AD) included in the microarray analysis are summarized in Table 1. There were no significant differences in age, gender balance, years of education, or postmortem interval (PMI). The AD group performed significantly poorer on the MMSE compared to the NCI and MCI groups (p < 0.0001), whereas GCS z-scores significantly declined throughout the transition from NCI to MCI to AD (p < 0.0001). Distribution of Braak scores was significantly different across the clinical groups. The NCI cases displayed significantly lower Braak scores than MCI or AD (p = 0.004). NCI cases were classified as Braak stages I/II (40%) or III/IV (60%). The MCI cases met the criteria for Braak stages I/II (20%), III/IV (50%), and V/VI (30%), and the AD cohort was classified as Braak stages I/II (12.5%), III/IV (25%), and V/VI (62.5%). The NIA-Reagan diagnosis also significantly differentiated NCI cases from MCI and AD subjects (p = 0.002). CERAD scores were significantly higher in the AD compared to NCI and MCI groups (p = 0.01).
NBM neuronal tau phenotypes and characteristics
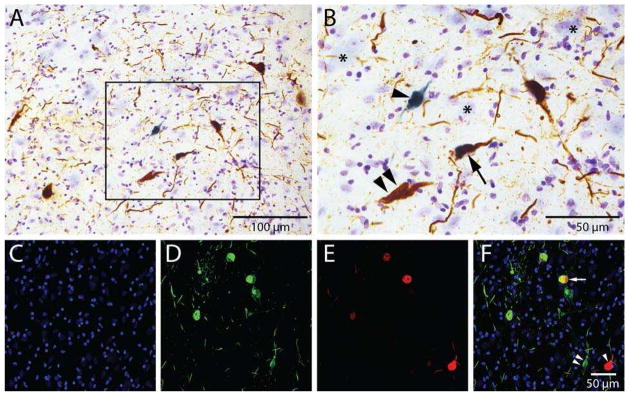
The progression of NBM tau pathology was marked by the site-specific tau antibodies pS422 and TauC3 [19]. Neuronal cytoplasm and processes singly immunolabeled with pS422 visualized with DAB appeared reddish-brown (Fig. 1A, B). TauC3 immunoreactivity produced a dark blue reaction product that discretely filled the cytoplasm (Fig. 1A, B). Confocal analysis revealed three discrete populations of NBM neurons expressing early, intermediate, or late stage NFT pathology (Fig. 1C–F), similar to that reported by our group [19]. Single pS422, TauC3, and dual-labeled NBM neurons were microdissected. Nissl cytochemistry was used to identify tau-negative NBM magnocellular neurons (Fig. 1B).
Figure 1. Identification of markers corresponding to early and late stage tangle pathology in distinct neurons within the NBM.
Tissue from the NBM of an AD case was stained with antibodies directed against tau phosphorylated at S422 (pS422) and C-terminal truncation of tau at D421 (TauC3) to identify early and late stage events in tangle evolution, respectively. Representative photomicrographs are shown in panels A–F. Low power (A) bright-field photomicrographs of NBM neurons labeled with the pS422 (reddish-brown) and TauC3 (blue) tau epitopes. Panel B is a higher magnification image of the box area in panel A showing single pS422+ NBM (double arrowheads) and TauC3+ NBM (arrowheads) neurons, as well as dual pS422+/TauC3+ ( arrow) NBM neurons. Nissl counterstain was used to identify NBM neurons lacking tau pathology (B; asterisks). Confocal immunofluorescence was used to confirm the presence of three discrete populations of NBM neurons displaying DAPI (blue, C), pS422 (green, D), TauC3 (red, E), and pS422/TauC3 (yellow in merged image, F) immunoreactivity. Scale bars: A) 100 μm; B) 50 μm; F) 50 μm and also applies to panels C–E.
Relationship of protein homeostasis gene expression to neuronal tau phenotypes
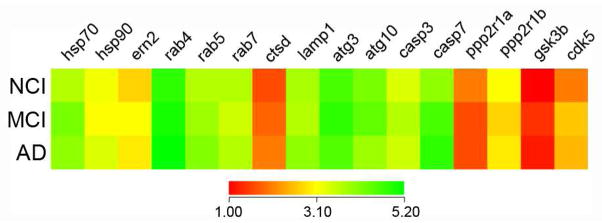
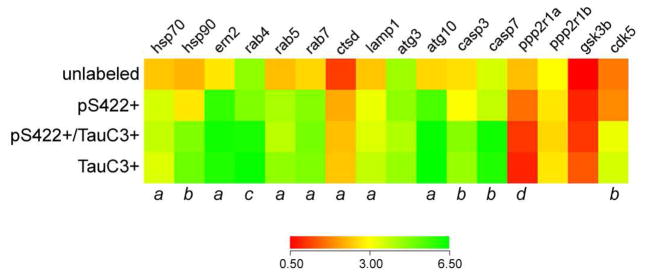
Expression profiling was performed on approximately 250 custom-designed microarrays following TC RNA amplification. Quantitative analyses compared the signal intensities of transcripts either between disease stages or between tau neuronal phenotypes. Comparison of transcript levels in pS422+ NBM neurons aspirated from each clinical group revealed no statistical differences (Fig. 2). However, when analyzed independent of clinical diagnosis, expression levels of key transcripts regulating protein homeostasis (e.g., hsp70, see below) and cell survival (e.g., casp3), were altered by NBM neuron phenotype transitioning from unlabeled to pS422+ to pS422+/TauC3+ (Fig. 3). The expression profile in single-labeled TauC3+ NBM neurons was equivalent to dual-labeled pS422+/TauC3+ NBM neurons, suggesting no additional alterations in transcript expression levels at this late pathological stage related to the genes examined (Fig. 3). These results suggest that it is the sequence of events underlying NFT evolution within NBM neurons, and not clinical disease status, drives changes in gene expression in this selectively vulnerable neuronal population.
Figure 2. Gene expression in pS422-immunopositive NBM neurons is equivalent throughout the progression of AD.
Color-coded heatmaps of relative expression profiles of select transcripts in pS422+ NBM neurons aspirated from NCI, MCI, and AD cases (red to green = increasing mRNA levels). Quantitative analysis revealed no statistical differences in the expression levels of the transcripts examined in pS422+ NBM neurons derived from MCI or AD subjects as compared to NCI cases. This observation suggests that tangle evolution, and not disease status, may drive changes in gene expression. Therefore, in the present analysis we compared mRNA levels in individual pS422+ and pS422+/TauC3+ NBM neurons independent of clinical diagnosis. Abbreviations: Molecular chaperone heat shock proteins 70, 90 (hsp70, -90) and ER-to-nucleus protein 2 (ern2), endosomal GTPases rab4-7, lysosomal markers cathepsin D (ctsd) and lysosome-associated membrane protein 1 (lamp1), autophagy-related E2-like enzymes atg3 and atg10, caspases 3, 7 (casp3, -7), protein phosphatase 2 regulatory subunits 1α, 1β (ppp2r1α, -1β), glycogen synthase kinase β (gsk3β), and cyclin-dependent kinase 5 (cdk5).
Figure 3. Select protein homeostasis genes are dysregulated in pS422+ NBM neurons as compared to unlabeled, tau pathology-negative NBM neurons.
Heatmap of expression levels of transcripts mediating protein homeostasis in NBM neurons (red to green = increasing mRNA levels). Quantitative analysis revealed that 7 of 16 genes involved in protein homeostasis were significant upregulated in pS422+ neurons as compared to unlabeled control neurons. One gene, ppp2r1a protein phosphatase 2 regulatory subunit was significantly downregulated. These alterations could indicate early mechanistic changes preceding NBM neurofibrillary degeneration. The appearance of the tau neoepitope TauC3 revealed upregulation of 5 additional transcripts. a, (pS422+, pS422+/tauC3+, tauC3+) > unlabeled, p < 0.01; b, (pS422+/tauC3+, tauC3+) > (unlabeled, pS422+), p < 0.01; c, (pS422+/tauC3+, tauC3+) > (unlabeled, pS422+), p < 0.05; d, unlabeled > pS422+ > (pS422+/tauC3+, tauC3+), p < 0.01. Abbreviations are as defined in Fig. 2.
Tau pathology and protein quality control-related mRNAs in NBM neurons during disease progression
Combining single neuron expression profiling with tau site-specific antibodies allowed for the characterization of the temporal molecular events associated with NFT formation within NBM neurons during the progression of AD. Quantitative analysis revealed differential expression within three classes of gene transcripts: protein quality control, protein trafficking and turnover, and protein degradation related to apoptosis or cell death (Fig. 3, 4). Of particular interest was the sequential upregulation of specific molecular chaperone mRNAs. Expression levels of heat shock protein 70 (hsp70) was significantly different among the cell phenotypes (F3,225 = 19.15, p < 0.0001), with pretangle pS422- containing NBM neurons displayed increased expression of hsp70 (1.5-fold; unlabeled < pS422+; p < 0.01; Fig. 3), whereas those positive for the late stage tau neoepitope, TauC3, continued to express high levels of hsp70, similar to pS422+ neurons. By contrast, heat shock protein 90 (hsp90) was also differentially regulated among the cell phenotypes (F3,224 = 8.73, p < 0.0001), but increased only with the appearance of the TauC3 epitope (1.8-fold; pS422+ < pS422+/TauC3+; p < 0.01; Fig. 3).
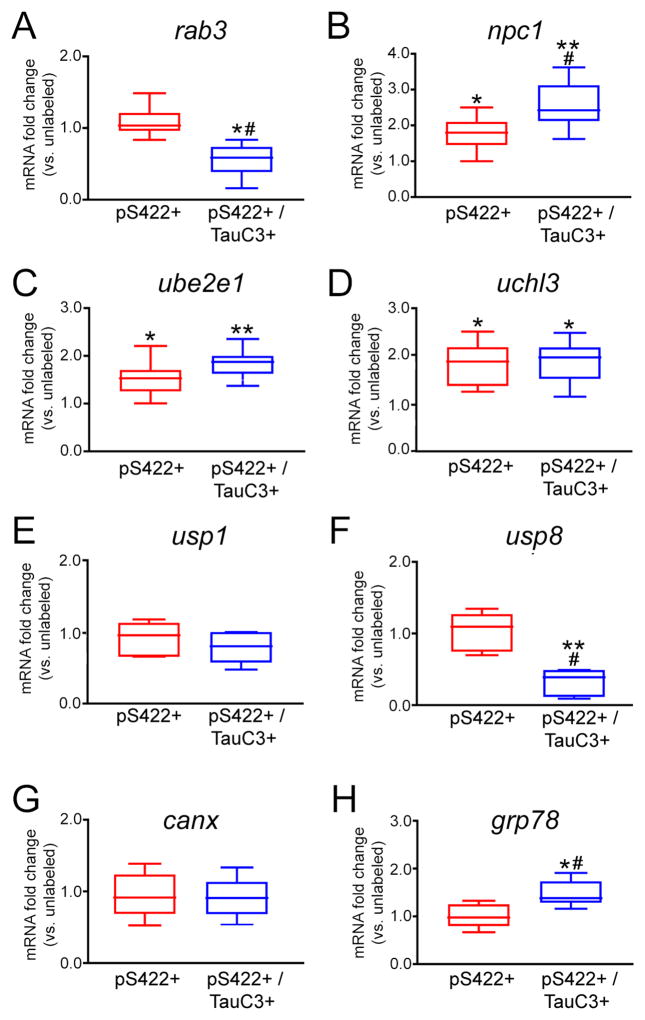
Figure 4. mRNAs regulating protein trafficking and metabolism are altered in NBM neurons containing pS422+/− TauC3 immunoreactivity.
Box plots demonstrate differential expression profiles of select transcripts encoding regulators of protein trafficking and metabolism is pS422+ and pS422+/TauC3+ NBM neurons relative to expression levels in unlabeled NBM neurons. *p < 0.05 vs. unlabeled, **p < 0.01 vs. unlabeled, # p < 0.05 vs. pS422. Abbreviations: Ras-associated protein-3 (rab3), Niemann-Pick disease, type C1 (npc1), ubiquitin-conjugating enzyme E2E 1 (ube2e1), ubiquitin carboxylterminal esterase L3 (uchl3), ubiquitin specific protease 1 (usp1), ubiquitin-specific protease 8 (usp8), calnexin (canx), glucose-regulated protein, kDa78 (grp78).
Effect of tau tangle evolution on endosomal-lysosomal protein trafficking-related mRNAs in NBM neurons
Prefibrillar pS422+ tau pathology was marked by a significant upregulation of four mRNAs encoding proteins involved in the endosomal-lysosomal pathway (Fig. 3), including the GTPases rab5 (F3,234 = 18.93, p < 0.0001; 1.5-fold increase; unlabeled < pS422+, p < 0.01) and rab7 (F3,241 = 10.88, p < 0.0001; 1.6-fold increase, unlabeled < pS422+, p < 0.001), which associate with early and late endosomes, respectively; the lysosomal aspartyl protease cathepsin D (ctsd; F3,238 = 7.88, p < 0.0001; 2-fold increase, unlabeled < pS422+, p < 0.01); and the lysosomal-associated membrane protein 1 (lamp1; F3,224 = 23.88, p = 0.0013; 1.4-fold increase, unlabeled < pS422+, p < 0.01). Cleavage at D421 and NFT pathology was associated with upregulation of the early endosomal GTPase rab4 (F3,224 = 14.70, p = 0.0031; 1.3-fold increase, pS422+ < pS422+/TauC3+, p < 0.05; Fig. 3). Finally, the transcript encoding Niemann-Pick disease type C1 (npc1), which delivers low-density lipoprotein to late endosomal/lysosomal compartments and modulates cholesterol metabolism, was significantly dysregulated across the cell phenotypes (F3,218 = 13.11, p = 0.0025), revealing a progressive upregulation in NBM neurons during the transition from unlabeled to pS422+ to pS422+/TauC3+ (p < 0.05; Fig. 4B).
Effect of tau pathology on protein turnover and tau processing mRNAs in NBM neurons
Impaired protein turnover was also evidenced by the differential regulation of mRNAs encoding enzymes involved in protein ubiquitination (Fig. 4). pS422 positivity was associated with the upregulation of ubiquitin conjugating enzyme E2E 1 (ube2e1; F3,230 = 9.73, p < 0.0001; 1.5-fold increase, unlabeled < pS422+; p < 0.01) and ubiquitin carboxy-terminal enzyme L3 (uchl3; F3,235 = 27.26, p < 0.0001; 1.8-fold increase, unlabeled < pS422+; p < 0.05), both of which remained significantly elevated in pS422+/TauC3+ and TauC3+ NBM neurons. By contrast, appearance of the TauC3 epitope was marked by downregulation of ubiquitin specific peptidase 8 (usp8; F3,224 = 19.45, p = 0.0011; 2-fold decrease, unlabeled, pS422+ > pS422+/TauC3; p < 0.05), a hydrolase that removes conjugated ubiquitin and inhibits protein degradation. The appearance of the TauC3 epitope within NBM neurons was also associated with an upregulation of two mRNAs encoding aspartyl proteases that mediate tau cleavage (Fig. 3), caspase-3 (casp3; F3,235 = 9.80, p < 0.0001; 1.4-fold increase, pS422+ < pS422+/TauC3+; p < 0.01) and caspase-7 (casp7; F3,234 = 13.47, p < 0.0001; 1.5-fold increase, pS422+ < pS422+/TauC3+; p < 0.01). Both caspase-3 and -7 mediate truncation of tau at D421 [35], and their coincident upregulation with the appearance of the TauC3 epitope suggests a role in C-terminal tau cleavage.
Effect of tau tangle evolution on the expression of canonical AD-related genes
There were no significant alterations of the more classic AD-related genes across the neuronal phenotypes or clinical groups examined. For example, no expression level changes were found for amyloid-β precursor protein (App), amyloid-β precursor-like protein 1 (Aplp1), amyloid-β precursor-like protein 2 (Aplp2), β-site APP-cleaving enzyme 1 (Bace1), presenilin 1 (Psen1), presenilin 2 (Psen2), low density lipoprotein-related protein 1 (Lrp1), high density lipoprotein binding protein (Hdlbp), α-2- macroglobulin (A2m), or β-2-microglobulin (B2m) (data not shown). In contrast, other transcripts more specifically associated with tau pathology were altered. For instance, cyclin-dependent kinase 5 (cdk5), which is known to mediate aberrant tau phosphorylation [46], was significantly dysregulated among the cell phenotypes (F3,224 = 12.56, p < 0.0001), displaying upregulation during the transition from pS422+ to pS422+/TauC3+ (1.5-fold; pS422+ < pS422+/TauC3+; p < 0.01; Fig. 3).
Microarray validation
qPCR and immunoblot validation analyses of frozen tissue samples were not conducted as previously described [10,37,47]. Therefore, changes associated with these phenotypes at the single neuron level would be masked by regional gene expression patterns from admixed neuronal and non-neuronal cell types. On the other hand, the upregulation of cathepsin D in NFT-bearing neurons in AD has been reported by combined in situ hybridization and immunocytochemical approaches [48,49]. Future studies employing single population RNA-sequencing, Fluidigm, and/or Nanostring nCounter analyses are warranted when transcriptomic technologies become more standardized and economical [50–52] for human postmortem tissue use.
Correlations with clinical pathologic variables
Correlation of levels of select transcripts within different tau phenotypic NBM neurons and demographic data, antemortem cognitive test scores and postmortem neuropathologic variables revealed no significant associations (Table 2). This was not surprising given our observation that transcript level alterations were dependent on the tau phenotype of the NBM neuron and not disease status. Hence, cognitive decline is likely related to the accumulation of dysfunctional NFT-bearing NBM neurons and subsequent cholinergic deficits in cortical target fields. Interestingly, we have previously demonstrated that the number of pS422-bearing NBM neurons and accumulation of pS422+ neurites in the basal forebrain is associated with the transition from NCI to MCI [19].
Table 2.
Clinical pathologic correlations of select transcripts in pS422-immunoreactive NBM neurons
| gene | MMSE | Braak | NIA-Reagan | CERAD |
|---|---|---|---|---|
| hsp70 | r = 0.17 (p = 0.4) | r = 0.20 (p = 0.3) | r = −0.11 (p = 0.5) | r = −0.27 (p = 0.2) |
| ern2 | r = −0.26 (p = 0.2) | r = 0.11 (p = 0.5) | r = 0.05 (p = 0.7) | r = 0.10 (p = 0.5) |
| rab5 | r = 0.18 (p = 0.4) | r = −0.04 (p = 0.7) | r = −0.22 (p = 0.3) | r = 0.09 (p = 0.6) |
| ctsd | r = 0.13 (p = 0.5) | r = 0.23 (p = 0.2) | r = −0.02 (p = 0.7) | r = 0.12 (p = 0.4) |
| atg10 | r = −0.22 (p = 0.2) | r = −0.31 (p = 0.2) | r = 0.10 (p = 0.5) | r = 0.15 (p = 0.4) |
Discussion
The present analysis characterizes alterations in the expression of select functional classes of genes that coincide with specific tau epitopes related to NFT formation in NBM neurons during the progression of AD (Table 3). Specifically, an early event in tangle evolution, phosphorylation at S422, is associated with upregulation of select gene transcripts subserving protein quality control and turnover, which are likely initiated to manage and degrade misfolded proteins, including tau. By contrast, a later tau truncation event at D421 involved in NFT formation is associated with the upregulation of additional mRNAs involved in protein trafficking, degradation, and apoptosis. Notably, these alterations were associated with specific stages of tau pathology and not clinical disease status, supporting the concept that the accumulation of NBM neurons bearing these tau epitopes over time drives the clinical progression of the disease [3,22]. In this regard, we and others have previously reported gene expression changes within individual NBM and hippocampal neurons during the progression from NCI to MCI to AD [10,37,40,53,54], as well as differences in the genetic signature of tangle-free neurons compared to NFT-containing hippocampal and entorhinal cortex neurons in control and AD cases [48,55–57]. Notably, Dunckley and colleagues revealed that thioflavine S (TS)-negative neurons in entorhinal cortex layer II displayed altered gene expression changes in AD cases compared to TS-negative neurons in control cases [55]. Likewise, Liang and colleagues isolated TS-negative neurons from several neocortical regions as well as entorhinal cortex and hippocampus revealing gene expression changes in AD compared to control cases [56]. Based on the present findings, it is possible that in both of these datasets, TS-negative AD neurons may have harbored early/pretangle tau epitopes that resulted in the observed altered gene expression patterns despite the absence of TS-positivity. Hence, to our knowledge, the present data are the first to explicitly demonstrate that progressive tau pathological epitopes are associated with the dysregulation of distinct functional gene classes and molecular pathways during disease progression.
Table 3.
Summary of gene expression changes associated with the progression of tau pathology in NB neurons
| gene | pS422 | pS422 + TauC3* |
|---|---|---|
| hsp70 |

|
|
| hsp90 |

|
|
| ern2 |

|
|
| rab4 |

|
|
| rab5 |

|
|
| rab7 |

|
|
| ctsd |

|
|
| lamp1 |

|
|
| atg3 | ||
| atg10 |

|
|
| casp3 |

|
|
| casp7 |

|
|
| ppp2r1a |
|
|
| ppp2r1b | ||
| gsk3b | ||
| cdk5 |

|
|
| rab3 |
|
|
| npc1 |

|

|
| ube2e1 |

|
|
| uchl3 |

|
|
| usp1 | ||
| usp8 |
|
|
| canx | ||
| grp78 |

|
|
| app | ||
| aplp1 | ||
| aplp2 | ||
| bace1 | ||
| psen1 | ||
| psen2 | ||
| lrp1 | ||
| hdlbp | ||
| a2m | ||
| b2m |
Expression profiles of TauC3 neurons not significantly different from pS422+TauC3 neurons
Prominent among these gene classes were those expressing heat shock proteins, underscoring the role of proteostatic stress in the development of neurofibrillary degeneration. For instance, Hsp70 and Hsp90 play essential roles in stabilizing proteins against improper protein folding and aggregation [58] and are upregulated in the AD brain [59,60]. Interestingly, Hsp70 is a potent inhibitor of tau aggregation [61], and attenuates tau toxicity by preferentially associating with soluble, monomeric and prefibrillar, oligomeric tau species [62]. Upregulation of the hsp70 molecular chaperone in pretangle pS422-positive neurons may be a compensatory mechanism initiated to abate toxicity associated with misfolded tau in NBM neurons early in the disease process. On the other hand, inhibitors of constitutively expressed hsp90 and its co-chaperones reduce tau pathology [63–65]. Hence, the observed induction of the hsp90 gene with later stage tau epitopes may represent an aberrant chaperone response that activates rather than inhibits tau aggregation. These data suggest that either hsp70/co-chaperone activators or hsp90/co-chaperone inhibitors affect tau pathology in various stage of tangle evolution.
In addition to an upregulation in protein chaperone markers during the progression of tau pathology, the endosomal markers rab5 and rab7, as well as the lysosomal markers ctsd and lamp1, are upregulated in NBM neurons displaying the pretangle marker, pS422. These results are in line with several observations that endosomal-lysosomal protein trafficking dysfunction is one of the earliest cellular disturbances in AD [37,66,67]. For example, enlarged rab5-positive endosomes occur in pyramidal cells of the neocortex prior to NFT formation [66], and an upregulation of rab5 and cathepsin D in the NBM and hippocampus is observed in MCI compared to NCI [47,53,68]. Whether protein trafficking and degradation facilitate tangle formation or are downstream consequences of tau fibrillization is unclear, but these data strengthen the hypothesis that endosomal-lysosomal abnormalities are an upstream molecular pathogenic event in AD pathophysiology that may be amenable to therapeutic targeting.
We also observed that select ubiquitination-related transcripts are differentially regulated in pS422-positive (e.g., an upregulation of ube2e1 and uchl3) and pS422/TauC3-positive (e.g., a downregulation of usp8) NBM neurons, suggesting dysfunctional protein metabolism and ubiquitin-proteasomal cascade activation during disease progression. These results have particular implications for regulation of tau protein turnover. The ubiquitin ligase for tau has been identified as the carboxyl terminus of Hsp70-interacting protein (CHIP) [69–71], which interacts with heat shock proteins to regulate tau degradation [64]. CHIP-mediated ligation of ubiquitin to its substrate requires interaction with an E2 conjugating enzyme and only a select subset of E2 conjugating enzymes bind and facilitate CHIP function, including Ube2E1 [72]. Therefore, upregulation of both ube2e1 and hsp70 in pS422+ NBM neurons suggests that a degradatory pathway for pathogenic tau is activated early in the disease process. Additionally, the upregulation of caspases casp3 and casp7 with the appearance of the TauC3 epitope may play an intrinsic cellular role in the mediation of apoptosis [73]. Once cleaved from their pro-forms by initiator caspases, activated caspase-3 and -7 induce apoptosis by dismantling key structural proteins within the neuron. Hence, the appearance of the TauC3 truncation epitope may signal that NBM neurons are already undergoing degeneration and that therapeutic interventions to prevent the conversion of pretangles to frank NFTs may have disease-modifying properties. However, the specific pathways that trigger caspase activation remain unclear.
In summary, these results suggest that pathways regulating protein homeostasis are altered prior to the onset of frank NFT pathology and that their temporal progression may confer pathogenicity in NBM neurons. While these human tissue based findings are inherently correlative and cannot address causality, they can point the way for more detailed mechanistic approaches for understanding selective vulnerability in preclinical models. Targeting these systems may form the basis for novel pharmacological treatment approaches for disease modification during the prodromal stage of AD.
Supplementary Material
HIGHLIGHTS.
The molecular pathways associated with neuronal tangle evolution in AD are unclear
Nucleus basalis neurons with pretangles show protein homeostasis gene changes
Gene changes depend on the stage of neuronal tau pathology, not clinical status
Dysregulation of protein homeostasis prior to frank NFTs may drive cell vulnerability
Targeting these systems may modify the progression of AD
Acknowledgments
This manuscript is dedicated to the memory of our late colleague, mentor, and friend, Dr. Lester I. “Skip” Binder. We thank Muhammad Nadeem and John Beck for excellent technical assistance. We are indebted to the altruism of the RROS participants. This work was supported by NIH grants AG014449 (CTT, SDG, NMK, EJM, SEC), AG043375 (EJM, SDG), AG044712 (SEC), AG107617 (Rush ADC), the Alzheimer’s Association (SDG), the Saint Mary’s Foundation (SEC), and the Barrow Neurological Foundation Barrow Beyond (EJM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Price JL, Davis PB, Morris JC, White DL. The Distribution of Tangles, Plaques and Related Immunohistochemical Markers in Healthy Aging and Alzheimers-Disease. NBA. 1991;12:295–312. doi: 10.1016/0197-4580(91)90006-6. [DOI] [PubMed] [Google Scholar]
- 2.Giannakopoulos P, Hof PR, Giannakopoulos AS, Herrmann FR, Michel JP, Bouras C. Regional distribution of neurofibrillary tangles and senile plaques in the cerebral cortex of very old patients. Arch Neurol. 1995;52:1150–1159. doi: 10.1001/archneur.1995.00540360028012. [DOI] [PubMed] [Google Scholar]
- 3.Mesulam M, Shaw P, Mash D, Weintraub S. Cholinergic nucleus basalis tauopathy emerges early in the aging-MCI-AD continuum. Ann Neurol. 2004;55:815–828. doi: 10.1002/ana.20100. [DOI] [PubMed] [Google Scholar]
- 4.Mufson EJ, Ma SY, Cochran EJ, Bennett DA, Beckett LA, Jaffar S, Saragovi HU, Kordower JH. Loss of nucleus basalis neurons containing trkA immunoreactivity in individuals with mild cognitive impairment and early Alzheimer’s disease. J Comp Neurol. 2000;427:19–30. doi: 10.1002/1096-9861(20001106)427:1<19::aid-cne2>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 5.Mufson EJ, Ma SY, Dills J, Cochran EJ, Leurgans S, Wuu J, Bennett DA, Jaffar S, Gilmor ML, Levey AI, Kordower JH. Loss of basal forebrain P75NTR immunoreactivity in subjects with mild cognitive impairment and Alzheimer’s disease. J Comp Neurol. 2002;443:136–153. doi: 10.1002/cne.10122. [DOI] [PubMed] [Google Scholar]
- 6.Mesulam MM, Mufson EJ, Levey AI, Wainer BH. Cholinergic innervation of cortex by the basal forebrain: cytochemistry and cortical connections of the septal area, diagonal band nuclei, nucleus basalis (substantia innominata), and hypothalamus in the rhesus monkey. J Comp Neurol. 1983;214:170–197. doi: 10.1002/cne.902140206. [DOI] [PubMed] [Google Scholar]
- 7.Mesulam MM, Geula C. Nucleus basalis (Ch4) and cortical cholinergic innervation in the human brain: observations based on the distribution of acetylcholinesterase and choline acetyltransferase. J Comp Neurol. 1988;275:216–240. doi: 10.1002/cne.902750205. [DOI] [PubMed] [Google Scholar]
- 8.Pappas BA, Bayley PJ, Bui BK, Hansen LA, Thal LJ. Choline acetyltransferase activity and cognitive domain scores of Alzheimer’s patients. Neurobiol Aging. 2000;21:11–17. doi: 10.1016/s0197-4580(00)00090-7. [DOI] [PubMed] [Google Scholar]
- 9.Bierer LM, Haroutunian V, Gabriel S, Knott PJ, Carlin LS, Purohit DP, Perl DP, Schmeidler J, Kanof P, Davis KL. Neurochemical correlates of dementia severity in Alzheimer’s disease: relative importance of the cholinergic deficits. J Neurochem. 2002;64:749–760. doi: 10.1046/j.1471-4159.1995.64020749.x. [DOI] [PubMed] [Google Scholar]
- 10.Ginsberg SD, Che S, Wuu J, Counts SE, Mufson EJ. Down regulation of trk but not p75 NTRgene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem. 2006;97:475–487. doi: 10.1111/j.1471-4159.2006.03764.x. [DOI] [PubMed] [Google Scholar]
- 11.Mufson EJ, Kordower JH. Nerve growth factor and its receptors in the primate forebrain: Alterations in Alzheimer’s disease and potential use in experimental therapeutics. In: Mattson MP, editor. Neuroprotective signal transduction. Humana Press; Clifton, NJ: 1997. pp. 23–59. [Google Scholar]
- 12.Mufson EJ, Kordower JH. Nerve growth factor in Alzheimer’s disease. In: Peter AA, Morrison JH, editors. Cerebral cortex. Kluwer Academic/Plenum Press; New York: 1999. pp. 681–731. [Google Scholar]
- 13.Counts SE, Mufson EJ. The role of nerve growth factor receptors in cholinergic basal forebrain degeneration in prodromal Alzheimer disease. J Neuropathol Exp Neurol. 2005;64:263–272. doi: 10.1093/jnen/64.4.263. [DOI] [PubMed] [Google Scholar]
- 14.Candy JM, Perry RH, Perry EK, Irving D, Blessed G, Fairbairn AF, Tomlinson BE. Pathological changes in the nucleus of meynert in Alzheimer“s and Parkinson”s diseases. J Neurol Sci. 1983;59:277–289. doi: 10.1016/0022-510x(83)90045-x. [DOI] [PubMed] [Google Scholar]
- 15.Geula C, Mesulam MM, Saroff DM, Wu CK. Relationship between plaques, tangles, and loss of cortical cholinergic fibers in Alzheimer disease. J Neuropathol Exp Neurol. 1998;57:63–75. doi: 10.1097/00005072-199801000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Wood JG, Mirra SS, Pollock NJ, Binder LI. Neurofibrillary Tangles of Alzheimer-Disease Share Antigenic Determinants with the Axonal Microtubule-Associated Protein Tau (Tau) Proc Natl Acad Sci U S A. 1986;83:4040–4043. doi: 10.1073/pnas.83.11.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal Phosphorylation of the Microtubule-Associated Protein Tau (Tau) in Alzheimer Cytoskeletal Pathology. Proc Natl Acad Sci U S A. 2004;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee VM, Balin BJ, Otvos LJ, Trojanowski JQ. A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 19.Vana L, Kanaan NM, Ugwu IC, Wuu J, Mufson EJ, Binder LI. Progression of Tau Pathology in Cholinergic Basal Forebrain Neurons in Mild Cognitive Impairment and Alzheimer’s Disease. AJPA. 2011;179:2533–2550. doi: 10.1016/j.ajpath.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillozet-Bongaarts AL, Garcia-Sierra F, Reynolds MR, Horowitz PM, Fu Y, Wang T, Cahill ME, Bigio EH, Berry RW, Binder LI. Tau truncation during neurofibrillary tangle evolution in Alzheimer’s disease. Neurobiol Aging. 2005;26:1015–1022. doi: 10.1016/j.neurobiolaging.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Sierra F, Ghoshal N, Quinn B, Berry RW, Binder LI. Conformational changes and truncation of tau protein during tangle evolution in Alzheimer’s disease. J Alzheimers Dis. 2003;5:65–77. doi: 10.3233/jad-2003-5201. [DOI] [PubMed] [Google Scholar]
- 22.Vana L, Kanaan NM, Hakala K, Weintraub ST, Binder LI. Peroxynitrite-Induced Nitrative and Oxidative Modifications Alter Tau Filament Formation. Biochemistry. 2011;50:1203–1212. doi: 10.1021/bi101735m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez SE, Getova DP, He B, Counts SE, Geula C, Desire L, Coutadeur S, Peillon H, Ginsberg SD, Mufson EJ. Rac1b Increases with Progressive Tau Pathology within Cholinergic Nucleus Basalis Neurons in Alzheimer’s Disease. AJPA. 2012;180:526–540. doi: 10.1016/j.ajpath.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillozet-Bongaarts AL, Cahill ME, Cryns VL, Reynolds MR, Berry RW, Binder LI. Pseudophosphorylation of tau at serine 422 inhibits caspase cleavage: in vitro evidence and implications for tangle formation in vivo. J Neurochem. 2006;97:1005–1014. doi: 10.1111/j.1471-4159.2006.03784.x. [DOI] [PubMed] [Google Scholar]
- 25.Bennett DA, Wilson RS, Schneider JA, Evans DA, Beckett LA, Aggarwal NT, Barnes LL, Fox JH, Bach J. Natural history of mild cognitive impairment in older persons. Neurology. 2002;59:198– 205. doi: 10.1212/wnl.59.2.198. [DOI] [PubMed] [Google Scholar]
- 26.Mufson EJ, Chen EY, Cochran EJ, Beckett LA, Bennett DA, Kordower JH. Entorhinal cortex beta-amyloid load in individuals with mild cognitive impairment. Exp Neurol. 1999;158:469–490. doi: 10.1006/exnr.1999.7086. [DOI] [PubMed] [Google Scholar]
- 27.Counts SE, Nadeem M, Lad SP, Wuu J, Mufson EJ. Differential expression of synaptic proteins in the frontal and temporal cortex of elderly subjects with mild cognitive impairment. J Neuropathol Exp Neurol. 2006;65:592–601. doi: 10.1097/00005072-200606000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Perez SE, Bin He, Nadeem M, Wuu J, Scheff SW, Abrahamson EE, Ikonomovic MD, Mufson EJ. Archival Report. BPS. 2015;77:693–703. doi: 10.1016/j.biopsych.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer“s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer”s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 30.Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- 31.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 32.Hyman BT, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Carrillo MC, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Thies B, Trojanowski JQ, Vinters HV, Montine TJ. National Institute on Aging–Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimer’s & Dementia. 2012;8:1–13. doi: 10.1016/j.jalz.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ. The Consortium to Establish a Registry for Alzheimer“s Disease (CERAD) Part II. Standardization of the neuropathologic assessment of Alzheimer”s disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 34.Ginsberg SD, Crino PB, Lee VM, Eberwine JH, Trojanowski JQ. Sequestration of RNA in Alzheimer’s disease neurofibrillary tangles and senile plaques. Ann Neurol. 1997;41:200–209. doi: 10.1002/ana.410410211. [DOI] [PubMed] [Google Scholar]
- 35.Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, Lu M, Fu Y, Garcia-Sierra F, LaPointe N, Miller R, Berry RW, Binder LI, Cryns VL. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2003;100:10032– 10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Counts SE, Alldred MJ, Che S, Ginsberg SD, Mufson EJ. Neuropharmacology. Neuropharmacology. 2014;79:172–179. doi: 10.1016/j.neuropharm.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ginsberg SD, Alldred MJ, Counts SE, Cataldo AM, Neve RL, Jiang Y, Wuu J, Chao MV, Mufson EJ, Nixon RA, Che S. Microarray Analysis of Hippocampal CA1 Neurons Implicates Early Endosomal Dysfunction During Alzheimer’s Disease Progression. Biol Psychiatry. 2010;68:885–893. doi: 10.1016/j.biopsych.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mufson EJ, Counts SE, Ginsberg SD. Gene expression profiles of cholinergic nucleus basalis neurons in Alzheimer’s disease. Neurochem Res. 2002;27:1035–1048. doi: 10.1023/a:1020952704398. [DOI] [PubMed] [Google Scholar]
- 39.Alldred MJ, Che S, Ginsberg SD. Terminal continuation (TC) RNA amplification without second strand synthesis. J Neurosci Meth. 2009;177:381–385. doi: 10.1016/j.jneumeth.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Counts SE, He B, Che S, Ikonomovic MD, DeKosky ST, Ginsberg SD, Mufson EJ. alpha 7 nicotinic receptor up-regulation in cholinergic basal forebrain neurons in Alzheimer disease. Arch Neurol. 2007;64:1771–1776. doi: 10.1001/archneur.64.12.1771. [DOI] [PubMed] [Google Scholar]
- 41.Counts SE, He B, Che S, Ginsberg SD, Mufson EJ. Galanin fiber hyperinnervation preserves neuroprotective gene expression in cholinergic basal forebrain neurons in Alzheimer’s disease. J Alzheimers Dis. 2009;18:885–896. doi: 10.3233/JAD-2009-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Che S, Ginsberg SD. Amplification of RNA transcripts using terminal continuation. Lab Invest. 2004;84:131–137. doi: 10.1038/labinvest.3700005. [DOI] [PubMed] [Google Scholar]
- 43.Ginsberg SD. RNA amplification strategies for small sample populations. Methods. 2005;37:229–237. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 45.Alldred MJ, Lee SH, Petkova E, Ginsberg SD. Expression profile analysis of vulnerable CA1 pyramidal neurons in young-Middle-Aged Ts65Dn mice. J Comp Neurol. 2015;523:61–74. doi: 10.1002/cne.23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noble W, Olm V, Takata K, Casey E, Mary O, Meyerson J, Gaynor K, LaFrancois J, Wang L, Kondo T, Davies P, Burns M, Veeranna, Nixon R, Dickson D, Matsuoka Y, Ahlijanian M, Lau L-F, Duff K. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron. 2003;38:555–565. doi: 10.1016/s0896-6273(03)00259-9. [DOI] [PubMed] [Google Scholar]
- 47.Ginsberg SD, Mufson EJ, Counts SE, Wuu J, Alldred MJ, Nixon RA, Che S. Regional selectivity of rab5 and rab7 protein upregulation in mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis. 2010;22:631–639. doi: 10.3233/JAD-2010-101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ginsberg SD, Hemby SE, Lee V, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 2000 [PubMed] [Google Scholar]
- 49.Callahan LM, Vaules WA, Coleman PD. Quantitative decrease in synaptophysin message expression and increase in cathepsin D message expression in Alzheimer disease neurons containing neurofibrillary tangles. J Neuropathol Exp Neurol. 1999;58:275–287. doi: 10.1097/00005072-199903000-00007. [DOI] [PubMed] [Google Scholar]
- 50.Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNAsequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 51.Kim T, Lim C-S, Kaang B-K. Cell type-specific gene expression profiling in brain tissue: comparison between TRAP, LCM and RNA-seq. BMB Rep. 2015;48:388–394. doi: 10.5483/BMBRep.2015.48.7.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ginsberg SD, Mufson EJ, Alldred MJ, Counts SE, Wuu J, Nixon RA, Che S. Upregulation of select rab GTPases in cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Chem Neuroanat. 2011;42:102–110. doi: 10.1016/j.jchemneu.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Counts SE, Che S, Ginsberg SD, Mufson EJ. Gender differences in neurotrophin and gluatmate receptor expression in cholinergic nucleus basalis neurons during the progression of Alzheimer’s disease. J Chem Neuroanat. 2011;42:111–117. doi: 10.1016/j.jchemneu.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunckley T, Beach TG, Ramsey KE, Grover A, Mastroeni D, Walker DG, LaFleur BJ, Coon KD, Brown KM, Caselli R, Kukull W, Higdon R, McKeel D, Morris JC, Hulette C, Schmechel D, Reiman EM, Rogers J, Stephan DA. Gene expression correlates of neurofibrillary tangles in Alzheimer’s disease. Neurobiol Aging. 2006;27:1359–1371. doi: 10.1016/j.neurobiolaging.2005.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA. Alzheimer’s Disease Is Associated with Reduced Expression of Energy Metabolism Genes in Posterior Cingulate Neurons. Proc Natl Acad Sci U S A. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callahan LM, Selski DJ, Martzen MR, Cheetham JE, Coleman PD. Preliminary Evidence: Decreased GAP-43 message in tangle-bearing neurons relative to adjacent tangle-free neurons in Alzheimer’s disease parahippocampal gyrus. Neurobiol Aging. 1994;15:381–386. doi: 10.1016/0197-4580(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 58.Brown IR. Heat Shock Proteins and Protection of the Nervous System. Ann N Y Acad Sci. 2007;1113:147–158. doi: 10.1196/annals.1391.032. [DOI] [PubMed] [Google Scholar]
- 59.Dou F, Netzer WJ, Tanemura K, Li F, Hartl FU, Takashima A, Gouras GK, Greengard P, Xu H. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci U S A. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahara N, Maeda S, Yoshiike Y, Mizoroki T, Yamashita S, Murayama M, Park JM, Saito Y, Murayama S, Takashima A. Molecular chaperone-mediated tau protein metabolism counteracts the formation of granular tau oligomers in human brain. J Neurosci Res. 2007;85:3098–3108. doi: 10.1002/jnr.21417. [DOI] [PubMed] [Google Scholar]
- 61.Voss K, Combs B, Patterson KR, Binder LI, Gamblin TC. Hsp70 Alters Tau Function and Aggregation in an Isoform Specific Manner. Biochemistry. 2012;51:888–898. doi: 10.1021/bi2018078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patterson KR, Ward SM, Combs B, Voss K, Kanaan NM, Morfini G, Brady ST, Gamblin TC, Binder LI. Heat shock protein 70 prevents both tau aggregation and the inhibitory effects of preexisting tau aggregates on fast axonal transport. Biochemistry. 2011;50:10300–10310. doi: 10.1021/bi2009147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blair LJ, Zhang B, Dickey CA. Potential synergy between tau aggregation inhibitors and tau chaperone modulators. Alzheimer’s Res Ther. 2013;5:1–8. doi: 10.1186/alzrt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, Patterson C, Dickson DW, Nahman NS, Jr, Hutton M, Burrows F, Petrucelli L. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carman A, Kishinevsky S, Koren J, Lou W, Chiosis G. Chaperone-dependent Neurodegeneration: A Molecular Perspective on Therapeutic Intervention. J Alzheimers Dis Parkinsonism. 2013 doi: 10.4172/2161-0460.S10-007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. AJPA. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nixon RA, Yang D-S, Lee J-H. Neurodegenerative lysosomal disorders: A continuum from development to late age. Autophagy. 2014;4:590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- 68.Perez SE, He B, Nadeem M, Wuu J, Ginsberg SD, Ikonomovic MD, Mufson EJ. Hippocampal Endosomal, Lysosomal, and Autophagic Dysregulation in Mild Cognitive Impairment: Correlation With A beta and Tau Pathology. J Neuropathol Exp Neurol. 2015;74:345–358. doi: 10.1097/NEN.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hatakeyama S, Matsumoto M, Kamura T, Murayama M, Chui D-H, Planel E, Takahashi R, Nakayama KI, Takashima A. U-box protein carboxyl terminus of Hsc70-interacting protein (CHIP) mediates poly-ubiquitylation preferentially on four-repeat Tau and is involved in neurodegeneration of tauopathy. J Neurochem. 2004;91:299–307. doi: 10.1111/j.1471-4159.2004.02713.x. [DOI] [PubMed] [Google Scholar]
- 70.Petrucelli L. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Gen. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 71.Shimura H. CHIP-Hsc70 Complex Ubiquitinates Phosphorylated Tau and Enhances Cell Survival. J Biol Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 72.Soss SE, Yue Y, Dhe-Paganon S, Chazin WJ. E2 Conjugating Enzyme Selectivity and Requirements for Function of the E3 Ubiquitin Ligase CHIP. J Biol Chem. 2011;286:21277–21286. doi: 10.1074/jbc.M111.224006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McIlwain DR, Berger T, Mak TW. Caspase Functions in Cell Death and Disease. Cold Spring Harb Perspect Biol. 2013;5:a008656–a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.