Abstract
Mechanical forces generated by nuclear-cytoskeletal coupling through the LINC (linker of nucleoskeleton and cytoskeleton) complex, an evolutionarily conserved molecular bridge in the nuclear envelope (NE), are critical for the execution of wholesale nuclear positioning events in migrating and dividing cells, chromosome dynamics during meiosis, and mechanotransduction. LINC complexes consist of outer (KASH (Klarsicht, ANC-1, and Syne homology)) and inner (SUN (Sad1, UNC-84)) nuclear membrane proteins. KASH proteins interact with the cytoskeleton in the cytoplasm and SUN proteins in the perinuclear space of the NE. In the nucleoplasm, SUN proteins interact with A-type nuclear lamins and chromatin-binding proteins. Recent structural insights into the KASH-SUN interaction have generated several questions regarding how LINC complex assembly and function might be regulated within the perinuclear space. Here we discuss potential LINC regulatory mechanisms and focus on the potential role of AAA+ (ATPases associated with various cellular activities) protein, torsinA, as a LINC complex regulator within the NE. We also examine how defects in LINC complex regulation by torsinA may contribute to the pathogenesis of the human neurological movement disorder, DYT1 dystonia.
INTRODUCTION
The LINC (linker of nucleoskeleton and cytoskeleton) complex is an evolutionarily conserved molecular bridge in the nuclear envelope (NE) that is responsible for physically coupling the nucleus to the cytoskeleton in most eukaryotic cells1. LINC complexes are composed of two type II membrane proteins: the ONM (outer nuclear membrane) KASH (Klarsicht, ANC-1, and Syne homology) proteins and the INM (inner nuclear membrane) SUN (Sad1 and UNC-84) proteins2. All KASH proteins contain a divergent cytoplasmic domain followed by a C-terminal trans-membrane domain and a conserved short (~10–32 residue) luminal KASH domain, which is necessary and sufficient for the targeting of their unconserved cytoplasmic domains to the ONM3. Within the cytoplasm, KASH proteins bind various cytoskeletal elements as well as signaling molecules3, 4. In addition, several KASH proteins also project into the nucleoplasm from the INM, where they interact with other INM proteins, A-type lamins, and chromatin-binding proteins5, 6. Within the perinuclear space, KASH proteins directly interact with the conserved SUN domain of SUN proteins through their lumenal KASH domain4. The divergent SUN protein C-terminus extends into the nucleoplasm, where it interacts with A-type lamins, chromatin-binding proteins, and other proteins1. It is through the KASH-SUN interaction that the LINC complex is able to transmit mechanical forces from the cytoplasm across the double membrane of the NE and into the nucleoplasm.
Over the past decade, the LINC complex has attracted much attention resulting in the identification of a growing list of KASH and SUN proteins, LINC complex accessory proteins, as well as LINC complex-dependent cellular functions including DNA damage repair, meiotic pairing of homologous chromosomes, mechanotransduction, and nuclear positioning1, 3, 7. In light of this wide range of cellular and developmental functions that rely upon LINC complex-dependent nuclear-cytoskeletal coupling, it should come as no surprise that mutations in the genes encoding KASH and SUN proteins are associated with an ever-expanding list of human diseases including ataxia, cancer, deafness, and muscular dystrophy.
While LINC complexes are fundamental force transducers across the NE, very little has been revealed about the regulatory mechanisms that control LINC complex assembly and function. These regulatory mechanisms must exist given the fact that the KASH domains of nesprins 1, 2, and 3 can promiscuously interact with the SUN domains of SUN1 and SUN2 within the perinuclear space with similar affinities8, 9. Furthermore, LINC complexes have to switch between different cellular functions and often reorganize into higher-ordered arrays within the NE10, 11. Here, we review recent structural insights into LINC complex regulation, discuss the potential role of the AAA+ protein torsinA as a candidate LINC complex regulator, and explore the possible relationship between defective LINC complex regulation by torsinA and the neurological movement disorder DYT1 dystonia.
STRUCTURAL INSIGHTS INTO LINC COMPLEX FUNCTION AND REGULATION
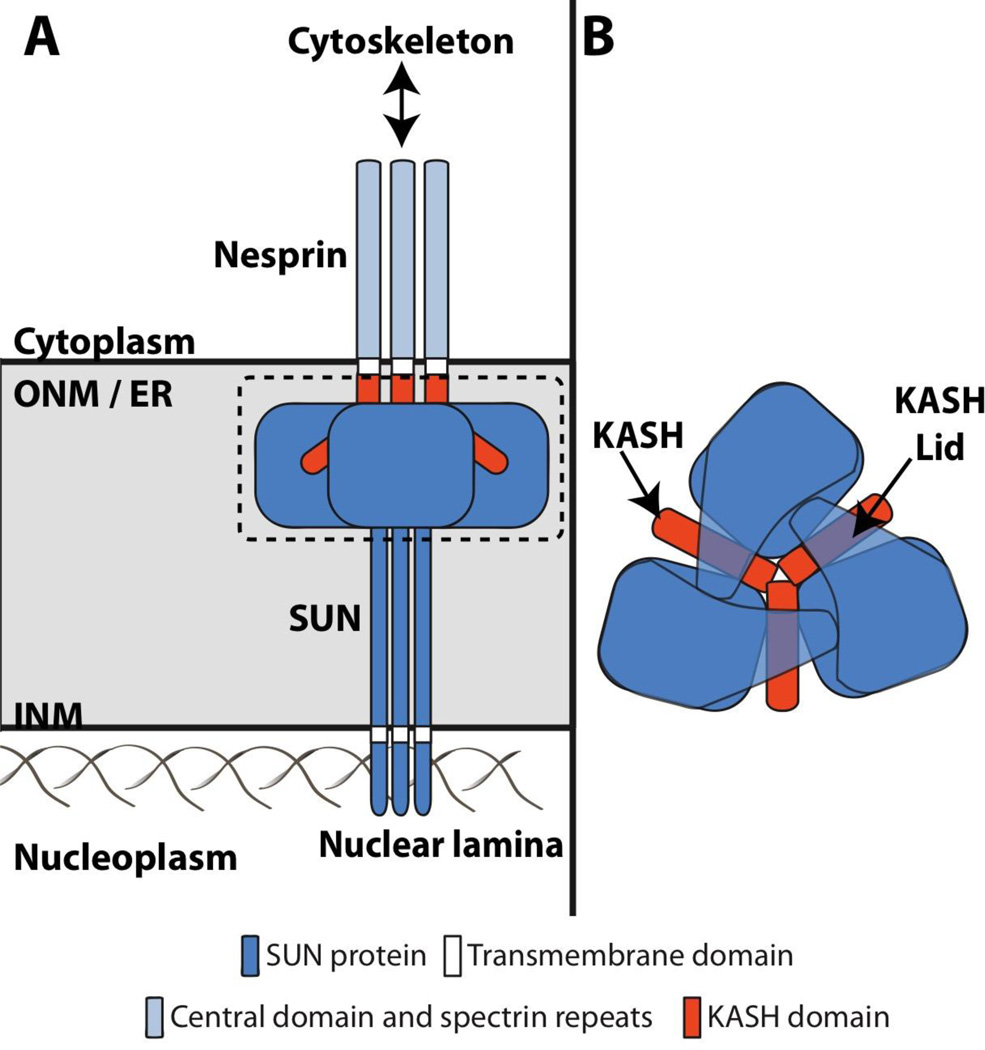
Recently, crystal structures of the human SUN2 bound to the C-terminal 29 residues (KASH2) from SYNE-2 (synaptic NE 2)/nesprin-2 (NE spectrin repeat protein 2) were independently solved and published by two groups12, 13. SUN2 is trimeric with the globular SUN domains sitting on top of a triple helical coiled-coil stalk (Figure 1A). KASH2 interacts with the SUN2 trimer within hydrophobic grooves formed at the interface of adjacent SUN domains (Figure 1). SUN2 further interacts with KASH2 through a “KASH-lid”, which conceals part of the KASH domain (Figure 1B). While SUN2 trimers exist in the absence of the KASH2, the KASH-lid assumes a random, non-ordered conformation, suggesting that it may function to stabilize the KASH-SUN interaction. In addition, two interesting structural features were identified in the SUN2-KASH2 complex: a loop that surrounds and coordinates a bound cation and a conserved intermolecular disulfide bond between KASH2 and the SUN domain. While the KASH2-SUN2 interaction was shown to require the cation loop, the disulfide bond appeared to be dispensable for binding12. However, molecular dynamics modeling recently demonstrated that this intermolecular disulfide bond was crucial for the stability of the KASH-SUN interaction as well as force transmission through the LINC complex14. The crystal structure of SUN2 in a complex with the KASH domain from SYNE-1/nesprin-1 was overall quite similar to the SUN2-KASH2 complex potentially explaining the conservation of the KASH domain12. Together the multiple non-covalent and single covalent interactions between KASH and SUN domains may explain how LINC complexes resist and perhaps transmit cytoskeletal-derived mechanical forces.
Figure 1. LINC complexes are heterohexamers of KASH and SUN proteins.
A) Assembled LINC complexes physically couple the nucleus to the cytoskeleton via the assembly of KASH-SUN heterohexamers within the perinuclear space12, 13. B) Top-down view of the KASH-SUN heterohexamer in A based on recently published crystal structures of the SUN-KASH interaction12, 13.
Highlighting the importance of SUN domain interfaces for KASH peptide binding, SUN2 trimerization through the triple helical coiled-coil is essential for KASH domain binding, as fusion of individual SUN domains of SUN2 to an unrelated trimeric coiled-coil can restore this interaction12. SUN protein trimerization may further allow efficient force transmission across the NE and the ability to endure the high loads necessary for movement of the nucleus or chromosomes within the nucleoplasm. Intriguingly, assembly of SUN protein trimers may also promote the formation of higher-order LINC complex arrays. Moving forward, it will be important to determine the existence of SUN protein trimers or LINC complexes with a SUN3:KASH3 stoichiometry within the NE of living cells. This knowledge will provide a useful intellectual framework for the future investigation of the kinetics and regulation of LINC complex assembly. While there is little question that these findings have significantly advanced our understanding of LINC complex structure and function, it is clear that they are just a preview of the increased mechanistic insights that will come in the future.
TORSINA AS A CANDIDATE LINC COMPLEX REGULATORY FACTOR
An often-discussed potential regulator of the LINC complex from within the perinuclear space is the evolutionarily conserved ATP-binding protein, torsinA12, 15, 16. Torsin proteins have been identified in all metazoans examined to date, but none have yet been found in unicellular eukaryotes or prokaryotic genomes17, 18. Recently, torsin proteins have attracted much attention as they have been implicated in multiple fascinating and important NE-localized functions ranging from the maintenance of proper ER/NE morphology19–21, the regulation of nuclear pore complex (NPC)-mediated nuclear import22, as well as the execution of a nuclear export pathway through which large ribonucleoprotein granules and herpesvirus capsids exit the nucleus via budding through the NE23, 24. The significance of torsinA function is illustrated by the fact that mice lacking torsinA die shortly after birth19. TorsinA belongs to the AAA+ protein superfamily, members of which typically function as ring-shaped hexameric molecular chaperones that utilize energy derived from ATP-hydrolysis to remodel protein complexes25, 26. Since torsinA has been shown to interact with the KASH domains from several nesprins, it has been proposed that torsinA acts as a molecular chaperone to control LINC complex assembly and/or disassembly16. Importantly, torsinA and the three additional mammalian torsin proteins (torsinB, torsin2, and torsin3) are the only AAA+ proteins known to reside within the lumen of ER (endoplasmic reticulum) and the contiguous perinuclear space of the NE17. Accumulating evidence suggest that torsinA exhibits chaperone-like behaviors both in vitro and in vivo27, 28. Notwithstanding these insights, the identity of the substrate(s) potentially remodeled by torsinA, and thus its precise cellular function(s), remain unidentified.
TORSINA IS AN ATYPICAL AAA+ PROTEIN
AAA+ proteins are defined by the presence of a 200–250 amino acid ATP-binding AAA domain, which contains Walker-A and -B motifs, as well as other motifs that differentiate them from classic P-loop NTPases25. Walker-A motifs bind ATP while Walker-B motifs hydrolyze ATP25. Mutation of the glutamate residue in the Walker-B motif prevents ATP-hydrolysis while leaving ATP-binding unaffected. Since ATP is required for substrate binding by most AAA+ proteins this mutation generates a useful reagent known as a ‘substrate trap’29. AAA+ proteins possess additional motifs including sensor-1, sensor-2, and arginine fingers, all of which participate in ATP-hydrolysis. Furthermore, the sensor-2 motif is also necessary for ATP-binding while the arginine fingers contribute to AAA+ protein oligomerization25.
Sequence-based analysis places torsinA in the classic AAA+ protein family and reveals a close relation to the C-terminal AAA+ domain of ClpB30. Upon sequence comparison, torsinA differs structurally from other AAA+ proteins in five major ways. First, unlike most AAA+ proteins, which are typically soluble, the first 20 amino acids of torsinA encode a signal sequence that directs the translocation of torsinA into the lumen of the ER and the contiguous perinuclear space of the NE31, 32. Second, torsinA possess a hydrophobic NTD (N-terminal domain) that follows the signal sequence allowing torsinA to monotopically associate with membranes and be retained within the secretory pathway by exclusion from ER exit sites33. Third, torsinA harbors critical differences in two conserved AAA+ motifs important for ATPase activity including the ATP-binding Walker-A motif and the sensor-2 motif, which promotes ATP-binding and -hydrolysis. Whereas the Walker-A motif is usually defined by the sequence motif GxxGxGK(T/S), torsinA contains an asparagine instead of threonine or serine34. Substituting the Walker-A threonine with asparagine in the bacterial AAA+ chaperone ClpB, which serves to reactivate aggregated proteins, results in a partial inhibition of ATPase activity34. The sensor II motif of torsinA atypically lacks the conserved arginine that interacts with the γ-phosphate of ATP in other AAA+ proteins35. This motif is referred to as the RSS (redox-sensing sensor) because it contains a unique cysteine-containing redox-sensing sensor-2 motif that is critical for both nucleotide- and partner-binding35. Fourth, no arginine fingers are found in torsinA35. Fifth, unlike other AAA+ proteins, torsinA lacks an identifiable substrate-binding domain or other functional domains outside of its ATPase domain, making it unclear how torsinA selectively interacts with and remodels potential substrates17.
To address this fundamental gap in our understanding of torsinA function, several groups have focused on determining the molecular mechanism of torsinA ATPase activation. However, the fact that torsinA is found within the oxidizing environment of the ER lumen, is glycosylated, and contains 6 conserved cysteine residues has posed a considerable challenge for biochemical analysis36. Demonstrative of this difficulty are the inconsistent results obtained by different laboratories regarding the ATPase activity of purified torsinA37–39. The critical evaluation of these conflicting results is hindered by the use of different expression systems (bacterial vs. insect), dissimilar torsinA constructs, and different assay conditions. Recently, pioneering work from the Schlieker laboratory provided significant insight into the mechanism of torsinA ATPase activation. They identified the major torsinA-interacting proteins LAP1 (lamina-associated polypeptide 1) and LULL1 (lumenal domain like LAP1) as stimulatory co-factors for torsinA40.
MECHANISM OF TORSINA ATPASE ACTIVATION
LAP1, a type II membrane protein, was the first torsinA-interacting protein identified in the NE41 (Figure 2). LAP1 binds tightly to an ATP-hydrolysis defective ‘substrate trap’ mutant of torsinA (E171Q)41, 42, which accumulates within the NE in cells21, 43. The N-terminus of LAP1 localizes to the nucleoplasm where it binds nuclear lamins, while its C-terminus resides within the perinuclear space where it interacts with torsinA41. Like torsinA, mice lacking LAP1 also exhibit perinatal lethality44. BLAST analysis revealed the existence of a novel protein, LULL1, with 60% identity to the lumenal domain of LAP141. LULL1 is also a type II membrane protein, the C-terminus of which resides within the ER/NE lumen where it interacts with torsinA in an ATP-dependent manner41, 42 (Figure 2). However, the N-terminus of LULL1 directs its localization to the peripheral ER and/or ONM, excluding it from the INM42. While LULL1 knockout mice have yet to be reported, there is evidence that functional segregation exists between LAP1 and LULL1. For example, LULL1 is required for the efficient growth of herpes simplex virus 1 and works together with torsinB to maintain proper ER morphology through undefined mechanisms20, 45.
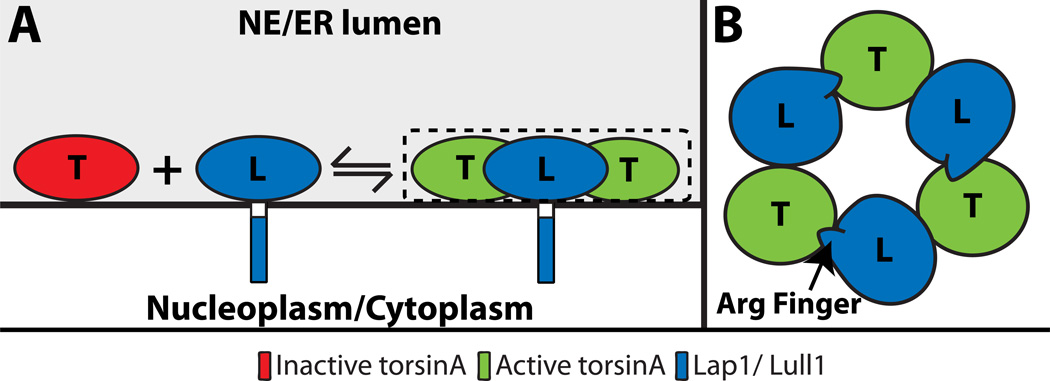
Figure 2. Stimulation of torsinA ATPase activity by LAP1 or LULL1.
A) The ATPase activity of torsinA is stimulated by LAP1 or LULL1 through the formation of heterohexamers on the INM and ER membranes, respectively18, 46. B) Top-down view of the alternating torsinA-LAP1/LULL1 heterohexamer in A18, 46.
Using torsinA purified from baculovirus, Zhao et al. demonstrate that this AAA+ protein has negligible ATPase activity40. Surprisingly though, when the lumenal domains of either LAP1 or LULL1 purified from bacteria were added to isolated torsinA, they found a significant stimulation of torsinA ATPase activity (Figure 2A). This ATPase activator function was also shown to control the activities of torsinB, torsin2, and torsin3, albeit to different extents. Substrates of AAA+ proteins often stimulate their ATPase activity25; however the lack of torsinA-mediated unfolding of LAP1 or LULL1 argues that these proteins most likely function as cofactors40. Two recent publications from the Schlieker and Schwartz laboratories further support this idea through experimental and computational evidence showing that the lumenal domains of LAP1 and LULL1 adopt a fold similar to an AAA+ domain18, 46. While neither protein possesses identifiable nucleotide-binding motifs, both contain a strictly conserved arginine finger that is dispensable for torsinA-binding but is required for the stimulation of torsinA ATPase activity18, 46 (Figure 2B).
Since torsins lack arginine fingers35, the authors proposed and provided evidence for a model whereby the lumenal domains of LAP1 or LULL1 activate torsinA through an active site complementation mechanism18, 46. In this model, these stimulatory cofactors interact with torsinA through a defined torsinA-cofactor interface that juxtaposes their arginine finger near the nucleotide bound in torsinA resulting in the assembly of an alternating hetero-hexameric (torsinA-LAP1/LULL1)3 holoenzyme (Figure 1). These important findings have perhaps generated more questions than they have answered. For example, the kinetic properties of torsinA ATPase activation by LAP1 and LULL1 are more similar to those observed for small GTPases where GAPs (GTPase-activating proteins) induce GTP hydrolysis above a negligible background47. This raises the possibility that torsinA functions mechanistically as a GTPase-like molecular switch rather than an AAA+ protein. In addition, the functional assembly state of torsinA remains undefined.
While the proposed active site complementation model requires the existence of an alternating hetero-hexameric (torsinA-LAP1/LULL1)3 holoenzyme (Fig. 2B), this may represent only one piece of a much more complicated puzzle as demonstrated by the conflicting published results from other groups studying torsinA oligomerization. For example, in vitro, isolated torsinA has been reported to be both monomeric and multimeric32, 39 while in cellulo experiments demonstrate that torsinA can self-associate into dimers and higher order oligomers consistent with the formation of a homo-hexamer48–52. These seemingly disparate results are not entirely mutually exclusive, as the oligomerization of HSP/Clp AAA+ protein family members are known to form homo- and/or hetero-hexameric assemblies53. Understanding torsinA oligomerization is further complicated by the fact that torsinB associates with and appears to be functionally redundant with torsinA in cells44, 54. To resolve these issues, it is important that a structure of torsinA alone and in complex with LAP1 or LULL1 be solved. Furthermore, structures of these proteins and complexes in the presence of transition state analogs would be helpful. It will also be interesting to investigate the potential role for the NTD during torsinA ATPase activation by LAP1 and LULL1, since a small fraction of torsinA has been shown to exist off the membrane in a soluble state33.
TARGETING OF TORSINA TO THE INM
Previously, a transient interaction between LULL1 and torsinA was shown to relocalize torsinA from the peripheral ER to the NE52. More recently, the Hanson laboratory demonstrated that the LULL1-torsinA interaction targets torsinA to the INM and destabilizes torsinA oligomers within the peripheral ER55. This finding is consistent with the fact that torsinA exists as a stable, slowly diffusing oligomer in the ER lumen that is unable to freely exchange between ER subdomains52. The targeting of torsinA to the INM by LULL1 was blocked by mutations in either protein that interfered with the stimulation of ATPase activity by active site complementation55. Remarkably, the forced dimerization of the N-terminus of LULL1 within the cytoplasm triggered this process, indicative of the existence of a signaling pathway within the cytoplasm that may control torsinA localization within the ER. These results suggest that LULL1 oligomerization leads to the transient disassembly of torsinA oligomers within the peripheral ER so that torsinA can reach LAP1 within the INM. Thus, while LULL1 and LAP1 both interact with and stimulate torsinA ATPase activity in vitro, the cellular consequence(s) of these interactions may be quite different. In the future, it will be interesting to further investigate the physical process responsible for the movement of torsinA from the peripheral ER to the INM following interaction with LULL1. The elucidation of the signaling pathway(s) that impinges upon the sub-organellar localization of torsinA will illuminate novel communication mechanisms that exist between the cytoplasm and ER.
EVIDENCE FOR TORSINA-MEDIATED LINC COMPLEX REGULATION
TorsinA was initially postulated to be a candidate regulator of the LINC complex because expression of a GFP-tagged torsinA construct encoding the ATP-hydrolysis-defective E171Q mutation in CHO cells resulted in the formation of NPC devoid stretches of NE with an abnormally narrow perinuclear space21. This close apposition of nuclear membranes was the opposite of what was observed in HeLa cells simultaneously depleted of SUN1 and SUN22. Therefore, since AAA+ proteins frequently remodel protein interactions, it was suggested that torsinA might modulate the interactions between nesprins and SUN proteins15. Consistent with this hypothesis are the following five results: 1) torsinA directly interacts with the KASH domains of several nesprins16; 2) torsinA is required for the localization of nesprin-3α to the NE16; 3) LULL1-mediated concentration of torsinA within the NE displaces nesprin-2G, nesprin-3, and SUN252; 4) SUN1 is involved in the proper localization of torsinA to the NE56; and 5) the Caenorhabditis elegans torsin protein OOC-5 is required for the NE localization of the KASH protein ZYG-12 in germ cells22.
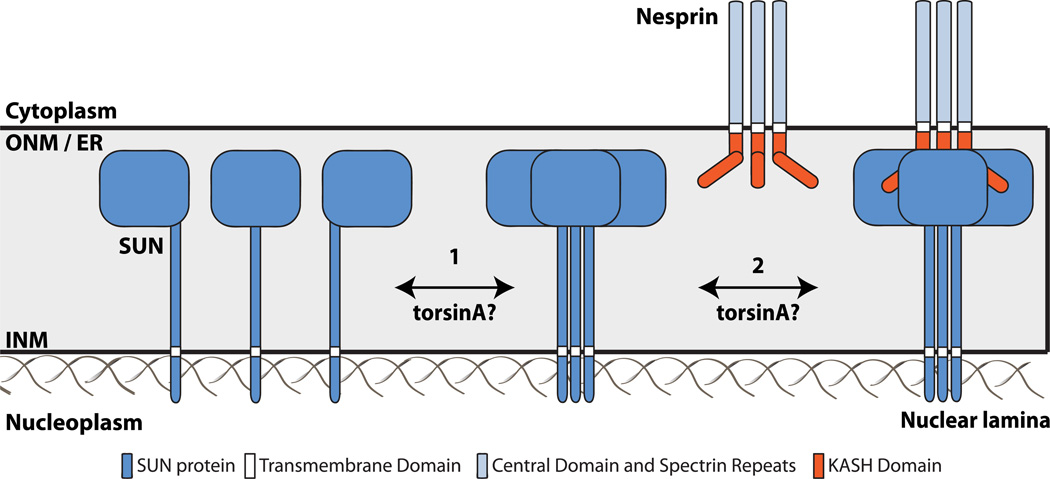
Although it is becoming increasingly clear that torsinA is involved in some aspect of LINC complex assembly and/or function, the mechanism responsible for this potential regulation is undefined. Based on the structural studies of the KASH-SUN interaction described above, we hypothesize that LINC complex assembly can be regulated by torsinA or other potential regulatory factors by influencing the ability of SUN proteins to homotrimerize and/or heterohexamerize with KASH proteins within the perinuclear space (Figure 3). An attractive potential mechanism for torsinA-mediated LINC complex regulation is provided by the fact that the C-terminus of torsinA is involved in binding the KASH domain16. Within this region of torsinA resides the previously discussed RSS motif that is important for nucleotide and partner binding35. When taken together with the existence of the KASH-SUN disulfide bond12, this raises the prospect that torsinA may use the RSS motif to break apart the SUN-KASH interaction in a fashion similar to a protein disulfide isomerase (PDI)57. However, the feasibility of this potential mechanism is complicated by the fact that KASH peptides bind to the domain interfaces of trimeric SUN proteins12, 13. Effectively, this shields the KASH domain, making it unclear how homo- or hetero-hexameric torsinA might access the KASH domain to regulate LINC complexes following their assembly. This gives rise to the alternative mechanism whereby torsinA may interact with and remodel the KASH domain of nesprins, preparing them for engagement with SUN proteins.
Figure 3. Working model of torsinA-mediated LINC complex regulation.
A) The two hypothesized steps of LINC complex assembly that may be regulated by torsinA within the perinuclear space are 1) homotrimerization of SUN proteins, and 2) heterohexamerization of KASH-SUN proteins. SUN proteins form homotrimers in the absence of KASH proteins12, 13, 83. While torsinA may positively and/or negatively regulate one or both of these steps, the precise regulatory mechanism remains unclear.
Another perplexing mechanistic issue regarding how torsinA could regulate the LINC complex is that torsinA is targeted to the INM in cells55. Since the width of the perinuclear space is ~30–50 nm58, it is unclear how torsinA would access the KASH domains of nesprins on the ONM (Figure 3). Models of the LINC complex suggest that the KASH domain extends no farther into the NE lumen than the SUN domain of SUN2, which represents only a fraction of the distance between the outer and inner nuclear membranes 12. Perhaps torsinA is involved in the proper ONM targeting of nesprins from the INM where several nesprin isoforms reside, projecting their N-terminus into the nucleoplasm to interact with the INM protein emerin and nuclear lamins59. Interestingly, LAP1 was recently demonstrated to interact with emerin within the nucleoplasm, and this interaction was shown to be essential for skeletal muscle maintenance in mice60. Emerin has often been considered a LINC complex component due to its ability to interact with both nesprins and SUN proteins61; thus, torsinA may regulate LINC complex assembly through modulating nesprin-emerin and/or SUN-emerin interactions at the INM. While interactions between LAP1 and LINC complex components have yet to be reported, they remain a formal possibility. It is also possible, although quite speculative, that the interaction between torsinA and LAP1 at the INM dislodges torsinA from the membrane allowing it to diffuse across the perinuclear space to interact with KASH domains on the ONM. Future experimental efforts should be focused on the biophysical, biochemical, and cellular mechanisms of LINC complex regulation by torsinA. It will also be interesting to investigate the role of redox potential and PDIs during LINC complex remodeling.
LINC COMPLEX DYSFUNCTION AND DYT1 DYSTONIA
TorsinA was first identified by the Breakefield laboratory in 1997 as the protein encoded by the DYT1/Tor1a gene, mutations of which cause DYT1 dystonia62. Dystonia is the third most frequent human neurological movement disorder behind essential tremor and Parkinson’s disease63. It is characterized by repetitive muscle contractions that result in involuntary twisting of the extremities and abnormal posturing64, 65. People afflicted with dystonia often experience serious disruptions in their ability to perform routine tasks including walking and sitting. Despite its prevalence, the pathogenesis of dystonia remains poorly understood. DYT1 dystonia is the most common and severe form of inherited dystonias, with autosomal-dominant inheritance and a penetrance of 30–40%66. The symptoms of DYT1 dystonia first appear at a mean age of 12.5 years67. Our understanding of DYT1 dystonia pathogenesis is complicated by the fact that torsinA is ubiquitously expressed, yet only the central nervous system appears to be affected in patients suffering from this disease68. Intriguingly, several other tissue-specific movement disorders (i.e. muscular dystrophies and cerebellar ataxias) are caused by mutations in widely expressed proteins that reside within the NE69. Furthermore, the lack of gross morphological defects in the brains of DYT1 patients does not provide obvious clues regarding disease mechanisms64; though recently a role for sensorimotor circuit neurodegeneration has been identified70. It is important to note that mutations in the TOR1AIP1 gene, which encodes LAP1, are also associated with severe dystonia, further highlighting the significance of the torsinA-LAP1 interaction71.
DYT1 dystonia results from the deletion of a single glutamic acid codon (ΔE302/303 or ΔE) in DYT1/Tor1a, the gene that encodes torsinA26. While the precise mechanism through which the ΔE mutation causes DYT1 dystonia is unclear, increasing evidence suggests that it is a loss-of-function mutation. For example, torsinA knockout mice are phenocopied by mice homozygous for the ΔE mutation 19, which impairs the ability of torsinA to interact with either LAP1 or LULL1, thereby disrupting its ATPase activity40, 42. This is surprising since the ΔE mutation causes torsinA to accumulate within the NE where LAP1 resides21, 43. Additionally, the efficiency of torsinA-mediated SUN2 displacement following LULL1-dependent NE targeting was significantly reduced by the presence of the disease mutation33. However, a gain-of-function mechanism has also been proposed since the ΔE mutation promotes an association between torsinA and SUN1, which is different from the interaction of ATP-bound torsinA with LAP156. In addition, the interaction of nesprin-3 with torsinA bearing the ΔE mutation was shown to be stronger than for wild type torsinA16. Taken together, these results suggest that LINC complex dysfunction may contribute to DYT1 dystonia pathogenesis.
Though widely speculated, the relationship between DYT1 dystonia and LINC complex dysfunction remains undefined. Nevertheless, torsinA containing the ΔE mutation disrupts some LINC complex-dependent functions such as cell migration and polarization, which may be important for neuronal function. The orientation of the centrosome to a position between the nucleus and leading edge is a hallmark of cell polarity in several directionally migrating cell types including fibroblasts and some neurons72. LINC complex-dependent nuclear-cytoskeletal coupling is critical for centrosome orientation and the efficient migration of these cells72–74. Interestingly, mouse embryonic fibroblasts from torsinA knockout mice and fibroblasts derived from DYT1 patients are defective in centrosome orientation16, 75. Centrosome orientation in fibroblasts is achieved through the coordinated positioning of the centrosome and the nucleus. The centrosome is maintained at the cell center through a microtubule-dependent mechanism while the nucleus is moved towards the cell rear by retrograde flowing actin cables on the dorsal nuclear surface, which are harnessed by linear arrays of nesprin-2G/SUN2 LINC complexes or TAN (transmembrane actin-associated nuclear) lines73. Currently, it is unknown if torsinA controls the positioning of the centrosome or the nucleus. While the brains of torsinA knockout mice lack obvious developmental defects normally associated with impaired neuronal migration19, subtle alterations in neuronal migration in the embryonic forebrain and from explants of the medial ganglionic eminence were observed in the absence of torsinA76. Although controversial, a correlation between the position of the centrosome and axon initiation has been demonstrated in developing hippocampal neurons77. Since many of the cellular components and mechanisms required for axon initiation are similar to those required for centrosome orientation during directional cell migration78, torsinA might also affect the establishment of axon-dendrite polarity in neurons. The decreased number of projections observed by group tractography in the cerebellothalamocortical pathway of mice heterozygous for ΔE mutation further supports this idea79.
A more speculative connection between DYT1 dystonia and LINC complex dysfunction was recently uncovered in Drosophila. Groundbreaking work from the Budnik laboratory identified a new mechanism through which NE budding drives the export of large ribonucleoprotein particles (RNPs) from the nucleus80. Interestingly, in neurons from homozygous torsinA knockout or ΔE knock-in mice the INM blebs into the perinuclear space19. This neural-specific NE morphology defect was proposed to be the result of the higher levels of torsinB expression found in non-neuronal cells vs. neurons44, 50. Interestingly, when RNP budding is disrupted in Drosophila torsinA mutants, large RNPs accumulate in the perinuclear space and synaptic mRNAs fail to traffic efficiently to their proper sites of protein synthesis, thereby compromising synaptic development23. TorsinA appears to function in the nuclear envelope budding process at a step involving scission of the inner nuclear membrane23. The movement of RNPs from muscle nuclei to postsynaptic sites in flies was shown to require the nesprin-1 orthologue, dNesp181. This research raises several questions relevant to the etiology of DYT1 dystonia. For example, it will be critical to determine if the involvement of torsinA in RNP export by NE budding occurs in neurons. It will also be of interest to resolve how RNPs are chosen and then selected for torsinA-dependent export. Finally, the identification of signaling pathways that regulate this torsinA-dependent process may represent novel therapeutic targets for DYT1 dystonia.
CONCLUSIONS
Our understanding of the structure and function of KASH-SUN interactions has dramatically increased since their original description 13 years ago 82. Since then the list of cellular, developmental, and disease-related roles of LINC complexes continues to grow at an exciting rate. Despite this explosion of interest, it remains unclear how LINC complexes are assembled and disassembled in a regulated manner. The identification of torsinA as a potential regulator of LINC complexes within the perinuclear space of the NE represents significant progress toward this end. However, the exact mechanism of torsinA-mediated LINC complex regulation remains an important mystery, the solution of which promises significant insight into LINC complex biology as well as the pathogenesis of DYT1 dystonia. A comprehensive mechanistic understanding of torsinA function will lead to the development of potential treatments for dystonia and related disorders.
Acknowledgments
We thank Drs. Melissa Gardner, David Greenstein, Joachim Mueller, and Meg Titus for helpful discussions. The authors apologize to those whose work we were unable to cite and cover in proper depth due to the limitations of length for this review. Studies in the Luxton Lab are supported by start up funding from the University of Minnesota, a P30 Pilot and Feasibility Grant from the Paul and Sheila Wellstone Muscular Dystrophy Center, and funding from the NIH (R21 NS095109-01, 1R41DA037622, and AR57220). C.A.S. is supported by an NIH training grant (NIH 5T32AR007612-14).
Footnotes
CONFLICT OF INTEREST
Cosmo A. Saunders and G.W. Gant Luxton have no conflicts on interest.
ETHICAL STANDARDS
The authors for this article carried out neither animal nor human studies for this study.
REFERENCES
- 1.Meinke P, Schirmer EC. LINC’ing form and function at the nuclear envelope. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Crisp M, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luxton GW, Starr DA. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol. 2014;28:69–75. doi: 10.1016/j.ceb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol. 2015;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann S, Noegel AA. Nesprins in cell stability and migration. Adv Exp Med Biol. 2014;773:491–504. doi: 10.1007/978-1-4899-8032-8_22. [DOI] [PubMed] [Google Scholar]
- 6.Duong NT, et al. Nesprins: tissue-specific expression of epsilon and other short isoforms. PLoS One. 2014;9:e94380. doi: 10.1371/journal.pone.0094380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swartz RK, Rodriguez EC, King MC. A role for nuclear envelope-bridging complexes in homology-directed repair. Mol Biol Cell. 2014;25:2461–2471. doi: 10.1091/mbc.E13-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostlund C, et al. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Luxton GW, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2011;2:173–181. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, et al. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res. 2012;22:1440–1452. doi: 10.1038/cr.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahed Z, Shams H, Mofrad MR. A Disulfide Bond Is Required for the Transmission of Forces through SUN-KASH Complexes. Biophys J. 2015;109:501–509. doi: 10.1016/j.bpj.2015.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerace L. TorsinA and torsion dystonia: Unraveling the architecture of the nuclear envelope. Proc Natl Acad Sci U S A. 2004;101:8839–8840. doi: 10.1073/pnas.0402441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nery FC, et al. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121:3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breakefield XO, Kamm C, Hanson PI. TorsinA: movement at many levels. Neuron. 2001;31:9–12. doi: 10.1016/s0896-6273(01)00350-6. [DOI] [PubMed] [Google Scholar]
- 18.Sosa BA, et al. How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife. 2014;3:e03239. doi: 10.7554/eLife.03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Rose AE, Zhao C, Turner EM, Steyer AM, Schlieker C. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J Biol Chem. 2014;289:552–564. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naismith TV, Heuser JE, Breakefield XO, Hanson PI. TorsinA in the nuclear envelope. Proc. Natl Acad Sci U S A. 2004;101:7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanGompel MJ, Nguyen KC, Hall DH, Dauer WT, Rose LS. A Novel Function for the C. elegans Torsin OOC-5 in Nucleoporin Localization and Nuclear Import. Mol Biol Cell. 2015 doi: 10.1091/mbc.E14-07-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jokhi V, et al. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–995. doi: 10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maric M, et al. A functional role for TorsinA in herpes simplex virus 1 nuclear egress. J Virol. 2011;85:9667–9679. doi: 10.1128/JVI.05314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 26.Ozelius LJ, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 27.Burdette AJ, Churchill PF, Caldwell GA, Caldwell KA. The early-onset torsion dystonia-associated protein, torsinA, displays molecular chaperone activity in vitro. Cell Stress Chaperones. 2010;15:605–617. doi: 10.1007/s12192-010-0173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nery FC, et al. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- 30.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Callan AC, Bunning S, Jones OT, High S, Swanton E. Biosynthesis of the dystonia-associated AAA+ ATPase torsinA at the endoplasmic reticulum. Biochem J. 2007;401:607–612. doi: 10.1042/BJ20061313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kustedjo K, Bracey MH, Cravatt BF. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem. 2000;275:27933–27939. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- 33.Vander Heyden AB, Naismith TV, Snapp EL, Hanson PI. Static retention of the lumenal monotopic membrane protein torsinA in the endoplasmic reticulum. EMBO J. 2011;30:3217–3231. doi: 10.1038/emboj.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy M, Wu HC, Liu Z, Kedzierska-Mieszkowska S, Zolkiewski M. Walker-A threonine couples nucleotide occupancy with the chaperone activity of the AAA+ ATPase ClpB. Protein Sci. 2009;18:287–293. doi: 10.1002/pro.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu L, Millen L, Mendoza JL, Thomas PJ. A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J Biol Chem. 2010;285:37271–37280. doi: 10.1074/jbc.M110.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu L, Wrabl JO, Hayashi AP, Rose LS, Thomas PJ. The torsin-family AAA+ protein OOC-5 contains a critical disulfide adjacent to Sensor-II that couples redox state to nucleotide binding. Mol Biol Cell. 2008;19:3599–3612. doi: 10.1091/mbc.E08-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kustedjo K, Deechongkit S, Kelly JW, Cravatt BF. Recombinant expression, purification, and comparative characterization of torsinA and its torsion dystonia-associated variant Delta E-torsinA. Biochemistry. 2003;42:15333–15341. doi: 10.1021/bi0349569. [DOI] [PubMed] [Google Scholar]
- 38.Konakova M, Pulst SM. Dystonia-associated forms of torsinA are deficient in ATPase activity. J Mol Neurosci. 2005;25:105–117. doi: 10.1385/JMN:25:1:105. [DOI] [PubMed] [Google Scholar]
- 39.Pham P, Frei KP, Woo W, Truong DD. Molecular defects of the dystonia-causing torsinA mutation. Neuroreport. 2006;17:1725–1728. doi: 10.1097/WNR.0b013e3280101220. [DOI] [PubMed] [Google Scholar]
- 40.Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci U S A. 2013;110:E1545–E1554. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol. 2005;168:855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naismith TV, Dalal S, Hanson PI. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem. 2009;284:27866–27874. doi: 10.1074/jbc.M109.020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci U S A. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim CE, Perez A, Perkins G, Ellisman MH, Dauer WT. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc Natl Acad Sci U S A. 2010;107:9861–9866. doi: 10.1073/pnas.0912877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner EM, Brown RS, Laudermilch E, Tsai PL, Schlieker C. The Torsin activator LULL1 is required for efficient growth of HSV-1. J Virol. 2015 doi: 10.1128/JVI.01143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown RS, Zhao C, Chase AR, Wang J, Schlieker C. The mechanism of Torsin ATPase activation. Proc Natl Acad Sci U S A. 2014;111:E4822–E4831. doi: 10.1073/pnas.1415271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 48.Giles LM, Chen J, Li L, Chin LS. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum Mol Genet. 2008;17:2712–2722. doi: 10.1093/hmg/ddn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon KL, Gonzalez-Alegre P. Consequences of the DYT1 mutation on torsinA oligomerization and degradation. Neuroscience. 2008;157:588–595. doi: 10.1016/j.neuroscience.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum Mol Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- 51.Torres GE, Sweeney AL, Beaulieu JM, Shashidharan P, Caron MG. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc Natl Acad Sci U S A. 2004;101:15650–15655. doi: 10.1073/pnas.0308088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell. 2009;20:2661–2672. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–F19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hewett JW, et al. TorsinB--perinuclear location and association with torsinA. J Neurochem. 2004;89:1186–1194. doi: 10.1111/j.1471-4159.2004.02404.x. [DOI] [PubMed] [Google Scholar]
- 55.Goodchild RE, et al. Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J Cell Sci. 2015;128:2854–2865. doi: 10.1242/jcs.167452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burke B. The nuclear envelope: filling in gaps. Nat Cell Biol. 2001;3:E273–E274. doi: 10.1038/ncb1201-e273. [DOI] [PubMed] [Google Scholar]
- 57.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 58.Cain NE, Starr DA. SUN proteins and nuclear envelope spacing. Nucleus. 2014 doi: 10.4161/19491034.2014.990857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris GE, Randles KN. Nesprin isoforms: are they inside or outside the nucleus. Biochem Soc Trans. 2010;38:278–280. doi: 10.1042/BST0380278. [DOI] [PubMed] [Google Scholar]
- 60.Shin JY, et al. Lamina-associated polypeptide-1 interacts with the muscular dystrophy protein emerin and is essential for skeletal muscle maintenance. Dev Cell. 2013;26:591–603. doi: 10.1016/j.devcel.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meinke P, Nguyen TD, Wehnert MS. The LINC complex and human disease. Biochem Soc Trans. 2011;39:1693–1697. doi: 10.1042/BST20110658. [DOI] [PubMed] [Google Scholar]
- 62.Ozelius LJ, et al. The TOR1A (DYT1) gene family and its role in early onset torsion dystonia. Genomics. 1999;62:377–384. doi: 10.1006/geno.1999.6039. [DOI] [PubMed] [Google Scholar]
- 63.Epidemiological SODIEESDECG. A prevalence study of primary dystonia in eight European countries. J Neurol. 2000;247:787–792. doi: 10.1007/s004150070094. [DOI] [PubMed] [Google Scholar]
- 64.Albanese A, et al. Phenomenology and classification of dystonia: a consensus update. Mov Disord. 2013;28:863–873. doi: 10.1002/mds.25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fahn S. Concept and classification of dystonia. Adv Neurol. 1988;50:1–8. [PubMed] [Google Scholar]
- 66.Bressman SB, et al. Idiopathic dystonia among Ashkenazi Jews: evidence for autosomal dominant inheritance. Ann Neurol. 1989;26:612–620. doi: 10.1002/ana.410260505. [DOI] [PubMed] [Google Scholar]
- 67.Ozelius LJ, Bressman SB. Genetic and clinical features of primary torsion dystonia. Neurobiol Dis. 2011;42:127–135. doi: 10.1016/j.nbd.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Breakefield XO, et al. The pathophysiological basis of dystonias. Nat Rev Neurosci. 2008;9:222–234. doi: 10.1038/nrn2337. [DOI] [PubMed] [Google Scholar]
- 69.Worman HJ, Dauer WT. The nuclear envelope: an intriguing focal point for neurogenetic disease. Neurotherapeutics. 2014;11:764–772. doi: 10.1007/s13311-014-0296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang CC, Tanabe LM, Jou S, Chi F, Dauer WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J Clin Invest. 2014;124:3080–3092. doi: 10.1172/JCI72830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dorboz I, et al. Severe dystonia, cerebellar atrophy, and cardiomyopathy likely caused by a missense mutation in TOR1AIP1. Orphanet J Rare Dis. 2014;9:174. doi: 10.1186/s13023-014-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23:579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nery FC, et al. Microfluidic platform to evaluate migration of cells from patients with DYT1 dystonia. J. Neurosci Methods. 2014;232:181–188. doi: 10.1016/j.jneumeth.2014.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCarthy DM, Gioioso V, Zhang X, Sharma N, Bhide PG. Neurogenesis and neuronal migration in the forebrain of the TorsinA knockout mouse embryo. Dev Neurosci. 2012;34:366–378. doi: 10.1159/000342260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Anda FC, et al. Centrosome localization determines neuronal polarity. Nature. 2005;436:704–708. doi: 10.1038/nature03811. [DOI] [PubMed] [Google Scholar]
- 78.Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 79.Ulug AM, et al. Cerebellothalamocortical pathway abnormalities in torsinA DYT1 knock-in mice. Proc Natl Acad Sci U S A. 2011;108:6638–6643. doi: 10.1073/pnas.1016445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Speese SD, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Packard M, et al. Nucleus to Synapse Nesprin1 Railroad Tracks Direct Synapse Maturation through RNA Localization. Neuron. 2015;86:1015–1028. doi: 10.1016/j.neuron.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 83.Zhou Z, et al. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem. 2012;287:5317–5326. doi: 10.1074/jbc.M111.304543. [DOI] [PMC free article] [PubMed] [Google Scholar]