Abstract
Hypothalamic hamartoma (HH) with gelastic epilepsy is a well-recognized drug-resistant epilepsy syndrome of early life.1 Surgical resection allows limited access to the small deep-seated lesions that cause the disease. Here, we report the results of a search for somatic mutations in paired hamartoma- and leukocyte-derived DNA samples from 38 individuals which we conducted by using whole-exome sequencing (WES), chromosomal microarray (CMA), and targeted resequencing (TRS) of candidate genes. Somatic mutations were identified in genes involving regulation of the sonic hedgehog (Shh) pathway in 14/38 individuals (37%). Three individuals had somatic mutations in PRKACA, which encodes a cAMP-dependent protein kinase that acts as a repressor protein in the Shh pathway, and four subjects had somatic mutations in GLI3, an Shh pathway gene associated with HH. In seven other individuals, we identified two recurrent and three single brain-tissue-specific, large copy-number or loss-of-heterozygosity (LOH) variants involving multiple Shh genes, as well as other genes without an obvious biological link to the Shh pathway. The Shh pathway genes in these large somatic lesions include the ligand itself (SHH and IHH), the receptor SMO, and several other Shh downstream pathway members, including CREBBP and GLI2. Taken together, our data implicate perturbation of the Shh pathway in at least 37% of individuals with the HH epilepsy syndrome, consistent with the concept of a developmental pathway brain disease.
Main Text
Some brain malformation syndromes are due to inherited germline mutations or germline or post-zygotic de novo mutations (reviewed in Poduri et al.2). Yet the cause of most brain malformations, with their frequent consequences of refractory epilepsy and intellectual disability, remains unknown. Recently, exome sequencing studies in privileged situations in which brain tissue is available have highlighted the role of somatic mutations confined to the brain. For example, somatic mutations in mTOR pathway genes (e.g., PIK3CA [MIM: 171834], AKT3 [MIM: 611223], and MTOR [MIM: 601231]) have been found to be important for a variety of malformations of cortical development, ranging from large hemispheric malformations to small focal cortical dysplasias.3, 4, 5, 6 Outside these rare disorders, the role of somatic mutations in drug-resistant epilepsies is largely unexplored.7, 8 Because the very rare dominant disorder of Pallister-Hall syndrome (MIM: 146510), comprising hypothalamic hamartomas (HHs [MIM: 241800]) and various other congenital anomalies, is due to germline truncation mutations in GLI3 (MIM: 165240),9 we previously searched for somatic GLI3 mutations in hamartoma tissue. We established that a few cases have de novo somatic point mutations or copy-number variants (CNVs) at this locus,10, 11 a finding that has now been independently confirmed.12 These early observations motivated a genome-wide search for somatic mutations via our unique access to hamartoma tissue and venous blood from individuals with HH. Surgical removal of these lesions was once regarded as hazardous. The development of innovative surgical techniques13 led to a relatively large series of this rare disorder being available at The Royal Children’s Hospital and The Barrow Institute.
Herein, we studied DNA extracted according to standard protocols from freshly frozen or formalin-fixed paraffin-embedded hamartoma tissue and leukocytes of 38 individuals with HH to identify somatic mutations. The human research ethics committees of the Austin Hospital and The Royal Children’s Hospital in Melbourne and the institutional review board of St. Joseph’s Hospital and Medical Center in Phoenix approved this study. Informed consent was obtained from affected individuals or their parents or legal guardians in the case of minors, those with intellectual disability, or deceased individuals. There were 11 females and 27 males, all with intractable epilepsy (Table S1). Epilepsy began in the first year of life in 30/38 of these indivdiuals, and all had gelastic (laughing) seizures. Additional features included intellectual disability in 24 individuals and central precocious puberty in 14 individuals; however, none had additional syndromic features of digital, oro-facial abnormalities or visceral malformations and none had a family history of HH. Samples were subjected to whole-exome sequencing (WES) as described previously14, 15, chromosomal microarray (CMA; Figure S1), and targeted resequencing (TRS) of 50 genes in the Shh pathway (Roche SeqCap EZ); the methodology selected depended on the quality and quantity of DNA from the brain samples. Due to limited DNA, nine hamartoma DNA samples were whole-genome amplified (QIAGEN Repli-g Single Cell) prior to WES.
Our first experiment was to subject a subset of paired DNA samples from hamartomas and leukocytes to WES (n = 15). Somatic single-nucleotide variants (sSNVs) were called from the aligned BAM files in both VarScan-2 and Mutect.16 VarScan-2 was used to call somatic insertion-deletion variants (sindels).17 sSNVs were removed from consideration if they were called in regions where there was less than 10-fold sequencing coverage in either the hamartoma or leukocytes, if the variant was present in less than three sequencing reads in the hamartoma, and if the variant was present in more than 5% of reads from leukocytes. Sindels with strand bias were filtered out with the Phred-scaled strand bias score. A somatic variant was classified as a candidate variant if it was predicted to change or truncate the amino acid sequence (Ensembl Variant Effect Predictor); this included missense (possibly- or probably-damaging or unknown according to PolyPhen-2), nonsense (frameshift and stop), and splice-site variants that were not present in controls sequenced in house (Institute for Genomic Medicine), in the Exome Variant Server (EVS), or in the Exome Aggregation Consortium (ExAC) database. The number of sSNVs and sindels detected in surgically resected tissue from the individuals with HH varied, despite high average coverage across exons (∼100 fold; Table S2). A total of 374 sSNVs were called on average per sample, including 12 candidate variants on average per sample (Tables S2 and S3). An average of 254 sindels were detected per sample, of which four on average per sample were candidate variants (Tables S2 and S3). Not unexpectedly, the nine samples for which the hamartoma DNA was whole-genome amplified due to very limited amounts of starting material prior to the preparation of the sequencing library produced larger numbers of sSNVs and sindels, most likely due to amplification artifacts. Among unamplified samples, 175 ± 51 and 186 ± 36 sSNVs and sindels, respectively, were called, versus 507 ± 479 sSNVs and 300 ± 211 sindels in the amplified samples (Table S2).
We performed a pathway analysis to determine whether candidate variants detected through WES were statistically significantly enriched in the Shh signaling pathway or any other Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (n = 301)18 by comparing the observed per base rate of candidate variants within and outside the pathway for each individual to the null expectation, estimated from the total number of candidate variants observed in the exome, the relative proportion of exonic bases within and outside the pathway that were sufficiently sequenced in the hamartoma and leukocyte DNA to call a candidate variant, and the relative mutability of the sequences within and outside of the pathway (see Supplemental Note). Two pathways of the 301 tested were enriched for candidate variants: the Shh pathway (p = 3 × 10−5) and the salivary secretion pathway (p = 0.017).
We next evaluated whether any genes in these two pathways were significantly enriched for candidate variants. GLI3 and PRKACA ([MIM: 601639], see Supplemental Note), both of which are involved in the Shh signaling pathway (defined by KEGG), were found to have two and three candidate variants, respectively, all in different individuals (Table 1). The uncorrected p values of seeing two and three candidate variants in GLI3 and PRKACA were 0.0025 and 2 × 10−7, respectively. Correcting for the ∼18,000 protein-coding genes genome wide, PRKACA is significantly enriched for candidate variants, implicating PRKACA as a HH-associated gene. We note that PRKACA encodes a protein that is named in both the Shh signaling and salivary secretion pathways (defined by KEGG). Given the existing evidence for the Shh signaling pathway in HH pathophysiology,10, 11, 12 we believe somatic variants in PRKACA lead to HH through disruption of the Shh signaling pathway. In addition to PRKACA, one additional gene, ZNF362, harboring a recurrent somatic mutation found in three individuals (Table S3), was found to be significantly enriched genome wide for candidate variants. These variants were found to be selectively observed in DNA specimens that were exome sequenced after whole-genome amplification in this study and in DNA from 12 out of 20 other individuals (without HH) that were also sequenced after whole-genome amplification as part of other genetic studies and therefore were not pursued further.
Table 1.
Study Participants with Confirmed Somatic Mutations in or Linked to the Shh Pathway
| Subject |
Genomic Analysis Method(s) |
Annotation of Candidate Mutations | Shh Gene(s) (KEGG) | VAF Brain | VAF Blood | Method of Detection | ||
|---|---|---|---|---|---|---|---|---|
| WES | CMA | TRS | ||||||
| hht25086 | × | − | − | c.984dupT (p.Asp329Ter) [GenBank: NM_002730.3] | PRKACA | 25% | 0.5% | WES, confirmed by Sanger |
| hht238a | × | − | − | c.983_984delTT (p.Phe328Ter) [GenBank: NM_002730.3] | PRKACA | 23% | 0% | WES, confirmed by Sanger |
| hht1198c | × | × | − | chr7q LOH (chr7:58,814,064–159,138,663) | SHH, SMO, WNT16, WNT2 | NA | NA | WES, confirmed by CMA |
| hht735 | × | × | × | chr16p LOH (chr16:0–31,543,619) | CREBBPa | NA | NA | WES, confirmed by CMA |
| hht209 | × | − | − | c.2989dupG (p.Ala997GlyfsTer87) [GenBank: NM_000168.5] | GLI3 | 57% | 0% | WES, confirmed by Sanger |
| hht26139 | × | − | − | c.3442C>T (p.Gln1148Ter) [GenBank: NM_000168.5] | GLI3 | 24% | 0% | WES, confirmed by Sanger |
| hht880 | − | × | − | chr2q12.1–q37.3 LOH (chr2:103,856,408–243,199,373) | GLI2, IHH, LRP2, STK36, WNT10A, WNT6 | NA | NA | CMA |
| hht953 | − | × | − | chr14q11.2–q32.33 LOH (chr14:24,419,118–106,072,470) | BMP4, AKT1 | NA | NA | CMA |
| hht25057 | − | × | − | chr16p11.2–p13.3 LOH (chr16:0–31543619) | CREBBPa | NA | NA | CMA |
| hht25063 | − | × | − | chr7p22.1–q36.3 CNG, CNL (chr7:986211–60069242, 58814064–159138663) | GLI3, SHH, SMO, WNT16, WNT2 | NA | NA | CMA |
| hht25094 | − | × | − | chr11q12.3–q25 LOH (chr11:64879188–135006516) | WNT11 | NA | NA | CMA |
| hht25077 | − | × | × | c.3172C>T (p.Arg1058Ter) [GenBank: NM_000168.5] | GLI3 | 18% | 0% | TRS, confirmed by Sanger |
| hht31536 | − | × | × | c.2071C>T (p.Gln691Ter) [GenBank: NM_000168.5] | GLI3 | 37% | 0.8% | TRS, confirmed by Sanger |
| hht25085 | − | − | − | c.226-231dup (p.Asp76_Lys77dup) [GenBank: NM_002730.3] | PRKACAb | – | – | Sanger |
Shh, sonic hedgehog; VAF, variant allele frequency; LOH, loss of heterozygosity (might be copy-number neutral LOH); CNG, copy-number gain; CNL, copy-number loss; WES, whole-exome sequencing; CMA, chromosomal microarray; TRS, targeted resequencing; ×, used; −, not used; NA, not applicable. All coordinates correspond to the UCSC Genome Browser reference genome (GRCH37/hg19).
Transcriptional regulator of the Shh pathway.
Identified by Sanger sequencing.
To screen for additional somatic mutations in PRKACA and GLI3, we then performed Sanger sequencing of the protein-coding exons by using standard protocols in hamartoma and blood DNA of 20 individuals with HH from our cohort that had not already been subjected to WES and identified a frameshift indel in PRKACA in an additional individual (hht25085; Table 1).
In addition to sSNVs and sindels, two large somatic loss-of-heterozygosity (sLOH) events were observed in the 15 samples analyzed by exome sequencing (Figure S2). One individual had sLOH across the p arm of chromosome 16, and another had sLOH across the q arm of chromosome 7 (Table 1; Figure 1). Interestingly, both regions contain genes connected to the Shh pathway; both SHH (MIM: 600725) and SMO (MIM: 601500), as well as WNT16 (MIM: 606267) and WNT2 (MIM: 147870), are located in the sLOH region on the q arm of chromosome 7; a transcriptional co-activator of the pathway, CREBBP (MIM: 600140), is located in the sLOH region on chromosome 16p.
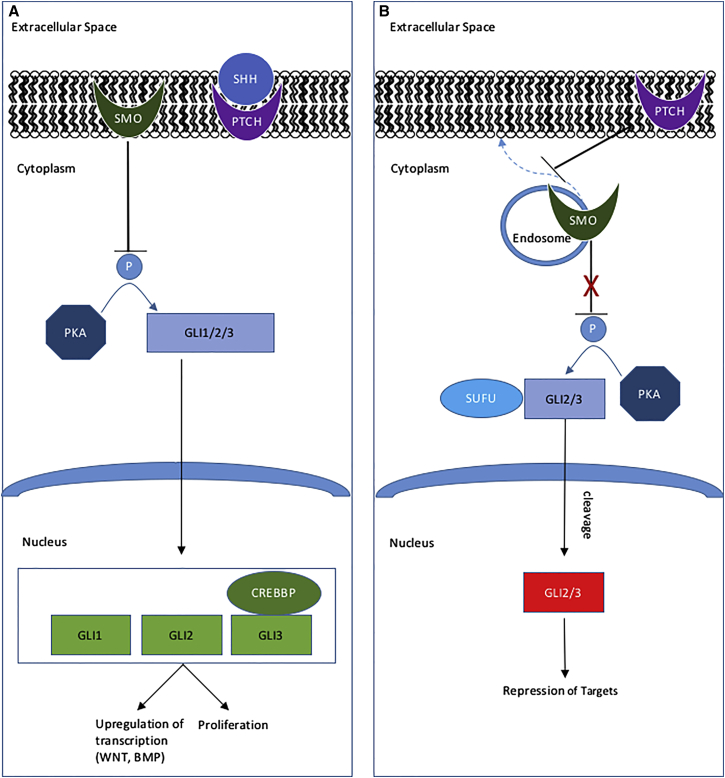
Figure 1.
Shh Pathway Showing Some of the Proteins Implicated in HH
(A) In the presence of SHH, PTCH no longer inhibits SMO from translocating to the membrane, where it inhibits phosphorylation of GLI2 and GLI3 by PKA (PRKACA). Full-length GLI2 and GLI3 translocate to the nucleus and, together with GLI1 and the transcriptional coactivator CREBBP, promote transcription of downstream target genes9
(B) In the absence of SHH, SMO remains localized to endosomes in the cytoplasm. GLI2 and GLI3 are bound by the SUFU complex and phosphorylated by PKA,15 leading to their cleavage. The cleaved protein translocates to the nucleus, where it acts to repress target gene transcription.
14 out of 38 individuals with HH had confirmed somatic mutations in the Shh pathway; an additional five individuals had unconfirmed findings in this pathway. Four individuals had confirmed somatic point mutations in GLI3, and an additional individual had a CNV that encompassed this gene. Three individuals had confirmed somatic mutations in PRKACA. Other proteins in the Shh pathway implicated in HH because of their occurrence in CNVs are SHH, SMO, CREBBP, GLI2, and STK36 (or, FU). Not shown on this simplified diagram are IHH, LRP2, LRP5, AKT1, and specific WNT and BMP pathway members that encode proteins in the Shh pathway, or that closely interact with the pathway, and are located within CNVs found in individuals with HH.16, 17, 18, 19
Because four of the five candidate variants were putatively protein-truncating mutations and because we observed two large LOH events with WES, we then used CMA to screen for somatic genomic deletions and other CNVs genome wide in 28 HH cases with adequate tumor DNA. We included the two exome-sequenced samples harboring large LOH regions on chromosomes 7q or 16p to confirm the detected somatic events. Analysis revealed a large somatic event in seven of the 28 individuals with HH, including confirmation of the two LOH lesions found by WES (Table 1; Figures S1 and S2). The lesions at 7q and 16p deletions were recurrent, given that one additional individual was found with copy-number neutral LOH (CNNLOH) events at each of these loci (Table 1). Three non-recurrent CNV or LOH events were also identified. One was found on chromosome 2q and included the Shh and/or WNT pathway members IHH (MIM: 600726), STK36 (MIM: 607652), GLI2 (MIM: 165230), LRP2 (MIM: 600073), WNT10A (MIM: 606268), and WNT6 (MIM: 604663). The second was on chromosome 14q and encompassed BMP4 (MIM: 112262) and AKT1 (MIM: 164730), a gene encoding a protein known to interact with the Shh pathway19 and that, when mutated, can result in a common type of primary brain tumor (meningioma).20 A third was discovered on chromosome 11q and included the Shh and/or WNT pathway members WNT11 (MIM: 603699) and LRP5 (MIM: 603506). The WNT pathway has parallel roles during development to the Shh signaling pathway21 and is also critical for axon guidance.22 A simulation analysis to assess the probability of observing this number of Shh genes in randomly selected (see Supplemental Note), similarly sized CNV or LOH events throughout the genome was significant (p = 0.046). We also performed a simulation analysis for the salivary secretion pathway but did not observe a significant enrichment for genes in this pathway (p = 0.716).
Given the preponderance of candidate somatic mutations in Shh genes, we then performed TRS of 50 genes in the Shh signaling pathway, as defined by KEGG,18 as well as of the transcriptional coactivator, CREBBP, in 15 individuals with HH to further screen for candidate causal mutations (see Supplemental Note for further methodological details). This analysis revealed somatic mutations in two additional individuals, both nonsense mutations in GLI3, confirmed by Sanger sequencing (Table 1). It is noteworthy that two of eight mutations in GLI3 and PRKACA detected by WES or TRS were present in a small fraction of sequencing reads from leukocyte-derived DNA, albeit a much smaller fraction than in tumors, and not confirmed to be present with another genotyping method (Table 1). This suggests that the mutations might be diffusely expressed in the brain, rather than just in the hamartoma per se. This is consistent with the observation of germline mutations in GLI3 in Pallister-Hall syndrome, in which HH is usually the only lesion in the brain.9 Although the biological significance of the observed enrichment of candidate variants in the salivary secretion pathway genes in HH is unclear, we did not further pursue these variants given that we detected no enrichment of genes in this pathway in somatic CNVs. Additionally, we note that Saitsu et al., who also recently screened for somatic mutations in HH, further support the association of the Shh pathway with HH with their work and did not report somatic mutations in the salivary secretion pathway genes.12
We predict that the somatic mutations in the pathway could lead to reduced Shh signaling. GLI3 can act as both a transcriptional activator and repressor of downstream targets.23 In the absence of Shh, protein kinase A (PKA) phosphorylates GLI3, causing its cleavage into a repressor form of the protein (Figure 123). This repressor then translocates to the nucleus and suppresses transcription of target genes. The somatic mutations we discovered in GLI3 all occurred in the same region as germline mutations in Pallister-Hall syndrome. The mutant, truncated protein in Pallister-Hall syndrome has been shown to localize to the nucleus and repress expression of target genes, leading to less Shh signaling.24 Two of three confirmed PRKACA mutations were located in the C-terminal region of the catalytic subunit; these truncating mutations could impair binding of the regulatory subunit of PKA, leading to a constitutively active form of the protein and subsequent repression of target genes due to increased phosphorylation and cleavage of GLI3. Interestingly, in adrenocortical tumors, a recurrent somatic mutation (encoding p.Leu206Arg) in PRKACA has been shown to inhibit binding of the regulatory subunit of PKA, leading to constitutive activation of the catalytic subunit of PKA and increased phosphorylation activity.25, 26
In summary, this comprehensive analysis revealed 14 somatic mutations (four nonsense changes, two frameshift indels, one in-frame insertion, and seven large CNV or LOH events; Table 1; Figure 1) in Shh pathway genes. PRKACA is now securely implicated as a HH-associated gene, in addition to the previously associated Shh gene, GLI3.10, 11, 12 An additional fifteen genes (AKT1, BMP4, CREBBP, GLI2, IHH, LRP2, LRP5, SHH, SMO, STK36, WNT2, WNT6, WNT10A, WNT11, and WNT16) associated with the Shh tumor suppressor pathway and the related WNT pathway are potentially implicated, as well as, possibly, other genes (within the large CNV or LOH regions) that do not have an obvious biological link to the Shh pathway. We did not identify somatic mutations in OFD1 (MIM: 300170), a recently reported candidate HH-associated gene identified in a mix of sporadic and syndromic cases from the Japanese population.12 Five cases had “unconfirmed findings” because there was insufficient DNA to complete the validation of all variants detected, whereas for 19 cases, no variants in genes encoding proteins directly or indirectly related to Shh were discovered (Table 2). Thus, we identified potentially pathogenic mutations in the Shh pathway in at least 37% (14/38) of individuals with HH, and possibly as many as 50% (19/38). This aggregation to a particular pathway is reminiscent of the remarkable contribution of various mTOR pathway gene mutations to malformations of cortical development, realized through similar clinico-molecular studies with privileged brain tissue over the last few years.2, 3, 27 The intense interest in Shh signaling in a variety of cancers has led to the development of potentially therapeutic agents acting on this pathway that could also be explored in treating this syndrome.28, 29
Table 2.
Study Participants with No Candidate or Unconfirmed Candidate Somatic Mutations in or Linked to the Shh Pathway
| Subject |
Genomic Analysis Method(s) |
Annotation of Candidate Mutations | Shh Gene(s) (KEGG) | VAF Brain | VAF Blood | Method of Detection | ||
|---|---|---|---|---|---|---|---|---|
| WES | CMA | TRS | ||||||
| hht929 | × | × | × | c.494dupG (p.Cys168LeufsTer4) [GenBank: NM_057168.1] | WNT16a | 31% | 4% | WES, TRS |
| hht25093 g | × | − | − | c.248T>C (p.Leu83Pro) [GenBank: NM_002730.3] | PRKACA | 14% | 1% | WES |
| hht20138 | − | × | × | c.984dupT (p.Asp329Ter) [GenBank: NM_002730.3] | PRKACA | 25% | 0% | TRS |
| hht25060 | − | × | × | c.1025dupG (p.Ala343ArgfsTer35) [GenBank: NM_003393.3] | WNT8B | 28% | 0% | TRS |
| c.394C>T (p.Gln132Ter) [GenBank: NM_182948.2] | PRKACB | 25% | 0% | |||||
| c.5293dupC (p.Gln1765ProfsTer201) [GenBank: NM_004380.2] | CREBBPb | 14% | 0% | |||||
| hht25186 | − | − | × | c.6858dupT (p.Glu2287Ter) [GenBank: NM_004525.2] | LRP2 | 17% | 3% | TRS |
| c.4230dupT (p.Gly1411TrpfsTer10) [GenBank: NM_004380.2] | CREBBPb | 14% | 0% | |||||
| hht25064 | − | − | × | ND | NA | NA | NA | |
| hht1276d | × | − | − | ND | NA | NA | NA | |
| hht322b | × | × | − | ND | NA | NA | NA | |
| hht25132h | × | × | − | ND | NA | NA | NA | |
| hht786 | × | × | × | ND | NA | NA | NA | |
| hht25080 | × | × | × | ND | NA | NA | NA | |
| hht25099 | × | × | × | ND | NA | NA | NA | |
| hht25059 | − | × | × | ND | NA | NA | NA | |
| hht25082 | × | − | × | ND | NA | NA | NA | |
| hht25050 | − | × | − | ND | NA | NA | NA | |
| hht25054 | − | × | − | ND | NA | NA | NA | |
| hht25066 | − | × | − | ND | NA | NA | NA | |
| hht25072 | − | × | − | ND | NA | NA | NA | |
| hht25079 | − | × | − | ND | NA | NA | NA | |
| hht25089 | − | × | − | ND | NA | NA | NA | |
| hht25097 | − | × | − | ND | NA | NA | NA | |
| hht25098 | − | × | − | ND | NA | NA | NA | |
| hht25052 | − | × | × | ND | NA | NA | NA | |
| hht25056 | − | × | × | ND | NA | NA | NA | |
Shh, Sonic hedgehog; VAF, variant allele frequency; WES, whole-exome sequencing; CMA, chromosomal microarray; TRS, targeted resequencing; ×, used; −, not used; NA, not applicable; ND, none detected. All coordinates correspond to the UCSC Genome Browser reference genome (GRCH37/hg19).
Candidate mutation found in WNT16 was detected with both next-generation sequencing technologies but not confirmed with Sanger sequencing because of insufficient DNA.
Transcriptional regulator of the Shh pathway.
Conflicts of Interest
I.E.S. discloses payments from UCB Pharma and Athena Diagnostics and Transgenomics for lectures and educational presentations. D.B.G. is an equity holder in Pairnomix, a company focused on modeling epilepsy mutations. S.F.B. discloses payments from UCB Pharma, Novartis Pharmaceuticals, Sanofi-Aventis, and Jansen Cilag for lectures and educational presentations and a patent for SCN1A testing held by Bionomics Inc. and licensed to various diagnostic companies.
Acknowledgments
We thank the families for their participation in this study, and we value the advocacy from the Hope for Hypothalamic Hamartoma Foundation (http://hopeforhh.org). Elena Aleksoska (Epilepsy Research Centre) performed genomic DNA extractions from tissues. Brett Copeland, Joshua Bridgers, and Sitharthan Kamalakaran (Institute for Genomic Medicine) provided bioinformatics support. Dr. Paul Lockhart, Greta Gillies, and Kate Pope (Murdoch Childrens Research Institute) assisted with collection of tissue, and Jeffrey Rosenfeld and Wirginia Maixner (Royal Children’s Hospital, Melbourne) performed surgeries. This study was supported by a National Health and Medical Research Council (NHMRC) Program Grant (628952) to S.F.B. and I.E.S., a Practitioner Fellowship (1006110) to I.E.S, and a Career Development Fellowship (1063799) to M.S.H. I.E.S. was also funded by grants from the NIH, Australian Research Council, Health Research Council of New Zealand, CURE, American Epilepsy Society, US Department of Defense Autism Spectrum Disorder Research Program, March of Dimes, and Perpetual Charitable Trustees. E.L.H. was supported by a NIH grant (R21-NS078657). N.C.J. was supported by an Australian Research Council Future Fellowship (13100100). Acknowledgments of individuals and funding sources responsible for the samples used in this study as controls are provided in the Supplemental Data.
Published: July 21, 2016
Footnotes
Supplemental Data include a Supplemental Note, two figures, three tables, and supplemental acknowledgments and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.05.031.
Contributor Information
Samuel F. Berkovic, Email: s.berkovic@unimelb.edu.au.
Erin L. Heinzen, Email: eh2682@cumc.columbia.edu.
Web Resources
Consensus Coding Sequence, https://www.ncbi.nlm.nih.gov/CCDS/
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Ingenuity Pathway Analysis, http://www.qiagen.com/ingenuity
Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/
NCBI Gene, http://www.ncbi.nlm.nih.gov/gene
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
Variant Effect Predictor, http://useast.ensembl.org/Homo_sapiens/Tools/VEP
Supplemental Data
References
- 1.Berkovic S.F., Arzimanoglou A., Kuzniecky R., Harvey A.S., Palmini A., Andermann F. Hypothalamic hamartoma and seizures: a treatable epileptic encephalopathy. Epilepsia. 2003;44:969–973. doi: 10.1046/j.1528-1157.2003.59102.x. [DOI] [PubMed] [Google Scholar]
- 2.Poduri A., Evrony G.D., Cai X., Walsh C.A. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J.H., Huynh M., Silhavy J.L., Kim S., Dixon-Salazar T., Heiberg A., Scott E., Bafna V., Hill K.J., Collazo A. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim J.S., Kim W.I., Kang H.C., Kim S.H., Park A.H., Park E.K., Cho Y.W., Kim S., Kim H.M., Kim J.A. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat. Med. 2015;21:395–400. doi: 10.1038/nm.3824. [DOI] [PubMed] [Google Scholar]
- 5.Nakashima M., Saitsu H., Takei N., Tohyama J., Kato M., Kitaura H., Shiina M., Shirozu H., Masuda H., Watanabe K. Somatic Mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann. Neurol. 2015;78:375–386. doi: 10.1002/ana.24444. [DOI] [PubMed] [Google Scholar]
- 6.Poduri A., Evrony G.D., Cai X., Elhosary P.C., Beroukhim R., Lehtinen M.K., Hills L.B., Heinzen E.L., Hill A., Hill R.S. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helbig I., Lowenstein D.H. Genetics of the epilepsies: where are we and where are we going? Curr. Opin. Neurol. 2013;26:179–185. doi: 10.1097/WCO.0b013e32835ee6ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindhout D. Somatic mosaicism as a basic epileptogenic mechanism? Brain. 2008;131:900–901. doi: 10.1093/brain/awn056. [DOI] [PubMed] [Google Scholar]
- 9.Kang S., Graham J.M., Jr., Olney A.H., Biesecker L.G. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat. Genet. 1997;15:266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- 10.Wallace R.H., Freeman J.L., Shouri M.R., Izzillo P.A., Rosenfeld J.V., Mulley J.C., Harvey A.S., Berkovic S.F. Somatic mutations in GLI3 can cause hypothalamic hamartoma and gelastic seizures. Neurology. 2008;70:653–655. doi: 10.1212/01.wnl.0000284607.12906.c5. [DOI] [PubMed] [Google Scholar]
- 11.Craig D.W., Itty A., Panganiban C., Szelinger S., Kruer M.C., Sekar A., Reiman D., Narayanan V., Stephan D.A., Kerrigan J.F. Identification of somatic chromosomal abnormalities in hypothalamic hamartoma tissue at the GLI3 locus. Am. J. Hum. Genet. 2008;82:366–374. doi: 10.1016/j.ajhg.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitsu H., Sonoda M., Higashijima T., Shirozu H., Masuda H., Tohyama J., Kato M., Nakashima M., Tsurusaki Y., Mizuguchi T. Somatic mutations in GLI3 and OFD1 involved in sonic hedgehog signaling cause hypothalamic hamartoma. Ann Clin Transl Neurol Epub. 2016 doi: 10.1002/acn3.300. Mar 24th. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenfeld J.V., Harvey A.S., Wrennall J., Zacharin M., Berkovic S.F. Transcallosal resection of hypothalamic hamartomas, with control of seizures, in children with gelastic epilepsy. Neurosurgery. 2001;48:108–118. doi: 10.1097/00006123-200101000-00019. [DOI] [PubMed] [Google Scholar]
- 14.EuroEPINOMICS-RES Consortium. Epilepsy Phenome/Genome Project. Epi4K Consortium De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epi4K Consortium; Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kebenko M., Drenckhan A., Gros S.J., Jücker M., Grabinski N., Ewald F., Grottke A., Schultze A., Izbicki J.R., Bokemeyer C. ErbB2 signaling activates the Hedgehog pathway via PI3K-Akt in human esophageal adenocarcinoma: identification of novel targets for concerted therapy concepts. Cell. Signal. 2015;27:373–381. doi: 10.1016/j.cellsig.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Clark V.E., Erson-Omay E.Z., Serin A., Yin J., Cotney J., Ozduman K., Avşar T., Li J., Murray P.B., Henegariu O. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulloa F., Martí E. Wnt won the war: antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Dev. Dyn. 2010;239:69–76. doi: 10.1002/dvdy.22058. [DOI] [PubMed] [Google Scholar]
- 22.Zou Y. Wnt signaling in axon guidance. Trends Neurosci. 2004;27:528–532. doi: 10.1016/j.tins.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 24.Shin S.H., Kogerman P., Lindström E., Toftgárd R., Biesecker L.G. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proc. Natl. Acad. Sci. USA. 1999;96:2880–2884. doi: 10.1073/pnas.96.6.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh G., Scholl U.I., Healy J.M., Choi M., Prasad M.L., Nelson-Williams C., Kunstman J.W., Korah R., Suttorp A.C., Dietrich D. Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors. Nat. Genet. 2014;46:613–617. doi: 10.1038/ng.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato Y., Maekawa S., Ishii R., Sanada M., Morikawa T., Shiraishi Y., Yoshida K., Nagata Y., Sato-Otsubo A., Yoshizato T. Recurrent somatic mutations underlie corticotropin-independent Cushing’s syndrome. Science. 2014;344:917–920. doi: 10.1126/science.1252328. [DOI] [PubMed] [Google Scholar]
- 27.Jamuar S.S., Walsh C.A. Somatic mutations in cerebral cortical malformations. N. Engl. J. Med. 2014;371:2038. doi: 10.1056/NEJMc1411784. [DOI] [PubMed] [Google Scholar]
- 28.Scales S.J., de Sauvage F.J. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Sheikh A., Alvi A.A., Aslam H.M., Haseeb A. Hedgehog pathway inhibitors - current status and future prospects. Infect. Agent. Cancer. 2012;7:29. doi: 10.1186/1750-9378-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.