Abstract
Rapid-Eye Movement (REM) sleep correlates with neuronal activity in the brainstem, basal forebrain and lateral hypothalamus (LH). LH melanin-concentrating hormone (MCH)-expressing neurons are active during sleep, however, their action on REM sleep remains unclear. Using optogenetic tools in newly-generated Tg(Pmch-Cre) mice, we found that acute activation of MCH neurons (ChETA, SSFO) at the onset of REM sleep extended the duration of REM, but not non-REM sleep episode. In contrast, their acute silencing (eNpHR3.0, ArchT) reduced the frequency and amplitude of hippocampal theta rhythm, without affecting REM sleep duration. In vitro activation of MCH neuron terminals induced GABAA-mediated inhibitory post-synaptic currents (IPSCs) in wake-promoting histaminergic neurons of the tuberomammillary nucleus (TMN), while in vivo activation of MCH neuron terminals in TMN or medial septum also prolonged REM sleep episodes. Collectively, these results suggest that activation of MCH neurons maintains REM sleep, possibly through inhibition of arousal circuits in the mammalian brain.
Introduction
Rapid Eye Movement (REM) sleep, or paradoxical sleep1, is characterized by rapid eye movements, muscle atonia and prominent theta rhythm in the hippocampus and cortex2. Pioneer studies initially located the REM sleep “generator(s)” within the pons1, where reciprocal inhibition between cholinergic neurons and serotonergic neurons from raphe nuclei and noradrenergic neurons from locus coeruleus were originally hypothesized to gate REM sleep3–7. More recently, neurons producing the neurotransmitter GABA (γ-aminobutyric acid) and glutamate were integrated to the brainstem circuitry critical for the onset of REM sleep8–11. In addition, the activity of neurons outside the brainstem also correlates with REM sleep state12, including neurons located in the anterior13 and lateral14–16 parts of the hypothalamus. Collectively, these studies suggested a role for the lateral hypothalamus (LH) networks in regulating REM sleep states, however, causal evidence are missing.
In this study, we investigated the role of neurons producing the melanin-concentrating hormone (MCH) peptide in regulating REM sleep state. Pharmacological infusion of the MCH peptide15,17 and selective activation of a subset of MCH neurons18 induced both NREM and REM sleep. MCH neurons are strongly activated during a homeostatic sleep rebound induced by a prolonged sleep deprivation 14,15,19,20, and discharge maximally during REM sleep, but were silent during NREM and wake episodes21. However, mice with a genomic deletion of the MCH22 and the MCH receptor 1 (MCH-R1)23 gene showed discrepant phenotype. Thus, it remains unclear whether MCH neurons are causally involved in the regulation of REM sleep.
Therefore, to characterize the role of the MCH neurons in regulating REM sleep state, we generated a new transgenic Tg(PMCH-Cre) driver mouse model and took advantages of recently-developed activating and silencing opsins to manipulate the activity of LH MCH neurons with high temporal resolution in behaving mice.
Results
Genetic targeting of LH MCH neurons
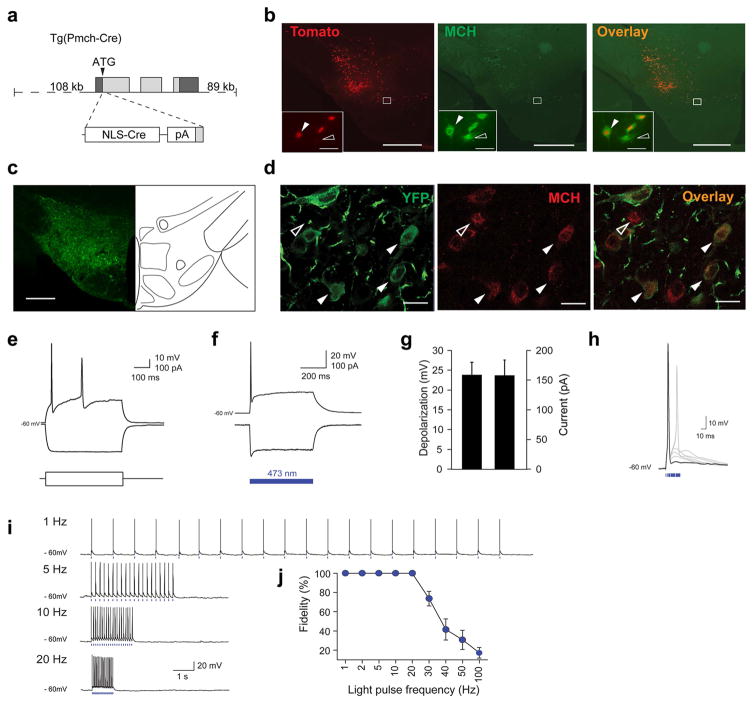
To target optogenetic actuators to MCH neurons located in the lateral hypothalamus and the zona incerta area (LH-ZI), we generated a Tg(Pmch-Cre) mouse using bacterial artificial chromosome (BAC) technology in which the Cre recombinase gene is driven by a ~ 108 kb MCH gene promoter (Fig. 1a). To assess the selectivity of this transgene, Tg(Pmch-Cre) mice were crossed with cre-dependent tdTomato reporter (Rosa-CAG-LSL-tdTomato-WPRE) and 83.6 ± 4.1 % of MCH-immuno-positive cells were found to express TdTomato in the LH, whereas 97.9 ± 0.4 % of TdTomato cells expressed MCH peptide (Fig. 1b and Online Methods). After stereotactic injection of Cre-inducible adeno-associated virus AAVdj-EF1-DIO-ChETA-EYFP24–26 into the LH-ZI of Tg(Pmch-Cre) mice, we found that 87.67 ± 2.96 % of MCH-immunopositive cells co-expressed ChETA-EYFP (1672/1912 cells, 3 sections per animal, N = 6 animals), and MCH immunoreactivity could be detected in 89.65±1.08% of ChETA-EYFP expressing cells (1672/1852 cells, 3 sections per animal, N = 6 animals) (Fig. 1d). No EYFP fluorescence was observed in wildtype (Cre-negative) animals following virus transduction, confirming that the expression was selective for MCH-cre neurons. ChETA-EYFP expression persisted as long as six months, and we observed dense ChETA-EYFP-containing MCH terminals within the hypothalamus, septum, raphe nucleus, locus coeruleus and cortex, consistent with previous characterizations27 (Supplementary Fig. S1).
Figure 1. Selective targeting and functional expression of optogenetic tools in MCH neurons.
a, Generation Tg(Pmch-Cre) mice using BAC technology. b, Photomicrograph of a brain section from [Tg(Pmch-Cre) X R26tom] mouse showing colocalization of tdTomato-positive cells (red) with MCH immuno-positive cells (green). Scale bar: 500 μm. Inset, magnification of hypothalamus area. White arrowheads represent MCH+/Cre+ cells. Open arrowhead represent MCH+/Cre– cell. Scale bar: 50 μm. c, Photomicrograph of ChETA-EYFP expression in the hypothalamus. Scale bar: 500 μm. d, Photomicrographs of ChETA-EYFP expression (green) within MCH neurons (red). White arrowheads represent MCH-positive cell expressing ChETA-EYFP. Open arrowhead represents non-transfected MCH neurons. Scale bar: 20 μm. e, MCH neurons transfected with AAVdj-ChETA-EYFP show typical firing-rate adaptation of wild-type MCH neurons in response to excitatory current injection (100 pA)18. f, ChETA-expressing MCH neurons show robust depolarization and spiking (top) upon 500-ms illumination (473 nm light) in current-clamp mode. This depolarization coincided with inward current in voltage-clamp mode (bottom). g, Quantification of voltage depolarization and inward currents of ChETA-expressing MCH neurons upon blue light illumination (n = 7 cells in 7 different slices, n=6 animals). h, Brief pulses of 473-nm light evoke single action potentials in ChETA-expressing MCH neurons. Note that pulse width > 20 ms typically resulted in spike doublets. i, Voltage responses of MCH cell shown in (e) to 20 pulses of blue light delivered at various frequencies (1–20 Hz). Blue bars represent 5-ms light pulses. j, Group data showing ChETA-expressing MCH neurons fidelity response to light pulses at frequencies up to 20 Hz (n = 7cells in 7 different slices, n=6 animals).
To test the response of ChETA-expressing MCH neurons to optical stimuli, we performed whole-cell patch-clamp recordings in acute hypothalamic slices (Fig. e–j). As previously reported for wild-type MCH neurons 28,29, ChETA-EYFP-expressing MCH neurons were not spontaneously active and retained typical resting membrane potential and input resistance (-61.7 ± 1.4 mV and 428.0 ± 63.1 MΩ, respectively; Fig. 1e). Blue light illumination (473 nm) evoked inward photocurrents (−158.2 ± 25.8 pA; Fig. 1f, g), and brief pulses of light (1–10 ms) evoked single action potentials (Fig. 1h) at frequencies up to 20 Hz with high fidelity (Fig. 1i, j). Extracellular recording in anesthetized animals confirmed that optogenetic stimulation of LH MCH neurons evoked spike waveforms similar to spontaneous waveforms (Supplementary Fig. S2).
Activation of MCH neurons stabilizes REM sleep
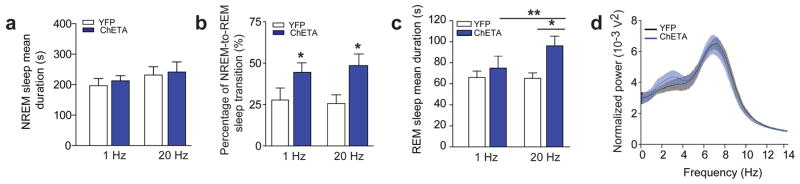
In mammals, sleep is not homogeneous, but comprises of a progression through various states, including NREM and REM sleep. After the onset of NREM sleep, animals switch to either wake or REM sleep states7. REM sleep episodes typically terminate by a transition to wakefulness. To probe the role of MCH neurons in different stages of sleep, we transduced Tg(Pmch-Cre) mice with AAVdj-EF1-DIO-ChETA-EYFP (ChETA) or AAVdj-EF1-DIO-EYFP (control) viruses and implanted electroencephalographic (EEG) and electromyographic (EMG) electrodes, as well as bilateral optical fibers above the LH-ZI area (Online Methods). Genetic and viral manipulations did not disrupt spontaneous sleep-wake cycles (Supplementary Fig. S3) and experiments were conducted during the second half of the light phase (12–7 PM), where REM sleep episodes are most frequent. To investigate the effects of MCH cell activity on NREM and REM sleep, optical stimuli were started at the onset of each sleep state, and stopped upon termination of each sleep state, as detected by real-time EEG/EMG analysis in freely-moving animals (Supplementary Fig. S4 for timeline and representative behavioral responses). Note that optogenetic stimulation patterns (1–20 Hz) were consistent with spontaneous discharge of MCH neurons during REM sleep21. We found that optogenetic stimulation of MCH neurons at 20 Hz or 1 Hz at the onset of NREM sleep episodes did not affect NREM sleep episode duration (p = 0.59 and 0.82 respectively; Fig. 2a), but increased the probability of NREM-to-REM sleep transitions (by ~80% and ~71%, respectively, in ChETA compared to control animals; p= 0.048 and 0.039 respectively; Fig. 2b). In contrast, we found that optical 20 Hz stimulation of MCH neurons at the onset of REM sleep episodes significantly extended their duration (+ 47 %) in ChETA compared to control animals (p =0.014; Fig. 2c), while 1 Hz stimulations had no effect (p = 0.49; Fig. 2c). Importantly, the EEG power spectrum of REM sleep (p = 0.45; Fig. 2d) and the muscular tone during NREM and REM sleep remained unchanged during optical activation of MCH neurons (YFP: 0.44 ± 0.007 mV2 vs. ChETA: 0.42 ± 0.006 mV2; p = 0.07 and YFP: 0.45 ± 0.020 mV2 vs. ChETA: 0.40 ± 0.013 mV2; p = 0.13, respectively).
Figure 2. Optogenetic activation of MCH neurons extends REM, but not NREM sleep duration.
a, Mean duration of NREM sleep upon optogenetic stimulation at 1 Hz and 20 Hz of control (white) and ChETA-EYFP group (blue) animals (n = 6 per group; > 15 stimulations per frequency and per animal). b, Percentage of successful NREM-to-REM sleep transitions upon optogenetic stimulation during NREM sleep shown in (a). Data are shown as a percentage of total number of NREM-to-REM sleep transitions on the total number of stimulation during NREM sleep (n = 6 animals per group). c, Mean duration of REM sleep episodes upon optogenetic stimulation at 1 Hz and 20 Hz of control (white) and ChETA-EYFP (blue) animals (n = 6 per group). Data analysis is based on an average of at least 15 stimulations per frequency and per animal during REM sleep episodes. d, Power spectrum analysis of REM sleep episodes upon 20 Hz optogenetic stimulation of control (black) and ChETA-EYFP (blue) animals (n = 6 per group).
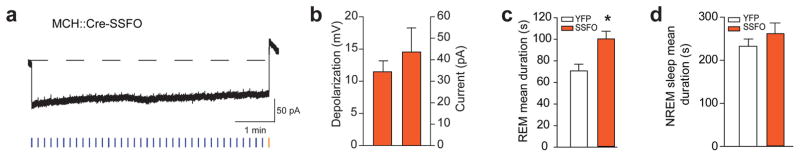
To rule out the possibility that the observed effects of MCH neurons on REM sleep were due to potentially unnatural synchrony among ChETA-driven MCH neurons, we utilized the ChR2 mutant step-function opsin (SSFO)30. Activation of SSFO with a single blue light pulse increases excitability of targeted cells through sustained depolarization, until SSFO is turned off (typically by a yellow light pulse)30. Animals were transduced with AAVdj-DIO-SSFO-EYFP in the LH-ZI area of Tg(Pmch-Cre) with similar efficiency as for ChETA virus (See Online Methods). Using whole-cell voltage-clamp recordings, we found that SSFO-mediated activation of MCH neurons (50-ms light pulses width at 0.1 Hz) induced prolonged inward currents that were rapidly terminated by a single pulse of yellow light (50 ms-pulse width; Fig. 3a). This produced a significant membrane depolarization that could be maintained for several minutes (> 5 min; Fig. 3a, b). To further assess the hyperexcitability of MCH neurons upon SSFO activation, we use intracellular current injection to “replay” a previously recorded current trace while recording the spontaneous synaptic inputs to MCH neurons in whole-cell current-clamp mode. We found that upon SSFO activation, MCH neurons significantly increased their firing activity by ~ 4.5 fold as compared to the de-activated state (p = 0.0008; Supplementary Fig. S5). Importantly, SSFO-induced spikes waveforms were indistinguishable from spontaneously occurring spikes (Supplementary Fig. S5b). Next, we used SSFO to study whether increasing excitability of MCH cells would extend REM sleep duration in freely-moving animals (Online Methods and Supplementary Fig. S4a). Indeed, we found that optical activation of SSFO-expressing MCH neurons during REM sleep significantly increased REM duration (+ 41 %, p =0.022; Fig. 3c), while it had no significant effect during NREM sleep (p = 0.40; Fig. 3d). Together with our previous results (Fig. 2c), these experiments provide evidence that activation of MCH selectively extends the duration of REM, rather than NREM sleep episodes.
Figure 3. SSFO activation of MCH neurons extends REM sleep duration.
a, Whole-cell voltage clamp recording from a MCH neuron expressing SSFO-EYFP shows a prolonged inward current upon blue light pulse (50 ms delivered every 10 s) that is terminated by a 50-ms yellow pulse. b, Quantification of membrane depolarization and inward currents of SSFO-expressing MCH neurons upon optogenetic stimulation (n = 8 cells in 5 slices, n = 3 animals). c, d, Quantification of REM (g) and NREM (h) sleep mean duration upon optogenetic stimulation (50 ms blue pulse delivered every 10 s; termination: 50 ms yellow pulse, see Methods) of EYFP and SSFO-EYFP animals (n = 4 per group). Data analysis is based on an average of at least 10 stimulations per frequency in each mouse during REM and NREM sleep episodes. Mean duration are represented as mean ± SEM. *, p<0.05, **, p<0.01 two-way mixed factorial ANOVA between stimulation condition and viral transduction, followed by Tukey post-hoc test or unpaired two tailed t test.
Silencing of MCH neurons slows REM sleep theta rhythm
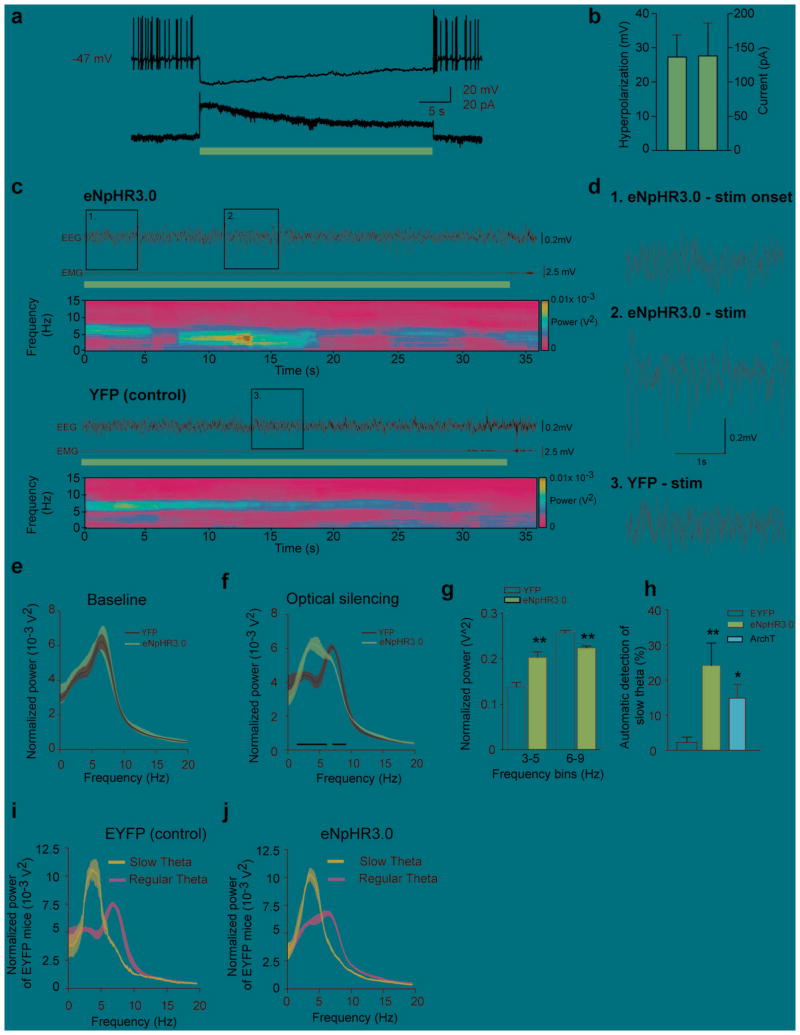
To assess whether activity of MCH neurons is necessary for natural REM sleep, we employed halorhodopsin (eNpHR3.0)31–33 to transiently and reversibly silence MCH neurons. We targeted the expression of eNpHR3.0-EYFP to MCH neurons by injecting AAVdj-DIO- eNpHR3.0-EYFP in the LH-ZI area of Tg(Pmch-Cre) (Online Methods). Using patch-clamp recordings performed in acute hypothalamic slices, we found that yellow illumination (593 nm) resulted in outward photocurrents (steady-state mean: 138.2 ± 48.4 pA; Fig. 4a, b) and a strong hyperpolarization (27.4 ± 6.4 mV; Fig. 4a, b) in eNpHR3.0-expressing MCH neurons. To study the effect of a rapid and reversible silencing of MCH neurons on REM sleep, we bilaterally delivered continuous yellow illumination (593 nm) to the LH-ZI area at the onset of REM sleep episodes (Supplementary Fig. S4a for timeline of the experiment). We found that optogenetic stimulation of MCH neurons during REM sleep did not significantly reduce its duration (eNpHR3.0: 67,2 ± 4.4 s vs EYFP: 60,2 ± 5.2 s, p = 0.337), but rather induced a shift in the dominant theta peak frequencies towards slower oscillations (Fig. 4c,d for representative traces). This was characterized by a significant reduction of 6–9 Hz theta power concomitant to an increase of 3–5 Hz slow theta power (p = 0.003; Fig. 4g), that occurred 8.31 ± 3.72 s after the light onset. Delta rhythms (0.5 – 4 Hz) remained unchanged during silencing of MCH neurons (eNpHR3.0: 0.28 ± 0.015 vs EYFP: 0.24 ± 0.016, p = 0.094). Similar to ChETA and SSFO experiments, muscle tone was not affected by optogenetic stimulation of MCH neurons (eNpHR3.0: 0.15 ± 0.06 vs EYFP: 0.24 ± 0.12, p = 0.481). These results were further confirmed with the use of archaerhodopsin (ArchT)34, a light-driven proton pump (p = 0.017; Fig. 4h, see Supplementary Fig. S6). Indeed, we found that the number of slow theta oscillations was significantly increased (by ~ 10 times) during optogenetic silencing of MCH neurons in eNpHR3.0 compared to control animals (p = 0.010;Fig. 4h). Importantly, the observed slow theta oscillations were not artificial because they occurred during spontaneous REM sleep, although less frequently, in all recorded animals, as detected using an unbiased waveform recognition algorithm (Fig. 4i, j,see Online Methods for criteria). Taken together, these results show that silencing of MCH neurons decreases theta peak frequency (i.e., REM sleep quality), rather than duration, of REM sleep episodes.
Figure 4. Optogenetic silencing of MCH neurons induced shift in the dominant theta peak frequencies towards slower oscillations.
a, eNpHR3.0-expressing MCH neurons show a persistent hyperpolarization (top) and outward current (bottom) upon 30-s constant yellow light illumination in current and voltage clamp, respectively. b, Quantification of membrane hyperpolarization and outward currents of eNpHR3.0-expressing MCH neurons upon optogenetic silencing (n= 8 cells in 5 slices, n=3 animals). c, Representative EEG/EMG recordings and EEG power spectrum during optogenetic silencing (horizontal yellow bars) in eNpHR3.0-YFP (top) and EYFP (bottom) animals during REM sleep. d, Magnification of the boxes in figure (c). Note the high amplitude slow theta oscillations few seconds after the onset of optogenetic silencing in eNpHR3.0 animals. e-f, Average spectral distribution of relative cortical EEG power densities during baseline (e) and optogenetic silencing (f) in EYFP and eNpHR3.0-YFP animals (n = 5 per group; > 15 stimulations per frequency per animal). g, Quantification of slow theta oscillations shown in (f) upon optogenetic silencing of control and eNpHR3.0-YFP animals (n = 5 per group). Normalized power densities are represented as mean ± SEM. *, p<0.05, **p<0.01, unpaired two-tailed t test. h, automatic detection of slow theta oscillations during optogenetic silencing of MCH neurons in control, eNpHR3.0 and ArchT animals. Percentage of detected events are represented as mean ± SEM. *, p<0.05 **, p<0.01, unpaired two-tailed t test between control and eNpHR3.0 or ArchT animals. i, j, Average spectral distribution of slow and regular theta oscillations during optogenetic silencing of MCH neurons in control (i) and eNpHR3.0 (j) animals. Note the similar power spectrum profile between control and eNpHR3.0 animals.
MCH neurons release GABA
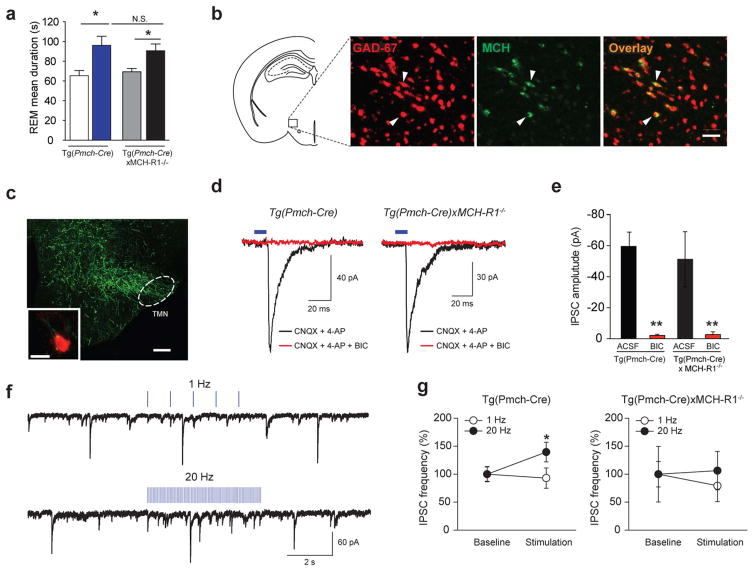
How do MCH neurons modulate downstream sleep-wake circuits? Subpopulations of MCH neurons express the vesicular GABA transporter, cocaine and amphetamine regulated transcript (CART), Nesfatin-1 and/or MCH-related peptides16,20,35–37, however, it remains unclear whether the firing of MCH neurons releases GABA and neuropeptides. First, to determine the role of MCH peptide in sleep, we optogenetically stimulated MCH neurons in mice lacking the MCH receptor 1 [Tg(Pmch-Cre) X MCH-R1−/−] (see Supplementary Fig. S3b for spontaneous sleep-wake cycle). Note that MCH-R1 is the only MCH receptor expressed in mice38. We found that optical activation of MCH neurons at 20 Hz extended REM sleep episodes similarly in both [Tg(Pmch-Cre) X MCH-R1−/−] and Tg(Pmch-Cre) transduced animals (p = 0.020; Fig. 5a), suggesting that GABA and other neurotransmitters/modulators, rather than MCH peptide, are important for acute extension of REM sleep by MCH cell firing. Using fluorescence in situ hybridization (FISH) labeling of GAD-67 mRNA combined with immuno-labeling of MCH (Fig. 5b, Online Methods), we found that 84.86 ± 2.18 % (315/386 cells, N = 9 slices, N = 4 animals) of MCH positive cells were also positive for GAD-67 signal and 17.09 ± 2.06 % of MCH cells that are negative for the GAD-67 signal, suggesting that a large majority of MCH neurons may release GABA.
Figure 5. MCH neurons release the inhibitory transmitter GABA.
a, Mean duration of REM sleep episodes upon 20 Hz optogenetic stimulation of Tg(Pmch-Cre) and [Tg(Pmch-Cre) X MCH-R1−/−] animals transduced with YFP or ChETA-EYFP viruses (n=6 per group; > 10 stimulations per animal). Data are represented as mean ± SEM. *, p<0.05 two-way ANOVA between genotype and viral transduction, followed by Tukey post-hoc test. b, Photomicrograph showing colocalization of GAD-67 transcripts (red) with MCH peptide (green). Scale bar: 100 μm. c, Photomicrograph showing ChETA-EYFP-expressing MCH terminals in TMN area. Scale bar: 200 μm. Inset, ChETA-EYFP-expressing MCH terminals (green) contacting histamine cell (red). Scale bar: 10 μm. d, IPSCs (black) were recorded in TMN histamine and non-histamine neurons (n=3 and 13, respectively) from Tg(Pmch-Cre) (left trace) and [Tg(Pmch-Cre) X MCH-R1−/−] animals (right trace) transduced with ChETA-EYFP viruses. Optically-evoked responses (black traces) were blocked by bicuculline (BIC, red traces). e, Mean amplitude of evoked IPSCs from Tg(Pmch-Cre) (n =9 cells in 7 slices, n=6 animals, left) and [Tg(Pmch-Cre) X MCH-R1−/−] (n=5 cells in 4 slices, n=3 animals, right) before and after bath application of BIC. f, Optogenetic stimulation of ChETA-expressing MCH axons at 20 Hz induced IPSC frequency in histamine neurons in Tg(Pmch-Cre), whereas 1-Hz stimulation did not. g, Mean IPSC frequency upon 20 Hz stimulation in Tg(Pmch-Cre) (left panel, n=10 cells in 7 slices, n=7 animals) and Tg(Pmch-Cre)xMCH-R1−/− (right panel, n= 5 cells in 4 slices, n=3 animals) animals. Mean IPSC amplitudes, latencies, and frequencies are represented as mean ± SEM. *, p<0.05, **, p<0.01, paired samples two tailed t-test.
We then functionally assessed whether MCH neurons release GABA. Anatomical mapping revealed dense ChETA-expressing MCH terminals in the tuberomammillary nucleus (TMN), in close apposition to the wake-promoting histaminergic (HA) neurons (Fig. 5c)39. We functionally mapped this LHMCH→TMN circuit using optogenetics in brain slices from control Tg(Pmch-Cre) and [Tg(Pmch-Cre) X MCH-R1−/−] animals transduced with ChETA virus. In both animal models, optogenetic stimulation of LHMCH→TMN circuit routinely evoked bicuculline-sensitive IPSCs in postsysynaptic HA and non-HA neurons (p < 0.001; Fig. 5d, e; Supplementary Fig. S7). IPSCs latencies were typically short, consistent with monosynaptic connections25,40, and did not differ between Tg(Pmch-Cre) and [Tg(Pmch-Cre) X MCH-R1−/−] slices (Tg(Pmch-Cre) ; 2.16 ± 1.61 ms and [Tg(Pmch-Cre) X MCH-R1−/−] : 5.36 ± 2.90 ms; p = 0.58).
The release of neuropeptides is thought to require the temporal proximity of multiple action potentials41. During optogenetic stimulation, not all light flashes evoked an IPSC, consistent with the low release probability at central synapses40. Nevertheless, in Tg(Pmch-Cre) mice, 20 Hz stimulation increased IPSC frequency relative to baseline (p < 0.05; Fig. 5f and 5g, left panel). However, when repeated in [Tg(Pmch-Cre) X MCH-R1−/−] mice, the effect of 20 Hz stimulation on IPSC frequency was no longer significant (p = 0.16; Fig. 5g, right panel). Because amplitudes of optically-evoked IPSCs were similar in Tg(Pmch-Cre) and Tg(Pmch-Cre)xMCH-R1−/− mice (p > 0.05; Fig. 5g), the effect of MCH-R1 deletion on IPSC frequency suggests a possible presynaptic facilitation of GABAergic transmission by MCH peptide activation of its receptor.
MCH neurons modulate multiple targets
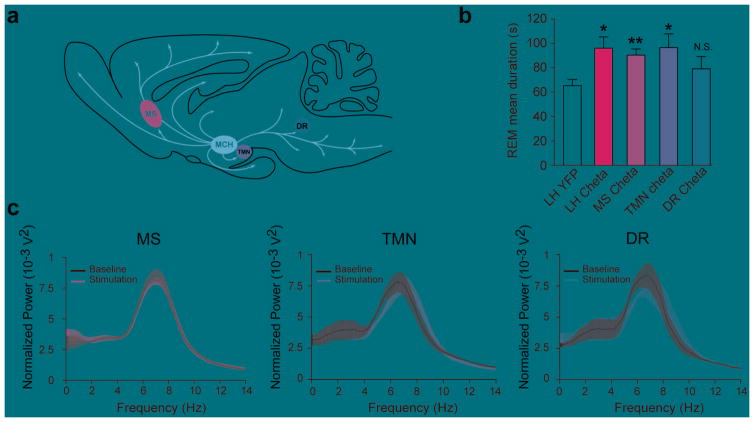
What are the downstream targets of MCH neurons for modulation of REM sleep? Results from this study and others suggest that MCH neurons exert an inhibitory action on postsynaptic targets, including the wake-promoting HA neurons located in the TMN (Fig. 5), as well as other targets located in the septum and dorsal raphe27,42. To test this hypothesis, we optogenetically activated ChETA-expressing MCH terminals in the tuberomammillary nuclei (LHMCH→TMN), medial septum (LHMCH→MS) or dorsal raphe (LHMCH→DR; Fig. 6a, b; see Supplementary Fig. S8 for placement of optical fibers). We showed that optogenetic stimulation of both LHMCH→TMN and LHMCH→MS circuits significantly increased the duration of REM sleep (TMN: + 48% ; p = 0.022, MS: + 38%; p = 0.006, respectively; Fig. 6b) similarly to optical activation of MCH cell bodies in the LH. In contrast, optogenetic stimulation of the LHMCH→DR circuit had no significant effect on REM sleep duration (REM sleep mean duration: 79.14±1 s, p = 0.22; Fig. 6b). The EEG power spectrum of REM sleep episodes remained unchanged during these experiments (MS: p = 0.79; TMN: p = 0.27; DR: p = 0.73; Fig. 6c). These results reveal two distinct downstream circuits through which MCH neurons modulate REM sleep episode.
Figure 6. MCH neurons control REM sleep duration trough multiple pathways.
a, Schematic of the MCH neuron projections in the rodent brain (based on Bittencourt et al (1992). Note the presence of MCH projection to the MS, TMN and DR. b, Mean REM sleep duration of animals stimulated at 20Hz during REM sleep in the TMN (n = 4 animals; bilateral), the DR (n = 4 animals; unilateral) or the MS (n = 6 animals; unilateral). Results of LH stimulation (LH ChETA) from Fig. 2c were reported for comparison. c, Average spectral distribution of relative cortical EEG power density during baseline (black) compared to stimulation (blue) of ChETA-containing MCH terminals within MS (left), TMN (center) and DR (right). Data analysis is based on an average of at least 10 stimulations per frequency for each mouse during REM sleep episodes. Mean duration are represented as mean ± SEM. *, p<0.05, **, p<0.01, unpaired two-tailed t test between YFP animal and ChETA animals in a given target.
Discussion
In this study, we show that acute activation of MCH neurons selectively modulates REM sleep state, using two distinct activating opsins (ChETA and SSFO) for stimulation of MCH neuron firing rates21. Consistent with the neuroanatomical distribution of MCH neurons (see Fig. 6a)27 and the MCH receptor27,43,44, our results suggest that LHMCH→TMN and LHMCH→MS circuits represent two distinct pathways mediating MCH neuron control of REM sleep state. None of the optogenetic manipulations affected the duration of NREM sleep episode, which is consistent with the discharge of MCH neuron during spontaneous REM, rather than NREM, sleep21.
To functionally map the underlying circuit, we first confirmed that MCH neurons express a vesicular GABA transporter16. Then, we showed that electrical activity in MCH neurons elicits GABAA-mediated IPSCs in post-synaptic neurons in vitro, revealing a local inhibitory LHMCH→TMN connection in the hypothalamus. Notably, TMN HA neurons are silent during both NREM and REM sleep but display arousal-specific neuronal activity39, in particular during REM sleep to wake transitions. Thus, we could speculate that activation of LHMCH→TMN circuit extends the duration of REM sleep episode by maintaining an inhibitory tone that delays the reactivation of histamine neurons39, as well as other hypothalamic45,46 and extra-hypothalamic2–6 arousal centers shown to be quiescent during REM sleep.
In agreement with previous studies47,48, we further found that activation of the LHMCH→MS, but not LHMCH→DR, circuit was sufficient to extend the duration of REM sleep episode, suggesting that MCH neurons and provides a functional input to septo-hippocampal circuits where they participate to the stabilization of hippocampus theta rhythm and cortical oscillations (around ~ 7 Hz) during REM sleep (as shown in Fig. 4). This hypothesis is further supported by our silencing experiment showing that rapid and reversible silencing of MCH neurons only during REM sleep episodes reduced the frequency and amplitude of theta oscillations in the hippocampus, which are a hallmark of REM sleep in mammals. Note that the silencing of MCH neurons did not reduced the duration of REM sleep episodes, suggesting that the termination of REM sleep episodes requires additional intra- and/or extra-hypothalamic circuits.
Experimental evidence show that activation of the MCH-R1 enhances cognitive processing during wakefulness49 and facilitates long-term plasticity at CA1 glutamatergic synapse in the hippocampus50, suggesting that MCH neurons may affect sleep-dependent cognitive processing through direct modulation of theta rhythm during REM sleep. However, additional circuits are likely involved and further experiments are now required to identify the mechanisms underlying the regulation of NREM and REM sleep states and their direct relevance in learning and memory.
The persistence of extended REM sleep episodes upon MCH neuron activation in animals lacking known MCH receptors suggests that the MCH peptide may have a minor role in acute regulation of REM sleep, in comparison to long-lasting activation of MCH neurons or MCH-R1. Indeed, previous pharmacological15,17 and optogenetic18 studies reported an increase of both NREM and REM sleep episode duration after cerebral infusion of MCH peptide or chronic optogenetic activation of MCH neurons. In contrast to our results, Konadhode and collaborators also observed an increase of NREM, as well as delta power, upon optogenetic activation of MCH neurons18. This is likely to result from different optogenetic stimulation paradigms or genetic targeting strategies. Based on in vivo data showing that MCH neurons are active mainly during REM sleep21, we used a temporal and sleep state-specific optogenetic stimulation – optical stimulation (1–20 Hz) were delivered at the onset of NREM or REM sleep and terminated at the following behavioral transition. In contrast, the previous study18 used a chronic random stimulation paradigm where optical stimulations were delivered at fixed interval (10 Hz, 1 min every 5 min for 24 h) independently of the behavioral state (i.e., wake, NREM or REM sleep) of the animals. Although we did not observe an extension of NREM sleep episode in our acute experiment, it is possible that chronic activation of MCH neurons results in a long-lasting activation of MCH or GABAA receptors, as well as receptors for transmitters/modulators produced by MCH neurons (Nesfatin-1, CART)20,35,37, that eventually modulate NREM sleep, as reported in pharmacological experiments15,17. Whether this reflects a physiological condition occurring during spontaneous sleep remains to be determined. Furthermore, we generated a new Tg(Pmch-Cre) driver mouse to target the entire MCH neuron population in the LH-ZI area, whereas they used a promoter-specific AAV that limited the transduction of opsin to half of the MCH neuron population (dorsal to the LH). Thus, it is likely that this partial targeting limited their optogenetic modulation of REM sleep, however, we cannot rule out the existence of anatomical subpopulation of MCH neurons with distinct modulatory function on NREM sleep.
Overall, our experiments and the results of others14–16, together with the neuroanatomical distribution of MCH neuron terminals, suggest that MCH neurons represent a subset of inhibitory cells outside the brainstem that stabilize REM sleep state, possibly through a distributed inhibition of arousal centers located in hypothalamus, forebrain and brainstem structures7,12. Further functional dissections of NREM and REM sleep circuits in the mammalian brain will undoubtedly support the identification of selective targets that protect the integrity, the structure and the function of sleep in healthy subjects and pathological conditions.
Online methods
Generation of Pmch-Cre transgenic mice
To restrict Cre expression to MCH neurons, we used a BAC clone containing the full-length pro-melanin-concentrating hormone (Pmch) gene (RP23-129A21) with upstream and downstream flanking sequences of 108kb and 89kb, respectively. BAC DNA was prepared and electroporated into E.coli strain SW102 as required51. An NLS-Cre PolyA construct (pML78, Mark Lewandowski, National Cancer Institute) was targeted to replace the ATG translational start codon of Pmch exon 1 and correct insertion was verified by PCR and sequencing. 5′ recombineering homology: TGAAAGTTTTCATCCAATGCACTCTTGTTTGGCTTTATGCAAGCATCAAA
3′ recombineering homology: CTGCAGAAAGATCCGTTGTCGCCCCTTCTCTGGAACAATACAAAAACGAC (See Fig. 1a). All DNA fragments used for recombineering were generated with the FastStart High Fidelity PCR System (Roche). The modified BAC insert was excised by NotI digestion, purified and used for pronuclear injection.
Heterozygous Tg(Pmch-Cre) mice were maintained on a C57BL/6 genetic background by intercross breeding. Tg(Pmch-Cre) male were used for sleep recordings and optogenetic stimulations, whereas both sex were used for in vitro electrophysiology. No detectable effects on phenotype were found. Mice were housed in individual custom-designed polycarbonate cages at constant temperature (22 ± 1 °C), humidity (30–50%), and circadian cycle (12 h light-dark cycle, starting at 8 AM). Food and water were available ad libitum. Animals were treated according to protocols and guidelines approved by McGill University and the Canadian Council of Animal Care.
Characterization of Pmch-Cre transgenic mice
To determine the specificity of the Cre recombinase expression, Tg(Pmch-Cre) were crossed with Rosa-CAG-LSL-td Tomato-WPRE (line Ai9, Jackson Laboratories). Quantification of co-localization was performed on adjacent sections containing the LH as described below. Some ectopic expression was found in the septum, the cortex and the cerebellum.
Plasmid and viral targeting
Ef1α-DIO-eNpHR3.0-EYFP, Ef1α-DIO-ChETA-EYFP, Ef1α-DIO-SSFO-EYFP or control Ef1α-DIO-EYFP plasmids were kindly provided by Dr K. Deisseroth and packaged in adeno-associated (AAV) DJ serotype52 at Vollum Vector Core. As reported previously3, viruses were stereotactically infused (0.8 μl total at 50 nl/min) bilaterally in the LH (anteroposterior (AP), −1.50 mm; mediolateral (ML), ± 0.9 mm; dorsoventral (DV), 5.35 mm) of Tg(Pmch-Cre) mice anesthetized with isoflurane (5% induction, 1% maintenance). Tg(Pmch-Cre) mice were randomly assigned to viral injection. Viral transduction efficiencies of MCH neurons were similar for all viruses.
In vitro electrophysiology
Slice preparation
P15–18 Tg(Pmch-Cre) were stereotactically injected into the LH (AP, −1.45 mm; ML, +/− 0.5 mm; DV, 5.35 mm). 3–5 weeks after virus transduction, mice were deeply sedated using isoflurane, decapitated, and the brain was rapidly removed and cooled in ice-cold high-sucrose cutting solution containing (in mM): 252 Sucrose, 26 NaHCO3, 2.5 KCl, 4 MgCl2, 1.25 KH2PO4, 0.1 CaCl2, 10 Glucose. All animals were sacrificed during the light phase and the slices were used during the afternoon. Coronal slices were cut using a vibratome (VT1000S, Leica), and allowed to recover for ~1.5 h at room temperature in artificial cerebrospinal fluid (ACSF) containing (in mM): 124 NaCl, 5 KCl, 1.25 NaH2PO4, 2 MgSO4, 2 CaCl2, 26 NaHCO3, and 10 dextrose saturated with 95% O2 and 5% CO2 (pH ~7.3; 300–310 mOsm). Coronal brain slices were transferred to the recording chamber, and perfused continuously with oxygenated ACSF (1.5–2 ml/min) maintained at 22 ± 0.5°C. Cellular morphology and fluorescence were visualized with with an upright fluorescent microscope (BX51WI, Olympus) equipped with a 40× water-immersion objective, differential interference contrast optics, IR DIC and a near-infrared fluorescence camera (EXi Blue, QImaging). Histaminergic neurons were identified based on previously published electrophysiological characteristics5 and immunohistochemical analyses (see below).
Electrophysiological recordings and data analysis
Micropipettes were prepared from borosilicate glass capillaries (1.0 mm OD, 0.58 mm ID) using a horizontal puller (P-97, Sutter Instr.), and had tip resistances between 3–8 MΩ. Somatic whole-cell current and voltage clamp recordings from MCH neurons were obtained using patch recording pipettes containing (in mM): 120 K-gluconate, 20 KCl, 10 N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), 7 phosphocreatine di-Tris, 2 MgCl2, 0.2 ethylene glyco-bis (β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 4 Na2+-ATP, 0.3 GTP-Tris (pH adjusted to 7.20 – 7.26 using KOH; 275–285 mOsm). In TMN experiments, voltage clamp recordings were obtained in patch recording pipettes containing (in mM): 70 K-gluconate, 70 KCl, 2 NaCl, 2 MgCl2, 10 HEPES, 5 QX-314, 1 EGTA, 4 MgATP, 0.3 GTP, and 0.1% biocytin (pH: 7.2–7.26, 275–285 mOsm). Inhibitory postsynaptic currents were routinely recorded in the presence of the AMPAR antagonist, CNQX (20 μM), and low concentrations of 4-AP (10 μM) to increase the reliability of the evoked IPSC4. In vitro electrophysiological data were discarded if intrinsic cell properties (resting membrane potential, input resitance) were more than 3SDs outside group mean, and/or if access resistance changed by >15%, as monitored by series resistance measurements throughout the course of the experiment (mean series resistance was 17 ± 1 MΩ).. Axopatch 700B amplifier was used for all current-clamp and voltage-clamp recordings, and signals were digitized (Axon Instr., Digidata 1322A) and sampled at 20 KHz for storage on hard-disk using pClamp 10.2 software package. Current-clamp recordings were filtered at 10 kHz, and voltage-clamp data were filtered at 2 kHz.
For optogenetic stimulation, square pulses of blue (473 nm) or yellow (593 nm) light, respectively, were delivered using lasers (Laserglow) connected to a 200 μm optical fiber and triggered via a built-in TTL circuit controlled by Clampex 10.3 software package. Light intensity was tested before each experiment, and was calibrated to emit 30–40 mW from the tip of the optical fiber.
Electrophysiological characteristics of MCH neurons were quantified using Clampfit 10.3 software package. Action potential measurements were derived from the first spike in response to a depolarizing intracellular current injection (typically 40–60 pA), and action potential amplitude was calculated from resting membrane potential. Action potential duration and afterhyperpolarizations were calculated from action potential threshold, and input resistance and rectification ratio was calculated in response to a −100 pA and −200 pA hyperpolarizing current pulse, respectively, from a holding voltage of −60 mV. Inhibitory postsynaptic current frequency and amplitude analyses were quantified in Clampfit 10.3 using event threshold detection using predefined criteria (acrit > 7 pA) over three non-consecutive 5-s samples of baseline and stimulation conditions, and all detected events were subsequently visually validated. Membrane potentials reported here were corrected off-line for the liquid junction potential, estimated at ~7.6 mV using JpCalc (Axon Instruments).
In vivo optrode recording
Simultaneous optogenetic stimulation and extracellular recording of LH neurons from Tg(Pmch-Cre) mice transduced with AAVdj-ChETA-YFP were carried out using glass pipette recording, as described elsewhere6. Briefly, recordings were conducted while gradually lowering the pipette (loaded with 0.5 KCl) in small increments to the LH area. The optical fiber was coupled to a 473 nm laser (~ 40 mW). Single unit recordings were recorded in urethane-anesthetized animals. Data were band-pass filtered at 300 Hz low/3 kHz high using an extracellular amplifier (Cygnus technologies) and acquired on a computer using Spike 2 software.
Pharmacological manipulations
Drugs were stored in frozen stock solution, and diluted to final concentrations in ACSF prior to recordings. In TMN experiments, bicuculline methiodide (10 μM) was used to block GABAA receptors. All salts and powders were purchased from Sigma-Aldrich, except for 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), DL-(±)-2-amino-5-phosphonopentanoic acid (AP-5), and bicuculline methiodide (10 μM) (Ascent Scientific).
In vivo polysomnographic recordings
Surgery
As described previously53, 9-weeks Tg(Pmch-Cre) and [Tg(Pmch-Cre) X MCH-R1−/−] animals were chronically implanted with bilateral fiber implants7 above the LH (Doric Lenses; AP, 1.40; ML, 0.95 mm; DV, 4.6 mm) and EEG/EMG connector secured to the skull with C&B Metabond (Patterson dental) and dental cement (Patterson dental). EEG and EMG signals were recorded from 2 pairs of electrodes inserted on the skull and the neck muscle, respectively. After 2 weeks of recovery from surgical procedure and 1 additional week of habituation to the EEG-EMG recording set up, optical patch cords and zirconia sleeves (ID=1.25; Doric Lenses) were connected permanently to the fiber implants. Black nail polish and black furcation tubing were used to blackout light refraction from the implant and the patch cord, respectively, during the optogenetic stimulation.
For circuit mapping experiments, a bilateral optical fiber implant was implanted above the TMN (AP, −2.18mm, ML,+/−1.12 mm and DV, −5.25mm; n = 4). A unilateral optical fiber implant with a 6° angle was implanted above the medial septum (AP,0.86 mm, ML,0.60 mm and DV, −3.62mm ; n = 6). A unilateral optical fiber implant with a 12° angle was implanted above the dorsal raphe (AP, −4 mm, ML,1 mm and DV, −3.40 mm; n = 4). Position of optical fibers was verified for each animals included in this study (see Supplementary Fig. S8). Animals with no viral expression, abnormal sleep-wake cycle (3SD outside group mean) or optical fiber implants outside the target area were discarded from the study. Based on these criteria, 4 ChETA, 2 EYFP, 2 NpHR3.0 transduced Tg(Pmch-Cre) animals and 1 ChETA transduced [Tg(Pmch-Cre) X MCH-R1−/−] animals were excluded from the study.
Polysomnographic Recording
All sleep recordings took place during the second part of the light phase (12–7 PM; light onset: 8 AM). EEG and EMG signals from electrodes were amplified (Grass Instruments) and digitized at 512 Hz (Vital Recorder, Kissei Comtec) as previously described3. Polysomnographic recordings were scored and analyzed using sleep analysis software (SleepSign for Animal, Kissei Comtec). All scoring was performed manually by 5-s epochs as previously described53, 58. Briefly, we defined wakefulness as desynchronized low-amplitude EEG and high tonic EMG activity with phasic bursts; NREM sleep as synchronized, high-amplitude, low-frequency (0.5–4 Hz) EEG and highly reduced EMG activity compared with wakefulness with no phasic bursts; REM sleep as having a pronounced theta rhythm (6–9 Hz) and a flat EMG. As previously described3, polysomnographic scorings were tested by two independent scorers and was found to lie within a 95% confidence interval.
Spectral EEG/EMG analysis
EEG power spectra were computed for 6 to 15 stimulated events per animal (> 20 s duration). Power spectra and time-frequency power spectra were calculated in Matlab with the Chronux toolbox59 using a 5-s sliding window (0.062 s increments). To normalize the data, all power spectral densities at the different frequency ranges, that is, delta (0.5–4 Hz), slow theta (3–5 Hz) or theta (6–9 Hz) were expressed as relative values to the total power of the same event. EMG power was calculated based on a 30–100 Hz range frequency.
Unbiased automatic detection of an increase of power in the 3–5 Hz frequency band during REM sleep
We automatically detected epochs when the EEG power in the ‘slow theta’ range was increased based on a detection threshold established during the baseline recording period (Fig. 4h–j). The analysis was performed as follows:
‘theta’ = 6–9 Hz and ‘slow theta’= 3–5 Hz were defined and kept constant for every mouse included in analysis.
For each animal, the average ‘slow theta’/’theta’ power ratio was calculated for all REM epochs (5 s duration each) occurring during baseline recording at the corresponding circadian time point (12–7 PM). Spectral power of each 5-s REM epoch was calculated in Matlab using the Chronux toolbox59 and the ‘slow theta/’theta’ ratio for each epoch was obtained from the summed spectral power from the specified ‘slow theta’ frequency range divided by that of the ‘theta’ frequency range. The average ‘slow theta/’theta’ power ratio and standard deviation for all baseline REM epochs was then calculated. The threshold ‘slow theta’/’theta’ power ratio for each mouse used for subsequent analysis during REM inhibition experiments (below) was defined as the average baseline REM ‘slow theta/’theta’ power ratio + 2 SDs.
For REM silencing experiments, the ‘slow theta’/’theta’ power ratio was calculated for each REM 5-s epoch as described for baseline recordings. Each epoch was identified as having a ‘slow theta’/’theta’ power ratio above or below the defined threshold calculated as described in step 2. Thus, for each mouse the percentage of detected relative to non-detected epochs was quantified and average power spectra values were calculated from all non-detected or detected epochs.
Optogenetic stimulation
All optogenetic stimulations (5-ms pulses; 1–20 Hz) were generated by a waveform generator (Master 9) that triggers two blue-diode lasers (473 nm, LaserGlow). Acute optogenetic stimulation consistently started ~10 s after the onset of a stable NREM or REM sleep episodes and terminated at the next behavioral transitions (See Supplementary Fig. S4c). During the experiment, frequencies of optogenetic stimulations (1 and 20 Hz) were randomly delivered at either NREM or REM sleep onset in Tg(Pmch-Cre) and control mice. For the stimulation of MCH projections in the MS, TMN or DR, 20 Hz stimulations were used. For NREM sleep experiments, we chose to discard micro-arousals (< 1s) during NREM sleep while stimulating and terminated stimulation only after the onset of a REM or a wake event > 5 s. To circumvent overstimulation of MCH neurons during prolonged NREM sleep period, 5-ms light pulses (1 Hz or 20 Hz) were delivered for 5 s every 10 s and this cycle was repeated until the animal exhibited a state transition (See Supplementary Fig. S4b). For SSFO experiment, we applied 50-ms blue light pulses every 10 s over the entire NREM or REM sleep episode and a single 50-ms yellow light pulse was used to deactivate SSFO at the end of the epsiode (See Fig. 3a–d). For silencing experiment, constant light delivery was provided by a yellow-light laser (593 nm; LaserGlow) split into two patch cords.
Immunohistochemistry
Mice were deeply anesthetized with ketamine/xylazine/acepromazide (100, 16 and 3 mg per kg, respectively, intraperitoneal injection) and perfused transcardially with 1× PBS-heparine 0.1%, pH 7.4, followed by 4% paraformaldehyde in PBS. Brains were extracted, postfixed overnight in the same fixative at 4 °C, and cryoprotected in PBS- Azide 0.1 %/30 % sucrose for an additional 24 h at 4 °C.
For double immunolabelling (Fig. 1b, d), 25-μm thick brain sections from virally-transduced mice were washed (PBST, 10 min, room temperature), incubated in a blocking solution (PBST/4% bovine serum albumin (BSA), 1 h, room temperature) and then in a goat anti-proMCH (SC14509, Santa Cruz Biotech.) at 1:1000 and in a rabbit anti-YFP (Ab290, Abcam) at 1:5000 in block solution (BSA 4%) at 4 °C for ~48 h. After 3 × 10-min washes in PBST, sections were incubated in a solution of Alexa 555 donkey anti-goat IgG secondary antibody (1:1000; A21432, Invitrogen) or Alexa 488 Donkey anti-rabbit IgG secondary antibody (1:1000, A21206, Invitrogen) in blocking solution (BSA 4%, 1 h, room temperature). Sections were washed 3 × 10min in PBST, mounted and coverslipped with Fluoromount-G (Southern Biotech.) and quantified for MCH-YFP colocalization in adjacent LH sections.
To verify that histamine was expressed in recorded TMN neurons, patch pipettes were filled with 0.1% biocytin (Sigma). Coronal brain slices were transferred to 4% PFA in PBS for >24 hrs and incubated for 30 min in 0.3% PBST for 1 h, rinsed 5× (5 min/wash) in 0.1% PBST, then further incubated in 0.1 % PBST with 1 % BSA for 1h. Slices were then transferred to the same solution containing 1% BSA with rabbit anti-histamine decarboxylase (1:500, Acris EUD2601) at 4º C. Slices were then washed (5× 10 min in 0.1% PBST), and incubated in a secondary antibody (anti-rabbit Alexa 555 and Strepdavidin-B conjugated with Alexa 649) for 1.5 h at room temperature, washed (5 × 10 min in PBS), and mounted on microscope glass.
In situ hybridization and immunocytochemistry
Tg(Pmch-Cre) mice were deeply anesthetized with 5% isoflurane and decapitated. Brains were extracted, quickly frozen in isopentane at −30° C, and stored at −80° C for at least 24h. Coronal brain sections (16 μm) were mounted onto glass slides and stored at −80° C. LH sections were transferred into cold 4% PFA for 20 min, followed by 5 × 5 min in 0.1 M PBS, and then incubated in PBS containing 0.3 % H2O2 for 10 min at room temperature, followed by 5 × 5 min washes in PBS. Acetylation was induced using 0.1M TEA buffer containing 0.25% acetic anhydride for 10 min, followed by 5 × 5 min wash in PBS. Sections were then dehydrated dried for 10 min, and placed in a humid chamber saturated with formamide. Sections were incubated in a pre-hybridization buffer (40 % formamide, 10 mM Tris-HCl, pH 8.0, 200 μg/ml yeast tRNA, 10 % dextran sulfate, 1× Denhardt’s solution, 600 mM NaCl, 1 mM EDTA, pH8.0) for 2 h at 60° C, and subsequently transferred to a hybridization buffer including the antisense- or sense- GAD67 riboprobe (1:1000) for 12h at 60° C, followed by SSC buffer washes under gentle agitation: 5 min in 5X SSC; 1 min in 2X SSC; 30 min in 0.2X SSC buffer containing 40 % formamide at 60° C; 5 min in 0.2X SSC at room temperature, and 5 min in 0.1M PBS. Sections were then blocked with 4% BSA and 0.5% Blocking Reagent (Roche) in PBS for 1 h at room temperature, and subsequently incubated in PBS containing goat anti-proMCH at 1:1000 for 72 h at 4° C, followed by 5 × 5 min washes in PBS. Slides were subsequently incubated in anti-DIG-POD (Roche) at 1:1000 overnight at 4° C, followed by 5 × 5 min washes in PBST. Antibodies were detected by incubation of sections using the TSA-Plus-cy5-fluorescein system (Perkin Elmer) for 10 min, followed by 5 × 5 min washes with PBS, and subsequent incubation in 0.1 M PBS containing 1:200 anti-goat Alexa Flour 488 for 1 h. Sections were then washed 5 × 5 min in PBS, coverslipped with Fluoromount-G before visualization with a fluorescent microscope. Quantification of colocalization was performed on sections containing LH-MCH cells.
Microscopy
Non-confocal images were collected on Axio Observer Carl Zeiss fluorescent microscope using fluorescent reflected light (Fig. 1b, c Fig. 5b, c). Confocal images (Fig. 1d, Supplementary Fig. S1) were collected on a LSM 710 Carl Zeiss confocal microscope. Digital images were minimally processed using Image J, Adobe Photoshop CS3 software or Zen to enhance brightness and contrast. All digital images were processed in the same way between experimental conditions to avoid artificial manipulation between different datasets.
Statistical analysis
Statistical analyses of electrophysiological properties and synaptic responses were assessed using repeated measures ANOVAs, two-tailed paired t-tests, and significant effects were investigated using pairwise multiple comparisons using Student-Newman-Keuls method for parametric data, unless otherwise indicated. In vitro data were tested for normality of the distribution using Kolmogorov-Smirnov test (with a Lilliefors’ correction). The typical alpha level, which was set at 0.050, was used to determine whether or not the data fit normal parametric distributions. Statistical analyses of optogenetic stimulations during sleep were assessed using repeated ANOVAs or two-tailed t test as mentioned throughout the text. Experimental sample size were defined based on previous studies53, 55. No statistical methods were used to pre-determine sample sizes. In vivo data distribution was assumed to be normal but this was not formally tested. Data collection and analysis were not performed blind to the conditions of the experiments. Data are presented as mean ± SEM. All data were analyzed using Prism 5.0 (GraphPad Software), Clampfit 10.3 (Axon Instruments), and Sigmaplot 11 (Systat).
Supplementary Material
Acknowledgments
We thank Drs B. Jones, M.E. Carter and L. de Lecea for helpful comments on a previous version of the manuscript. We thank the Tidis Lab members for their technical help and comments. Optogenetic plasmids were kindly provided by Dr K. Deisseroth. S. D. G. was supported by the Fonds de Recherche du Québec - Santé (FRQS). D. B. and A. A. were supported by the Human Frontier Science Program. A. A. was supported by the Douglas Foundation, McGill University, Canadian Fund for Innovation (CFI), Canadian Research Chair (CRC Tier 2), Canadian Institute for Health Research (CIHR) and the Natural Science and Engineering Council of Canada (NSERC).
Footnotes
Author contribution. All authors designed the experiments. S.J., S.D.G., C.G., M.E., and S. J. R. collected data and performed analysis. M.E and J.F. generated the transgenic mouse model. All authors discussed the results and S. J., S.D.G., D.B. and A.A. wrote the manuscript.
References
- 1.JOUVET M. Research on the neural structures and responsible mechanisms in different phases of physiological sleep. Arch Ital Biol. 1962;100:125–206. [PubMed] [Google Scholar]
- 2.Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep state switching. Neuron. 2010;68:1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGinty DJ, Harper RM. Dorsal raphe neurons: depression of firing during sleep in cats. Brain Res. 1976;101:569–575. doi: 10.1016/0006-8993(76)90480-7. [DOI] [PubMed] [Google Scholar]
- 4.Aston-Jones G, Bloom FE. Activity of norepinephrine-containing locus coeruleus neurons in behaving rats anticipates fluctuations in the sleep-waking cycle. J Neurosci. 1981;1:876–886. doi: 10.1523/JNEUROSCI.01-08-00876.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarley RW, Hobson JA. Neuronal excitability modulation over the sleep cycle: a structural and mathematical model. Science. 1975;189:58–60. doi: 10.1126/science.1135627. [DOI] [PubMed] [Google Scholar]
- 6.Hobson JA, McCarley RW, Wyzinski PW. Sleep cycle oscillation: reciprocal discharge by two brainstem neuronal groups. Science. 1975;189:55–58. doi: 10.1126/science.1094539. [DOI] [PubMed] [Google Scholar]
- 7.Pace-Schott EF, Hobson JA. The neurobiology of sleep: genetics, cellular physiology and subcortical networks. Nat Rev Neurosci. 2002;3:591–605. doi: 10.1038/nrn895. [DOI] [PubMed] [Google Scholar]
- 8.Sapin E, et al. Localization of the brainstem GABAergic neurons controlling paradoxical (REM) sleep. PLoS ONE. 2009;4:e4272. doi: 10.1371/journal.pone.0004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boissard R, Fort P, Gervasoni D, Barbagli B, Luppi PH. Localization of the GABAergic and non-GABAergic neurons projecting to the sublaterodorsal nucleus and potentially gating paradoxical sleep onset. Eur J Neurosci. 2003;18:1627–1639. doi: 10.1046/j.1460-9568.2003.02861.x. [DOI] [PubMed] [Google Scholar]
- 10.Verret L, Fort P, Gervasoni D, Leger L, Luppi PH. Localization of the neurons active during paradoxical (REM) sleep and projecting to the locus coeruleus noradrenergic neurons in the rat. J Comp Neurol. 2006;495:573–586. doi: 10.1002/cne.20891. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Sherman D, Devor M, Saper CB. A putative flip-flop switch for control of REM sleep. Nature. 2006;441:589–594. doi: 10.1038/nature04767. [DOI] [PubMed] [Google Scholar]
- 12.Luppi P-H, Clément O, Fort P. Paradoxical (REM) sleep genesis by the brainstem is under hypothalamic control. Current Opinion in Neurobiology. 2013 doi: 10.1016/j.conb.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, et al. Selective activation of the extended ventrolateral preoptic nucleus during rapid eye movement sleep. Journal of Neuroscience. 2002;22:4568–4576. doi: 10.1523/JNEUROSCI.22-11-04568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clément O, et al. The lateral hypothalamic area controls paradoxical (REM) sleep by means of descending projections to brainstem GABAergic neurons. Journal of Neuroscience. 2012;32:16763–16774. doi: 10.1523/JNEUROSCI.1885-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verret L, et al. A role of melanin-concentrating hormone producing neurons in the central regulation of paradoxical sleep. BMC Neurosci. 2003;4:19. doi: 10.1186/1471-2202-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sapin E, et al. A Very Large Number of GABAergic Neurons Are Activated in the Tuberal Hypothalamus during Paradoxical (REM) Sleep Hypersomnia. PLoS ONE. 2010;5:e11766. doi: 10.1371/journal.pone.0011766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monti JM, Torterolo P, Lagos P. Melanin-concentrating hormone control of sleep-wake behavior. Sleep Med Rev. 2013;17:293–298. doi: 10.1016/j.smrv.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Konadhode RR, et al. Optogenetic stimulation of MCH neurons increases sleep. Journal of Neuroscience. 2013;33:10257–10263. doi: 10.1523/JNEUROSCI.1225-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modirrousta M, Mainville L, Jones BE. Orexin and MCH neurons express c-Fos differently after sleep deprivation vs. recovery and bear different adrenergic receptors. Eur J Neurosci. 2005;21:2807–2816. doi: 10.1111/j.1460-9568.2005.04104.x. [DOI] [PubMed] [Google Scholar]
- 20.Hanriot L, et al. Characterization of the melanin-concentrating hormone neurons activated during paradoxical sleep hypersomnia in rats. J Comp Neurol. 2007;505:147–157. doi: 10.1002/cne.21482. [DOI] [PubMed] [Google Scholar]
- 21.Hassani OK, Lee MG, Jones BE. Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep-wake cycle. Proc Natl Acad Sci USA. 2009;106:2418–2422. doi: 10.1073/pnas.0811400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willie JT, Sinton CM, Maratos-Flier E, Yanagisawa M. Abnormal response of melanin-concentrating hormone deficient mice to fasting: hyperactivity and rapid eye movement sleep suppression. Neuroscience. 2008;156:819–829. doi: 10.1016/j.neuroscience.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adamantidis A, et al. Sleep architecture of the melanin-concentrating hormone receptor 1-knockout mice. Eur J Neurosci. 2008;27:1793–1800. doi: 10.1111/j.1460-9568.2008.06129.x. [DOI] [PubMed] [Google Scholar]
- 24.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX Switch Targets Channelrhodopsin-2 to Multiple Cell Types for Imaging and Long-Range Circuit Mapping. Journal of Neuroscience. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunaydin LA, et al. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 27.Bittencourt JC, et al. The melanin-concentrating hormone system of the rat brain: an immuno- and hybridization histochemical characterization. J Comp Neurol. 1992;319:218–245. doi: 10.1002/cne.903190204. [DOI] [PubMed] [Google Scholar]
- 28.van den Pol AN, Acuna-Goycolea C, Clark KR, Ghosh PK. Physiological properties of hypothalamic MCH neurons identified with selective expression of reporter gene after recombinant virus infection. Neuron. 2004;42:635–652. doi: 10.1016/s0896-6273(04)00251-x. [DOI] [PubMed] [Google Scholar]
- 29.Burdakov D, Gerasimenko O, Verkhratsky A. Physiological changes in glucose differentially modulate the excitability of hypothalamic melanin-concentrating hormone and orexin neurons in situ. J Neurosci. 2005;25:2429–2433. doi: 10.1523/JNEUROSCI.4925-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 31.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 32.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS ONE. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elias CF, et al. Characterization of CART neurons in the rat and human hypothalamus. J Comp Neurol. 2001;432:1–19. doi: 10.1002/cne.1085. [DOI] [PubMed] [Google Scholar]
- 36.Del Cid-Pellitero E, Jones BE. Immunohistochemical evidence for synaptic release of GABA from melanin-concentrating hormone containing varicosities in the locus coeruleus. Neuroscience. 2012;223:269–276. doi: 10.1016/j.neuroscience.2012.07.072. [DOI] [PubMed] [Google Scholar]
- 37.Fort P, et al. The satiety molecule nesfatin-1 is co-expressed with melanin concentrating hormone in tuberal hypothalamic neurons of the rat. Neuroscience. 2008;155:174–181. doi: 10.1016/j.neuroscience.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 38.Tan CP, et al. Melanin-concentrating hormone receptor subtypes 1 and 2: species-specific gene expression. Genomics. 2002;79:785–792. doi: 10.1006/geno.2002.6771. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K, Lin JS, Sakai K. Neuronal activity of histaminergic tuberomammillary neurons during wake-sleep states in the mouse. J Neurosci. 2006;26:10292–10298. doi: 10.1523/JNEUROSCI.2341-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schöne C, et al. Optogenetic probing of fast glutamatergic transmission from hypocretin/orexin to histamine neurons in situ. Journal of Neuroscience. 2012;32:12437–12443. doi: 10.1523/JNEUROSCI.0706-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Pol AN. Neuropeptide Transmission in Brain Circuits. Neuron. 2012;76:98–115. doi: 10.1016/j.neuron.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagos P, Torterolo P, Jantos H, Monti JM. Immunoneutralization of melanin-concentrating hormone (MCH) in the dorsal raphe nucleus: effects on sleep and wakefulness. Brain Res. 2011;1369:112–118. doi: 10.1016/j.brainres.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 43.Saito Y, Cheng M, Leslie FM, Civelli O. Expression of the melanin-concentrating hormone (MCH) receptor mRNA in the rat brain. J Comp Neurol. 2001;435:26–40. doi: 10.1002/cne.1191. [DOI] [PubMed] [Google Scholar]
- 44.Chee MJS, Pissios P, Maratos-Flier E. Neurochemical characterization of neurons expressing melanin-concentrating hormone receptor 1 in the mouse hypothalamus. J Comp Neurol. 2013;521:2208–2234. doi: 10.1002/cne.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MG, Hassani OK, Jones BE. Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle. Journal of Neuroscience. 2005;25:6716–6720. doi: 10.1523/JNEUROSCI.1887-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mileykovskiy BY, Kiyashchenko LI, Siegel JM. Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron. 2005;46:787–798. doi: 10.1016/j.neuron.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu ZH, et al. Melanin concentrating hormone induces hippocampal acetylcholine release via the medial septum in rats. Peptides. 2013;44:32–39. doi: 10.1016/j.peptides.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 48.Wu M, Dumalska I, Morozova E, van den Pol A, Alreja M. Melanin-concentrating hormone directly inhibits GnRH neurons and blocks kisspeptin activation, linking energy balance to reproduction. Proc Natl Acad Sci USA. 2009;106:17217–17222. doi: 10.1073/pnas.0908200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamantidis A, et al. Disrupting the melanin-concentrating hormone receptor 1 in mice leads to cognitive deficits and alterations of NMDA receptor function. Eur J Neurosci. 2005;21:2837–2844. doi: 10.1111/j.1460-9568.2005.04100.x. [DOI] [PubMed] [Google Scholar]
- 50.Pachoud B, et al. Major impairments of glutamatergic transmission and long-term synaptic plasticity in the hippocampus of mice lacking the melanin-concentrating hormone receptor-1. Journal of Neurophysiology. 2010;104:1417–1425. doi: 10.1152/jn.01052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic acids research. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimm D, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagode DA, Tang AH, Karson MA, Klugmann M, Alger BE. Optogenetic release of ACh induces rhythmic bursts of perisomatic IPSCs in hippocampus. PLoS ONE. 2011;6:e27691. doi: 10.1371/journal.pone.0027691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schöne C, et al. Optogenetic probing of fast glutamatergic transmission from hypocretin/orexin to histamine neurons in situ. Journal of Neuroscience. 2012;32:12437–12443. doi: 10.1523/JNEUROSCI.0706-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sparta DR, et al. Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits. Nat Protoc. 2012;7:12–23. doi: 10.1038/nprot.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franken P, Chollet D, Tafti M. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21:2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bokil H, Andrews P, Kulkarni JE, Mehta S, Mitra PP. Chronux: a platform for analyzing neural signals. J Neurosci Methods. 2010;192:146–151. doi: 10.1016/j.jneumeth.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.