Abstract
Vascular malformations are non-neoplastic expansions of blood vessels that arise due to errors during angiogenesis. They are a heterogeneous group of sporadic or inherited vascular disorders characterized by localized lesions of arteriovenous, capillary, or lymphatic origin. Vascular malformations that occur inside bone tissue are rare. Herein, we report loss-of-function mutations in ELMO2 (which translates extracellular signals into cellular movements) that are causative for autosomal-recessive intraosseous vascular malformation (VMOS) in five different families. Individuals with VMOS suffer from life-threatening progressive expansion of the jaw, craniofacial, and other intramembranous bones caused by malformed blood vessels that lack a mature vascular smooth muscle layer. Analysis of primary fibroblasts from an affected individual showed that absence of ELMO2 correlated with a significant downregulation of binding partner DOCK1, resulting in deficient RAC1-dependent cell migration. Unexpectedly, elmo2-knockout zebrafish appeared phenotypically normal, suggesting that there might be human-specific ELMO2 requirements in bone vasculature homeostasis or genetic compensation by related genes. Comparative phylogenetic analysis indicated that elmo2 originated upon the appearance of intramembranous bones and the jaw in ancestral vertebrates, implying that elmo2 might have been involved in the evolution of these novel traits. The present findings highlight the necessity of ELMO2 for maintaining vascular integrity, specifically in intramembranous bones.
Introduction
Embryonic development of blood vessels involves two sequential processes: vasculogenesis and angiogenesis. Vasculogenesis is the de novo vascular growth from mesoderm-derived angioblasts and the subsequent formation of the primary capillary plexus in vascular islets.1 Angiogenesis refers to the subsequent process whereby the primary capillary plexus undergoes remodeling and extension via endothelial cell (EC) proliferation and vascular smooth muscle cell (vSMC) recruitment to form mature blood vessels.2 Errors in molecular control of angiogenesis cause vascular anomalies—localized lesions of arteriovenous, capillary, or lymphatic origin. The International Society for the Study of Vascular Anomalies (ISSVA) classifies vascular anomalies into two major categories: tumors and malformations, according to their clinical behavior and endothelial properties.3, 4, 5 Vascular tumors are characterized by actively proliferating neoplastic ECs, whereas vascular malformations are non-neoplastic abnormal expansions of vascular tissue, without prominent endothelial proliferation.3 The most common vascular tumor—infantile hemangioma—is a rapidly growing benign tumor that regresses during the first decade of life. In contrast, vascular malformations never regress and grow proportionally with age.
Both sporadic and inherited forms of vascular malformations have been described.5 Hereditary vascular malformations are caused by dominant mutations in several genes, with some indication of the contribution of a second-hit, whose protein products regulate EC-vSMC communication, as well as recruitment and migration of vSMCs to the vascular bed.6, 7, 8 Mutations in TEK (MIM: 600221) cause multiple cutaneous and mucosal venous malformations (MIM: 600195), mutations in GLML (MIM: 601749) cause glomuvenous malformation (MIM: 138000), mutations in RASA1 (MIM: 139150) cause capillary malformation-arteriovenous malformation (MIM: 608354) and Parkes-Weber syndrome (MIM: 608355), mutations in ENG (MIM: 131195) and ACVRL1 (MIM: 601284) cause hereditary hemorrhagic telangiectasia type 1 (MIM: 187300) and type 2 (MIM: 600376), respectively, and mutations in KRIT1 (MIM: 604214), CCM2 (MIM: 607929), and PDCD10 (MIM: 609118) cause cerebral cavernous malformations type 1 (MIM: 116860), type 2 (MIM: 603284), and type 3 (MIM: 603285), respectively.
Intraosseous vascular malformations are rare abnormalities that account for <0.2% of all bony tumors.9 They are almost exclusively described in sporadic cases involving the skull and vertebral column; however, we identified the first two autosomal-recessive families with primary intraosseous vascular malformation (VMOS [MIM: 606893]), which severely affected cranial bones.10, 11 The malformation is characterized by severe and progressive blood vessel expansion within the craniofacial bones, variably accompanied by midline abnormalities such as diastasis recti and supraumbilical raphe.10 Prior to the onset of puberty, the vascular malformation and bone enlargement is restricted to the mandibular and maxillary region; thereafter, rapid expansion occurs with extension to all cranial bones. Clinically, the gradual increase in intracranial pressure or massive bleeding—either spontaneous or induced by surgery—can be life threatening. Treatment is extremely challenging, because embolization, sclerotherapy, or surgical manipulations are effective only for slowing disease progression. The facial appearance of individuals with VMOS and their CT images resemble cherubism, as mandibular bone is replaced by excessive amounts of fibrous tissue.10, 12, 13 Pathological findings are critical for differential diagnosis, because there are no pathognomonic radiographic findings for VMOSs.11, 13, 14
Various terms have been used to describe malformations similar to VMOSs, including intraosseous cavernous hemangioma, extraspinal osseous hemangioma, central hemangioma, cavernous angiomata of skull, and cystic angiomatosis.12, 14, 15, 16 Recently, the nomenclature of vascular bone lesions was updated and earlier cases were re-classified according to the new ISSVA classification.4, 5, 17 With this reclassification, the diverse and sometimes contradictory terminology used for intraosseous vascular lesions was largely standardized; however, because no mode of inheritance for the aforementioned malformations were addressed, it remains unclear whether VMOS and similar conditions reported in the literature have a common genetic etiology.18
The present study used homozygosity mapping and massively parallel sequencing in combination to identify four distinct germline mutations in ELMO2 (MIM: 606421), which encodes Engulfment and cell motility protein 2 (ELMO2), in five consanguineous families affected by VMOS. Functional testing using affected cells showed that there was a significant reduction in ELMO2 transcript and loss of ELMO2 with concomitant downregulation of DOCK1. Overexpressed ELMO2 mutant proteins derived from an affected individual’s mutant transcripts were unable to stably recruit interacting protein DOCK1 or fully enhance downstream RAC1 activation. The fibroblasts from an affected individual showed a deficiency in cell migration that could be partially rescued by exogenous ELMO2. Attempts to model VMOS in zebrafish by generating CRISPR/Cas9-elmo2 null alleles showed that unlike in humans, elmo2 might not be essential in zebrafish for proper vasculature development and homeostasis.
Material and Methods
Study Participants
The study included 21 individuals, of which 8 were affected with VMOS, from two original Turkish families (families A and B) plus one Saudi Arabian (family C), one Iraqi (family D), and one North American (family E) family (Figure 1).10 Skin biopsies were obtained from three members of family A (individuals I:1, II:2, and II:3) and bone biopsies were obtained for pathological examination from A-II:3 during surgical treatment. Genomic DNA was extracted via standard protocols from peripheral blood of the affected individuals and other available family members. The study protocol was approved by the Hacettepe University Ethics Committee (FON 00/9-4) and the National University of Singapore Institutional Review Board (NUS IRB 10-051). The study was conducted in accordance with the Declaration of Helsinki and written informed consent was obtained from the participants.
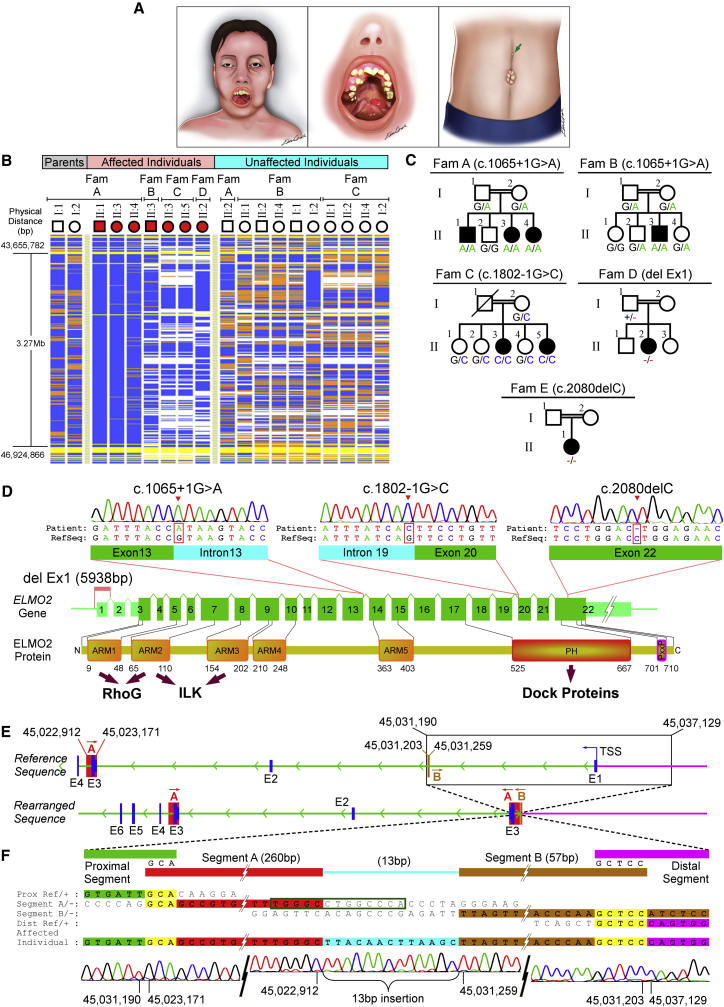
Figure 1.
Identification of the ELMO2 Mutations that Cause VMOS
(A) Illustration of the morphological VMOS findings. Maxillomandibular enlargement is evident, both extraorally and intraorally. Facial asymmetry due to bone enlargement is shown on the left. Exophthalmos and loss of vision usually accompany the disease in later stages. Ectopic eruption, impaction, and displacement of the teeth, as well as severe alveolar bone expansion are common findings in intraoral examination (middle). Supraumbilical raphe (green arrow) and umbilical hernia are the primary extraosseous findings in VMOS (right).
(B) Homozygosity mapping via VIGENOS showed a 3.27 Mbp candidate region in chromosome 20q13. Homozygous genotypes identical to the genotype data obtained from the affected individual A-II:1 are shown in blue. Contrasting homozygote genotypes are shown in white, whereas heterozygous genotypes appear in orange. Non-informative genotypes resulting from heterozygous SNPs in parent-child trios are shown in yellow. Note that all affected individuals are homozygous for the candidate region; however, each of the four families have a different haplotype for this interval.
(C) Pedigrees of families A–E. Mutations found in ELMO2 are shown for each family and the genotypes for the corresponding mutation are indicated below each individual whose DNA samples were available.
(D) Schematic representation of homozygous ELMO2 mutations co-segregating with VMOS in the five families. ELMO2 contains a total of 22 exons (green boxes). The untranslated regions of the exons are denoted with smaller light green boxes. In families A and B, the c.1065+1G>A mutation substitutes the first nucleotide of the 13th intron in the splice donor site. In family C, the c.1802−1G>C mutation substitutes the last nucleotide of the 19th intron in the splice acceptor site. In family D, a complex rearrangement involving a 5,938 bp deletion removes the first exon of ELMO2. In family E, the c.2080delC mutation deletes one cytosine, which leads to a frameshift predicted to produce a longer protein. Below is a schematic representation of the ELMO2. ELMO2 protein domains are linked by arrows to their interacting proteins. Abbreviations are as follows: ARM, Armadillo repeat; PH, pleckstrin homology domain; PxxP, proline-rich motif.
(E) Complex rearrangement schematized on the genomic sequence. The upper line illustrates the 5′ end of wild-type ELMO2. The exons are shown as blue boxes, and segments A and B, which represent inserted segments in the complex rearrangement (see Figure S1A), are shown as red and light brown boxes, respectively. The green arrowheads indicate the direction of transcription. The black rectangle shows the deleted portion of the genome. Abbreviation: TSS, transcription start site. The bottom line shows the rearranged sequence. Note that inserted sequences A and B are in reverse orientation (inverted). Genomic positions are indicated where applicable.
(F) Enlarged view of the complex rearrangement region. The upper part represents the sequence alignment of the breakpoint junctions, showing the homology with four genomic regions, namely proximal segment (green) (centromeric end of the breakpoint), inverted segment A (red), inverted segment B (light brown), and distal segment (purple) (telomeric end of the breakpoint). Two of the breakpoint junctions share 3-bp- and 5-bp-long microhomology sequences (shown in yellow), whereas a 13-bp-long joining segment shown in blue joins segments A and B. Nucleotide sequences homologous to RefSeq are depicted in bold. The green box is a 13-bp-long inverted repeat sequence at the breakpoint junction of segment A. The lower part shows the corresponding Sanger sequences for the three breakpoint junctions. The genomic positions of the nucleotides in RefSeq are indicated below the electropherogram.
Also see Figure S2 for details.
Identification of Causative Mutations
Genome-wide genotyping was performed with the Affymetrix Human Mapping 250K NspI Array, according to the manufacturer’s protocol, in 7 affected and 12 unaffected individuals from families A–D (Figures 1B and 1C). Genotype files (.chp files) were generated with GTYPE software (Affymetrix) and were transferred to VIGENOS (Visual Genome Studio) (Hemosoft) to construct genome-wide haplotypes.19, 20 Haplotype blocks indicating homozygosity-by-descent were compared between affected and unaffected individuals. Because homozygosity mapping indicated a single candidate region on chromosome 20q13.12, targeted massive parallel sequencing of the critical interval (hg19: chr20: 43,655,782–46,924,866) was performed using four affected individuals, one from each family (A-II:3, B-II:3, C-II:3, and D-II:2). This sequencing service was purchased from the University of Wisconsin-Madison Biotechnology Center, Genome Center of Wisconsin. In brief, the Illumina Paired-End Sample Preparation Kit was used to construct genomic libraries. The genomic fragments within the critical interval were captured from this library using the Agilent Custom SureSelect Kit. The enriched genomic fragments were sequenced via Illumina Genome Analyzer IIx as 75 bp paired-end reads.
Whole-exome sequencing was employed independently for the detection of variants in family members and to confirm the targeted sequencing results. High-quality DNA available from 14 individuals in 3 families (family A: I:1, I:2, II:2, II:3, and II:4; family B: I:1, I:2, II:3, and II:4; family C: I:2, II:1, II:2, II:3, and II:5) were analyzed via whole-exome sequencing in the deep sequencing core facility of the TUBİTAK Advanced Genomics and Bioinformatics Research Center (IGBAM), Kocaeli, Turkey. Exome enrichment was performed by the TruSeq Exome Enrichment Kit (Illumina) and sequenced as 100 bp paired-end reads using an Illumina HiSeq 2000 system. Reads from both targeted sequencing and exome sequencing were aligned to the human reference genome (GRCh37/hg19) via BWA read mapper.21 Genome Analysis Toolkit (GATK) v.1.6 and ANNOVAR were used for variant calling and functional annotation.22, 23 A prioritization scheme was adopted to determine the causative mutation for VMOS (Tables S1 and S2). Integrated Genome Viewer (IGV) was used for visual inspection of targeted sequencing data (Figure S1).24 For de novo assembly of 75 bp paired-end reads from targeted sequencing of individual D-II:1, Exact DE Novo reads Assembler (Edena v.110920) was used (Figure S2).25 The IGBAM in-house exome database (n = 279 individuals) was screened for the defined mutations. Confirmation and segregation of identified ELMO2 variants were performed via Sanger sequencing of all available family members. The sequence of human ELMO2 and variants presented herein refer to the transcript GenBank: NM_133171.4. All primers that were used are listed in Table S3.
Pathological Examination of Human Tissue Samples
Standard 4-μm-thick sections of formalin-fixed, paraffin-embedded tissues were stained with hematoxylin and eosin (H&E) or immunostained. Decalcification of bone specimens was performed using 10% formic acid after fixation. Antigen retrieval was performed using Epitope Retrieval Solution 2 (Leica) prior to all immunostaining procedures, except for anti-smooth muscle actin (SMA) and anti-desmin. Tissue sections were stained with anti-SMA (Thermo, MS113P, 1/800), anti-desmin (Biocare, CM036C, 1/50), anti-h-caldesmon (Thermo, MS1169P1, 1/50), anti-CD31 (DBS, MOB034, 1/20), and anti-Ki-67 (Leica, NCLLKi67MM1, 1/200), according to the manufacturers’ protocol, for 15 min at 37°C using a Leica Bond Polymer Refine Detection Kit and Leica Bond Max Autostainer. Antibody concentrations were adjusted with appropriate negative and positive controls. Images were obtained with a SPOT Insight Camera attached to an Olympus BX51 microscope, and were processed by SPOT v.4.1 software (Diagnostic Instruments) and ACDSee Photo Editor 2008 v.291 (ACD Systems).
Tissue and Cell Culture
Primary dermal fibroblast cultures were established from skin biopsies obtained from members of family A. In brief, biopsy samples were grown in DMEM (Biological Industries) supplemented with 10% fetal bovine serum (Biological Industries) and 2 mM L-glutamine (Biological Industries). Subsequent fibroblast passages as well as HEK293T cells were maintained in high-glucose DMEM (HyClone, SH30243.01) supplemented with 10% fetal bovine serum (HyClone, SH30070.03) and 1% Pen Strep (GIBCO, 15140-122). Human iPSCs were generated according to published protocols (Figure S3).26
RT-PCR and Quantitative PCR
RNA from fibroblasts and iPSCs was extracted with the RNeasy Mini Kit (QIAGEN). cDNA was synthesized with the iScript cDNA Synthesis Kit (Bio-Rad). RT-PCR of splice variants obtained from an affected individual was performed with primers flanking the mutated splice site (see Table S3 for primer sequences). qRT-PCR was performed with FastStart Universal SYBR Green Master (Roche) and the ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems). Gene expression was normalized to GAPDH for human, and actin for zebrafish samples.
Constructs and Transfections
The open reading frame (ORF) of wild-type ELMO2 was amplified from control cDNA and cloned between the BamHI and XbaI sites of pCS2+. The same method was used to amplify all aberrantly spliced ELMO2 ORFs from cDNA obtained from an affected individual. More than 40 colonies were sequenced and yielded a final tally of 4 mutant constructs—ELMO2675, ELMO2444, ELMO2363, and ELMO2364 (see Figures 2 and S4). Five ELMO2 constructs, including the four mutant and one wild-type ELMO2, were subcloned between the HindIII and NotI sites of pXJ40-mCherry. The open reading frame of DOCK1 was cloned between the XhoI and NotI sites of pXJ40-GFP using a pCAGGS-DOCK1 construct as the template (gift from Raymond Birge). These constructs were overexpressed in HEK293T cells using FuGENE HD Transfection Reagent (Promega), according to the manufacturer’s protocol, in Opti-MEM I Reduced Serum Medium (GIBCO).
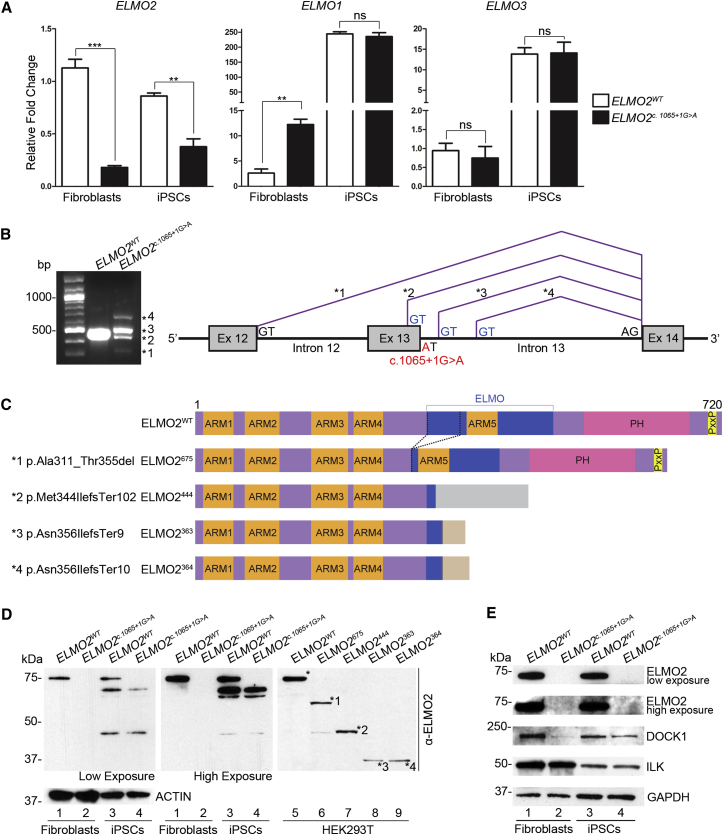
Figure 2.
The ELMO2 c.1065+1G>A Splice Mutation Behaves as a Loss-of-Function Allele
(A) By quantitative PCR, total ELMO2 transcript levels were significantly lower in the fibroblasts and iPSCs of affected individual A-II:3 than in those of control subject A-II:2. ELMO1 transcript levels were significantly upregulated in the affected individual’s fibroblasts, as compared to control subject, but not in iPSCs. ELMO3 transcript levels were the same in fibroblasts and iPSCs of affected and control subjects. Data are shown as mean ± SEM, unpaired two-tailed t test. ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ns: p ≥ 0.05.
(B) RT-PCR analysis of ELMO2 cDNA flanking the splice mutation site shows the presence of at least four alternatively spliced transcripts in affected individual A-II:3’s fibroblasts, as compared to control subject A-II:2 (left). Schematic diagram of corresponding activated cryptic splice donor sites deduced via cloning and sequencing of ELMO2 ORF from affected individual’s cDNA (right).
(C) Schematic diagram of mutant ELMO2 proteins derived from affected individual A-II:3’s alternatively spliced transcripts, as compared to full-length wild-type ELMO2 (720 amino acids). Abbreviations are as follows: ARM, Armadillo repeat; ELMO, ELMO domain; PH, pleckstrin homology domain; PxxP, proline-rich motif.
(D) Western blot analysis shows that affected individual A-II:3’s fibroblasts and iPSCs do not express wild-type ELMO2 (75 kDa), as compared to control cells. Mutant ELMO2 proteins are not detected, either. Lanes 1 to 4: endogenous protein from affected and control fibroblasts and iPSCs were probed using a mouse monoclonal antibody against ELMO2, with high and low exposures shown. Lanes 3 and 4: multiple smaller bands in iPSC lysates are probably non-specific proteins. Lanes 6 to 9: cloned affected individual’s transcripts were overexpressed in HEK293T cells to show the size of the mutant ELMO2 in comparison to wild-type ELMO2 (lane 5). Asterisks (∗) on the western blot indicate wild-type and mutant ELMO2 (numbered asterisks). The same α-ELMO2 antibody was used for detection.
(E) Endogenous protein (ELMO2, DOCK1, and ILK) levels in affected individual A-II:3’s and control subject A-II:2’s fibroblasts and iPSCs. The DOCK1 level was significantly reduced in affected fibroblasts and to a lesser extent in iPSCs. ILK levels were the same between affected and control subjects. GAPDH was used as the loading control.
Western Blot and Antibodies
Cells were lysed in RIPA extraction buffer supplemented with 1 mM dithiothreitol (DTT) and 1× Protease Inhibitor Cocktail (Roche). Protein concentrations of cleared lysates were measured using the Pierce BCA Protein Assay Kit and equal quantities of protein were loaded on precast 7% or 10% SDS-polyacrylamide gels (Bio-Rad). Transferred PVDF membranes were blotted with the following antibodies: mouse anti-ELMO2 (Santa Cruz cat# sc-365739; RRID: AB_10846327), goat anti-DOCK1 (Santa Cruz cat# sc-6167; RRID: AB_2261864), rabbit anti-ILK (Millipore cat# 04-1149; RRID: AB_1977290), mouse anti-GAPDH (Santa-Cruz cat# sc-47724; RRID: AB_627678), and mouse anti-ACTIN (Millipore cat# MAB1501R, RRID: AB_11214116). Secondary anti-mouse-HRP, anti-rabbit-HRP, and anti-goat-HRP were used at 1:4,000 dilution before visualization on X-ray film using SuperSignal West Pico or Dura Chemiluminescent Substrate (ThermoScientific).
Fluorescence Lifetime Imaging Microscopy
Time-domain FLIM experiments were performed using a Time-Correlated Single Photon Counting (TCSPC) system (PicoQuant) attached to an Olympus FV-1000 confocal microscope (Olympus) with a 60 × 1.2 W objective. The excitation light source was a 485 nm pulsed diode laser controlled by a Sepia II (PicoQuant) driver with a dichroic mirror of 488/559 and a 520/530 emission filter. The arrival of individual photons was detected using an SPAD detector and events were recorded by a PicoHarp 300 TCSPC module. Lifetime analysis was performed using SymPhoTime 200 software. Mono- and bi-exponential fittings were applied.
RAC1 Activation Assay
RAC1-GTP levels were measured using a luminescence-based G-LISA Rac1 Activation Assay Biochem Kit (Cytoskeleton), according to the manufacturer’s protocol. In brief, 750,000 HEK293T cells were plated onto 6-cm cell culture dishes pre-coated with poly-L-lysine (Sigma) and then transfected with the indicated constructs. After 24 hr of transfection, the cells were lysed and protein concentrations were equalized across samples to 1 mg/mL via quantification by Precision Red Advanced Protein Assay Reagent. The luminescence signal was measured using a SpectraMax M5 multi-mode microplate reader running SoftMax Pro v.5.3.
Scratch-Wound Assay
Primary fibroblasts from affected individual A-II:3 and control individual A-II:2 were seeded onto 24-well plates and allowed to reach confluency. For each experiment, 4–6 wells were used as technical replicates. In each well, a P200 yellow pipette tip was used to make a central vertical scratch. An orthogonal line perpendicular to the vertical scratch was drawn on the underside of the plate to demarcate the area of measurement. At the indicated time points after scratch, brightfield images were obtained at 10× objective and the wound area was measured by pixel area using ImageJ. Cell migration was evaluated via calculation of the percent decrease in the wound area over time.
Electroporation of Fibroblasts
Transient overexpression in primary fibroblasts was achieved via electroporation of endotoxin-free constructs, as indicated, using the NEPA21 electroporator (Nepa Gene). In brief, 1 million cells were resuspended in 100 μL of Opti-MEM I Reduced Serum Medium (GIBCO). The indicated amount of construct was added, mixed with the cells, and then transferred to a Nepagene 2.0-mm gap electroporation cuvette. The cells were electroporated with the following parameters: Poring pulse: 200 V, 5 ms, 50-ms interval, 2 pulses, 10% D. Rate, + polarity; transfer pulse: 20 V, 50 ms, 50-ms interval, 5 pulses, 40% D. Rate, +/– polarity. Cells were then immediately plated onto 6 cm dishes with media for recovery.
CRISPR-Mediated Zebrafish Knockout
The protocols used for zebrafish fertilization, microinjections, and whole-mount in situ hybridization are available at the Reversade lab protocol website (see Web Resources). Zebrafish were maintained and used according to the Singapore National Advisory Committee on Laboratory Animal Research Guidelines. The Tg(fli1a:EGFP) and Tg(acta2:mCherry) transgenic fish lines were gifts from Nathan Lawson.
Custom gBLOCKs (Integrated DNA Technologies) were designed incorporating a guide RNA-targeting sequence preceded by a T7 promoter sequence. Two guide RNAs were used independently and the targeting sequences on the elmo2 were 5′-GGAGACGCAGCAGAACCCAG-3′ on the forward strand and 5′-GGCCGCCGCTTTCCTCTTCT-3′ on the reverse strand (Figure S5). Guide RNAs were synthesized using a MEGA shortscript Kit (Applied Biosystems) according to the manufacturer’s protocol and were purified with an RNeasy Mini Kit (QIAGEN). Cas9 mRNA was synthesized with an mMESSAGE mMACHINE Kit (SP6) and a NotI-linearized zebrafish codon-optimized Cas9 construct in pCS2+ (gift from Tom Carney). The gRNA and Cas9 mRNA were mixed together to a concentration of 150 ng/μL each, and 2 nL of RNA was injected into the yolk of 1-cell AB zebrafish embryos.
Zebrafish Jaw Assay
Using 0.017% tricaine (3-amino benzoic acidethylester, Sigma), 2-month-old zebrafish were anesthetized. Individual fish were then mounted on a petri dish and observed under a Leica stereomicroscope. Surgical scissors were used to make a complete fracture in the most anterior part of lower jaw (Movie S1). Fish were then immediately placed in clean fish water with methylene blue for recovery and kept isolated for 2 days without food to prevent prophylaxis. Next, the fish were returned to the system and assayed for jaw healing and morphology 1 month later. Images were obtained with a Leica M205 FA stereomicroscope.
Comparative Genomics
The ELMO protein or deduced amino acid sequences from nine species (human, Homo sapiens; mouse, Mus musculus; chicken, Gallus gallus; frog, Xenopus tropicalis; zebrafish, Danio rerio; elephant shark, Callorhinchus milii; Japanese lamprey, Lethenteron japonicum; Florida lancelet, Branchiostoma floridae; and fruit fly, Drosophila melanogaster) were aligned with ClustalX2.27 The sequences were then trimmed and phylogenetic analysis of amino acid sequences was performed with the neighbor-joining method implemented in ClustalX2, with outputs displayed using TreeView (Figure S6).28 Confidence in the phylogeny was assessed via bootstrap re-sampling of the data.
Results
Clinical Features of Intraosseous Vascular Malformation
In total, eight children born to five different consanguineous families from Turkey, Saudi Arabia, Iraq, and the United States were diagnosed with VMOS (Table 1 and Figure 1). Detailed clinical and radiological features of families A and B have been described previously.10, 11 All eight affected individuals appeared to be unaffected at birth, but painless swelling of the mandible began in early childhood and progressed with age.
Table 1.
Clinical Features of Affected Individuals in Five VMOS-Affected Families
| Clinical Findings |
Family A |
Family B |
Family C |
Family D |
Family E |
|||
|---|---|---|---|---|---|---|---|---|
| II:1 | II:3 | II:4 | II:3 | II:3 | II:5 | II:2 | II:1 | |
| Sex | male | female | female | male | female | female | female | male |
| Country | Turkey | Turkey | Turkey | Turkey | Saudi Arabia | Saudi Arabia | Iraq | USA |
| First symptom | enlargement of facial bones | enlargement of facial bones | enlargement of facial bones | enlargement of facial bones | ND | ND | gingival bleeding | gingival bleeding |
| Course | progressive | progressive | progressive | progressive | progressive | progressive | progressive | progressive |
| Facial asymmetry | yes | yes | yes | yes | yes | yes | yes | yes |
| Craniofacial bone involvement | mandible, maxilla, nasal bones, calvaria, sphenoid, and clivus | mandible, maxilla, nasal bones, calvaria, sphenoid, and clivus | mandible, maxilla, nasal bones, calvaria, sphenoid, and clivus | mandible, maxilla, nasal bones, calvaria, sphenoid, and clivus | mandible, maxilla | mandible, maxilla | mandible, maxilla, nasal bones, sphenoid | mandible, maxilla, nasal bones, sphenoid, calvaria |
| Clavicular involvement | yes | yes | yes | NA | ND | ND | no | no |
| Rib involvement | no | no | no | yes | ND | ND | ND | yes |
| Vertebral involvement | no | no | no | yes | ND | ND | no | no |
| History of gingival bleeding | yes | yes | yes | yes | yes | yes | yes | yes |
| Ectopic tooth eruption | yes | yes | yes | yes | yes | yes | yes | yes |
| Exophthalmos | yes | yes | yes | yes | yes | yes | no | yes |
| Vision loss | yes | yes | yes | yes | ND | ND | no | yes |
| Increased intracranial pressure | yes | yes | yes | NA | ND | ND | no | no |
| Anemia | yes | yes | yes | yes | ND | ND | ND | yes |
| Alkaline phosphatase | elevated | elevated | elevated | elevated | ND | ND | ND | normal |
| Angiography findingsa | abnormal staining and late phase capillary pooling | abnormal staining and late phase capillary pooling | abnormal staining and late phase capillary pooling | abnormal staining and late phase capillary pooling and internal carotid aneurysm | ND | ND | abnormal staining and late phase capillary pooling | expansile hypervascular lesions of the calvarial, facial, and orbital bones |
| Treatment of acute bleeding episodes | endovascular embolization and percutaneous sclerotherapy with ethanol | endovascular embolization and percutaneous sclerotherapy with ethanol | endovascular embolization and percutaneous sclerotherapy with ethanol | endovascular embolization and percutaneous sclerotherapy with ethanol | ND | ND | endovascular embolization and percutaneous sclerotherapy with ethanol | endovascular embolization |
| Treatment | interferon alpha-2a treatment (no improvement), full-mouth tooth extraction | mandibulectomy, mandibular reconstruction with fibular graft, ventriculo-peritoneal shunt | extraction of some teeth | full-mouth tooth extraction, obliteration of internal carotid aneurysm | NA | NA | extraction of some teeth, endovascular obliteration of interosseous vascular lakes | full-mouth tooth extraction |
| Supraumbilical raphe, diastasis recti, and umbilical hernia | yes | yes | yes | yes | yes | yes | yes | yes (no umbilical hernia) |
| Other | died due to brain stem herniation | – | – | cryptorchidism, hydrocele, paraplegia, died due to massive bleeding | – | – | – | enlarged aortic arch, repaired ASD, marfanoid habitus, pectus carinatum, petechiae on feet |
Abbreviation is as follows: ND, no data.
Angiographic images were obtained via percutaneous angiography or magnetic resonance angiography.
The first symptom observed by the families was enlargement of facial bones (n = 4) and gingival bleeding (n = 2). Follow-up of the affected individuals in families A and B showed that enlargement of facial bones accelerated during early puberty and caused severe facial asymmetry (Figure 1A). The affected bones included the mandible, maxilla, nasal bones, calvaria, sphenoid, and clivus; the mandible and maxilla were involved in all affected individuals. Less frequently was the involvement of the clavicles (n = 4), ribs (n = 2), and vertebrae (n = 1). Alkaline phosphatase (ALP) levels were elevated in four of the five affected individuals, indicating increased bone turnover. Ectopic tooth eruption also occurred in all affected individuals. Removal of teeth and facial bones slowed disease progress and provided temporary symptomatic relief.
All the affected individuals had a history of gingival bleeding, which required surgical intervention, such as endovascular embolization and percutaneous sclerotherapy. Additionally, five affected individuals from families A, B, and E had chronic microcytic hypochromic anemia and suffered from repeated bleeding episodes. Individual B-II:3 had a chronically elevated lactate dehydrogenase (LDH) level, indicating intravascular hemolysis, probably due to malformed vessels. Unfortunately, spontaneous massive bleeding in individual B-II:3 led to hemorrhagic shock and subsequent death at the age of 27. Radiological imaging findings in family A were described in detail previously.11 In five of the six affected individuals, angiography showed abnormal organization of vessels and late-phase capillary pooling in affected bones, indicating that they are slow-flow lesions of capillary or venous origin. MR angiography in affected individual E-II:1 showed hypervascular lesions of the facial bones.
Growth of the vascular lesions caused bone deformation and enlargement, which led to compression of organ compartments and resulted in various complications. Exophthalmos due to deformation of the orbit was present in seven of the affected individuals, of which five had some loss of vision (Table 1). Lesions of vascular origin extending to the spinal canal resulted in spinal cord compression and paraplegia in affected individual B-II:3. In three affected individuals, compression of the calvarial base and brain caused variably severe increases in intracranial pressure. Increased intracranial pressure resulted in brain herniation and subsequent death in affected individual A-II:1 at 36 years and required a ventriculoperitoneal shunt in affected individual A-II:3. Primary extraosseous findings were supraumbilical raphe, diastasis recti, and umbilical hernia.
Homozygous Mutations in ELMO2 Cause VMOS
Affected individual A-II:1 was chosen for the construction of a genome-wide haplotype (Figure 1B). Haplotypes indicating homozygosity-by-descent were compared with homozygous haplotypes of other affected and unaffected individuals in families A, B, C, and D. This analysis showed that there was a single homozygous region spanning 3.27 Mb between DNA markers rs6065774 and rs401976 on chromosome 20q13.12. This critical interval resides at chr20: 43,655,782–46,924,866 (GRCh37/hg19). One affected individual each from families A, B, C, and D were selected and massive parallel sequencing was performed, targeting the critical homozygous region. The variant filtering and prioritization scheme used is presented in Table S1. dbSNP138 and IGBAM in-house exome databases (n = 279) were used to determine minor allele frequencies in the general population. After filtering out the variants that were of low quality, low coverage, and occurred at a high frequency (minor allele frequency > 0.01) in the general population, we focused on single nucleotide or small deletion/insertion variants that were homozygous and likely to be deleterious (i.e., missense, stop gain/loss, splice site, insertion, deletion, or indel variants). ELMO2 was the only gene with potentially deleterious variants in families A and C (Table S1).
A homozygous c.1065+1G>A mutation affecting an intronic splice donor site was detected in affected individual A-II:4 (Figures 1C and 1D). The same homozygous c.1065+1G>A mutation was also present in family B. SNP-based haplotype comparison did not show a common haplotype for the critical region in families A and B; however, intragenic variant analysis showed a consensus haplotype of approximately 16 kb spanning exons 10–21 of ELMO2 and harboring the c.1065+1G>A mutation. As such, a very distant founder mutation might explain the common mutation in these Turkish families, although recurrent mutations cannot be ruled out.
In family C a homozygous intronic splice acceptor site mutation c.1802−1G>C in ELMO2 caused VMOS (Figures 1C and 1D). This mutation is predicted to disrupt the pleckstrin homology (PH) domain that mediates binding to DOCK proteins, which then activate the small GTPase RAC1 (Figure 1D).29 Family E was included later in this study and Sanger sequencing of ELMO2 in affected individual E-II:1 showed a homozygous single-base deletion, c.2080delC in the last exon, which results in a frameshift (Figures 1C and 1D). The resulting mutant protein p.Leu694Trpfs∗127 is predicted to be longer by an extra 99 amino acids and would lack the carboxy-terminal proline-rich (PxxP) motif as well as the critical Trp713 residue that is a target for phosphorylation by the upstream factor Axl (Figure 1D).30
Whole-exome sequencing in 14 individuals in families A, B, and C also confirmed the results obtained via targeted sequencing (Table S2). Sanger sequencing of the respective mutations in families A, B, C, and E showed that all available parents were heterozygous and siblings were either heterozygous for the respective ELMO2 mutation in the family or homozygous for the wild-type allele (Figure 1C). In family D no single nucleotide or small deletion/insertion variant in ELMO2 was found. Instead, missense variants in OCSTAMP (GenBank: NM_080721) and NCOA3 (MIM: 601937) were detected. Because no small-scale homozygous mutations were found for ELMO2 in family D, unlike in the other four families, copy-number variations (CNVs) in ELMO2 were investigated.
A visual inspection of ELMO2 via Integrated Genomics Viewer (IGV) showed that the region around the first exon of ELMO2 was deleted as part of a homozygous complex rearrangement that was also confirmed and delineated by de novo assembly of the targeted sequencing reads (Figures 1D and S1).24 In brief, 5,172 contigs of the whole region, with a mean length of 600 bp, were obtained when a minimum size of the overlaps was taken as 50% of the read lengths. When focused on chr20: 45,027,684–45,037,538 region, we confirmed a 5,938 bp deletion and a 330 bp insertion, resulting in g.45031191_45037128del5938ins330 mutation (GenBank: KU680992) (Figure S2). This complex rearrangement was confirmed by PCR and the father D-I:1 was heterozygous for this complex rearrangement (Figure S1D). The mother’s DNA and RNA samples from members of family D were unavailable for further analysis.
This rearrangement results in the deletion of exon 1, along with surrounding promoter and intronic sequences, as well as insertion of portions of exon 3, intron 3, and intron 1 in reverse orientation (Figures 1E and S1). In both ELMO2 alleles, the deleted sequence includes the transcription start site and is enriched in transcription factor binding sites as well as histone H3K27Ac marks commonly found in active enhancer sites (ENCODE project at UCSC), likely diminishing ELMO2 transcription substantially. Further delineation of the complex rearrangement showed that the breakpoint junctions of the inserted sequences (260 bp long segment A and 57 bp long segment B shown in Figures 1F and S1) shared microhomology with the breakpoints of the deleted sequence (Figure 1F), which indicates that microhomology-mediated replicative mechanisms may underlie this complex rearrangement.31 It is notable that an inverted repeat is present, overlapping the breakpoint of segment A in event 2 (Figure 1F). This is predicted to result in a naturally occurring strong hairpin structure, also noted in the non-B database, which might serve as a catalyst for event 2, as well as the entire complex rearrangement presented here.32, 33
The c.1065+1G>A Splice Mutation in ELMO2 Behaves as a Loss-of-Function Allele
Functional investigations focused on the c.1065+1G>A allele, due to availability of material and because it was present in four of the eight individuals with VMOS. Endogenous RNA levels of ELMO2, along with ELMO1 and ELMO3, were quantified in affected individual A-II:3 and her unaffected non-carrier brother A-II:2 (control subject) (Figure 2A). Total ELMO2 RNA was significantly reduced in the affected individual’s primary fibroblasts and iPSCs, relative to control cells, probably indicative of nonsense-mediated decay (Figure 2A). The levels of ELMO2 in the unaffected heterozygous father A-I:1 were between those of affected individual A-II:3 and the non-carrier brother (Figure S4A). Total ELMO1 RNA was significantly upregulated in affected individual A-II:3’s fibroblasts, but not in iPSCs, relative to control cells, whereas ELMO3 RNA levels remained unchanged (Figure 2A). The upregulation of ELMO1 mRNA is probably a compensatory response to the loss of ELMO2, in a manner resembling the upregulation of Elmo2 in Elmo1−/− mice.34
To examine sequence changes in ELMO2 transcripts, the open reading frame was amplified from cDNA in affected individual A-II:3 and control A-II:2 fibroblasts. Unlike in control A-II:2 fibroblasts, at least four aberrantly spliced ELMO2 transcripts (labeled ∗1 to ∗4) were detected via RT-PCR in the fibroblasts of affected individual A-II:3 (Figure 2B). Transcript ∗1 had a complete loss of exon 13, resulting in the putative ELMO2 mutein p.Ala311_Thr355del (referred to as ELMO2675). Transcripts ∗2, ∗3, and ∗4 resulted from the activation of cryptic splice donor sites in exon 13 and intron 13, each of which lead to a premature stop codon (Figures 2B, 2C, and S4B), and are predicted to generate p.Met344IlefsTer102 (ELMO2444), p.Asn356IlefsTer9 (ELMO2363), and p.Asn356IlefsTer10 (ELMO2364) muteins, respectively (Figure 2C). These findings indicate that the affected individual A-II:3’s cells cannot express wild-type ELMO2, but instead produce low levels of abnormally spliced transcripts that are likely targeted for degradation.
We next determined whether these aberrant transcripts could be translated into ELMO2 mutant proteins. Via western blotting, endogenous ELMO2 (75 kDa) was readily detected in control subject A-II:2’s cells, but not in affected individual A-II:3’s cells, even after high exposure (Figures 2D and 2E). The four aberrantly spliced ELMO2 transcripts were cloned and overexpressed in HEK293T cells and were observed to be recognized by this antibody (Figure 2D). There was no detectable ELMO2 signal in affected individual A-II:3’s fibroblasts or iPSCs that corresponded to the sizes of ELMO2675 (lane 6), ELMO2444 (lane 7), ELMO2363 (lane 8), and ELMO2364 (lane 9) muteins (Figure 2D), suggesting that these truncated ELMO2 muteins could be unstable. We next searched for interacting proteins DOCK1 and ILK.35, 36 Endogenous DOCK1 levels were significantly lower in the affected individual A-II:3’s fibroblasts than those of control A-II:2’s fibroblasts, whereas ILK levels were the same (Figure 2E). These findings show that the c.1065+1G>A splice mutation not only knocks out endogenous ELMO2 at the protein level, but may also reduce the stability of DOCK1, similar to ELMO1’s role in inhibiting ubiquitylation and degradation of DOCK1.37
Although mutant ELMO2 is not detected in fibroblasts and the skin is spared from disease, craniofacial bones may still express ELMO2 muteins. To rule out this possibility and gain insight into the effects of ELMO2 mutations on cellular processes, we carried out biochemical and cellular assays. First, the interaction between ELMO2 and DOCK1, which is the initiating event for downstream RAC1 activation, was investigated using fluorescence lifetime imaging microscopy (FLIM) in HEK293T cells co-expressing GFP-tagged DOCK1 (GFP-DOCK1) and mCherry-tagged ELMO2 (mCherry-ELMO2) constructs (Figure 3A).36 As expected, wild-type mCherry-ELMO2 caused a significant and stable decrease in the fluorescence lifetime of GFP-DOCK1, indicating a positive interaction (Figure 3A). In contrast, the C-terminally truncated mCherry-ELMO2444 and mCherry-ELMO2363 mutants did not alter the fluorescence lifetime of GFP-DOCK1, indicating abrogated interaction (Figure 3A). The internally truncated mCherry-ELMO2675 also decreased GFP-DOCK1 fluorescence lifetime; however, measurements fluctuated over a relatively wide range (Figure 3A), suggesting an unstable interaction. These results show that three ELMO2 mutant proteins tested can no longer interact stably with DOCK1 and, therefore, are unlikely to initiate downstream signaling.
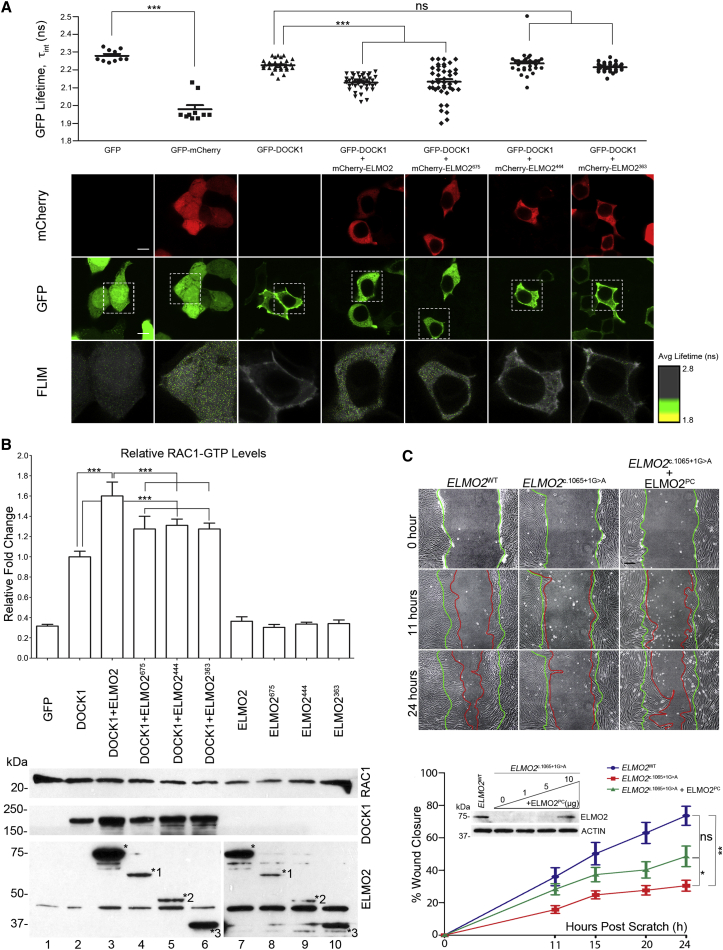
Figure 3.
Functional Analysis of ELMO2 Mutant Proteins
(A) FLIM analysis shows that wild-type ELMO2 interacts stably with DOCK1, but the mutants ELMO2444 and ELMO2363 do not. The mutant ELMO2675 can also interact with DOCK1, but does so in an unstable fashion. The GFP-mCherry fusion protein served as a positive control. Data represented as a scatter dot plot, with mean ± SEM, one-way ANOVA with Bonferroni’s multiple comparison test, ∗∗∗p ≤ 0.001. Dotted white box in each GFP image represents an enlarged version of the corresponding FLIM image. Scale bar represents 10 μm. Bottom: Representative immunofluorescence images show co-localization of DOCK1 and ELMO2 proteins at the plasma membrane and in the cytoplasm.
(B) G-LISA RAC1 activation assay showed that wild-type ELMO2 significantly enhanced RAC1 activation in the presence of DOCK1. Muteins ELMO2675, ELMO2444, and ELMO2363 significantly decreased this activation but did not abrogate it. Data are shown as mean ± SD, one-way ANOVA with Bonferroni’s multiple comparison test. ∗∗∗p ≤ 0.001. Asterisks (∗) on the western blot indicate wild-type and mutant ELMO2 (numbered asterisks).
(C) Affected individual A-II:3’s fibroblasts migrated significantly slower than control fibroblasts in scratch wound assays. This defect was partially rescued by re-expressing exogenous ELMO2 at a level equal to that in control A-II:2’s fibroblasts (western blot inset). Green lines on brightfield images represent wound edge at T0. Red lines represent new wound edge over time. Data are shown as mean ± SEM, two-way ANOVA with Bonferroni post-test. ∗p ≤ 0.05; ∗∗p ≤ 0.01. Scale bar represents 200 μm.
Subsequently, to determine whether ELMO2 muteins could synergize with DOCK1 to enhance RAC1 activation, untagged ELMO2 and DOCK1 were co-expressed in HEK293T cells and RAC1-GTP levels were measured via a G-LISA assay (Figure 3B). Overexpression of DOCK1 alone, but not ELMO2 or its mutants alone, was sufficient to activate RAC1 over basal levels (Figure 3B). This activation was further enhanced in the presence of wild-type ELMO2 (Figure 3B); however, co-expression of DOCK1 with any of the three ELMO2 mutants resulted in reduced enhancement of RAC1 activation but was nonetheless higher than DOCK1 alone (Figure 3B), indicating that these mutants may retain partial activity. Combined, these data indicate that ELMO2 mutant proteins, if stable, do not possess full functionality and are therefore likely to be hypomorphic.
Given the role of the ELMO/DOCK complex in regulating actin dynamics, we sought to determine whether affected individual A-II:3’s primary fibroblasts exhibit a deficiency in cellular migration by means of a scratch wound assay.29, 38 Relative to the control A-II:2’s fibroblasts, affected individual A-II:3’s fibroblasts, which lack wild-type ELMO2 and expressed less DOCK1, migrated twice as slowly toward the middle of the wound (Figure 3C). This delay could be partially rescued by re-expressing wild-type ELMO2 back into affected individual A-II:3’s fibroblasts (Figure 3C). These findings suggest that a specific loss of ELMO2, paralleled by a reduction in DOCK1 in human fibroblasts, significantly compromises cell migration.
Immature Perivascular Cells Surround Aberrant Vascular Channels in VMOS
To further characterize the nature of the vascular lesions, we examined affected individual A-II:3’s mandibular tissue sections. H&E staining showed the presence of irregular, thin-walled, dilated, and engorged vascular channels, accompanied by abundant adipose tissue among bone trabeculae whereas fibular tissue obtained during mandibular reconstruction was normal (Figures 4A and 4B). The abnormal vessels in the mandible were lined by a single layer of ECs highlighted by CD31 staining (Figure 4C). The absence of endothelial multi-layering and negative Ki-67 staining, an indicator of mitotic activity (Figure 4D), excluded a proliferative or neoplastic process, such as hemangioma. These morphological features are consistent with the diagnosis of vascular venous malformation and differ from fibrous dysplasia and other fibrous tumors of the bone in which abundant cellular fibrous tissue between bone trabeculae is diagnostic.
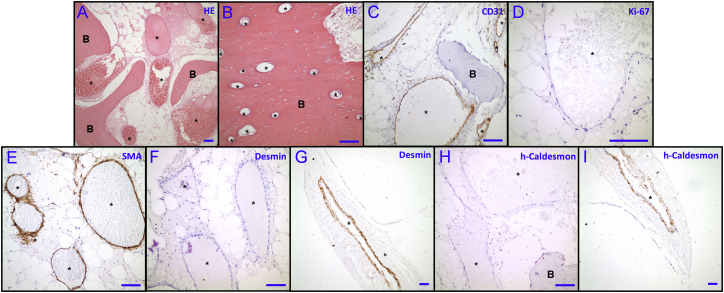
Figure 4.
Histopathological and Immunohistochemical Examination of Affected Tissues
(A and B) Cross-section H&E staining of (A) diseased mandible and (B) normal fibula tissue obtained from affected individual A-II:3.
(C–I) Cross-section of diseased mandible stained from same individual by (C) anti-CD31, (D) anti-Ki-67, (E) anti-SMA, (F, G) anti-desmin, and (H, I) anti-h-caldesmon.
(G and I) Cross-sections from the neighboring tissue with normal blood vessels and serve as positive controls for the immunohistochemistry in (F) and (H).
Symbols: B, bone; ∗, blood vessel lumens. Scale bars in each panel represent 100 μm.
Given that known genetic vascular malformations are often caused by deficiencies in vSMC function, VMOS tissue was stained with markers of mature vSMCs.8 Smooth muscle actin (SMA) expression showed the presence of a thin smooth muscle layer in malformed vessels (Figure 4E); however, desmin (Figures 4F and 4G) and h-caldesmon (Figures 4H and 4I) were absent—both proteins are considered markers for mature vSMCs.39, 40, 41 Similarly, myofibroblast-like desmin negativity in the mural layer of periorbital intraosseous vascular malformations has also been reported.42 Moreover, given that h-caldesmon tethers actin and myosin in smooth muscle cells for regulation of muscle tone, the vSMCs in VMOS are likely to form an immature smooth muscle layer incapable of withstanding blood pressure.43
Zebrafish elmo2 Mutants, Unlike Humans, Are Phenotypically Normal
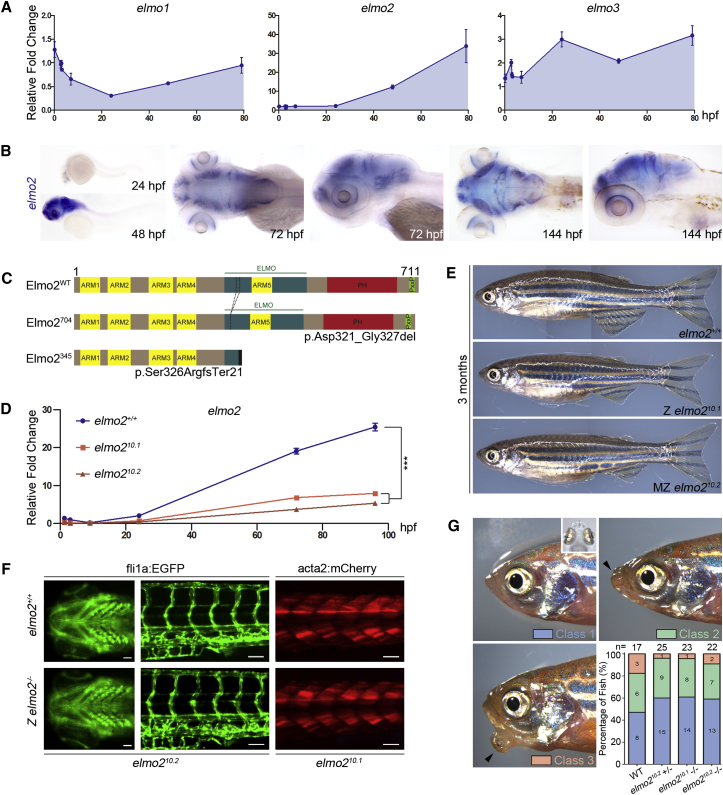
The zebrafish genome contains three paralogs—elmo1, elmo2, and elmo3. Both elmo1 and elmo3 are maternally and zygotically expressed (Figure 5), whereas elmo2 has no maternal contribution and is expressed only at 24 hr post fertilization (Figures 5A and 5B).44 Cells that express elmo2 are localized in the most anterior part of the larvae, most noticeably in the olfactory bulbs, optic tectum, retinal ganglion layer, mid-cerebral vein, upper rhombic lip, and the lower rhombic lip of the hindbrain (Figure 5B).
Figure 5.
elmo2 Knockout Zebrafish Develop Normally to Adulthood
(A) By quantitative PCR, elmo2 is expressed 24 hpf and onward, whereas elmo1 and elmo3 appear to be both maternally and zygotically expressed. Data are shown as mean ± SEM.
(B) By WISH, elmo2 is expressed predominantly in the brain and craniofacial structures, including the upper and lower rhombic lips of the hindbrain, the mid-cerebral vein, optic tectum, olfactory bulbs, and the retinal ganglion layer.
(C) Schematic diagram of zebrafish wild-type Elmo2 and mutants derived from allelic series of elmo2 knockouts. Elmo2704 (p.Asp321_Gly327del) corresponds to the elmo221 (c.961_981del) allele, and Elmo2345 (p.Ser326Argfs∗21) corresponds to both the elmo210.1 (c.975_984del) and elmo210.2 (c.969_978del) alleles. Abbreviations are as follows: ARM, Armadillo repeat; ELMO, ELMO domain; PH, pleckstrin homology domain; PxxP, proline-rich motif.
(D) Quantitative PCR shows significantly lower levels of elmo2 transcripts in MZ homozygous elmo210.1 and elmo210.2 fish, as compared to matched elmo2+/+ controls from 0 to 96 hpf. Data are shown as mean ± SEM, two-way ANOVA with Bonferroni posttest. ∗∗∗p ≤ 0.001.
(E) Both zygotic elmo210.1 and maternal-zygotic elmo210.2 homozygous fish develop normally to fertile adults, without a gross discernible phenotype.
(F) Normal blood vessel development in the pharyngeal arches and trunk at 5 dpf in elmo210.2 homozygous fish carrying the fli1a:EGFP transgene is shown. Fish homozygous for elmo210.1 carrying the acta2:mCherry transgene show normal visceral smooth muscle cells at 6 dpf. Scale bars represent 0.1 μm.
(G) Based on evaluation of jaw morphology in a jaw injury assay, elmo2 is not required for proper jaw healing in adult zebrafish. Class 1 is defined by complete jaw healing. Class 2 fish have slight jaw defects marked by small gaps (black arrowhead). Class 3 fish have gross jaw deformities and protrusions (black arrowhead). Top right-side inset on class 1 indicates axis of injury.
To gain insight into the pathogenesis of VMOS, we generated an allelic series of zebrafish elmo2 knockouts using the CRISPR/Cas9 system, targeting exon 12 of the elmo2 (Figure S5). In all, three distinct elmo2 alleles—elmo210.1, elmo210.2, and elmo221—were isolated, outcrossed, and propagated (Figure S5). The allele elmo221 is an in-frame 21 bp deletion in exon 12, resulting in an Elmo2 (p.Asp321_Gly327del) that is predicted to be missing seven amino acids from the ELMO domain (Figures 5C and S5). Alleles elmo210.1 and elmo210.2 both have 10 bp deletions in exon 12, resulting in transcripts that differ by only a single nucleotide encoding the same p.Ala323 residue. Both transcripts reach a pre-termination stop codon, resulting in the exact same C-terminal truncated protein, p.Ser326ArgfsTer21 (Figures 5C and S5). When bred to homozygosity, elmo210.1 and elmo210.2 zygotic null embryos were observed in expected Mendelian ratios. In contrast to the human disease phenotype, these embryos developed normally into fertile adults. To rule out a possible rescue from maternal Elmo2 or trace elmo2 mRNA deposited in the egg, zygotic elmo2–/– parents were incrossed to generate complete maternal-zygotic (MZ) knockout embryos. MZ elmo2-null embryos of the elmo210.1 and elmo210.2 genotypes were phenotypically unremarkable and could be maintained as such for several generations (Figure 5E). Developmental QPCR analysis of MZ knockout fish showed that mutant elmo210.1 and elmo210.2 transcripts were present at significantly lower levels than those of wild-type elmo2 in control fish (Figure 5D), which indicates that as in the individuals with VMOS, aberrantly spliced elmo2 transcripts are also subjected to nonsense-mediated decay, confirming that these fish mutations are likely to be loss-of-function alleles.
Assuming a potential role for ELMO2 in maintaining blood vessel integrity via vascular smooth muscle cells, the elmo210.1 allele was crossed into the Tg(fli1a:EGFP) line to assess vasculature development and the elmo210.2 allele was crossed into the Tg(acta2:mCherry) line to examine vascular smooth muscle cells. Pharyngeal arch development was identical in the wild-type and mutants, and gross vasculature was generally normal (Figure 5F). Visceral smooth muscle in the trunk was also similar in the wild-type and mutant elmo2 fish (Figure 5F). To unravel the possible role for elmo2 in maintaining integrity of the jaw vasculature upon injury, we further challenged fish of the wild-type and elmo2 heterozygous and homozygous genotypes. To mimic the dental trauma experienced by individuals with VMOS undergoing oral surgery for ectopic tooth eruption or mandibulectomy, juvenile fish were subjected to a complete non-union lower jaw bone fracture using surgical scissors (Figure 5G, inset, and Movie S1). After injury, the fish were allowed to rest for 1 month, and then jaw morphology was assessed. All injured fish, regardless of genotype, survived the procedure and were grouped according to phenotypic severity of the jaw (Figure 5G). Most fish healed completely (class 1) or had small defects (class 2). A few fish had a prominent jaw protrusion (class 3) that was not biased toward the loss of elmo2 (Figure 5G). These results indicate that the loss of elmo2 in zebrafish has no noticeable effect on development, adult physiology, or response to major jaw trauma. Despite high ELMO2 conservation between vertebrate species, we speculate that zebrafish have built-in redundancy systems to buffer against the loss of elmo2, whereas in humans ELMO2 is absolutely vital for maintaining blood vessel and bone homeostasis.
The Elmo2 Gene Family Originated upon Vertebrate Jaw Evolution
Phylogenetic analysis shows that all the vertebrate Elmo genes examined in this study form a monophyletic group derived from a single ancestral gene via gene duplications (Figure S6). The ELMO genes of human as well as other vertebrate model species (mouse, chicken, frog, and zebrafish) all clearly divide into three lineages deduced to be three paralogous genes: Elmo1, Elmo2, and Elmo3. The protein sequences from different species within each of the three Elmo sub-families are highly conserved (Figure S7). Moreover, this analysis places Elmo1 and Elmo2 closer to each other than to Elmo3 (Figure S6).
To further elucidate the early evolutionary history of vertebrate Elmo genes, we analyzed the genomes of two basal vertebrates—cartilaginous fish and lampreys—and an invertebrate chordate, the amphioxus. Chondrichthyes (cartilaginous fish) protein sequences from the elephant shark (Callorhinchus milii) included three different sequences (GenBank: XP_007887897, XP_007883848, and XP_007904034) that were clearly orthologous to the ELMO1/2/3 proteins of the model vertebrates, respectively, demonstrating that the vertebrate Elmo1/2/3 gene family was established before the divergence of cartilaginous fishes from all other jawed vertebrates. Analysis of genome sequences from lamprey (Lethenteron japonicum) showed that extant agnathans (jawless fish) have at least two homologous genes to the Elmo1/2/3 gene family of jawed vertebrates. One of the homologs (GenBank: KE993686) forms a sister group with the clade of Elmo1 and Elmo2 (Figure S6). This clearly indicates that the second gene duplication took place after lampreys were diverged from other jawless vertebrates. In other words, the Elmo2 gene family was established around the time when the “jaw” evolved but before cartilaginous fish and other jawed vertebrates diverged. A phylogenetic relationship between the other lamprey ELMO (GenBank: KE993714) and ELMO1/2/3 proteins was not clearly resolved. In amphioxus at least three Elmo proteins were identified (221852, 282737, and 121840 at JGI Genome Portal: Branchiostoma floridae genome), but these were not orthologous to vertebrate Elmo1/2/3. It is likely that three amphioxus Elmo genes were formed by lineage-specific duplications, independently of vertebrate duplications of Elmo1/2/3.
Discussion
Well-orchestrated migration of ECs and vSMCs is crucial for angiogenesis. Angiogenesis is stimulated by local signals that activate proliferation and directed migration of ECs to form vascular channels, followed by perivascular cell recruitment and vessel stabilization.45 ELMO2 encodes engulfment and cell motility protein 2, which is recruited to the plasma membrane and involved in the signaling cascade that controls cytoskeleton dynamics and cell migration. Herein, we describe four distinct homozygous mutations in ELMO2 that cause severe VMOS. All of the affected individuals in the five presented families have a common phenotype of expanding vascular malformations in craniofacial bones, which is progressive and potentially life threatening. Similar phenotypic features of VMOS have previously been described in several cases with no indication of family history; however, family history might be overlooked because such cases can appear as sporadic.4, 12, 46 As such, recessive ELMO2 mutations might be more common than previously thought.
A Spectrum of ELMO2 Mutations Is Responsible for VMOS
The ELMO2 mutation spectrum predicts severe or complete loss of function. The phenotype is strikingly similar among affected individuals with different ELMO2 mutations. One of the splice site mutations, c.1065+1G>A, was studied in detail, using primary cells and iPSCs from affected individual A-II:3, which shows that this allele behaves as a loss-of-function variant. The second splice site mutation, c.1802−1G>C, affects the splice acceptor site of intron 19 and is predicted to reduce ELMO2 function in a similar manner. The third mutation, c.2080delC, deletes 1 bp in the last exon, leading to a frameshift, and is predicted to delete the proline-rich motif and elongate the C terminus by 99 random amino acids. Such aberrantly elongated proteins skipping the canonical termination codon have been shown to be targeted for proteasomal degradation and, therefore, this is considered to be another loss-of-function variant.47, 48
The fourth mutation, a homozygous complex rearrangement, deletes the critical transcription initiation site, which is predicted to abolish ELMO2 transcription. We think that at least three successive microhomology-mediated events should have occurred to account for the insertion of two different sequences during rearrangement in a common ancestor (Figure S1C). Events 1 and 3 probably occurred via either microhomology-mediated break-induced replication (MMBIR) or fork stalling-template switch (FoSTeS).49, 50, 51 In contrast, the presence of a 13 bp joining sequence with several microhomologies suggests that >1 DNA synthesis and trimming event may have occurred in event 2 (Figures S1C and S2). In Drosophila melanogaster similar events in which DNA synthesis with replication slippage and microhomology-mediated end-joining (MMEJ) have been observed and are referred to as synthesis-dependent MMEJ (SD-MMEJ).52 Nonetheless, ≥2 SD-MMEJ events are required to explain two microhomologies in the joining segment observed in the present study.
Depletion of ELMO2 from VMOS-Affected Cells Impairs Cell Migration
The primary consequence of the c.1065+1G>A splice site mutation is loss of wild-type ELMO2 transcripts and protein due to aberrant splicing. Although no appreciable mutant ELMO2 was detected in primary fibroblasts or iPSCs (Figure 2D), it cannot be excluded that they may be translated and remain stable in disease-related tissues. Functional tests with overexpressed ELMO2 muteins showed that, if ever present, these muteins would not retain appreciable RAC1 signaling, further confirming that these rare ELMO2 variants causing VMOS are most likely behaving as loss-of-function alleles. In accordance with these findings, cell migration was impaired in ELMO2-depleted primary fibroblasts, which was partially rescued by wild-type ELMO2, indicating a specific role for ELMO2 in cell migration.
Six ELMO domain-containing proteins are described in humans, of which three are ELMOD proteins that interact with ARF to control intracellular trafficking.53 The other three are ELMO proteins—ELMO1, ELMO2, and ELMO3—and interact with RAC1 to regulate the cytoskeleton. Similar to other members of the ELMO family, ELMO2 has several ARM domains that interact with RHOG and ILK, a PH domain interacting with DOCK proteins, and a PxxP domain for which the interacting partners remain unknown (Figure 1D).35 RHOG is a bridge between ELMO2 and several upstream factors, including ephrin, ephexins, integrins, epidermal growth factor receptor (EGFR), and yet undescribed proteins.35, 54, 55 These effectors regulate the small GTPase RAC1 via the ELMO/DOCK bipartite guanine nucleotide exchange factor. This signaling pathway has been shown to control actin and microtubule cytoskeletal dynamics, cell adhesion, cell polarity, and cell migration in keratinocytes.29, 55, 56 Angiogenesis is primarily based on the dynamic migration of endothelial and perivascular cells, so it is not surprising that a human disorder with angiogenic malformation is caused by the lack of ELMO2.
The specific upstream elements interacting with ELMO2 in ECs and vSMCs are yet to be determined. However, ELMO2 has been shown to be phosphorylated by hepatocyte growth factor (HGF) in A549 adenocarcinoma cells.57 This may be an important event, because HGF, which is released by ECs upon angiopoetin-1 stimulation, is a key factor in vSMC recruitment.58 This may be especially important because mutations in angiopoetin-1 receptor, Tie2, are implicated in hereditary venous/capillary malformations. In addition, Rho GEFs are known to be key mediators in vessel function.59 We hypothesize that ELMO2, in complex with DOCK1, is a key GEF in vascular smooth muscle cells to maintain vessel integrity by an unknown mechanism, perhaps by maintaining cadherin-mediated adhesion.29
Zebrafish, Unlike Humans, Are Able to Compensate for the Loss of ELMO2
VMOS was modeled in vivo by creating elmo2 knockout alleles in zebrafish that would mimic the out-of-frame mutations observed in the individuals with VMOS. Mutant elmo2 fish did not phenocopy VMOS syndrome, even when maternal-zygotic null, but this model serves as a possible starting point to elucidate compensatory networks activated upon the loss of elmo2. One candidate compensatory protein is Elmo1, due to its high amino acid sequence and critical domain homology with Elmo2 (Figure S6). Consistent with a possible redundancy between Elmo1 and Elmo2 is the report that zygotic elmo1 knockout fish also have no phenotype.60 If this is the case, we suspect that a double elmo1/2 zebrafish knockout might be more inclined to recapitulate and model VMOS in zebrafish.
The requirement of ELMO2 in human development, physiology, and homeostasis might differ from that in zebrafish. The human mandible is a highly vascularized intramembranous bone, whereas in small fish like Danio rerio the equivalent Meckel’s cartilage is a non-vascularized endochondral bone. Although numerous vessels surround the zebrafish jaw, those in elmo2-null fish were structurally normal and did not cause enlargement of the cranium (Figure 5E and 5F). Larger fish such as salmon have vessels within bone, but no associated hematopoiesis (G. Crump, personal communication). The mouse mandible and associated vasculature is more comparable with that of humans and might, therefore, be a better model for recapitulating the VMOS phenotype.61
Lastly, the pathophysiology of VMOS might be human specific, despite close homology between human ELMO2 and zebrafish elmo2 (Figure S7). The carotid arteries in humans are subjected to relatively high pressure from blood exiting the aorta and flowing upward against gravity. The lack of a mature vascular smooth muscle layer decreases the resilience of these vessels and can lead to abnormal dilation. In zebrafish, the need to buffer against blood pressure is likely less crucial, given its horizontal and aquatic life.
What Can ELMO2 and VMOS Tell Us about Vertebrate Evolution?
To seek a wider implication from this study, we further explored our analysis by bringing a phylogenetic approach together with our findings. Invertebrates, such as Caenorhabditis and Drosophila, have only a single gene (ced-12; GenBank: NM_135704.3) homologous to the vertebrate Elmo1/2/3 gene family, whereas vertebrates possess three Elmo paralogs. These form a monophyletic group established by two gene duplications from a single ancestral invertebrate gene (Figure S6).62, 63 The first gene duplication separated the common ancestor of Elmo1 and Elmo2 (Elmo1/2) from Elmo3 during early vertebrate evolution. The next gene duplication established the current Elmo2 gene family from the Elmo1/2 common ancestor gene. The latter duplication event took place after lampreys were diverged from other jawless vertebrates, but before chondrichthyes (cartilaginous fishes like sharks) were diverged from all the other jawed vertebrates. Interestingly, this timing overlaps with the period when vertebrate ancestors acquired dermal (intramembranous) bones and the jaw, implying that Elmo2 may have been involved in the evolution of these novel traits in our ancestor.64
Vertebrates have two different types of bones: dermal and cartilaginous. Dermal bones are mainly formed by intramembranous ossification of cells derived from neural crest and are regarded as the first type of bone that appeared in vertebrate evolution. As described above, a common characteristic feature of individuals with VMOS is that the majority of vascular malformations are limited to dermal bones, including the mandible. Although little is known about the relationship between angiogenesis and intramembranous ossification, our results clearly demonstrate that ELMO2 is a key factor in the development of dermal bones.
Conclusion
We present loss-of-function mutations in ELMO2, via impaired RAC1 signaling and defective vascular smooth muscles, as the causative factor for autosomal-recessive VMOS. Abnormally enlarged blood vessels are localized specifically to intramembranous bone, resulting in dysregulated bone remodeling. Based on these findings, we consider ELMO2 to be a crucial intermediate signaling molecule in RAC1-dependent control of vascular stability in dermal bones.
Acknowledgments
We are indebted to the five study families for their kind participation. We thank Dr. Gen Nishimura, Tokyo Metropolitan Children’s Medical Center, Japan, for his help with radiographic diagnosis of individual E-II:1, Tom Carney, Nathan Lawson, and Raymond Birge for sharing relevant materials and zebrafish transgenic lines, Atilla Göknur and Lütfiye Mesci for performing targeted massive parallel sequencing, Afsin Oktay for his early laboratory work that inspired to the design of this study, and Ebru Orallı Bircan for illustrations.
This study was supported by a Scientific and Technological Research Council of Turkey (TÜBİTAK) grant (108S420 to N.A.A.), under the framework of ERA-Net for Research on Rare Diseases, CRANIRARE consortium (R07197KS), a grant from the Hacettepe University Research Fund (00-02-101-009 and 03-D07-101-001 to N.A.A.), a grant from TUBİTAK-BİLGEM-UEKAE (K030-T439 to M.Ş.S. and B.Y.) under the framework of the Republic of Turkey Ministry of Development Infrastructure Grant to establish Advanced Genomics and Bioinformatics Research Center (2011K120020), and a grant from the Strategic Positioning Fund for Genetic Orphan Diseases, as well as an inaugural A∗STAR Investigatorship from the Agency for Science, Technology, and Research in Singapore. We are grateful to all members of the B.R. laboratory for their support. B.R. is a fellow of the Branco Weiss Foundation and an A∗STAR and EMBO Young Investigator. J.R.X. is funded by the A∗STAR Graduate Scholarship.
Published: July 28, 2016
Footnotes
Supplemental Data include seven figures, three tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.06.008.
Contributor Information
Bruno Reversade, Email: bruno@reversade.com.
Nurten Ayse Akarsu, Email: nakarsu@hacettepe.edu.tr.
Accession Numbers
The accession number for the g.45031191_45037128del5938ins330 sequence reported in this paper is GenBank: KU680992.
Web Resources
ENCODE, https://www.encodeproject.org/
Ensembl Genome Browser, http://www.ensembl.org/index.html
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Japanese Lamprey Genome Project, http://jlampreygenome.imcb.a-star.edu.sg/
JGI Genome Portal, http://genome.jgi.doe.gov/Brafl1/Brafl1.home.html
NCBI Gene, http://www.ncbi.nlm.nih.gov/gene
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
non-B DB, https://nonb-abcc.ncifcrf.gov
OMIM, http://www.omim.org/
Primer3, http://bioinfo.ut.ee/primer3
Protocols from the Reversade Laboratory, http://www.reversade.com-a.googlepages.com/protocols/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
A video clip depicting the zebrafish lower jaw fracture assay. Following anesthetization, fish were placed on petri dishes and the lower jaw was fractured using surgical scissors.
References
- 1.Helker C.S., Schuermann A., Pollmann C., Chng S.C., Kiefer F., Reversade B., Herzog W. The hormonal peptide Elabela guides angioblasts to the midline during vasculogenesis. eLife. 2015;4:e06726. doi: 10.7554/eLife.06726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conway E.M., Collen D., Carmeliet P. Molecular mechanisms of blood vessel growth. Cardiovasc. Res. 2001;49:507–521. doi: 10.1016/s0008-6363(00)00281-9. [DOI] [PubMed] [Google Scholar]
- 3.Mulliken J.B., Glowacki J. Hemangiomas and vascular malformations in infants and children: a classification based on endothelial characteristics. Plast. Reconstr. Surg. 1982;69:412–422. doi: 10.1097/00006534-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Bruder E., Perez-Atayde A.R., Jundt G., Alomari A.I., Rischewski J., Fishman S.J., Mulliken J.B., Kozakewich H.P. Vascular lesions of bone in children, adolescents, and young adults. A clinicopathologic reappraisal and application of the ISSVA classification. Virchows Arch. 2009;454:161–179. doi: 10.1007/s00428-008-0709-3. [DOI] [PubMed] [Google Scholar]
- 5.Wassef M., Blei F., Adams D., Alomari A., Baselga E., Berenstein A., Burrows P., Frieden I.J., Garzon M.C., Lopez-Gutierrez J.C., ISSVA Board and Scientific Committee Vascular anomalies classification: recommendations from the International Society for the Study of Vascular Anomalies. Pediatrics. 2015;136:e203–e214. doi: 10.1542/peds.2014-3673. [DOI] [PubMed] [Google Scholar]
- 6.Akers A.L., Johnson E., Steinberg G.K., Zabramski J.M., Marchuk D.A. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limaye N., Wouters V., Uebelhoer M., Tuominen M., Wirkkala R., Mulliken J.B., Eklund L., Boon L.M., Vikkula M. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat. Genet. 2009;41:118–124. doi: 10.1038/ng.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen H.L., Boon L.M., Vikkula M. Genetics of vascular malformations. Semin. Pediatr. Surg. 2014;23:221–226. doi: 10.1053/j.sempedsurg.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Dickins J.R. Cavernous hemangioma of the sphenoid wing. Arch. Otolaryngol. 1978;104:58–60. [PubMed] [Google Scholar]
- 10.Vargel I., Cil B.E., Er N., Ruacan S., Akarsu A.N., Erk Y. Hereditary intraosseous vascular malformation of the craniofacial region: an apparently novel disorder. Am. J. Med. Genet. 2002;109:22–35. doi: 10.1002/ajmg.10282. [DOI] [PubMed] [Google Scholar]
- 11.Vargel I., Cil B.E., Kiratli P., Akinci D., Erk Y. Hereditary intraosseous vascular malformation of the craniofacial region: imaging findings. Br. J. Radiol. 2004;77:197–203. doi: 10.1259/bjr/88438282. [DOI] [PubMed] [Google Scholar]
- 12.Hossein Mortazavi S., Khodayari A., Khojasteh A., Abbas F.M., Mehrdad L., Kiani M.T., Farman A.G. Massive craniofacial intraosseous vascular malformation resembling cystic angiomatosis: report of 2 cases and review of the literature. J. Oral Maxillofac. Surg. 2011;69:204–214. doi: 10.1016/j.joms.2010.07.072. [DOI] [PubMed] [Google Scholar]
- 13.Kim J.E., Yi W.J., Heo M.S., Lee S.S., Choi S.C., Huh K.H. An unusual case of intraosseous vascular malformation of the maxilla mimicking fibrous dysplasia: a case report and literature review on imaging features of intraosseous vascular anomalies of the jaw. Dentomaxillofac. Radiol. 2014;43:20130400. doi: 10.1259/dmfr.20130400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenger D.E., Wold L.E. Benign vascular lesions of bone: radiologic and pathologic features. Skeletal Radiol. 2000;29:63–74. doi: 10.1007/s002560050012. [DOI] [PubMed] [Google Scholar]
- 15.Goutoudi P.C., Sferopoulos N.K., Papavasiliou V., Konstantinidis A. Cystic angiomatosis of bone: a case report. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 1996;81:89–92. doi: 10.1016/s1079-2104(96)80155-7. [DOI] [PubMed] [Google Scholar]
- 16.Xu P., Lan S., Liang Y., Xiao Q. Multiple cavernous hemangiomas of the skull with dural tail sign: a case report and literature review. BMC Neurol. 2013;13:155. doi: 10.1186/1471-2377-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson S.E., Prayson R.A. Primary skull lesions in the pediatric population: a 25-year experience. Arch. Pathol. Lab. Med. 2007;131:761–766. doi: 10.5858/2007-131-761-PSLITP. [DOI] [PubMed] [Google Scholar]
- 18.Naraghi A., Keene M. Maxillary intraosseous vascular malformation: a case report and literature review. J. Laryngol. Otol. 2007;121:404–407. doi: 10.1017/S0022215107004562. [DOI] [PubMed] [Google Scholar]
- 19.Kayserili H., Uz E., Niessen C., Vargel I., Alanay Y., Tuncbilek G., Yigit G., Uyguner O., Candan S., Okur H. ALX4 dysfunction disrupts craniofacial and epidermal development. Hum. Mol. Genet. 2009;18:4357–4366. doi: 10.1093/hmg/ddp391. [DOI] [PubMed] [Google Scholar]
- 20.Uz E., Alanay Y., Aktas D., Vargel I., Gucer S., Tuncbilek G., von Eggeling F., Yilmaz E., Deren O., Posorski N. Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am. J. Hum. Genet. 2010;86:789–796. doi: 10.1016/j.ajhg.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez D., François P., Farinelli L., Osterås M., Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Page R.D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 29.Toret C.P., Collins C., Nelson W.J. An Elmo-Dock complex locally controls Rho GTPases and actin remodeling during cadherin-mediated adhesion. J. Cell Biol. 2014;207:577–587. doi: 10.1083/jcb.201406135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abu-Thuraia A., Gauthier R., Chidiac R., Fukui Y., Screaton R.A., Gratton J.P., Côté J.F. Axl phosphorylates Elmo scaffold proteins to promote Rac activation and cell invasion. Mol. Cell. Biol. 2015;35:76–87. doi: 10.1128/MCB.00764-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 2013;41:D94–D100. doi: 10.1093/nar/gks955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vissers L.E., Bhatt S.S., Janssen I.M., Xia Z., Lalani S.R., Pfundt R., Derwinska K., de Vries B.B., Gilissen C., Hoischen A. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum. Mol. Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. [DOI] [PubMed] [Google Scholar]
- 34.Elliott M.R., Zheng S., Park D., Woodson R.I., Reardon M.A., Juncadella I.J., Kinchen J.M., Zhang J., Lysiak J.J., Ravichandran K.S. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature. 2010;467:333–337. doi: 10.1038/nature09356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho E., Irvine T., Vilk G.J., Lajoie G., Ravichandran K.S., D’Souza S.J., Dagnino L. Integrin-linked kinase interactions with ELMO2 modulate cell polarity. Mol. Biol. Cell. 2009;20:3033–3043. doi: 10.1091/mbc.E09-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brugnera E., Haney L., Grimsley C., Lu M., Walk S.F., Tosello-Trampont A.C., Macara I.G., Madhani H., Fink G.R., Ravichandran K.S. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 37.Makino Y., Tsuda M., Ichihara S., Watanabe T., Sakai M., Sawa H., Nagashima K., Hatakeyama S., Tanaka S. Elmo1 inhibits ubiquitylation of Dock180. J. Cell Sci. 2006;119:923–932. doi: 10.1242/jcs.02797. [DOI] [PubMed] [Google Scholar]
- 38.Lu M., Ravichandran K.S. Dock180-ELMO cooperation in Rac activation. Methods Enzymol. 2006;406:388–402. doi: 10.1016/S0076-6879(06)06028-9. [DOI] [PubMed] [Google Scholar]
- 39.Fisher C. Myofibrosarcoma. Virchows Arch. 2004;445:215–223. doi: 10.1007/s00428-004-1038-9. [DOI] [PubMed] [Google Scholar]
- 40.Kilarski W.W., Jura N., Gerwins P. An ex vivo model for functional studies of myofibroblasts. Lab. Invest. 2005;85:643–654. doi: 10.1038/labinvest.3700255. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe K., Kusakabe T., Hoshi N., Saito A., Suzuki T. h-Caldesmon in leiomyosarcoma and tumors with smooth muscle cell-like differentiation: its specific expression in the smooth muscle cell tumor. Hum. Pathol. 1999;30:392–396. doi: 10.1016/s0046-8177(99)90113-2. [DOI] [PubMed] [Google Scholar]
- 42.Werdich X.Q., Jakobiec F.A., Curtin H.D., Fay A. A clinical, radiologic, and immunopathologic study of five periorbital intraosseous cavernous vascular malformations. Am. J. Ophthalmol. 2014;158:816–826.e1. doi: 10.1016/j.ajo.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y.H., Gallant C., Guo H., Li Y., Wang C.A., Morgan K.G. Regulation of vascular smooth muscle tone by N-terminal region of caldesmon. Possible role of tethering actin to myosin. J. Biol. Chem. 2000;275:3213–3220. doi: 10.1074/jbc.275.5.3213. [DOI] [PubMed] [Google Scholar]
- 44.Epting D., Wendik B., Bennewitz K., Dietz C.T., Driever W., Kroll J. The Rac1 regulator ELMO1 controls vascular morphogenesis in zebrafish. Circ. Res. 2010;107:45–55. doi: 10.1161/CIRCRESAHA.109.213983. [DOI] [PubMed] [Google Scholar]
- 45.Ucuzian A.A., Gassman A.A., East A.T., Greisler H.P. Molecular mediators of angiogenesis. J. Burn Care Res. 2010;31:158–175. doi: 10.1097/BCR.0b013e3181c7ed82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh C.H., Wu Y.M., Chen Y.L., Wong H.F. Contralateral de novo intraosseous arteriovenous malformation in a child with arteriovenous malformation of mandible treated by endovascular embolotherapy. A case report. Interv. Neuroradiol. 2012;18:484–489. doi: 10.1177/159101991201800415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patronas Y., Horvath A., Greene E., Tsang K., Bimpaki E., Haran M., Nesterova M., Stratakis C.A. In vitro studies of novel PRKAR1A mutants that extend the predicted RIα protein sequence into the 3′-untranslated open reading frame: proteasomal degradation leads to RIα haploinsufficiency and Carney complex. J. Clin. Endocrinol. Metab. 2012;97:E496–E502. doi: 10.1210/jc.2011-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata N., Ohoka N., Sugaki Y., Onodera C., Inoue M., Sakuraba Y., Takakura D., Hashii N., Kawasaki N., Gondo Y., Naito M. Degradation of stop codon read-through mutant proteins via the ubiquitin-proteasome system causes hereditary disorders. J. Biol. Chem. 2015;290:28428–28437. doi: 10.1074/jbc.M115.670901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 50.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verdin H., D’haene B., Beysen D., Novikova Y., Menten B., Sante T., Lapunzina P., Nevado J., Carvalho C.M., Lupski J.R., De Baere E. Microhomology-mediated mechanisms underlie non-recurrent disease-causing microdeletions of the FOXL2 gene or its regulatory domain. PLoS Genet. 2013;9:e1003358. doi: 10.1371/journal.pgen.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu A.M., McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.East M.P., Bowzard J.B., Dacks J.B., Kahn R.A. ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for Arf protein family GTPases. J. Biol. Chem. 2012;287:39538–39553. doi: 10.1074/jbc.M112.417477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiramoto-Yamaki N., Takeuchi S., Ueda S., Harada K., Fujimoto S., Negishi M., Katoh H. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J. Cell Biol. 2010;190:461–477. doi: 10.1083/jcb.201005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho E., Dagnino L. Epidermal growth factor induction of front-rear polarity and migration in keratinocytes is mediated by integrin-linked kinase and ELMO2. Mol. Biol. Cell. 2012;23:492–502. doi: 10.1091/mbc.E11-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson B.C., Ivanova I.A., Dagnino L. An ELMO2-RhoG-ILK network modulates microtubule dynamics. Mol. Biol. Cell. 2015;26:2712–2725. doi: 10.1091/mbc.E14-10-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammond D.E., Hyde R., Kratchmarova I., Beynon R.J., Blagoev B., Clague M.J. Quantitative analysis of HGF and EGF-dependent phosphotyrosine signaling networks. J. Proteome Res. 2010;9:2734–2742. doi: 10.1021/pr100145w. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi H., DeBusk L.M., Babichev Y.O., Dumont D.J., Lin P.C. Hepatocyte growth factor mediates angiopoietin-induced smooth muscle cell recruitment. Blood. 2006;108:1260–1266. doi: 10.1182/blood-2005-09-012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kather J.N., Kroll J. Rho guanine exchange factors in blood vessels: fine-tuners of angiogenesis and vascular function. Exp. Cell Res. 2013;319:1289–1297. doi: 10.1016/j.yexcr.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 60.Kok F.O., Shin M., Ni C.W., Gupta A., Grosse A.S., van Impel A., Kirchmaier B.C., Peterson-Maduro J., Kourkoulis G., Male I. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Percival C.J., Richtsmeier J.T. Angiogenesis and intramembranous osteogenesis. Dev. Dyn. 2013;242:909–922. doi: 10.1002/dvdy.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gumienny T.L., Brugnera E., Tosello-Trampont A.C., Kinchen J.M., Haney L.B., Nishiwaki K., Walk S.F., Nemergut M.E., Macara I.G., Francis R. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 63.Geisbrecht E.R., Haralalka S., Swanson S.K., Florens L., Washburn M.P., Abmayr S.M. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 2008;314:137–149. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu M., Yu X., Ahlberg P.E., Choo B., Lu J., Qiao T., Qu Q., Zhao W., Jia L., Blom H., Zhu Y. A Silurian placoderm with osteichthyan-like marginal jaw bones. Nature. 2013;502:188–193. doi: 10.1038/nature12617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A video clip depicting the zebrafish lower jaw fracture assay. Following anesthetization, fish were placed on petri dishes and the lower jaw was fractured using surgical scissors.