Abstract
In ∼30% of families affected by colorectal adenomatous polyposis, no germline mutations have been identified in the previously implicated genes APC, MUTYH, POLE, POLD1, and NTHL1, although a hereditary etiology is likely. To uncover further genes with high-penetrance causative mutations, we performed exome sequencing of leukocyte DNA from 102 unrelated individuals with unexplained adenomatous polyposis. We identified two unrelated individuals with differing compound-heterozygous loss-of-function (LoF) germline mutations in the mismatch-repair gene MSH3. The impact of the MSH3 mutations (c.1148delA, c.2319−1G>A, c.2760delC, and c.3001−2A>C) was indicated at the RNA and protein levels. Analysis of the diseased individuals’ tumor tissue demonstrated high microsatellite instability of di- and tetranucleotides (EMAST), and immunohistochemical staining illustrated a complete loss of nuclear MSH3 in normal and tumor tissue, confirming the LoF effect and causal relevance of the mutations. The pedigrees, genotypes, and frequency of MSH3 mutations in the general population are consistent with an autosomal-recessive mode of inheritance. Both index persons have an affected sibling carrying the same mutations. The tumor spectrum in these four persons comprised colorectal and duodenal adenomas, colorectal cancer, gastric cancer, and an early-onset astrocytoma. Additionally, we detected one unrelated individual with biallelic PMS2 germline mutations, representing constitutional mismatch-repair deficiency. Potentially causative variants in 14 more candidate genes identified in 26 other individuals require further workup. In the present study, we identified biallelic germline MSH3 mutations in individuals with a suspected hereditary tumor syndrome. Our data suggest that MSH3 mutations represent an additional recessive subtype of colorectal adenomatous polyposis.
Keywords: familial colorectal cancer, adenomatous polyposis, candidate genes, mismatch repair, exome sequencing, massive parallel sequencing, hereditary tumor syndromes
Introduction
Adenomatous polyposis syndromes of the colorectum are precancerous conditions characterized by the presence of dozens to thousands of adenomatous polyps, which, unless detected early and removed, invariably result in colorectal cancer (CRC). The phenotypic spectrum ranges from an early-onset manifestation with high numbers of adenomas and a positive family history to isolated late-onset cases with a low polyp burden.
To date, two major inherited monogenic forms of colorectal adenomatous polyposis can be delineated by molecular genetic analyses: (1) autosomal-dominant familial adenomatous polyposis (FAP [MIM: 175100]), caused by heterozygous germline mutations in the tumor-suppressor gene APC (APC, WNT signaling pathway regulator [MIM: 611731]);1, 2 and (2) autosomal-recessive MUTYH-associated polyposis (MAP [MIM: 608456]), caused by biallelic germline mutations in the base-excision-repair (BER) gene MUTYH (mutY DNA glycosylase [MIM: 604933]).3, 4
Currently, high-throughput sequencing approaches, in particular whole-exome sequencing (WES), are considered the most powerful tools for detecting causative variants in genes in as yet unexplained Mendelian conditions.5, 6 Very recent WES investigations have identified two rare forms of colorectal adenomatous polyposis: (1) autosomal-dominant polymerase-proofreading-associated polyposis (PPAP [MIM: 612591]), caused by specific germline missense mutations in the polymerase genes POLE (DNA polymerase epsilon, catalytic subunit [MIM: 174762]) and POLD1 (DNA polymerase delta 1, catalytic subunit [MIM: 174761]);7, 8, 9 and (2) another very rare autosomal-recessive colorectal adenomatous polyposis (MIM: 616415), caused by biallelic mutations in NTHL1 (nth-like DNA glycosylase 1 [MIM: 602656]).10 The WES approach has also detected ZSWIM7 (zinc finger SWIM-type containing 7 [MIM: 614535]) and PIEZO1 (piezo type mechanosensitive ion channel component 1 [MIM: 611184]) as promising candidate genes carrying variants causing colorectal adenomatous polyposis.11
However, in around 30% of polyposis cases, no underlying germline mutation has been identified, although a genetic basis is likely. Here, classic approaches to gene identification, such as linkage analysis, are not feasible, given that most of these cases are either sporadic or characterized by an uncertain family.12, 13, 14, 15 Over the past two decades, a number of candidate-gene studies have been performed without convincing results.16, 17, 18, 19 Neither loss-of-heterozygosity (LOH) analyses nor profiling of somatic mutations has contributed to the identification of promising novel genetic causes. A fraction of cases might be explained by deep intronic APC mutations,20 APC mutational mosaicism,21 rare APC missense mutations,22 or other genes predisposing to cancer.23, 24, 25, 26 In addition, rare germline copy-number variants (CNVs) and low-penetrant variants might contribute to the genetic predisposition for the formation of colorectal adenomas.15, 27, 28
Another hereditary CRC syndrome, Lynch syndrome, is not accompanied by a florid colorectal polyposis and is characterized by microsatellite instable tumors. The underlying cause is a heterozygous germline mutation in one of the mismatch-repair (MMR) genes MLH1 (mutL homolog 1 [MIM: 120436]), MSH2 (mutS homolog 2 [MIM: 609309]), MSH6 (mutS homolog 6 [MIM: 600678]), PMS2 (PMS1 homolog 2, mismatch repair system component [MIM: 600259]), or EPCAM (epithelial cell adhesion molecule [MIM: 185535]).29, 30 Biallelic mutations in these genes lead to constitutional MMR deficiency (CMMRD [MIM: 276300]) with multiple tumors and childhood onset.31, 32 In contrast, familial CRC without polyposis or microsatellite instability is etiologically very heterogeneous. In some of these families, germline mutations in FAN1 (FANCD2/FANCI-associated nuclease 1 [MIM: 613534]), encoding a nuclease involved in DNA repair, were recently identified by a WES approach.33
To uncover additional genes with high-penetrance mutations causing colorectal polyposis, we sequenced the germline exomes of 102 unrelated individuals with unexplained adenomatous polyposis. The identification of further genetic causes will extend the knowledge of disease mechanisms, biological pathways, and potential therapeutic targets.
Material and Methods
Cohort and Data Collection
All 102 individuals were determined to have unexplained colorectal adenomatous polyposis, i.e., no germline mutation in APC or MUTYH was identified by Sanger sequencing of the coding regions or deletion and duplication analysis by multiplex ligation-dependent probe amplification (MLPA).34 All participants were screened for APC mutations in the mosaic state.21 All persons were examined for pathogenic deep intronic APC mutations: 67 persons were screened via transcript analysis, and 35 were tested for known intronic mutations.20 Furthermore, the two hotspot mutations in POLE and POLD1 were excluded.9 In addition, a SNP-array-based CNV analysis was performed in all individuals, as described elsewhere.28
For all 102 persons included in this study, a hereditary cause of the disease was considered highly likely. The inclusion criteria were the presence of at least 20 synchronous, or 40 metachronous, histologically confirmed colorectal adenomas, irrespective of inheritance pattern or extraintestinal lesions. All participants were of central European origin according to family name and self-report. Relatives were only considered to be affected if their medical records confirmed fulfilment of the inclusion criteria. The study was approved by the local ethics review board (Medical Faculty of the University of Bonn, board no. 224/07), and all participants provided written informed consent prior to inclusion.
High-Throughput Sequencing and Bioinformatics Workflow
Genomic DNA was extracted from peripheral EDTA-anticoagulated blood samples by the standard salting-out procedure. WES was performed at the Yale Center for Genome Analysis via capture by the NimbleGen 2.1M Human Exome Array, and then paired-end sequencing was performed on a HiSeq 2000 instrument (Illumina) as described elsewhere.35 Targeted bases were covered by a mean of 67 independent reads, and an average of 94% of all bases were covered eight or more times (Table S1). Reads were aligned to the hg19 human reference genome (UCSC Genome Browser) with ELAND (Illumina). SAMtools software was used for marking duplicated reads, performing local realignment around short indels, recalibrating base quality scores, and calling single-nucleotide variants (SNVs) and short indels.
Variant call quality was assessed with SAMtools. A minimum quality score of 100 and a minimum coverage of 10× were required. Synonymous or intronic variants other than those affecting consensus splice sites were excluded from further analysis.
The resulting variants were filtered for (1) rare truncating (loss-of-function [LoF]) alterations (nonsense mutations, frameshift indels, and mutations at highly conserved splice sites) and (2) missense variants located in highly conserved nucleotide positions and predicted to be disease causing, damaging, or deleterious by at least two of three in silico analysis tools (PolyPhen-2, MutationTaster, and SIFT).
The variants were selected according to a recessive (presumed biallelic mutations) or dominant (heterozygous mutations) mode of inheritance and an estimated disease frequency of 0.01% in the population. In the dominant and recessive disease models, variants with a minor allele frequency (MAF) of ≥0.01% and ≥1%, respectively, were considered benign polymorphisms or low-penetrance variants and excluded from further analysis. In addition, recurrent dominant (heterozygous) variants were selected with a less stringent frequency threshold (MAF = 1%). Population allele frequencies are based on data from dbSNP, the 1000 Genomes Project (TGP), the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) Exome Variant Server (EVS), the Exome Aggregation Consortium (ExAC) Browser, and a large in-house exome database containing all germline variants identified in 2,816 exomes of individuals without known tumor disease and sequenced under similar conditions. To exclude obvious sequencing artifacts, we performed a detailed visual inspection of the remaining variants with a read browser (Integrative Genomics Viewer [IGV]).
We considered only genes affected by potentially pathogenic variants in at least two alleles of the cohort (heterozygous in at least two individuals or homozygous or compound heterozygous in at least one individual). Finally, we inspected all genes carrying the remaining rare variants for the presence of rare, non-polymorphic heterozygous CNVs of ≥10 kb in order to identify additional recurrently mutated genes or biallelic, compound-heterozygous variants.28
Splicing efficiencies of the normal and mutant sequences were calculated with the following splice prediction programs: Human Splicing Finder,36 GeneSplicer (University of Maryland Center for Bioinformatics and Computational Biology), MaxEntScan,37 and NNSPLICE 0.9 (Berkeley Drosophila Genome Project).
The etiological relevance of the mutations was further explored by evaluation of their genetic intolerance to functional variation, as measured by the Residual Variation Intolerance Score (RVIS),38 and the likelihood of haploinsufficiency, as measured by haploinsufficiency scores (from dataset S2, including imputed values).39 The expression of candidate genes was determined with the expressed-sequence-tag profiles of human colon tissue and protein detection data from human colon tissue (glandular cells) provided by UniGene and the Human Protein Atlas.
Frequency of Colorectal Tumors with Somatic Mutations in Candidate Genes
Data concerning the frequency (percentage) of colorectal tumors with somatic mutations in candidate genes were obtained from the exome database of The Cancer Genome Atlas (TCGA). Somatic variants identified in exome data from colonic (n = 273) and rectal (n = 116) adenocarcinomas were downloaded from the TCGA data portal. To correct the data for the presence of passenger mutations, we excluded hypermutated tumors from the dataset. Therefore, the distribution of somatic variants in the TCGA exomes was analyzed, and all tumors with >200 variants (24% of the tumors) were excluded. We used the remaining 295 exomes (76% of tumors) to calculate the frequency of tumors with somatic mutations in candidate genes.28
Sanger Sequencing and Validation
The identified truncating variants were validated via Sanger sequencing of the corresponding region according to standard protocols. We used genomic leukocyte-derived DNA to amplify the genomic region of the respective variant. PCR products were purified and sequenced on an ABI 3500xl Genetic Analyzer (Applied Biosystems). To avoid pseudogene amplification, as described previously,40 we based Sanger sequencing of PMS2 on long-range PCR with primers specific to PMS2.
Transcript Analysis
Venous blood samples were collected into PAXgene blood RNA tubes (Becton Dickinson). Total RNA was extracted with the PAXgene Blood RNA Kit (QIAGEN) in accordance with the manufacturer’s protocol. First-strand cDNA was synthesized from 2–3 μg of total RNA by random hexamer-primed reverse transcription and the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen GmbH) in accordance with the manufacturer’s protocol. RT-PCR fragments were obtained according to standard PCR protocols, and different primers were used for generating the appropriate fragments. RT-PCR products were separated on 2% agarose gel and visualized with ethidium bromide with an UV imaging system (Bio-Rad). Individual bands were excised from the gel and eluted with the High Pure PCR Product Purification Kit (Roche Diagnostics GmbH). Eluted DNA was re-amplified with the same primer pairs and sequenced as described above.
Analysis of Altered MSH3 Products
MSH3 (mutS homolog 3 [MIM: 600887]) frameshift mutations, c.1148delA and c.2760delC, were generated via site-directed mutagenesis (QuikChange II Kit, Stratagene; primers: 5′-GTTAGGGACAAAAAAAGGGCAACATT-3′ and 5′-AATGTTGCCCTTTTTTTGTCCCTAAC-3′ [c.1148delA] and 5′-GGCTCAGATTGGCTCTATGTTCCTGCAGAAG-3′ and 5′-CTTCTGCAGGAACATAGAGCCAATCTGAGCC-3′ [c.2760delC]) with the wild-type pcDNA3.1−/MSH3-WT vector (kindly provided by Grazia Graziani, Italy).41 Plasmids were confirmed by sequencing. To mimic the splice-site variants MSH3 c.3001−2A>C and c.2319−1G>A, resulting cDNAs including the premature stop codons were synthesized (Gene Art) and subcloned into a pcDNA3.1+ expression vector.
Transient transfection was carried out with HEK293T cells as described previously.42 In brief, HEK293T cells were transfected at 50%–70% confluence with expression plasmid pcDNA3.1−/MSH3-WT, pcDNA3.1−/MSH3-c.1148delA, pcDNA3.1−/MSH3-c.2760delC, pcDNA3.1+/MSH3-c.3001−2A>C, or pcDNA3.1+/MSH3-c.2319−1G>A (0.5 μg/ml, respectively) with the use of 2 μl/ml of the cationic polymer polyethylenimine (Polysciences; stock solution 1 mg/ml). 48 hr after transfection, cell extracts were prepared for western blot analysis with anti-MSH3 (H-300, Santa Cruz Biotechnologies) and anti-β-actin (Sigma). Fluorescence signals (680 and 800, Li-Cor) were detected in a FLA-9000 (Fujifilm).
The effect of whole exon deletions on protein structure was illustrated in silico. MSH3 structure was obtained from the Protein Data Bank (PDB: 3THY; MutSβ complexed with an indel loop of two bases and ADP).43 We mapped the amino acids coded by exons 17 and 22 to the MSH3 structure by using the PyMOL Molecular Graphics System (version 1.7.0.0, Schrödinger).
Immunohistochemistry of MMR Proteins
Immunohistochemical (IHC) staining of formalin-fixed, paraffin-embedded (FFPE) tissue samples was performed according to established routine procedures on a fully automated Bond-III IHC stainer (Leica) according to the manufacturer’s protocol with the following primary antibodies: MLH1, MSH2, MSH6, and PMS2 (all purchased from Leica). The amount of protein staining in tumor cells was compared to that in normal tissue. The amount of MMR protein was considered deficient if the nuclei showed no or only very weak immunostaining in relation to normal tissue.
IHC of MSH3 was performed on 2–3 μm FFPE tissue specimens with an automated staining system (480 S Autostainer, Medac). For antigen retrieval, a pre-treatment module (Medac) was used. A rabbit polyclonal antibody for MSH3, raised against an NH2-terminal polypeptide comprising amino acids 1–200, was used at a dilution of 1:100.44, 45 The reaction was developed with a horseradish-peroxidase (HRP)-conjugated detection system (C-DPVB 500 HRP, Medac) and the 3,3′-diaminobenzidine system (495192F, Medac).
Targeted Sequencing of Tumor Tissue
DNA was extracted from 10 μm FFPE tissue sections. After deparaffinization, tumor tissue was macrodissected from unstained slides. A previously marked H&E-stained slide served as a reference. Extraction of FFPE tissue DNA was carried out with the BioRobot M48 robotic workstation and the corresponding MagAttract DNA Mini M48 Kit (QIAGEN) or the Maxwell 16 FFPE Tissue LEV DNA Purification Kit (Promega) in accordance with the manufacturer’s protocol. Analysis of microsatellite status was performed according to the previously described methods.46 For examination of somatic APC mutations, high-coverage targeted sequencing (read depths > 1,000) was performed with the FAP MASTR Kit (Multiplicom) on a MiSeq platform (Illumina) in one person (individual 1661.1). The results were analyzed with SeqPilot software (JSI Medical Systems).
Microsatellite Analysis
Microsatellite analysis was performed on matched tumor and normal DNA samples by conventional fragment analysis or next-generation-sequencing-based analysis as previously described.46 This involved using the National Cancer Institute panel of reference markers to evaluate microsatellite instability (MSI) in CRC. This panel consists of two mononucleotide (BAT25 and BAT26) and three dinucleotide (D2S123, D5S346, and D17S250) repeats.47, 48, 49 Tumor DNA was extracted from microdissected tumor tissue. Normal DNA was extracted from normal tissue or peripheral-blood leukocytes. Tumors were scored as highly instable (MSI-H) if two or more of these five markers exhibited additional alleles and as stable (MSS) if none of the five markers showed instability.
A second panel of five markers was complemented. This consisted of one tetranucleotide and four dinucleotide repeats (BAT40, D10S197, D13S153, MYCL1, and D18S58). The tumor was classified as MSI-H if two or more of the ten markers exhibited instability and as MSI-low (MSI-L) if only one marker exhibited additional alleles.
For detecting elevated microsatellite instability at selected tetranucleotide repeats (EMAST), DNA from tumor and normal tissue was analyzed with five more tetranucleotide repeat markers (D20S82, D2S443, D21S1436, D9S747, and UTS037) as described elsewhere.50
Results
To identify high-penetrance germline mutations causing colorectal adenomatous polyposis and located in genes not related to polyposis so far, we performed WES of leukocyte-derived DNA in 102 unrelated individuals with unexplained adenomatous polyposis. Most of the individuals presented with an attenuated colorectal phenotype (late-onset disease and/or <100 colorectal adenomas). The mean age at diagnosis was 44 years (range = 14–73 years). The majority of individuals in the whole cohort had no evidence of extracolonic lesions and were sporadic or isolated cases. The basic clinical features of the cohort are summarized in Tables S2 and S3.
The median coverage of mapped reads was 56× (66% on-target), and 84% of bases were covered at ≥20×. The overall performance of exome sequencing is described in Table S1. A principal-component analysis demonstrated that all but one of the participants were of central European origin (Figure S1). The outlier was excluded from further analysis. Two further persons were removed as a result of low coverage. A mean of 30,152 SNVs per sample was called in the coding and flanking intronic regions. Assuming a dominant or recessive disease model, we applied a number of stringent filter steps to select for rare, non-polymorphic, truncating (LoF) variants.
We identified two unrelated individuals each carrying two different mutations in MSH3 (Figures 1 and 2, and Figure S2). We also detected one unrelated person with two different mutations in PMS2. Furthermore, potentially pathogenic germline variants were identified in 14 additional protein-coding genes (Figure S3 and Table S4).
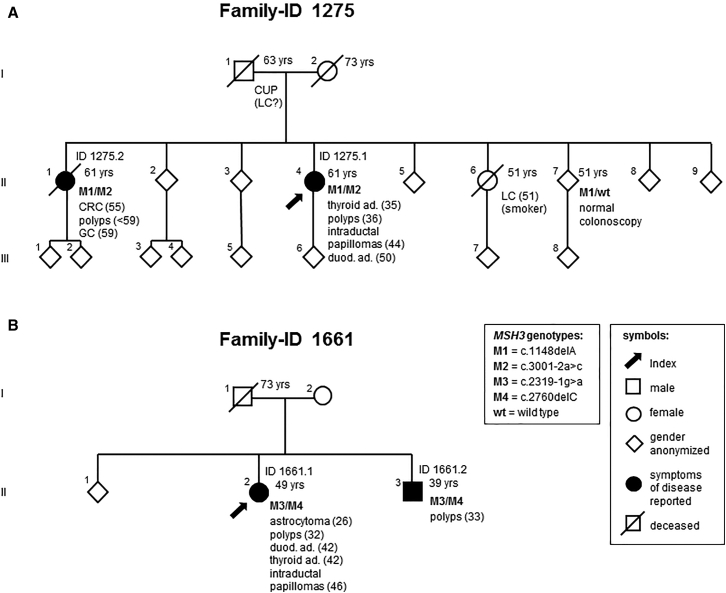
Figure 1.
Pedigrees of the Two Index Individuals with Biallelic MSH3 Germline Mutations
Pedigrees of family 1275 (A) and 1661 (B). The index persons are indicated by arrows (see main text for details). Above the symbols, identifiers are given for affected individuals. The number on the upper right side of a symbol displays the age at death, or in living persons, the age at last contact. On the lower right, genotype and phenotype information is displayed. The numbers following a disease represent the age at first diagnosis. Abbreviations are as follows: ad, adenomas; CRC, colorectal carcinoma; CUP, cancer of unknown primary; duod, duodenal; GC, gastric cancer; LC, lung cancer; polyps, multiple colorectal adenomatous polyps; and yrs, years.
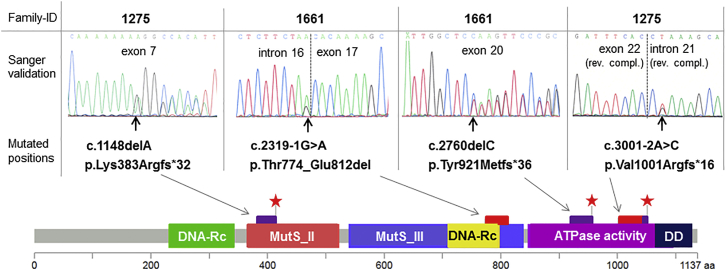
Figure 2.
Germline Mutations in MSH3
All four MSH3 mutations identified in the present study were validated by Sanger sequencing of the respective exons with flanking intronic regions. Their protein-level effects are depicted below in an adapted lollipop plot of MSH3 (created with MutationMapper). A premature stop is symbolized by a red star, a purple bar denotes altered amino acids, and a red bar denotes lost amino acids. Regions with conserved functional residuals43 are highlighted. Abbreviations are as follows: DD, dimerization domain; DNA-Rc, DNA-recognition; and MutS_II-III, PFAM domains of the MutS family.
Clinical Description of Individuals Carrying Biallelic MSH3 Mutations
Individual 1275.1 (II-4 in Figure 1A) is female and was diagnosed with colorectal adenomatous polyposis at age 36 years. She underwent a preventive sigmoidectomy at age 48 years and a right hemicolectomy at age 53 years. Histology results were available for >40 polyps, all of which were tubular or tubulovillous adenomas with low to intermediate dysplasia, often accompanied by inflammatory infiltration. Three distal hyperplastic polyps were also documented. In addition, this person has a history of proliferative disorders in other organs: thyroid adenoma at age 35 years, a small polyp of the corpus uteri and uterine leiomyomas at age 44 years, multiple small intraductal papillomas of (peripheral) mammary glands at age 44 years, and multiple adenomatous polyps in the duodenum at age 50 years. Hypertrophy of the retinal pigment epithelium was excluded by ophthalmological examination at age 50 years.
Individual 1661.1 (II-2 in Figure 1B) is also female and was diagnosed at age 32 years with colorectal tubular and tubulovillous adenomas with low-grade intraepithelial neoplasia. At age 42 years, she underwent proctocolectomy and excision of large duodenal adenomas. This individual has a striking past medical history: at age 26 years, a grade II astrocytoma was diagnosed and surgically treated. At age 27 years, she underwent oophorectomy for the presence of ovarian cysts, including one dermoid cyst. A hysterectomy was performed for a myoma at age 34 years, and a thyroidectomy was performed for follicular adenomas at age 42 years. At age 43 years, she showed a cutaneous fibrolipoma, and at age 46 years, a flat epithelial atypia, multiple peripheral small intraductal papillomas, usual ductal hyperplasias, and cysts with apocrine metaplasia were detected in the mammary glands.
Both index persons have one affected sibling, whereas their respective parents have no reported history of malignant gastrointestinal disease (Figure 1). A sister (individual 1275.2; II-1 in Figure 1A) of individual 1275.1 (II-4 in Figure 1A) was diagnosed with a rectal adenocarcinoma at age 56 years and with a signet cell gastric carcinoma at age 59 years. The available histology reports describe multiple tubulovillous adenomas of the entire colon and proximal duodenum, with up to high-grade intraepithelial neoplasia, and two hyperplastic polyps of the transverse colon. Small bilateral renal cysts were reported as a secondary finding. The brother (individual 1661.2; II-3 in Figure 1B) of individual 1661.1 (II-2 in Figure 1B) was diagnosed with colorectal polyps at age 33 years; he underwent colectomy at age 37 years.
Characterization of the MSH3 Mutations
In total, the two index persons harbor four different MSH3 variants, all with a putative LoF effect. MSH3 (GenBank: NM_002439.4) on 5q14.1 is one of the six MMR genes identified to date in eukaryotic cells.51 It consists of 24 exons and encodes a protein composed of 1,137 amino acids, including several functional domains (Figure 2). In both families, each affected individual carries one frameshift and one splice-site mutation (Figures 1 and 2). All mutations were validated by Sanger sequencing (Figure 2).
The frameshift mutation c.1148delA (p.Lys383Argfs∗32; chr5: g.79970921delA) in exon 7 in family 1275 is predicted to result in a premature stop codon after 31 amino acids. It corresponds to a known somatic cancer mutation located in a poly-A(8) tract of exon 7. The frameshift mutation c.2760delC (p.Tyr921Metfs∗36; chr5: g.80109507delC) in exon 20 in family 1661 is predicted to result in a premature stop codon after 35 amino acids. After transfection of MSH3 plasmids with each frameshift mutation in HEK293T cells, we demonstrated via western blot that the altered proteins are shortened by the expected length (Figures S4A and S4C).
The splice-site mutations c.2319−1G>A (chr5: g.80074538G>A) in intron 16 in family 1661 and c.3001−2A>C (chr5: g.80160630A>C) in intron 21 in family 1275 are both located in highly conserved splice acceptor sites and are predicted to alter splicing in four of four prediction tools. To demonstrate their functional effect, we performed a transcript analysis with primers located in flanking exons (Figures 3A and 3B). RT-PCR products obtained from cDNA of individual 1661.1, who carries the c.2319−1G>A mutation, showed two bands on an agarose gel. Sequencing of the shortened transcript confirmed a loss of exon 17, which is predicted to result in an in-frame loss of 39 amino acids (774–812) at the protein level. These amino acids are involved in DNA recognition (Figures 2 and S5).43 Sequencing of the short RT-PCR fragment from individual 1275.1 (II-4 in Figure 1A), who has the c.3001−2A>C mutation, confirmed a loss of exon 22. This is predicted to result in a frameshift mutation with a premature stop codon after 16 amino acids, altering the dimerization domain (Figures 2 and S5). In addition, we demonstrated via western blot that MSH3 cDNA variants lacking the respective exon lead to altered proteins shortened by the expected length (Figures S4B–S4C).
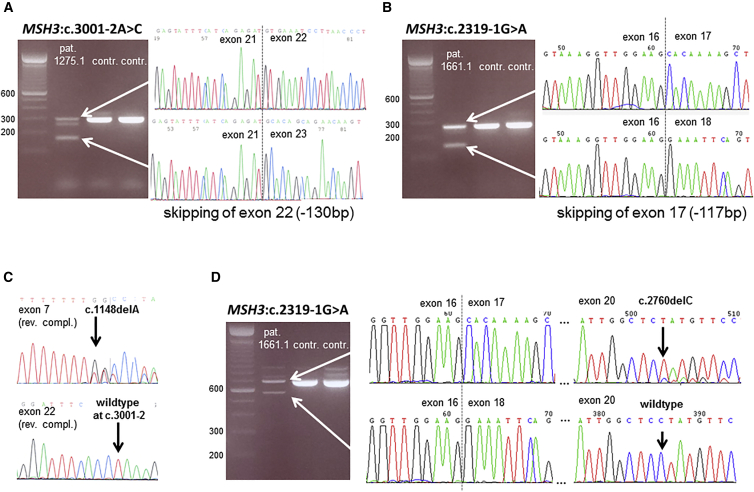
Figure 3.
Transcript Analyses of MSH3 Splice-Site Mutations and Confirmation of Compound Heterozygosity
(A) An agarose gel shows RT-PCR products obtained from mRNA of individual 1275.1 (lane 1) and control samples (lanes 2–3); primers are localized in exon 21 (forward) and exon 23 (reverse). In the affected individual’s sample, a shorter band of around 165 bp is visible in addition to the expected band of 295 bp. Sequencing of the shorter fragment demonstrated a loss of exon 22 caused by the c.3001−2A>C mutation.
(B) An agarose gel shows RT-PCR products obtained from mRNA of individual 1661.1 (lane 1) and control samples (lanes 2–3); primers are localized in exon 16 (forward) and exon 18 (reverse). In the affected individual’s sample, a shorter band of around 162 bp is visible in addition to the expected band of 279 bp. Sequencing of the shorter fragment demonstrated a loss of exon 17 caused by the c.2319−1G>A mutation.
(C) The mutation sites in family 1275 were amplified and sequenced from lymphocyte DNA of an unaffected sibling. Only one of the mutations was detectable in the heterozygous state, confirming that the two mutations are located on separate alleles.
(D) Agarose gel shows RT-PCR products obtained from mRNA of individual 1661.1 (lane 1) and control samples (lanes 2–3); primers are localized in exon 16 (forward) and exon 21 (reverse) in order to capture the effects of the splice-site mutation and the frameshift mutation within the same amplicon. Sequencing the shorter band proved that the product of aberrant splicing does not carry frameshift mutation c.2760delC in exon 20, whereas the frameshift mutation is present in the longer band. This confirms that the two mutations are located on distinct transcripts and thus separate alleles (compound-heterozygous state).
In control individuals (ExAC Browser), the variant c.1148delA is listed ten times (as “chr5:79970914CA/C”), and the variant c.2760delC is listed twice (as “chr5:80109505TC/T”) in a heterozygous state, corresponding to an allele frequency of 0.008% and 0.0016%, respectively. The two other MSH3 variants are not reported in the general population, and none of the variants are listed in the Human Gene Mutation Database (HGMD). To determine the frequency of MSH3 LoF mutations in the normal population, we queried large exome datasets from control individuals (TGP, EVS, and ExAC Browser). Although these datasets listed heterozygous LoF MSH3 mutations with a MAF of <0.2%, no homozygous mutation was reported.
We were able to obtain paraffin-embedded tumor and adjacent normal tissue from the affected sister 1275.2 (II-1 in Figure 1A) and a blood sample from the affected brother 1661.2 (II-3 in Figure 1B) and found the same two MSH3 mutations as those present in the index individual (data not shown).
To confirm compound heterozygosity, we gathered a leukocyte-derived DNA sample from one unaffected sibling (II-7 in Figure 1A) of individual 1275.1 (II-4 in Figure 1A). Examination of both regions mutated in the index individual revealed that the unaffected sibling carries only the frameshift mutation in the heterozygous state and not the splice-site mutation (Figures 1 and 3C). For individual 1661.1 (II-2 in Figure 1B), the biallelic genotype was confirmed via transcript analysis: by creating an amplicon spanning both mutated regions, we could demonstrate that the transcript of normal length carries the single-nucleotide deletion resulting in a frameshift mutation. In contrast, this single-nucleotide deletion was not detected in the shortened product transcribed from the allele carrying the splice-site mutation (Figure 3D).
To assess potential additional causal alterations, we performed supplementary sequencing and MLPA of genes with influence on MSH3: neither of the two index individuals showed a germline mutation in other MMR genes (MLH1, MSH2, MSH6, or PMS2), EPCAM, or TP53 (tumor protein p53 [MIM: 191170]), any evidence of APC mosaicism in leukocyte DNA, or somatic MSH2 or MSH6 mutations in tumor tissue. Furthermore, IHC staining of two adenomas per index person demonstrated a strong presence of the MMR proteins MLH1, MSH2, MSH6, and PMS2 (Figure S6).
Effect of MSH3 Deficiency on Colorectal Tumor Tissue
IHC staining with a rabbit polyclonal antibody proved complete nuclear loss of MSH3 in normal colon mucosa of individual 1275.1 and in adenomas of both index persons (Figure 4). In the control samples, non-tumorous mucosa of an independent person with CRC showed MSH3 predominantly located in the nuclei.
Figure 4.
IHC Staining of MSH3 in Tumor and Normal Tissue
MSH3 was stained with rabbit polyclonal antibody targeting N-terminal human MSH3.
(A) MSH3 was nearly undetectable in FFPE normal colon mucosa and colorectal adenoma samples from index person 1275.1 and a colorectal adenoma sample from index person 1661.1, who has a complete loss of nuclear MSH3.
(B) In contrast, control samples, which were taken from FFPE normal mucosa of an independent subject with colon cancer and were processed in parallel, show a strong nuclear MSH3 staining. The same results were obtained in an examination of a second independent colorectal adenoma from both index persons (data not shown).
Scale bars represent 100 μm.
In adenoma-derived DNA from both index persons, we found stability of mononucleotide repeats. Complementarily, we examined several dinucleotide and tetranucleotide markers in order to focus on lesions that are processed by MSH3. Individuals 1275.1 and 1661.1 exhibited instability of one of four (MSI-L) and three of four (MSI-H) dinucleotide markers, respectively. In addition, individuals 1275.1 and 1275.2 showed instability of three of six and three of five tetranucleotide markers, respectively, whereas individual 1661.1 displayed instability of four of six tetranucleotide markers (Figures 5 and S7). Thus, we demonstrated EMAST in tumors of all three examined individuals.
Figure 5.
Results of Microsatellite Analysis
Five tetranucleotide repeat markers (D20S82, D2S443, D21S1436, D9S747, and UT5037) were examined in normal (N) and tumor (T) tissue. In each tumor sample, two or three out of five markers showed clear instability and thus demonstrated elevated microsatellite instability at selected tetranucleotide repeats (EMAST). The x axes denote peak positions related to fragment lengths. Please note that only x axes within tumor-normal pairs allow direct comparison. The y axes quantifying peak heights were generally not standardized. The results of both index persons were confirmed in a second colorectal adenoma (data not shown).
In addition, we used the adenoma-derived DNA of individual 1661.1 to screen for somatic APC mutations. Using targeted deep sequencing, we compared four independent polyps with leukocyte DNA and found seven different somatic APC mutations (one to two per polyp) in 6%–36% of the reads (Figure S8). All mutations were small deletions of two to eight nucleotides. In four of seven mutations, the sequence context proved to be di- or trinucleotide repeats.
Identification of a Biallelic PMS2 Mutation
Individual 1138 harbors the PMS2 germline mutations c.2T>A (p.Met1?) in exon 1 and c.863delA (p.Gln288Argfs∗19) in exon 8 (GenBank: NM_000535.5) (Figure S9A). Both mutations were validated by Sanger sequencing (Figure S9B). Compound heterozygosity could be confirmed given that the healthy mother only carries mutation c.863delA in exon 8. The start-loss mutation c.2T>A (p.Met1?) was predicted to be pathogenic or damaging by two of three in-silico tools. In accordance with the assumed protein truncation caused by the two germline mutations, IHC staining showed complete loss of PMS2 in both tumor and normal tissue (Figure S9C).
Additional clinical information and careful re-evaluation of the medical history revealed that individual 1138 had been diagnosed with early-onset colorectal polyposis with 20–25 adenomas at age 14 years and had undergone proctocolectomy with pouch-anal anastomosis at age 16 years. In addition, a primitive neuroectodermal tumor of the cerebellum had been diagnosed at 4 years, and a history of a pilomatrixoma, thyroid cysts, and three café-au-lait spots had been reported. The family history was unremarkable.
Mutations in Further Candidate Genes
In the remaining individuals with unexplained polyposis, 29 different rare mutations in 14 protein-coding genes were found in 26 other individuals (Table S4 and Figure S3). All genes, apart from one (DNAJB7 [DnaJ heat shock protein family (Hsp40) member B7 (MIM: 611336)]), are reported to be expressed in colon tissue. Two genes (MAGT1 [magnesium transporter 1 (MIM: 300715)] and SLC27A5 [solute carrier family 27 member 5 (MIM: 603314)]) were affected by a homozygous LoF mutation in one individual each, and seven genes (BTBD9 [BTB domain containing 9 (MIM: 611237)], CD36 [CD36 molecule (MIM: 173510)], ECHDC3 [enoyl-CoA hydratase domain containing 3], SSC5D [scavenger receptor cysteine rich family member with 5 domains], UGGT2 [UDP-glucose glycoprotein glucosyltransferase 2 (MIM: 605898)], WDR35 [WD repeat domain 35 (MIM: 613602)], and ZC3H8 [zinc finger CCCH-type containing 8]) were recurrently affected by heterozygous LoF mutations. Of these, three genes (CD36, WDR35, and ZC3H8) have been implicated in cell adhesion or apoptosis. Five individuals were found to carry more than one heterozygous mutation. On the basis of the CNV data, we identified no further heterozygous or additional biallelic large duplication or deletion in these genes.
Discussion
Most cases of colorectal adenomatous polyposis are attributable to heterozygous germline mutations in the tumor-suppressor gene APC and are thus diagnosed as FAP. However, the few novel subtypes delineated in recent years are caused by genes involved in DNA repair. Whereas heterozygous mutations affecting the proofreading domain encoded by DNA polymerase genes POLE and POLD1 lead to the rare, dominantly inherited PPAP, recessive MUTYH-associated polyposis and NTHL1-associated polyposis are caused by biallelic germline mutations in BER genes.3, 7, 10 After causal variants in those genes were initially described in only a few families, identification of additional cases expanded the mutation spectrum and allowed refinement of the respective phenotypes.8, 9, 52, 53
In a number of individuals with colorectal adenomatous polyposis, however, no germline mutation in the established genes has been identified. Although the synchronous or metachronous occurrence of dozens to hundreds of adenomas is strongly suggestive of an underlying genetic basis, so far it remains unclear whether the predisposing genetic factors mainly act in a monogenic fashion or contribute as low- or moderate-penetrance variants to a more complex, oligo- or polygenic trait.
Interestingly, increasing evidence suggests that biallelic germline mutations in MMR genes can result in a phenotype with features overlapping those of colorectal polyposis. Typically, these conditions are designated as CMMRD or biallelic MMR deficiency and are characterized by early-onset CRC, brain tumors, hematological malignancies, and café-au-lait skin macules.31, 32 Nonetheless, several individuals with homozygous or compound-heterozygous PMS2, MSH2, or MSH6 germline mutations and an early-onset colorectal adenomatous polyposis in the second or first decade of life have been described;23, 31, 54 the majority of these cases were until then misclassified as mutation-negative FAP. In some individuals, particularly those with biallelic PMS2 mutations, however, the colorectal phenotype manifests not before the third or even fourth decade of life, resembling the clinical presentation in the present polyposis cohort.
To uncover further monogenic causes, we performed exome sequencing of leukocyte DNA in a cohort of 102 unrelated individuals with histologically confirmed, genetically unexplained adenomatous polyposis. The clinical and family characteristics of the participants are consistent with published data from other mutation-negative polyposis cohorts.14, 15, 19, 55 Using this approach, we identified two families affected by biallelic LoF germline mutations in the MMR gene MSH3, a genotype that has not yet been described as causative for a polyposis phenotype. In addition, we found one individual with biallelic PMS2 mutations and several persons harboring homo- and heterozygous LoF variants.
Biallelic MSH3 Germline Mutations
The genotypes and pedigrees of the two unrelated persons with compound-heterozygous MSH3 germline mutations are in full agreement with a recessively inherited trait. Interestingly, unlike the majority of the examined cohort, these two individuals do have affected siblings and documented extraintestinal neoplasias. Neither index person has a germline mutation in any of the known genes associated with gastrointestinal polyposis nor any further mutation in other MMR genes or EPCAM. In addition, IHC staining of MLH1, MSH2, MSH6, and PMS2 was normal. The haploinsufficiency score (0.486) of MSH3 indicated a rather low probability of haploinsufficiency (16.2%).39 In large control sets, none of the LoF MSH3 germline mutations were identified in the homozygous state, and the frequency of heterozygosity is compatible with a rare recessive disease.
Confirming compound heterozygosity is critical to demonstrating recessive inheritance; in both families, we have clearly shown that the two mutations are located on different alleles either by examination of an unaffected sibling who is heterozygous for just one mutation (family 1275) or by transcript analysis indicating that both mutations are located on different alleles (family 1661). Taken together, these data strongly support the hypothesis that deleterious MSH3 mutations follow a recessive mode of inheritance.
The MMR system is a critical pathway that corrects base-base and indel mispairs occurring as a result of replication errors, thus increasing the fidelity of DNA replication.56 Defects in the MMR system result in a mutator phenotype, which manifests as MSI in the DNA of affected cells. In tumors with MSI, microsatellite loci containing mono-, di-, tri-, and tetranucleotide repeats can be affected.57
During DNA repair, mispaired bases are recognized by two heterodimers of MutS homologs (DNA mismatch recognition complex)—MSH2 and MSH6 (MutSα) and MSH2 and MSH3 (MutSβ)—with partially overlapping mispair-recognition specificities.56 In humans, MutSα efficiently binds single-base substitutions and small (single-base) indel mispairs, whereas MutSβ has a stronger affinity for larger base-indel loops with up to ten unpaired nucleotides.57, 58 Thus, loss of MutSβ due to MSH3 inactivation in human cells not only results in MSI at loci containing dinucleotide repeats but also results in MSI at certain loci with tetranucleotide repeats, termed EMAST.57, 59 It is known that di- and tetranucleotide repeats are affected in the majority of CRC with MSI-L.58 MMR deficiency can also result from an imbalance in the relative amounts of MSH3 or MSH6.60
All four MSH3 germline mutations detected in the present cohort are strongly predicted to have a LoF effect. According to previous work, somatic MSH3 frameshift mutations at the (A)8 repeat in exon 7 result in a loss of MSH3.45, 61 To evaluate the pathophysiological consequences of the four mutations in more detail, we performed several experiments. Using transcript analysis, we confirmed aberrant splicing caused by the two mutations located within the conserved consensus splice motifs. This would affect regions relevant for dimerization and for DNA recognition, according to the MSH3 structure described by Yang’s group (Figure S5).43
Three of the four identified MSH3 mutations are predicted to result in premature stop codons and thus might lead to nonsense-mediated mRNA decay (NMD). However, mRNA analysis, performed with fresh blood samples not treated with NMD inhibitors, demonstrated that the affected transcripts are expressed. This might be due to NMD escape, a phenomenon that is known from several mutations in other polyposis and MMR genes and that was also described for MSH3. You et al. found that MSH3 transcripts with a frameshift mutation at the (A)8 repeat in exon 7 are not degraded by NMD but instead experience repression of protein translation.62
The western blot experiments illustrated that the altered MSH3 proteins are shortened by the expected length (Figure S4), which would lead to a loss of the conserved C-terminal dimerization domain (Figures 2 and S5).43 Because the stable proteins were obtained in vitro in a human embryonic kidney cell line and a strong promoter for high-level expression, this observation is not per se transferable to the in vivo situation. In fact, IHC staining clearly demonstrated loss of nuclear MSH3 in both normal and colorectal tumor tissues of the affected individuals, confirming the expected MSH3 deficiency. Different mechanisms such as repressed protein translation or blocked nuclear transport by hampered dimerization or changes in protein conformation might explain the nuclear absence of MSH3.
Microsatellite analysis of adenoma-derived DNA demonstrated EMAST, high and low instability at dinucleotide markers, and no instability at mononucleotide repeats in any of the examined tumors. These findings further confirm the functional relevance of the MSH3 mutations. In addition, presumed effects of the MSH3 deficiency are well reflected by the inflammatory infiltration, a characteristic feature of MSI colorectal tumors, and the somatic APC mutation spectrum observed in the adenomas.
Several lines of evidence support the causal relevance of MSH3 deficiency to the initiation of genetic instability and tumorigenesis. Around 50% of MSI tumors contain somatic frameshift mutations in the (A)8 tract in codons 381–383 of MSH3.57, 63, 64 The detection of LOH in some of these tumors supports the role of MSH3 and MSH6 as primary mutators. In CRC and human colon epithelial cells, MSH3 deficiency is associated with EMAST ([AAAG]n repeats) and MSI-L at dinucleotide repeats and results in the formation of double-strand breaks and significant changes in the proteome.57, 65
In yeast and extracts of Msh3−/− cells, Msh3 deficiency leads to a partial MMR defect and MSI.66, 67, 68 In mouse models, elimination of either Msh3 or Msh6 alone still maintains some functional MMR activity, which is consistent with the persistence of the MutSα or MutSβ heterodimer, respectively. Of all MMR-knockout mice, Msh3-deficient mice exhibited the lowest, yet still significantly elevated, mutation frequencies in comparison to wild-type mice.68
Whereas MLH1, MSH2, MSH6, and PMS2 are established genes associated with Lynch syndrome, the causal relevance of MSH3 germline variants in cancer predisposition has remained uncertain until now. To date, MSH3 mutations have neither been consistently linked to a Lynch-like phenotype nor described in polyposis cases. In several previous studies, common MSH3 polymorphisms were significantly associated with CRC and prostate cancer as low-penetrance risk alleles.69, 70, 71, 72 In contrast, a potentially high-penetrance pathogenic MSH3 germline mutation has very rarely been identified in persons with a suspected predisposition to cancer.73, 74 Msh3-deficient mice develop late-onset MSI gastrointestinal cancers. However, given the small number of reported tumors, the significance of this finding remains unclear, and survival did not differ significantly from that of wild-type control animals.67, 75, 76
In a Chinese cohort with suspected familial breast cancer, Yang et al. found three heterozygous MSH3 germline variants (two in-frame deletions and one frameshift mutation; Table S5). They examined eight tumor samples from three families, and all showed MSS in the standard marker panel (BAT25, BAT26, D2S123, D5S346, and D17S250) and, on average, two instable loci in nine additional dinucleotide or EMAST markers. Two individuals showed less MSH3 staining in tumor tissue (breast and ovary) than was shown in normal tissue.74 One family showed no evidence of Lynch syndrome; the MSH3 in-frame variant segregated incompletely with the disease, and MSH3 staining showed no relevant deficiency in the tumors. The second family met the clinical criteria for Lynch syndrome, and the MSH3 in-frame germline variant segregated well with the disease in three generations. However, the tumor spectrum was broad and included breast, ovarian, renal, and colon cancer. It was not reported whether genetic causes of Lynch syndrome had been excluded systematically. The third family carried the MSH3 frameshift variant, which incompletely segregated with the disease. A comparison with currently available frequency data in the general population (ExAC Browser) suggests that both in-frame deletions are likely to be polymorphisms (Table S5).
In a family with suspected but genetically unexplained Lynch syndrome, Duraturo et al. found that two brothers each with three metachronous CRCs had a compound-heterozygous MSH3 genotype comprising a potentially pathogenic missense variant and a silent variant.73 However, no functional data to confirm the pathogenicity of the variants was reported. Moreover, the silent MSH3 variant is meanwhile listed as a frequent polymorphism (rs1805355; Table S5).
In our study, all carriers of biallelic MSH3 mutations have attenuated colorectal and duodenal involvement and no or late-onset cancer. This is similar to the phenotype observed in persons with MAP or attenuated FAP and is consistent with the phenotype described in MSH3-knockout mice. Two of the four carriers are reported to have extraintestinal tumors: whereas various thyroid neoplasias also occur in FAP and MAP individuals, the early-onset astrocytoma fits well in the tumor spectrum observed in CMMRD.
A high frequency of EMAST was also observed in a wide range of extraintestinal sporadic malignancies, such as skin, bladder, kidney, lung, ovarian, head, and neck cancer,59, 77, 78 although the underlying mechanism remained unclear, and an association with MSH3 impairment was not proven. Recent studies, however, provide strong evidence that EMAST formation is driven by MSH3 deficiency either as a result of MSH3 mutations or, e.g., by a nuclear-to-cytosol shift induced by oxidative stress.79, 80 Thus, it can be speculated that MSH3-induced EMAST is more common than previously thought and might occur in different tumor types. Consequently, the tumor spectrum in individuals with biallelic MSH3 germline mutations might include a much broader extraintestinal tumor spectrum than observed in the persons identified in the present study.
Although the clinical information and underlying molecular changes point to a broader tumor spectrum and some degree of overlap with CMMRD, exploring the whole oncologic phenotype will require further individuals with biallelic MSH3 mutations.
Biallelic PMS2 Germline Mutations
The identification of one individual with a biallelic PMS2 mutation demonstrates that CMMRD is a rare but important cause of adenomatous polyposis. The c.2T>A (p.Met1?) start-loss mutation is listed twice in ClinVar and is considered pathogenic. According to data from the ExAC Browser, the allele frequency of this mutation in the European population is 0.003%. Apart from the individual reported here, we recently identified another person with a CMMRD phenotype (B cell lymphoma, acute lymphatic leukemia, carcinoma of the rectum, and a multifocal grade III-IV astrocytoma occurring between the ages of 9 and 15 years) in a multiple-tumor cohort. This person carries similar start-loss (c.1A>T [p.Met1?]) and frameshift (c.2117delA [p.Lys706Serfs∗19]) mutations in PMS2. A similar potential founder mutation in the start codon (c.1A>G) was found in three unrelated CMMRD individuals, all of whom have a compound-heterozygous PMS2 genotype and isolated loss of PMS2 on IHC staining.81 The PMS2 locus-specific database (Leiden Open Variation Database [LOVD]) lists a fourth family with the same genotype. Although the person identified in the present study has extracolonic features suggestive of CMMRD, these manifestations are often unspecific and could remain unreported or unrecognized (e.g., café-au-lait spots). This suggests that CMMRD is an underdiagnosed condition and should be included in the differential diagnosis of any unexplained early-onset case of adenomatous polyposis.
Further Candidate Genes
Under the assumption of a monogenic mode of inheritance with high penetrance, the frequency of causative germline mutations in the general population is expected to be low. In 26 of the 96 remaining individuals (excluding three samples after quality control and three resolved cases), we identified unique (i.e., not present in control individuals) or rare (i.e., frequency < 0.01% for the dominant or 1% for the recessive model in control individuals) potentially pathogenic germline variants in 14 protein-coding genes. The causative relevance of these interesting candidate genes awaits exploration in larger cohorts and via functional analysis.
In the present study, mutations might have been overlooked, e.g., in low-coverage regions or within repeat tracts in coding sequences. Moreover, some causative mutations might be located beyond the exome, e.g., in non-coding regions or in unannotated genes.
Conclusions
In conclusion, this study describes the identification of biallelic pathogenic MSH3 germline mutations as a cause of an inherited tumor syndrome. Specifically, biallelic LoF MSH3 germline mutations appear to cause an additional rare recessively inherited subtype of colorectal adenomatous polyposis, which was present in 2% of the study participants. Data from the present and previous studies consistently show that mutations in newly identified genes associated with inherited tumor predisposition syndromes are very rare (0.3%–0.5% in unexplained polyposis cohorts with familial cancer).7, 82 At least some of these syndromes appear to show extreme genetic heterogeneity, and identifying recurrently mutated genes will require large cohorts.
Preliminary experiments indicate that MSH3-deficient cells are more sensitive to cisplatin treatment or platinum-based adjuvant treatment for CRC.57 Thus, MSH3 deficiency might also be of therapeutic relevance for individuals with MSH3-associated polyposis.
Conflicts Of Interest
M.M.N. is managing scientific director of Life & Brain GmbH.
Acknowledgments
We thank the individuals and their families for participating in the study and Prof. Grazia Graziani (University of Rome Tor Vergata, Italy) for generously providing the pcDNA3.1−/MSH3-WT vector. This work was supported by the German Cancer Aid (grant no. 108421, Deutsche Krebshilfe, Bonn), the Gerok-Stipendium of the University Hospital Bonn (grant no. O-149.0098), and NIH Centers for Mendelian Genomics (5U54HG006504). R.C.B. and M.M.N. are members of the Excellence Cluster ImmunoSensation, funded by the German Research Foundation (Deutsche Forschungsgemeinschaft). The funding sources had no involvement in the study design; the collection, analysis, or interpretation of data; the writing of the report; or the decision to submit the paper for publication. The corresponding author (S.A.) had full access to all data in the study and had final responsibility for the decision to submit the manuscript for publication.
Published: July 28, 2016
Footnotes
Supplemental Data include nine figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.06.015.
Accession Numbers
The accession numbers for the variant data reported in this paper are LOVD: 00074677 and 00074678.
Web Resources
1000 Genomes, http://www.1000genomes.org
Berkeley Drosophila Genome Project NNSplice 0.9, http://www.fruitfly.org/seq_tools/splice.html
ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/
dbSNP, www.ncbi.nlm.nih.gov/SNP/
Ensembl (release 54), http://may2009.archive.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
HGMD, http://www.hgmd.cf.ac.uk
International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
LOVD 2.0, APC, http://www.lovd.nl/APC
LOVD 2.0, MUTYH, http://www.lovd.nl/MUTYH
LOVD 2.0, PMS2, http://www.lovd.nl/PMS2
LOVD 3.0, MSH3, http://databases.lovd.nl/shared/genes/MSH3
MutationMapper, http://www.cbioportal.org/mutation_mapper.jsp
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Primer3, http://bioinfo.ut.ee/primer3-0.4.0/primer3/input.htm
Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
SIFT, http://sift.jcvi.org/
The Human Protein Atlas, http://www.proteinatlas.org/
UCSC Genome Browser, http://genome.ucsc.edu
UMD-APC Mutations Database, http://www.umd.be/APC/
UMD-MUTYH Mutations Database, http://www.umd.be/MUTYH/
UniGene, http://www.ncbi.nlm.nih.gov/unigene/
Supplemental Data
References
- 1.Galiatsatos P., Foulkes W.D. Familial adenomatous polyposis. Am. J. Gastroenterol. 2006;101:385–398. doi: 10.1111/j.1572-0241.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 2.Grover S., Kastrinos F., Steyerberg E.W., Cook E.F., Dewanwala A., Burbidge L.A., Wenstrup R.J., Syngal S. Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. JAMA. 2012;308:485–492. doi: 10.1001/jama.2012.8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R., Cheadle J.P. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 4.Mazzei F., Viel A., Bignami M. Role of MUTYH in human cancer. Mutat. Res. 2013;743-744:33–43. doi: 10.1016/j.mrfmmm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Krawitz P.M., Schweiger M.R., Rödelsperger C., Marcelis C., Kölsch U., Meisel C., Stephani F., Kinoshita T., Murakami Y., Bauer S. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- 6.Gilissen C., Hoischen A., Brunner H.G., Veltman J.A. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palles C., Cazier J.B., Howarth K.M., Domingo E., Jones A.M., Broderick P., Kemp Z., Spain S.L., Guarino E., Salguero I., CORGI Consortium. WGS500 Consortium Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valle L., Hernández-Illán E., Bellido F., Aiza G., Castillejo A., Castillejo M.I., Navarro M., Seguí N., Vargas G., Guarinos C. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum. Mol. Genet. 2014;23:3506–3512. doi: 10.1093/hmg/ddu058. [DOI] [PubMed] [Google Scholar]
- 9.Spier I., Holzapfel S., Altmüller J., Zhao B., Horpaopan S., Vogt S., Chen S., Morak M., Raeder S., Kayser K. Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int. J. Cancer. 2015;137:320–331. doi: 10.1002/ijc.29396. [DOI] [PubMed] [Google Scholar]
- 10.Weren R.D., Ligtenberg M.J., Kets C.M., de Voer R.M., Verwiel E.T., Spruijt L., van Zelst-Stams W.A., Jongmans M.C., Gilissen C., Hehir-Kwa J.Y. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 11.Spier I., Kerick M., Drichel D., Horpaopan S., Altmüller J., Laner A., Holzapfel S., Peters S., Adam R., Zhao B. Exome sequencing identifies potential novel candidate genes in patients with unexplained colorectal adenomatous polyposis. Fam. Cancer. 2016;15:281–288. doi: 10.1007/s10689-016-9870-z. [DOI] [PubMed] [Google Scholar]
- 12.Knudsen A.L., Bisgaard M.L., Bülow S. Attenuated familial adenomatous polyposis (AFAP). A review of the literature. Fam. Cancer. 2003;2:43–55. doi: 10.1023/a:1023286520725. [DOI] [PubMed] [Google Scholar]
- 13.Renkonen E.T., Nieminen P., Abdel-Rahman W.M., Moisio A.L., Järvelä I., Arte S., Järvinen H.J., Peltomäki P. Adenomatous polyposis families that screen APC mutation-negative by conventional methods are genetically heterogeneous. J. Clin. Oncol. 2005;23:5651–5659. doi: 10.1200/JCO.2005.14.712. [DOI] [PubMed] [Google Scholar]
- 14.Thirlwell C., Howarth K.M., Segditsas S., Guerra G., Thomas H.J., Phillips R.K., Talbot I.C., Gorman M., Novelli M.R., Sieber O.M., Tomlinson I.P. Investigation of pathogenic mechanisms in multiple colorectal adenoma patients without germline APC or MYH/MUTYH mutations. Br. J. Cancer. 2007;96:1729–1734. doi: 10.1038/sj.bjc.6603789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hes F.J., Ruano D., Nieuwenhuis M., Tops C.M., Schrumpf M., Nielsen M., Huijts P.E., Wijnen J.T., Wagner A., Gómez García E.B. Colorectal cancer risk variants on 11q23 and 15q13 are associated with unexplained adenomatous polyposis. J. Med. Genet. 2014;51:55–60. doi: 10.1136/jmedgenet-2013-102000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venesio T., Balsamo A., Rondo-Spaudo M., Varesco L., Risio M., Ranzani G.N. APC haploinsufficiency, but not CTNNB1 or CDH1 gene mutations, accounts for a fraction of familial adenomatous polyposis patients without APC truncating mutations. Lab. Invest. 2003;83:1859–1866. doi: 10.1097/01.lab.0000106722.37873.8d. [DOI] [PubMed] [Google Scholar]
- 17.Fearnhead N.S., Wilding J.L., Winney B., Tonks S., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc. Natl. Acad. Sci. USA. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dallosso A.R., Dolwani S., Jones N., Jones S., Colley J., Maynard J., Idziaszczyk S., Humphreys V., Arnold J., Donaldson A. Inherited predisposition to colorectal adenomas caused by multiple rare alleles of MUTYH but not OGG1, NUDT1, NTH1 or NEIL 1, 2 or 3. Gut. 2008;57:1252–1255. doi: 10.1136/gut.2007.145748. [DOI] [PubMed] [Google Scholar]
- 19.Mongin C., Coulet F., Lefevre J.H., Colas C., Svrcek M., Eyries M., Lahely Y., Fléjou J.F., Soubrier F., Parc Y. Unexplained polyposis: a challenge for geneticists, pathologists and gastroenterologists. Clin. Genet. 2012;81:38–46. doi: 10.1111/j.1399-0004.2011.01676.x. [DOI] [PubMed] [Google Scholar]
- 20.Spier I., Horpaopan S., Vogt S., Uhlhaas S., Morak M., Stienen D., Draaken M., Ludwig M., Holinski-Feder E., Nöthen M.M. Deep intronic APC mutations explain a substantial proportion of patients with familial or early-onset adenomatous polyposis. Hum. Mutat. 2012;33:1045–1050. doi: 10.1002/humu.22082. [DOI] [PubMed] [Google Scholar]
- 21.Spier I., Drichel D., Kerick M., Kirfel J., Horpaopan S., Laner A., Holzapfel S., Peters S., Adam R., Zhao B. Low-level APC mutational mosaicism is the underlying cause in a substantial fraction of unexplained colorectal adenomatous polyposis cases. J. Med. Genet. 2016;53:172–179. doi: 10.1136/jmedgenet-2015-103468. [DOI] [PubMed] [Google Scholar]
- 22.Azzopardi D., Dallosso A.R., Eliason K., Hendrickson B.C., Jones N., Rawstorne E., Colley J., Moskvina V., Frye C., Sampson J.R. Multiple rare nonsynonymous variants in the adenomatous polyposis coli gene predispose to colorectal adenomas. Cancer Res. 2008;68:358–363. doi: 10.1158/0008-5472.CAN-07-5733. [DOI] [PubMed] [Google Scholar]
- 23.Will O., Carvajal-Carmona L.G., Gorman P., Howarth K.M., Jones A.M., Polanco-Echeverry G.M., Chinaleong J.A., Günther T., Silver A., Clark S.K., Tomlinson I. Homozygous PMS2 deletion causes a severe colorectal cancer and multiple adenoma phenotype without extraintestinal cancer. Gastroenterology. 2007;132:527–530. doi: 10.1053/j.gastro.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 24.Rio Frio T., Lavoie J., Hamel N., Geyer F.C., Kushner Y.B., Novak D.J., Wark L., Capelli C., Reis-Filho J.S., Mai S. Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N. Engl. J. Med. 2010;363:2628–2637. doi: 10.1056/NEJMoa1006565. [DOI] [PubMed] [Google Scholar]
- 25.Lefevre J.H., Bonilla C., Colas C., Winney B., Johnstone E., Tonks S., Day T., Hutnik K., Boumertit A., Soubrier F. Role of rare variants in undetermined multiple adenomatous polyposis and early-onset colorectal cancer. J. Hum. Genet. 2012;57:709–716. doi: 10.1038/jhg.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Voer R.M., Geurts van Kessel A., Weren R.D., Ligtenberg M.J., Smeets D., Fu L., Vreede L., Kamping E.J., Verwiel E.T., Hahn M.M. Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are risk factors for colorectal cancer. Gastroenterology. 2013;145:544–547. doi: 10.1053/j.gastro.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Tomlinson I.P., Webb E., Carvajal-Carmona L., Broderick P., Howarth K., Pittman A.M., Spain S., Lubbe S., Walther A., Sullivan K., CORGI Consortium. EPICOLON Consortium A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 28.Horpaopan S., Spier I., Zink A.M., Altmüller J., Holzapfel S., Laner A., Vogt S., Uhlhaas S., Heilmann S., Stienen D. Genome-wide CNV analysis in 221 unrelated patients and targeted high-throughput sequencing reveal novel causative candidate genes for colorectal adenomatous polyposis. Int. J. Cancer. 2015;136:E578–E589. doi: 10.1002/ijc.29215. [DOI] [PubMed] [Google Scholar]
- 29.Lynch H.T., Lynch P.M., Lanspa S.J., Snyder C.L., Lynch J.F., Boland C.R. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niessen R.C., Hofstra R.M., Westers H., Ligtenberg M.J., Kooi K., Jager P.O., de Groote M.L., Dijkhuizen T., Olderode-Berends M.J., Hollema H. Germline hypermethylation of MLH1 and EPCAM deletions are a frequent cause of Lynch syndrome. Genes Chromosomes Cancer. 2009;48:737–744. doi: 10.1002/gcc.20678. [DOI] [PubMed] [Google Scholar]
- 31.Durno C.A., Sherman P.M., Aronson M., Malkin D., Hawkins C., Bakry D., Bouffet E., Gallinger S., Pollett A., Campbell B., Tabori U., International BMMRD Consortium Phenotypic and genotypic characterisation of biallelic mismatch repair deficiency (BMMR-D) syndrome. Eur. J. Cancer. 2015;51:977–983. doi: 10.1016/j.ejca.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Lavoine N., Colas C., Muleris M., Bodo S., Duval A., Entz-Werle N., Coulet F., Cabaret O., Andreiuolo F., Charpy C. Constitutional mismatch repair deficiency syndrome: clinical description in a French cohort. J. Med. Genet. 2015;52:770–778. doi: 10.1136/jmedgenet-2015-103299. [DOI] [PubMed] [Google Scholar]
- 33.Seguí N., Mina L.B., Lázaro C., Sanz-Pamplona R., Pons T., Navarro M., Bellido F., López-Doriga A., Valdés-Mas R., Pineda M. Germline Mutations in FAN1 Cause Hereditary Colorectal Cancer by Impairing DNA Repair. Gastroenterology. 2015;149:563–566. doi: 10.1053/j.gastro.2015.05.056. [DOI] [PubMed] [Google Scholar]
- 34.Aretz S., Stienen D., Uhlhaas S., Pagenstecher C., Mangold E., Caspari R., Propping P., Friedl W. Large submicroscopic genomic APC deletions are a common cause of typical familial adenomatous polyposis. J. Med. Genet. 2005;42:185–192. doi: 10.1136/jmg.2004.022822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloğlu A., Ozen S., Sanjad S. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 38.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaughn C.P., Robles J., Swensen J.J., Miller C.E., Lyon E., Mao R., Bayrak-Toydemir P., Samowitz W.S. Clinical analysis of PMS2: mutation detection and avoidance of pseudogenes. Hum. Mutat. 2010;31:588–593. doi: 10.1002/humu.21230. [DOI] [PubMed] [Google Scholar]
- 41.Tentori L., Muzi A., Dorio A.S., Dolci S., Campolo F., Vernole P., Lacal P.M., Praz F., Graziani G. MSH3 expression does not influence the sensitivity of colon cancer HCT116 cell line to oxaliplatin and poly(ADP-ribose) polymerase (PARP) inhibitor as monotherapy or in combination. Cancer Chemother. Pharmacol. 2013;72:117–125. doi: 10.1007/s00280-013-2175-0. [DOI] [PubMed] [Google Scholar]
- 42.Brieger A., Plotz G., Zeuzem S., Trojan J. Thymosin beta 4 expression and nuclear transport are regulated by hMLH1. Biochem. Biophys. Res. Commun. 2007;364:731–736. doi: 10.1016/j.bbrc.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Gupta S., Gellert M., Yang W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2011;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleczkowska H.E., Marra G., Lettieri T., Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plaschke J., Krüger S., Jeske B., Theissig F., Kreuz F.R., Pistorius S., Saeger H.D., Iaccarino I., Marra G., Schackert H.K. Loss of MSH3 protein expression is frequent in MLH1-deficient colorectal cancer and is associated with disease progression. Cancer Res. 2004;64:864–870. doi: 10.1158/0008-5472.can-03-2807. [DOI] [PubMed] [Google Scholar]
- 46.Kloth M., Ruesseler V., Engel C., Koenig K., Peifer M., Mariotti E., Kuenstlinger H., Florin A., Rommerscheidt-Fuss U., Koitzsch U. Activating ERBB2/HER2 mutations indicate susceptibility to pan-HER inhibitors in Lynch and Lynch-like colorectal cancer. Gut. 2016;65:1296–1305. doi: 10.1136/gutjnl-2014-309026. [DOI] [PubMed] [Google Scholar]
- 47.Boland C.R., Thibodeau S.N., Hamilton S.R., Sidransky D., Eshleman J.R., Burt R.W., Meltzer S.J., Rodriguez-Bigas M.A., Fodde R., Ranzani G.N., Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 48.Suraweera N., Duval A., Reperant M., Vaury C., Furlan D., Leroy K., Seruca R., Iacopetta B., Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 49.Bellizzi A.M., Frankel W.L. Colorectal cancer due to deficiency in DNA mismatch repair function: a review. Adv. Anat. Pathol. 2009;16:405–417. doi: 10.1097/PAP.0b013e3181bb6bdc. [DOI] [PubMed] [Google Scholar]
- 50.Burger M., Burger S.J., Denzinger S., Wild P.J., Wieland W.F., Blaszyk H., Obermann E.C., Stoehr R., Hartmann A. Elevated microsatellite instability at selected tetranucleotide repeats does not correlate with clinicopathologic features of bladder cancer. Eur. Urol. 2006;50:770–775. doi: 10.1016/j.eururo.2006.04.010. discussion 776. [DOI] [PubMed] [Google Scholar]
- 51.Fujii H., Shimada T. Isolation and characterization of cDNA clones derived from the divergently transcribed gene in the region upstream from the human dihydrofolate reductase gene. J. Biol. Chem. 1989;264:10057–10064. [PubMed] [Google Scholar]
- 52.Rivera B., Castellsagué E., Bah I., van Kempen L.C., Foulkes W.D. Biallelic NTHL1 Mutations in a Woman with Multiple Primary Tumors. N. Engl. J. Med. 2015;373:1985–1986. doi: 10.1056/NEJMc1506878. [DOI] [PubMed] [Google Scholar]
- 53.Sieber O.M., Lipton L., Crabtree M., Heinimann K., Fidalgo P., Phillips R.K., Bisgaard M.L., Orntoft T.F., Aaltonen L.A., Hodgson S.V. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 54.Herkert J.C., Niessen R.C., Olderode-Berends M.J., Veenstra-Knol H.E., Vos Y.J., van der Klift H.M., Scheenstra R., Tops C.M., Karrenbeld A., Peters F.T. Paediatric intestinal cancer and polyposis due to bi-allelic PMS2 mutations: case series, review and follow-up guidelines. Eur. J. Cancer. 2011;47:965–982. doi: 10.1016/j.ejca.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 55.Tieu A.H., Edelstein D., Axilbund J., Romans K.E., Brosens L.A., Wiley E., Hylind L., Giardiello F.M. Clinical Characteristics of Multiple Colorectal Adenoma Patients Without Germline APC or MYH Mutations. J. Clin. Gastroenterol. 2016;50:584–588. doi: 10.1097/MCG.0000000000000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivatsan A., Bowen N., Kolodner R.D. Mispair-specific recruitment of the Mlh1-Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2-Msh3 complex. J. Biol. Chem. 2014;289:9352–9364. doi: 10.1074/jbc.M114.552190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haugen A.C., Goel A., Yamada K., Marra G., Nguyen T.P., Nagasaka T., Kanazawa S., Koike J., Kikuchi Y., Zhong X. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res. 2008;68:8465–8472. doi: 10.1158/0008-5472.CAN-08-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plaschke J., Preußler M., Ziegler A., Schackert H.K. Aberrant protein expression and frequent allelic loss of MSH3 in colorectal cancer with low-level microsatellite instability. Int. J. Colorectal Dis. 2012;27:911–919. doi: 10.1007/s00384-011-1408-0. [DOI] [PubMed] [Google Scholar]
- 59.Hile S.E., Shabashev S., Eckert K.A. Tumor-specific microsatellite instability: do distinct mechanisms underlie the MSI-L and EMAST phenotypes? Mutat. Res. 2013;743-744:67–77. doi: 10.1016/j.mrfmmm.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marra G., Iaccarino I., Lettieri T., Roscilli G., Delmastro P., Jiricny J. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc. Natl. Acad. Sci. USA. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miquel C., Jacob S., Grandjouan S., Aimé A., Viguier J., Sabourin J.C., Sarasin A., Duval A., Praz F. Frequent alteration of DNA damage signalling and repair pathways in human colorectal cancers with microsatellite instability. Oncogene. 2007;26:5919–5926. doi: 10.1038/sj.onc.1210419. [DOI] [PubMed] [Google Scholar]
- 62.You K.T., Li L.S., Kim N.G., Kang H.J., Koh K.H., Chwae Y.J., Kim K.M., Kim Y.K., Park S.M., Jang S.K., Kim H. Selective translational repression of truncated proteins from frameshift mutation-derived mRNAs in tumors. PLoS Biol. 2007;5:e109. doi: 10.1371/journal.pbio.0050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohmiya N., Matsumoto S., Yamamoto H., Baranovskaya S., Malkhosyan S.R., Perucho M. Germline and somatic mutations in hMSH6 and hMSH3 in gastrointestinal cancers of the microsatellite mutator phenotype. Gene. 2001;272:301–313. doi: 10.1016/s0378-1119(01)00517-0. [DOI] [PubMed] [Google Scholar]
- 64.Harrington J.M., Kolodner R.D. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol. Cell. Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campregher C., Schmid G., Ferk F., Knasmüller S., Khare V., Kortüm B., Dammann K., Lang M., Scharl T., Spittler A. MSH3-deficiency initiates EMAST without oncogenic transformation of human colon epithelial cells. PLoS ONE. 2012;7:e50541. doi: 10.1371/journal.pone.0050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Risinger J.I., Umar A., Boyd J., Berchuck A., Kunkel T.A., Barrett J.C. Mutation of MSH3 in endometrial cancer and evidence for its functional role in heteroduplex repair. Nat. Genet. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 67.Edelmann W., Umar A., Yang K., Heyer J., Kucherlapati M., Lia M., Kneitz B., Avdievich E., Fan K., Wong E. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res. 2000;60:803–807. [PubMed] [Google Scholar]
- 68.Hegan D.C., Narayanan L., Jirik F.R., Edelmann W., Liskay R.M., Glazer P.M. Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6. Carcinogenesis. 2006;27:2402–2408. doi: 10.1093/carcin/bgl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orimo H., Nakajima E., Yamamoto M., Ikejima M., Emi M., Shimada T. Association between single nucleotide polymorphisms in the hMSH3 gene and sporadic colon cancer with microsatellite instability. J. Hum. Genet. 2000;45:228–230. doi: 10.1007/s100380070031. [DOI] [PubMed] [Google Scholar]
- 70.Berndt S.I., Platz E.A., Fallin M.D., Thuita L.W., Hoffman S.C., Helzlsouer K.J. Mismatch repair polymorphisms and the risk of colorectal cancer. Int. J. Cancer. 2007;120:1548–1554. doi: 10.1002/ijc.22510. [DOI] [PubMed] [Google Scholar]
- 71.Hirata H., Hinoda Y., Kawamoto K., Kikuno N., Suehiro Y., Okayama N., Tanaka Y., Dahiya R. Mismatch repair gene MSH3 polymorphism is associated with the risk of sporadic prostate cancer. J. Urol. 2008;179:2020–2024. doi: 10.1016/j.juro.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jafary F., Salehi M., Sedghi M., Nouri N., Jafary F., Sadeghi F., Motamedi S., Talebi M. Association between mismatch repair gene MSH3 codons 1036 and 222 polymorphisms and sporadic prostate cancer in the Iranian population. Asian Pac. J. Cancer Prev. 2012;13:6055–6057. doi: 10.7314/apjcp.2012.13.12.6055. [DOI] [PubMed] [Google Scholar]
- 73.Duraturo F., Liccardo R., Cavallo A., De Rosa M., Grosso M., Izzo P. Association of low-risk MSH3 and MSH2 variant alleles with Lynch syndrome: probability of synergistic effects. Int. J. Cancer. 2011;129:1643–1650. doi: 10.1002/ijc.25824. [DOI] [PubMed] [Google Scholar]
- 74.Yang X., Wu J., Lu J., Liu G., Di G., Chen C., Hou Y., Sun M., Yang W., Xu X. Identification of a comprehensive spectrum of genetic factors for hereditary breast cancer in a Chinese population by next-generation sequencing. PLoS ONE. 2015;10:e0125571. doi: 10.1371/journal.pone.0125571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Wind N., Dekker M., Claij N., Jansen L., van Klink Y., Radman M., Riggins G., van der Valk M., van’t Wout K., te Riele H. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat. Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- 76.Edelmann L., Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: animal models for human hereditary nonpolyposis colorectal cancer. Am. J. Med. Genet. C. Semin. Med. Genet. 2004;129C:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- 77.Arai H., Okudela K., Oshiro H., Komitsu N., Mitsui H., Nishii T., Tsuboi M., Nozawa A., Noishiki Y., Ohashi K. Elevated microsatellite alterations at selected tetra-nucleotide (EMAST) in non-small cell lung cancers--a potential determinant of susceptibility to multiple malignancies. Int. J. Clin. Exp. Pathol. 2013;6:395–410. [PMC free article] [PubMed] [Google Scholar]
- 78.Danaee H., Nelson H.H., Karagas M.R., Schned A.R., Ashok T.D., Hirao T., Perry A.E., Kelsey K.T. Microsatellite instability at tetranucleotide repeats in skin and bladder cancer. Oncogene. 2002;21:4894–4899. doi: 10.1038/sj.onc.1205619. [DOI] [PubMed] [Google Scholar]
- 79.Tseng-Rogenski S.S., Chung H., Wilk M.B., Zhang S., Iwaizumi M., Carethers J.M. Oxidative stress induces nuclear-to-cytosol shift of hMSH3, a potential mechanism for EMAST in colorectal cancer cells. PLoS ONE. 2012;7:e50616. doi: 10.1371/journal.pone.0050616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carethers J.M., Koi M., Tseng-Rogenski S.S. EMAST is a Form of Microsatellite Instability That is Initiated by Inflammation and Modulates Colorectal Cancer Progression. Genes (Basel) 2015;6:185–205. doi: 10.3390/genes6020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Senter L., Clendenning M., Sotamaa K., Hampel H., Green J., Potter J.D., Lindblom A., Lagerstedt K., Thibodeau S.N., Lindor N.M. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.