Abstract
Cell division terminates with cytokinesis and cellular separation. Autosomal-recessive primary microcephaly (MCPH) is a neurodevelopmental disorder characterized by a reduction in brain and head size at birth in addition to non-progressive intellectual disability. MCPH is genetically heterogeneous, and 16 loci are known to be associated with loss-of-function mutations predominantly affecting centrosomal-associated proteins, but the multiple roles of centrosomes in cellular function has left questions about etiology. Here, we identified three families affected by homozygous missense mutations in CIT, encoding citron rho-interacting kinase (CIT), which has established roles in cytokinesis. All mutations caused substitution of conserved amino acid residues in the kinase domain and impaired kinase activity. Neural progenitors that were differentiated from induced pluripotent stem cells (iPSCs) derived from individuals with these mutations exhibited abnormal cytokinesis with delayed mitosis, multipolar spindles, and increased apoptosis, rescued by CRISPR/Cas9 genome editing. Our results highlight the importance of cytokinesis in the pathology of primary microcephaly.
Main Text
Primary microcephaly is characterized by reduced head size accompanied by variable degrees of intellectual, language, and motor-skill disability, but not progressive cognitive decline, spasticity, or epilepsy.1 In autosomal-recessive primary microcephaly (MCPH [MIM: 251200]), the cerebral cortex is particularly reduced in size, leading to an apparently “simplified gyral pattern” because mantle thickness is grossly preserved but surface area is dramatically reduced.2 MRI of an affected fetus with a prenatal diagnosis has shown the frontal lobes of the cerebral cortex to be affected as early as 30 weeks of gestation.3
MCPH is considered a disorder of neural progenitor cell (NPC) proliferation or survival during embryogenesis, predominantly of autosomal-recessive or X-linked inheritance. Mutations in 16 loci (MCPH1–MCPH16) have been described as associated with MCPH, and many of these loci encode proteins implicated in the biogenesis and function of the centrosome, an organelle critical for multiple cellular functions.4 MCPH-associated genes encode essential centrosomal-duplication proteins—SAS-6, STIL, CPAP, CEP63, CEP135, and CEP152—and pericentriolar-matrix proteins—WDR62, ASPM, and CEP215 (also known as CDK5Rap2). Most of these mutations generate premature stop codons that predominantly result in nonsense-mediated mRNA decay and absent proteins. ASPM (MIM: 605481) mutations, which result in alterations in mitotic-spindle regulation, account for approximately 40% of MCPH cases.1 Recently, a hierarchy of MCPH-associated proteins co-recruiting to the centrosome was described as mediating centrosomal duplication,5 suggesting a critical role for these MCPH-associated proteins in centrosomal biogenesis.
Because centrosomes play multiple roles in the cell (e.g., serving as microtubule-organizing centers, as spindle poles during mitosis, and as basal bodies in ciliogenesis), the cellular etiology of MCPH has remained unclear. Initial studies focused on the roles of MCPH-associated proteins at the spindle pole in determining mitotic-spindle orientation. The position of the centrosome in the dividing cell regulates mitotic orientation, which can, in turn, regulate neural stem cell fate decisions.6 Indeed, defects in the orientation of the mitotic cleavage plane contribute to neurogenesis defects in an MCPH model.7 Recent work has extended the roles of centrosomes in microcephaly to include a potential function in cilia. MCPH-associated proteins play essential roles in cilia, which are critical for cells to process Sonic hedgehog signals that can regulate neurogenesis.8 Furthermore, cells harboring mutations in microcephaly-associated genes can show disrupted ciliogenesis,9, 10 but classically, ciliopathy disorders do not demonstrate microcephaly. Thus, identifying non-centrosomal factors that lead to MCPH when mutated could help clarify mechanisms.
To identify additional mechanisms of microcephaly, we recruited individuals displaying MCPH in the setting of parental consanguinity. Subjects were enrolled according to protocols approved by institutional review boards at affiliated institutions. We performed whole-exome sequencing (WES) on at least one affected member per family and focused on the identification of potentially deleterious rare homozygous variants.11 Three independent consanguineous families whose children showed non-syndromic MCPH presented with homozygous missense variants in CIT (MIM: 605629), encoding citron rho-interacting kinase (CIT), which has established roles in cytokinesis. The variants were unique in our dataset of more than 5,000 geographically matched individuals sequenced by exome, were not represented in the Greater Middle Eastern Variome or the Exome Aggregation Consortium (ExAC) Browser, were fully segregated in the families according to a recessive mode of inheritance (Figure S1A), and were predicted to be damaging with altered evolutionarily conserved amino acids (Figure S1B and Table S1). Moreover, none of the families displayed functionally relevant variants in any of the previously established MCPH-associated genes in OMIM, and no other variants that met screening criteria showed segregation according to a recessive fully penetrant mode of inheritance (Table S1).
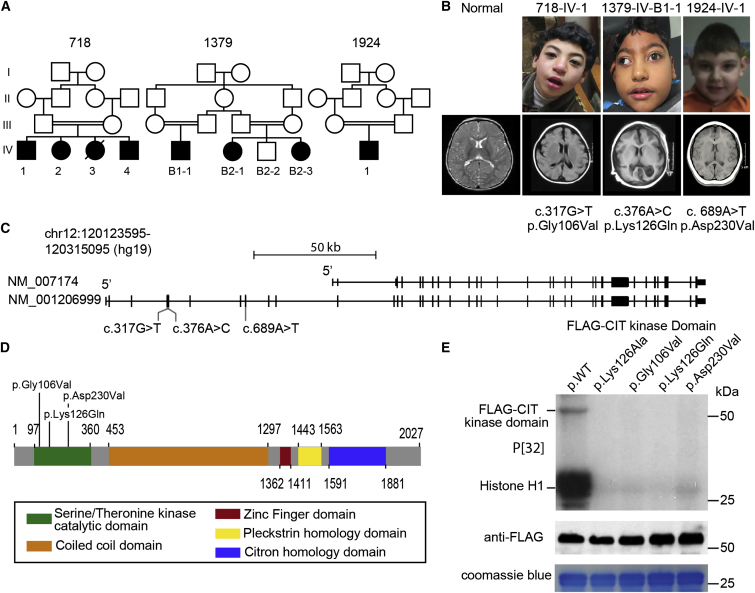
Family 718 from Egypt had four children affected by MCPH from a consanguineous marriage (Figure 1A and Table 1). Individual 718-IV-1 was born at full term with a reduced head circumference (HC) of 32.5 cm (−1 SD) and an average body weight. Developmental delay with motor, speech, and social impairment was noted at 3 years. At 9 years, HC was 44.8 cm (−5.6 SDs). He displayed a sloping forehead (Figure 1B), moderate intellectual disability, mild hypertonia, and brisk reflexes. His sister (718-IV-2) had a similar clinical course and had a HC of 43 cm (−7.4 SDs) at 8 years of age. Similarly, 718-IV-3 was noted to have microcephaly at birth but died on the first day of life as a result of unrelated causes, and 718-IV-4 had a HC of 41 cm (−5.6 SDs) at age 2 years and mild intellectual disability. Clinical features were not progressive, and no spasticity or epileptic seizures were described. Brain MRI of 718-IV-1, 718-IV-2, and 718-IV-4 revealed a simplified gyral pattern and a thin corpus callosum (Figure 1B). WES identified a homozygous missense variant (c.317G>T [p.Gly106Val]; hg19 chr12: g.120295424C>A; GenBank: NM_001206999.1) affecting the kinase domain of CIT (Figures 1C and 1D).
Figure 1.
Mutations in CIT Cause Primary Microcephaly
(A) Pedigrees of consanguineous families 718, 1379, and 1924. Symbols are as follows: filled, affected; empty, unaffected; circle, female; square, male; hash, deceased; B, branch.
(B) Faces (top) and axial MRI (bottom) of representative affected individuals from each family show reduced brain volume and a simplified gyral pattern, consistent with a diagnosis of MCPH.
(C) Exonic structure of CIT and the location of the identified mutations. The shorter CIT-N isoform, encoded by GenBank: NM_007174.2, lacks the N-terminal kinase domain. The longer CIT-K isoform, encoded by GenBank: NM_001206999.1, encodes the kinase domain where the CIT variants localize.
(D) Identified missense variants cluster within the kinase domain of full length CIT-K.
(E) Defective activity of the kinase domain with CIT variants. P[32] incorporation was detected in the wild-type only for histone H1 and FLAG-CIT autophosphorylation.
Table 1.
Clinical Features of the Affected Individuals in This Study
| 718-IV-1 | 718-IV-2 | 718-IV-4 | 1379-IV-B2-1 | 1379-IV-B2-3 | 1379-IV-B1-1 | 1924-IV-1 | |
|---|---|---|---|---|---|---|---|
| Gender | male | female | male | female | female | male | male |
| Ethnic origin | Egypt | Egypt | Egypt | Egypt | Egypt | Egypt | Turkey |
| Pregnancy duration | term | term | term | term | term | term | term |
| Weight at birth | 3.2 kg (−0.2 SD) | 3 kg (−0.4 SD) | 3.5 kg (+0.1 SD) | 2.5 kg (−1.9 SDs) | 2.5 kg (−1.9 SDs) | 2 kg (−2.4 SDs) | NA |
| Length at birth | 48 cm (−0.4 SD) | 49 cm | 50 cm (+0.4 SD) | 50 cm (+0.4 SD) | 49 cm | 49.5 cm (+0.2 SD) | NA |
| HC at birth | 31 cm | 31.5 cm | 32 cm | 31 cm | 30 cm | 30 cm | NA |
| HC at last examination | 44.8 cm (−5.6 SDs) | 43 cm (−7.4 SDs) | 41 cm (−5.6 SDs) | 41.2 cm (−7.8 SDs) | 40 cm (−8.4 SDs) | 39.5 cm (−8.3 SDs) | 45 cm (−6.5 SDs) |
| Diagnosis age | 3 years | 2 years | at birth | 5 years | 1 year | 1 year | 4 years |
| Intellectual disability | moderate | mild | mild | moderate | moderate | severe | moderate |
| Development | |||||||
| Gross motor | delayed | delayed | delayed | delayed | delayed | delayed | delayed |
| Fine motor | delayed | delayed | normal | delayed | delayed | absent | delayed |
| Language | delayed | delayed | delayed | delayed | delayed | absent | delayed |
| Social | delayed | delayed | delayed | delayed | delayed | delayed | delayed |
| Seizures | |||||||
| Present | − | − | − | − | − | − | − |
| Neurological Findings | |||||||
| Hypertonia | mild | mild | − | mild | mild | severe, acquired arthrogryposis | − |
| Hypotonia | − | − | − | − | − | − | − |
| Deep tendon reflexes | brisk | brisk | normal | brisk | brisk | brisk | normal |
| Spastic tetraplegia | − | − | − | − | − | + | − |
| Ataxia | − | − | − | − | − | − | − |
| Investigations | |||||||
| Metabolic | normal | normal | NA | NA | NA | normal | normal |
| VEP and ERG | normal | normal | normal | NA | NA | normal | normal |
| EEG | normal | normal | normal | NA | NA | generalized epileptogenic activity | normal |
| MRI | |||||||
| Simplified gyral pattern | + | + | + | NA | NA | + | + |
| Hypogenesis of corpus callosum | + | + | + | NA | NA | + | − |
| Cerebellar hypoplasia | − | − | − | NA | NA | − | − |
| Brainstem hypoplasia | − | − | − | NA | NA | − | − |
| White-matter abnormalities | − | − | − | NA | NA | − | + |
| Miscellaneous | |||||||
| Short stature | 125 cm (−1 SD) | 119 cm (−1.2 SDs) | 86 cm (mean) | 124 cm (−1.5 SDs) | 102 cm (−1.2 SDs) | 104 cm (−0.9 SD) | NA |
| Optic atrophy | − | − | − | − | − | − | NA |
| Autistic features | − | − | − | − | − | mild | NA |
| Dysmorphism | sloping forehead, tubular nose, full lips and cheeks, large ears | sloping forehead, tubular nose, full lips and cheeks, large ears | sloping forehead, tubular nose, full lips and cheeks, large ears | sloping forehead, tubular nose, full lips and cheeks, large ears | sloping forehead, tubular nose, full lips and cheeks, large ears | flat occiput, esotropia (R), large ears, retruded mandible, full lips and cheeks, tubular nose | low anterior hairline, full lips, large ears |
Abbreviations are as follows: EEG, electroencephalography; ERG, electroretinography; HC, head circumference; NA, not available; R, right; and VEP, visual evoked potential.
Family 1379 from Egypt had two first-cousin parental consanguineous branches and three children affected by MCPH. All exhibited reduced head size, intellectual disability, delayed motor and speech development, hypertonia, brisk reflexes, sloping foreheads, and relatively large ears. HC was −7 to −8.4 SDs in the first decade of life. MRI of individual 1379-IV-B1-1 demonstrated a simplified gyral pattern and a thin corpus callosum, similar to those in family 718. WES revealed a homozygous missense variant (c.376A>C [p. Lys126Gln]; hg19 chr12: g.120295365T>G) also affecting the kinase domain. We noticed that subject 1379-B1-1 was more severely impaired than other affected individuals from this family. Whether other variants from this individual (Table S2) contribute to his clinical symptoms requires further investigation.
Family 1924 from Turkey had a single male child affected with MCPH from a consanguineous marriage. He displayed a reduced HC of 45 cm (−6.5 SDs), intellectual disability, delayed motor and speech development, and large protruding ears. WES detected a homozygous variant (c.689A>T [p.Asp230Val]; hg19 chr12: g.120270639T>A) also affecting the encoded kinase domain. It is worth noting that another common feature of the MCPH-affected individuals from these three families is short stature, which is also seen in individuals with MCPH1 mutations12 but is not a common finding in other MCPH cases.
CIT was identified as a 183 kDa GTP-bound Rac and Rho binding protein.13 There are two cDNA isoforms: (1) one (GenBank: NM_001206999.1) encoding a 2,027 aa full-length CIT-K, which has N-terminal kinase, coiled-coil, zinc finger, Pleckstrin homology, and citron homology domains,14 and (2) another (GenBank: NM_007174.2) encoding a 1,545 aa CIT-N, which is produced by alternative transcriptional initiation and lacks the kinase domain but maintains the other C-terminal domains (Figures 1C and 1D).15 CIT-K mRNA is specifically expressed in proliferating neural precursors in the developing brain, whereas CIT-N mRNA is expressed by post-mitotic cells,16 suggesting a role for the kinase in neurogenesis. CIT-K localizes to the cleavage furrow and midbody, where it promotes cytokinesis during cell division.17
The CIT variants that we identified were notable for several reasons. First, variants were not observed in the ExAC Browser or in our collective ethnically matched database of 10,000 WES traces from families affected by neurodevelopmental disease. Second, all variants occurred within the kinase domain of the protein. Third, all variants altered highly conserved residues, not just in evolutionarily orthologs of CIT but also in other sequence-similar kinase family members, including ROCK1 and AKT1 (Figure S1C). In fact, p.Lys126Gln, the amino acid substitution detected in family 1379, was in the invariant lysine (K) residue in subdomain II, which is present in all known kinases and is involved in the phosphotransfer reaction.18 Fourth, using the resolved structure of the ROCK1 kinase domain to predict the function of the altered amino acids in CIT,19 we found that two variants (p.Gly106Val and p.Lys126Gln) were within the ATP binding pocket of the kinase domain, and one (p.Asp230Val) was within the DFG (Asp-Phe-Gly) motif, which positions ATP for phosphoryl transfer20 (Figure S1D). These findings suggest that the mutations impair kinase activity.
To test for impaired kinase activity, we FLAG tagged amino acids 1–480 of CIT-K, encoding the kinase domain, either as wild-type (WT) or with each identified CIT variant engineered. The tagged proteins were immunoprecipitated from transfected 293T cells with anti-FLAG antibody and then used in an in vitro kinase assay. We utilized a kinase-dead (p.Lys126Ala) version with an alanine substitution at the ATP donor site as a negative control.17 As reported, WT CIT-K exhibited kinase activity toward exogenous histone H1 substrate, as well as autophosphorylation activity.21 Neither of these kinase activities was observed from the clone harboring p.Lys126Ala. Furthermore, none of the clones with CIT variants showed phosphorylation of histone H1 or autophosphorylation on the basis of P[32] incorporation (Figure 1E), suggesting that all three variants impair kinase activity.
Cytokinesis refers to the final stage of the cell cycle, in which the physical separation of two daughter cells occurs during cell division. During cytokinesis, the contractile ring forms beneath the cell equatorial surface to form the cleavage furrow, and then ingression of the furrow results in the formation of an intercellular bridge called the midbody. The cell cycle completes as the midbody is resolved and the two daughter cells separate, a process known as abscission.22 Disrupted cytokinesis frequently leads to binucleated cells, aneuploidy, chromosomal instability, activation of p53, cell-cycle arrest, and apoptosis. CIT-K is localized in the midbody during abscission, and knockdown can result in failed abscission followed by re-fusion of the two daughter cells.23
To study the functional consequences of the CIT variants at the cellular level, we collected skin-derived fibroblasts from affected and carrier individuals from families 718 and 1379, from affected individuals from family 1924, and from a healthy unrelated control individual. Primary fibroblasts were unremarkable in culture and displayed no defects in cell proliferation or mitosis (data not shown). Therefore, we generated induced pluripotent stem cells (iPSCs) through reprograming by integration-free episomal methods.24 We excluded gross chromosomal post-reprogramming rearrangements. Furthermore, we found no defect in differentiation of iPSCs into mesodermal or endodermal lineages.
From iPSCs, we next generated NPCs by using a dual-SMAD inhibition protocol.25 As expected, amounts of PAX6 and CIT-K in mutant NPCs were comparable to those in related controls (Figures S2A and S2B). CIT-K was detected and localized to the midbody core in a manner indistinguishable from that of the wild-type during cytokinesis (Figure S2C), suggesting that the absence of kinase activity does not lead to protein mislocalization. Furthermore, midbody integrity was intact, as evidenced by indistinguishable localization of Aurora B to the midbody flank.26
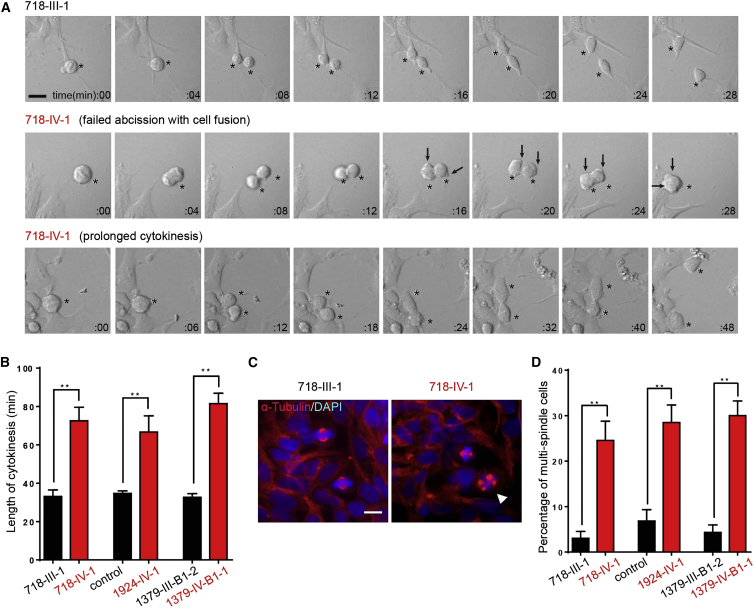
To assess for functional defects in cytokinesis, we used time-lapse imaging to monitor cell-cycle progression by comparing the entire duration of mitosis in NPCs. Previous work in HeLa cells showed that knockdown of CIT resulted in delayed cytokinesis and “blebbing” of the cellular membrane, followed by either “early fusion” or “late fusion” of the daughters with binucleated derivatives.23 In mutant NPCs, we found a dramatic increase in the length of cytokinesis and cellular blebbing in all mitotic cells recorded. This resulted in one of two outcomes. In about 25% of cells, blebbing was followed by failed cytokinesis and the formation of cellular fusions and binucleated cells (Figure 2A, middle). In the remainder, after the delay, cytokinesis completed with separation of the two daughter cells (Figure 2A, bottom). On average, the length of cytokinesis was doubled in mutant cells (mean ± SEM: 73.6 ± 4.1 min versus 33.5 ± 1.4 min).
Figure 2.
Cell-Division Defects in NPCs Derived from Individuals with CIT Variants
(A) Representative phase-contrast time-lapse images of NPCs from unaffected and affected individuals. 718-III-1 is a healthy father (black typeface), and 718-IV-1 is an affected member (red typeface). Asterisks mark the dividing cells analyzed in each series. Note that unaffected cells completed division within 28 min. Affected cells (top row) showed blebbing at 16 min and then incomplete cytokinesis with binucleated derivatives at 28 min. Affected cells (bottom row) showed minimal blebbing and then division by 48 min. Arrows mark cortical blebbing.
(B) Quantification of the length of cytokinesis (n = 15 cells per group).
(C and D) Representative images and quantification of the percentage of multipolar cells in NPCs from unaffected (C) and affected (D) individuals. Arrowheads mark multipolar cells (n = 7 cultures per group).
Bar graphs show the mean ± SEM. ∗∗p < 0.01 (Student’s t test). Scale bar represents 10 μm.
Failure to achieve cytokinesis can result in subsequent multipolar spindles as a result of duplicated centrosomes and nuclear material. To evaluate for multipolar spindles, we fixed and stained sub-confluent NPCs for α-tubulin and DAPI and then counted the percentage of mitotic figures with three or more spindle poles. Multipolar spindles were evident in about 28% of mitotic cells, whereas they were evident in fewer than 5% of control cells. We conclude that multipolar spindles are a frequent feature of CIT-mutated NPCs.
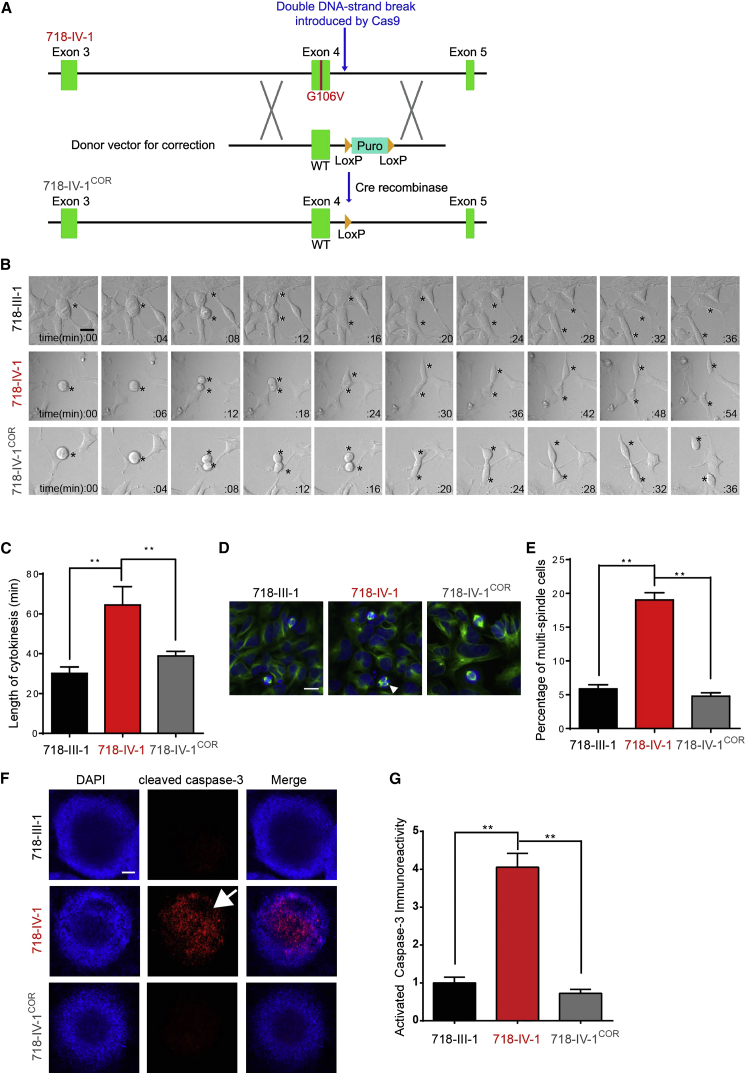
To test whether the mutation caused the observed cellular phenotypes, we corrected the c.317G>T (p.Gly106Val) mutation by using CRISPR/Cas9-based genome editing in iPSCs derived from affected individual 718-IV-1 by homologous recombination (Figure 3A). A genome-editing donor plasmid containing wild-type CIT exon 4, flanking genomic sequence, and a puromycin cassette (LoxP-Puro-LoxP) was electroporated together with Cas9- and gRNA-expressing plasmids into iPSCs. Individual iPSC clones were propagated after puromycin selection. After successful biallelic targeting (Figures S3A–S3C), the puromycin cassette was removed by transfection with a plasmid expressing Cre recombinase (Figure S3D). Of note, the expression level of CIT was indistinguishable between affected and wild-type cells after puromycin recombination in corrected lines (data not shown), suggesting that genome editing did not adversely affect expression. We then differentiated cells into NPCs and performed time-lapse microscopy to evaluate cytokinesis. Corrected cells showed no delay in the length of cytokinesis (Figures 3B and 3C). Furthermore, there was no accumulation above baseline of multiple spindle cells in corrected NPCs (Figures 3D and 3E). We conclude that the CIT variants identified from the affected individual were necessary for mediating defects in cytokinesis, given that cellular defects were corrected when the wild-type allele was genetically engineered into mutant cells.
Figure 3.
Correcting the CIT Variant Rescues Cellular NPC Phenotypes in Cells from Affected Individuals
(A) Schematic of gene-editing strategy for correcting the CIT variant in iPSCs. WT exon 4 and the floxed puromycin cassette were recombined at the mutant locus after Cas9 cleavage. Cre-mediated removal of the LoxP-Puro-LoxP cassette left only the single LoxP site.
(B) Representative differential interference contrast (DIC) time-lapse images from unaffected (black), affected (red), and mutation-corrected (gray) NPCs during cytokinesis. Scale bar represents 10 μm.
(C) Quantification of the length of cytokinesis (n = 15 cells for each group).
(D and E) Representative images and quantification of the percentage of multipolar cells in unaffected, affected, and mutation-corrected (COR) NPCs. Arrowheads mark multipolar cells (n = 4 cultures per group). Scale bar represents 10 μm.
(F and G) Representative images (F) and quantification (G) of active caspase-3 immunoreactivity in unaffected, affected, and mutation-corrected neurospheres (n = 6 cultures per group). Note the dramatically increased amount of cleaved caspase-3 in mutant cells (718-IV-1) prior to correction (F, arrow). Scale bar represents 150 μm.
(C, E, and G) One-way ANOVA was followed by Tukey’s multiple-comparison test.
Bar graphs show the mean ± SEM. ∗∗p < 0.01.
Both prolonged and incomplete cell division are associated with genotoxic stress and increased apoptotic cell death22, 27 and are proposed as mechanisms in mouse and zebrafish MCPH models.28, 29, 30 Furthermore, Cit-deficient mice display binucleated cells and increased apoptosis in the developing cerebral cortex.16 To test for apoptosis in a complex cellular milieu, we generated neurospheres from control and mutant cells, together with CIT-mutation-corrected cells, and then stained for cleaved caspase-3 as a marker of apoptosis. Dramatic evidence for increased apoptosis (an overall 4-fold increase in intensity) was detected in mutant but not control or mutation-corrected neurospheres (Figures 3F and 3G).
Here, we describe families affected by recessive MCPH caused by biallelic missense CIT alleles affecting the kinase domain and thus leading to undetectable kinase activity. These mutations were remarkable in that they did not alter the expression level of the mRNA or CIT localization in anaphase, suggesting that the kinase activity of CIT is required for function in controlling cytokinesis. Previous studies using overexpression of kinase-dead CIT in HeLa cells found largely retained effects, but these studies could not differentiate between loss-of-function and toxic gain-of-function effects.17 The fact that the parent and sibling carriers were healthy and had a normal HC suggests that our mutations did not act in a dominant-negative fashion, and because no individuals with truncating CIT mutations were detected with MCPH, it is tempting to speculate that CIT retains some biological activity even in the setting of kinase loss. This is further supported by the finding of the more severe microlissencephaly phenotype observed among individuals with truncating CIT mutations, shown in the accompanying paper by Li et al.31
CIT is classified as an AGC kinase (by homology with protein kinases A, G, and C), which are regulated by second messengers, such as cyclic AMP (PKA) and lipids (PKC).32 CIT binds activated Rho and Rac,13 and activated Rho correlates with the translocation of CIT to the cleavage furrow.33 Most AGC kinases require sequential phosphorylations of the “turn” and “hydrophobic” motifs to stabilize the active conformation,34 but it is still unclear whether other kinases regulate CIT’s kinase activity. Although CIT can function with proteins such as KIF14 and TUBB3,26, 35 direct phosphorylation targets of CIT have not yet been identified.
Several spindle-pole-localized MCPH-associated proteins, including ASPM, CENPJ, and CDK5RRAP2, are also present in the midbody during cytokinesis.36 Moreover, loss of ASPM in cultured cells leads to cytokinesis failure followed by apoptosis, in addition to misorientation of the mitotic spindle.37 Our results confirm that cytokinesis failure and subsequent apoptosis are underlying mechanisms for the genetic forms of MCPH. Interestingly, the C terminus of ASPM interacts with CIT-K, suggesting that CIT-K and ASPM might function together in regulating cytokinesis.36
Our data align with those of other studies arguing against MCPH as a primary defect of centrosomes for the following reasons. First, fruit flies without centrosomes still undergo neurogenesis,38 suggesting that centrosomes are not required for neuronal proliferation. Second, mutations in microcephaly-associated genes Sas4 and Cep63 in mice lead to genotoxic stress, activation of the p53 pathway, and apoptosis, probably independent of cleavage plane effects.39 Third, neurogenesis defects in microcephaly mouse models have been rescued at least in part by concurrent removal of p53.39, 40 The data support a model in at least some forms of microcephaly, whereby loss of mitotic integrity through an effect on centrosomes or cytokinesis can result in genome instability, genotoxic stress, apoptosis, and subsequently, reduced cerebral volume.
Acknowledgments
We thank Joseph LoTurco and Ferdinando Di Cunto for communicating unpublished results and Susan Taylor for providing suggestions on the project. We thank the subjects and their families for their contributions to this study. This work was supported by the NIH (R01NS041537, R01NS048453, R01NS052455, and P01HD070494 to N.C.), the Howard Hughes Medical Institute (J.G.G.), and the Druckenmiller Fellowship from the New York Stem Cell Foundation (to H.L). We thank the Broad Institute (U54HG003067 to E. Lander) and the Yale Center for Mendelian Disorders (U54HG006504 to R. Lifton) for sequencing support.
Published: July 21, 2016
Footnotes
Supplemental Data include three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.004.
Contributor Information
Murat Gunel, Email: murat.gunel@yale.edu.
Joseph G. Gleeson, Email: jogleeson@ucsd.edu.
Accession Numbers
The WES data for all study subjects who consented to data release have been deposited under accession number dbGaP: phs000288.
Web Resources
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
Greater Middle East Variome, http://igm.ucsd.edu/gme
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org/
SeattleSeq, http://snp.gs.washington.edu/SeattleSeqAnnotation137/
SIFT, http://sift.jcvi.org/
Supplemental Data
References
- 1.Faheem M., Naseer M.I., Rasool M., Chaudhary A.G., Kumosani T.A., Ilyas A.M., Pushparaj P., Ahmed F., Algahtani H.A., Al-Qahtani M.H., Saleh Jamal H. Molecular genetics of human primary microcephaly: an overview. BMC Med. Genomics. 2015;8(Suppl 1):S4. doi: 10.1186/1755-8794-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi Y., Poduri A., Kawaguch A., Yoon G., Salih M.A., Yamashita F., Walsh C.A., Barkovich A.J. Congenital microcephaly with a simplified gyral pattern: associated findings and their significance. AJNR Am. J. Neuroradiol. 2011;32:1123–1129. doi: 10.3174/ajnr.A2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desir J., Cassart M., David P., Van Bogaert P., Abramowicz M. Primary microcephaly with ASPM mutation shows simplified cortical gyration with antero-posterior gradient pre- and post-natally. Am. J. Med. Genet. A. 2008;146A:1439–1443. doi: 10.1002/ajmg.a.32312. [DOI] [PubMed] [Google Scholar]
- 4.Woods C.G., Basto R. Microcephaly. Curr. Biol. 2014;24:R1109–R1111. doi: 10.1016/j.cub.2014.09.063. [DOI] [PubMed] [Google Scholar]
- 5.Kodani A., Yu T.W., Johnson J.R., Jayaraman D., Johnson T.L., Al-Gazali L., Sztriha L., Partlow J.N., Kim H., Krup A.L. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. eLife. 2015;4:4. doi: 10.7554/eLife.07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaltschmidt J.A., Davidson C.M., Brown N.H., Brand A.H. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat. Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- 7.Fish J.L., Kosodo Y., Enard W., Pääbo S., Huttner W.B. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong C.K., Han Y.G., Shah J.K., Obernier K., Guinto C.D., Alvarez-Buylla A. Primary cilia are required in a unique subpopulation of neural progenitors. Proc. Natl. Acad. Sci. USA. 2014;111:12438–12443. doi: 10.1073/pnas.1321425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waters A.M., Asfahani R., Carroll P., Bicknell L., Lescai F., Bright A., Chanudet E., Brooks A., Christou-Savina S., Osman G. The kinetochore protein, CENPF, is mutated in human ciliopathy and microcephaly phenotypes. J. Med. Genet. 2015;52:147–156. doi: 10.1136/jmedgenet-2014-102691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W.F., Pomp O., Ben-Omran T., Kodani A., Henke K., Mochida G.H., Yu T.W., Woodworth M.B., Bonnard C., Raj G.S. Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron. 2014;84:1240–1257. doi: 10.1016/j.neuron.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon-Salazar T.J., Silhavy J.L., Udpa N., Schroth J., Bielas S., Schaffer A.E., Olvera J., Bafna V., Zaki M.S., Abdel-Salam G.H. Exome sequencing can improve diagnosis and alter patient management. Sci. Transl. Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neitzel H., Neumann L.M., Schindler D., Wirges A., Tönnies H., Trimborn M., Krebsova A., Richter R., Sperling K. Premature chromosome condensation in humans associated with microcephaly and mental retardation: a novel autosomal recessive condition. Am. J. Hum. Genet. 2002;70:1015–1022. doi: 10.1086/339518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madaule P., Furuyashiki T., Reid T., Ishizaki T., Watanabe G., Morii N., Narumiya S. A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe S., De Zan T., Ishizaki T., Narumiya S. Citron kinase mediates transition from constriction to abscission through its coiled-coil domain. J. Cell Sci. 2013;126:1773–1784. doi: 10.1242/jcs.116608. [DOI] [PubMed] [Google Scholar]
- 15.Camera P., da Silva J.S., Griffiths G., Giuffrida M.G., Ferrara L., Schubert V., Imarisio S., Silengo L., Dotti C.G., Di Cunto F. Citron-N is a neuronal Rho-associated protein involved in Golgi organization through actin cytoskeleton regulation. Nat. Cell Biol. 2003;5:1071–1078. doi: 10.1038/ncb1064. [DOI] [PubMed] [Google Scholar]
- 16.Di Cunto F., Imarisio S., Hirsch E., Broccoli V., Bulfone A., Migheli A., Atzori C., Turco E., Triolo R., Dotto G.P. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–127. doi: 10.1016/s0896-6273(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 17.Madaule P., Eda M., Watanabe N., Fujisawa K., Matsuoka T., Bito H., Ishizaki T., Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 18.Carrera A.C., Alexandrov K., Roberts T.M. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc. Natl. Acad. Sci. USA. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs M., Hayakawa K., Swenson L., Bellon S., Fleming M., Taslimi P., Doran J. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J. Biol. Chem. 2006;281:260–268. doi: 10.1074/jbc.M508847200. [DOI] [PubMed] [Google Scholar]
- 20.Kornev A.P., Haste N.M., Taylor S.S., Eyck L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Cunto F., Calautti E., Hsiao J., Ong L., Topley G., Turco E., Dotto G.P. Citron rho-interacting kinase, a novel tissue-specific ser/thr kinase encompassing the Rho-Rac-binding protein Citron. J. Biol. Chem. 1998;273:29706–29711. doi: 10.1074/jbc.273.45.29706. [DOI] [PubMed] [Google Scholar]
- 22.Green R.A., Paluch E., Oegema K. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- 23.Gai M., Camera P., Dema A., Bianchi F., Berto G., Scarpa E., Germena G., Di Cunto F. Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell. 2011;22:3768–3778. doi: 10.1091/mbc.E10-12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 25.Novarino G., El-Fishawy P., Kayserili H., Meguid N.A., Scott E.M., Schroth J., Silhavy J.L., Kara M., Khalil R.O., Ben-Omran T. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruneberg U., Neef R., Li X., Chan E.H., Chalamalasetty R.B., Nigg E.A., Barr F.A. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilaz L.J., McMahon J.J., Miller E.E., Lennox A.L., Suzuki A., Salmon E., Silver D.L. Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron. 2016;89:83–99. doi: 10.1016/j.neuron.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIntyre R.E., Lakshminarasimhan Chavali P., Ismail O., Carragher D.M., Sanchez-Andrade G., Forment J.V., Fu B., Del Castillo Velasco-Herrera M., Edwards A., van der Weyden L., Sanger Mouse Genetics Project Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genet. 2012;8:e1003022. doi: 10.1371/journal.pgen.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novorol C., Burkhardt J., Wood K.J., Iqbal A., Roque C., Coutts N., Almeida A.D., He J., Wilkinson C.J., Harris W.A. Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biol. 2013;3:130065. doi: 10.1098/rsob.130065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaindl A.M., Passemard S., Kumar P., Kraemer N., Issa L., Zwirner A., Gerard B., Verloes A., Mani S., Gressens P. Many roads lead to primary autosomal recessive microcephaly. Prog. Neurobiol. 2010;90:363–383. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Harding B.N., Moccia A., Soukarieh O., Tubeuf H., Drunat S., Chitty L.S., Verloes A., Gressens P., El Ghouzzi V., Joriot S. Mutations in Citron Kinase Cause Recessive Microlissencephaly with Multinucleated Neurons. Am. J. Hum. Genet. 2016;99:511–520. doi: 10.1016/j.ajhg.2016.07.003. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearce L.R., Komander D., Alessi D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 33.Eda M., Yonemura S., Kato T., Watanabe N., Ishizaki T., Madaule P., Narumiya S. Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J. Cell Sci. 2001;114:3273–3284. doi: 10.1242/jcs.114.18.3273. [DOI] [PubMed] [Google Scholar]
- 34.Hauge C., Antal T.L., Hirschberg D., Doehn U., Thorup K., Idrissova L., Hansen K., Jensen O.N., Jørgensen T.J., Biondi R.M., Frödin M. Mechanism for activation of the growth factor-activated AGC kinases by turn motif phosphorylation. EMBO J. 2007;26:2251–2261. doi: 10.1038/sj.emboj.7601682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sgrò F., Bianchi F.T., Falcone M., Pallavicini G., Gai M., Chiotto A.M., Berto G.E., Turco E., Chang Y.J., Huttner W.B., Di Cunto F. Tissue-specific control of midbody microtubule stability by Citron kinase through modulation of TUBB3 phosphorylation. Cell Death Differ. 2016;23:801–813. doi: 10.1038/cdd.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paramasivam M., Chang Y.J., LoTurco J.J. ASPM and citron kinase co-localize to the midbody ring during cytokinesis. Cell Cycle. 2007;6:1605–1612. doi: 10.4161/cc.6.13.4356. [DOI] [PubMed] [Google Scholar]
- 37.Higgins J., Midgley C., Bergh A.M., Bell S.M., Askham J.M., Roberts E., Binns R.K., Sharif S.M., Bennett C., Glover D.M. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 2010;11:85. doi: 10.1186/1471-2121-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 39.Marjanović M., Sánchez-Huertas C., Terré B., Gómez R., Scheel J.F., Pacheco S., Knobel P.A., Martínez-Marchal A., Aivio S., Palenzuela L. CEP63 deficiency promotes p53-dependent microcephaly and reveals a role for the centrosome in meiotic recombination. Nat. Commun. 2015;6:7676. doi: 10.1038/ncomms8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Insolera R., Bazzi H., Shao W., Anderson K.V., Shi S.H. Cortical neurogenesis in the absence of centrioles. Nat. Neurosci. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.