Abstract
Primary ciliary dyskinesia (PCD) is an autosomal-recessive disease due to functional or ultra-structural defects of motile cilia. Affected individuals display recurrent respiratory-tract infections; most males are infertile as a result of sperm flagellar dysfunction. The great majority of the PCD-associated genes identified so far encode either components of dynein arms (DAs), which are multiprotein-ATPase complexes essential for ciliary motility, or proteins involved in DA assembly. To identify the molecular basis of a PCD phenotype characterized by central complex (CC) defects but normal DA structure, a phenotype found in ∼15% of cases, we performed whole-exome sequencing in a male individual with PCD and unexplained CC defects. This analysis, combined with whole-genome SNP genotyping, identified a homozygous mutation in DNAJB13 (c.833T>G), a gene encoding a HSP40 co-chaperone whose ortholog in the flagellated alga Chlamydomonas localizes to the radial spokes. In vitro studies showed that this missense substitution (p.Met278Arg), which involves a highly conserved residue of several HSP40 family members, leads to protein instability and triggers proteasomal degradation, a result confirmed by the absence of endogenous DNAJB13 in cilia and sperm from this individual. Subsequent DNAJB13 analyses identified another homozygous mutation in a second family; the study of DNAJB13 transcripts obtained from airway cells showed that this mutation (c.68+1G>C) results in a splicing defect consistent with a loss-of-function mutation. Overall, this study, which establishes mutations in DNAJB13 as a cause of PCD, unveils the key role played by DNAJB13 in the proper formation and function of ciliary and flagellar axonemes in humans.
Keywords: DNAJB13, PCD, central complex, radial spoke, cilia, sperm flagellum
Main Text
Primary ciliary dyskinesia (PCD [MIM: 244400]) is a rare autosomal-recessive disease resulting from functional or ultra-structural defects of motile cilia. This disorder is characterized by chronic airway infections. Half of PCD cases are associated with situs inversus, thereby defining Kartagener syndrome. In addition, because motile cilia and sperm flagella share a conserved microtubule-based structure, most male individuals with PCD are infertile as a result of severe or total asthenozoospermia.1
The internal cytoskeleton of motile cilia and flagella, named the axoneme, consists of nine doublets of microtubules circularly arranged around the central complex (CC), which comprises a central pair of microtubules (CP) surrounded by a central sheath (“9 + 2” pattern). The beating of cilia and flagella is orchestrated by the outer and inner dynein arms (ODAs and IDAs, respectively); these are multiprotein ATPase complexes that provide the sliding force for motility.2 The structural organization of the axoneme is dependent on (1) the nexin-dynein regulatory complex that connects the peripheral microtubules and (2) the radial spokes (RSs), which are multiprotein T-shaped structures that link each peripheral microtubule doublet to the CC.3 Interactions between the RSs and the CC have been shown to locally control dynein-driven microtubule sliding.2, 4
Several genes encoding proteins involved in the structure or assembly of the axoneme are implicated in PCD. In most cases, the axonemal defects concern the dynein arms, but structural defects of the CC have also been reported in approximately 15% of cases.5 So far, among the 31 genes implicated in PCD with structural defects of the axoneme, only five have been identified with mutations inducing CC defects. These are RSPH1 (radial spoke head 1 homolog [MIM: 609314]), RSPH4A (radial spoke head 4 homolog A [MIM: 612647]), and RSPH9 (radial spoke head 9 homolog [MIM: 612648]), which encode proteins located in the spoke head, RSPH3 (radial spoke 3 homolog [MIM: 615876]), which encodes the main constituent of the spoke stalk,4 and HYDIN (HYDIN, axonemal central pair apparatus protein [MIM: 610812]), which encodes a component of the central sheath.6 The CC defects can be schematically subdivided into two groups: in the first one, which has been shown to result from mutations in RSPH1, RSPH4A, RSPH9, and RSPH3,7, 8, 9, 10 there is no central-sheath defect, and the main defect involves the CP: the CP is missing (“9 + 0” or “9 + 1” pattern), and a transposition (“8 + 1” pattern) is seen in a variable proportion of cilia.5 In the second group of CC defects, in which mutations in HYDIN have been identified,11 the main ultrastructural defect involves the central sheath; in that situation, the CP is rarely missing, and a microtubule transposition is rarely seen as well. Studies performed on Chlamydomonas reinhardtii flagella have shown that RSs are dimeric structures, which are partially assembled in the cytoplasm and contain at least 23 proteins per monomer;12, 13 mutations in these RS components are likely to explain unresolved cases of PCD with CC defects.
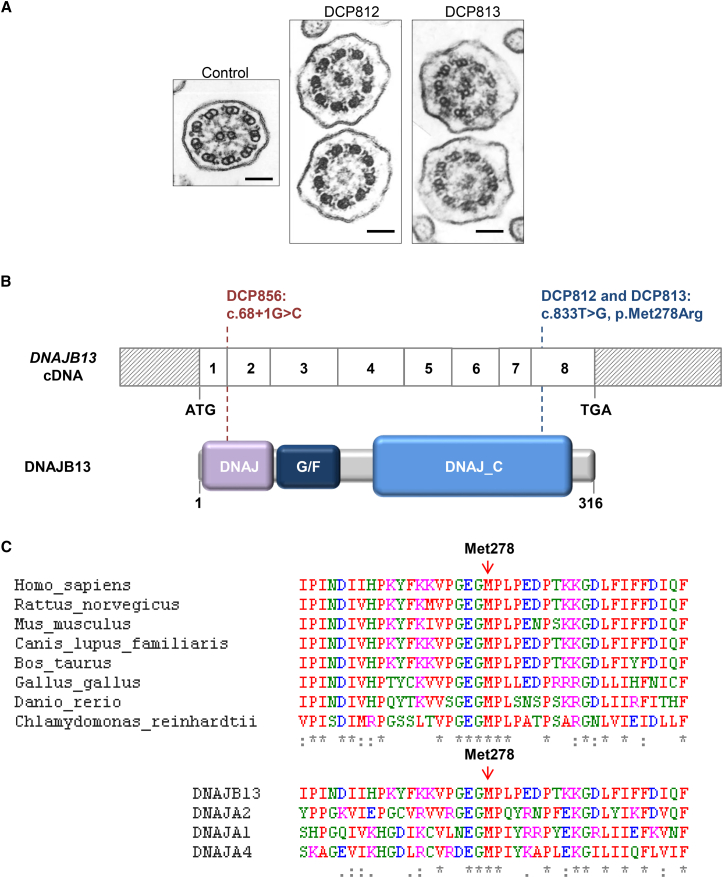
The present study was performed with the approval of the Comité de Protection des Personnes CPP Ile de France III (approval nos. CPP07729 and CPP02748), and written informed consent was obtained from all participating individuals. We first analyzed a family (DC387) in whom two siblings—a female individual (DCP812) and a male individual (DCP813)—present with a PCD phenotype characterized by ultrastructural abnormalities reminiscent of those reported in the first group of CC defects and have had no mutations identified by Sanger sequencing of all coding exons and flanking intronic sequences of the four genes already implicated in this phenotype (i.e., RSPH1, RSPH3, RSPH4A, and RSPH9). These individuals, who were aged 51 and 53 years at the time of diagnosis, have a sinopulmonary syndrome (bronchiectasis, rhinosinusitis, and otitis) that began in childhood without laterality defects (Table 1). In both affected siblings, transmission electron microscopy (TEM) analysis of the respiratory cilia obtained by nasal biopsy showed an abnormal percentage of cilia lacking central microtubule(s) (29% and 37% in individuals DCP812 and DCP813, respectively), including mainly cilia with a “9 + 0” pattern and some rare cilia with a “9 + 1” or a “8 + 1” pattern (Figure 1A and Table 2). In both individuals, nasal nitric oxide (NO) levels were found to be very low (16.5 and 17.3 nL/min in DCP812 and DCP813, respectively; normal values > 100 nL/min) (Table 2). High-speed videomicroscopy (HSV) performed on airway epithelial cells obtained by nasal biopsy showed that, although most cilia were motile (75% and 60% in individuals DCP812 and DCP813, respectively), the ciliary beating frequency (CBF) was lower (5 and 4.3 Hz in individuals DCP812 and DCP813, respectively) than in control individuals,15 and the beating pattern was abnormally characterized by movements of reduced amplitude (Table 2 and Movies S1, S2, S3, S4, S5, and S6); such a reduced amplitude is similar to what has been observed for RSPH1 or RSPH3 mutations via the same HSV technique.7 In addition, the male individual (DCP813) is infertile as a result of severe oligo-astheno-terato-zoospermia and necrozoospermia. Three semen evaluations performed between 2009 and 2015 indicated a complete absence of progressive sperm motility (normal value > 32%). Moreover, static motility (i.e., non-progressive motility) was displayed by 12% of spermatozoa in 2009 and was absent in the two following samples. The total sperm count was between 0.6 and 1.4 million (normal value > 39 million/ejaculate), and sperm viability was between 10% and 25% (normal value > 58%) (Table 3).
Table 1.
Phenotypic Features of Individuals DCP812, DCP813, and DCP856, in Whom DNAJB13 Mutations Were Identified
| Individual | Family (Origin) | Consanguinity | Gender (Age at Diagnosis) | Airway Disease | Laterality Defect | Fertility | Allele 1 | Allele 2 |
|---|---|---|---|---|---|---|---|---|
| DCP812 | DC387 (Brittany, France) | yes | female (51 years) | bronchitis, bronchiectasis, rhinosinusitis, otitis | no | NT | c.833T>G (p.Met278Arg) | c.833T>G (p.Met278Arg) |
| DCP813 | DC387 (Brittany, France) | yes | male (53 years) | NRD, bronchiectasis, rhinosinusitis, otitis | no | infertility | c.833T>G (p.Met278Arg) | c.833T>G (p.Met278Arg) |
| DCP856 | DC416 (Algeria) | yes | female (14 years) | bronchitis, bronchiectasis, rhinosinusitis, otitis | no | NT | c.68+1G>C (p.Tyr24∗) | c.68+1G>C (p.Tyr24∗) |
Abbreviations are as follows: NRD, neonatal respiratory distress; and NT, not tested.
Figure 1.
Ultrastructural Ciliary Defects and Identified DNAJB13 Mutations in Individuals with PCD
(A) Electron micrographs of cross-sections of cilia from a control individual and from individuals DCP812 and DCP813, who carry the mutation c.833T>G (p.Met278Arg). For each affected individual, one section with a well-organized configuration (9 + 2) shows the presence of a CP (top), and another shows an abnormal configuration (9 + 0) lacking the two central microtubules (bottom). Black scale bars represent 0.1 μm.
(B) DNAJB13 mutations and their expected impact at the protein level in individuals with PCD are shown in an exonic organization of the human DNAJB13 cDNA (top) and a domain-organization model of the corresponding protein (bottom).
(C) Top: partial sequence alignment of DNAJB13 proteins shows the evolutionary conservation of the region surrounding Met278, which is located in the DNAJ-C domain and is mutated in individuals DCP812 and DCP813 (red vertical arrow). Bottom: partial sequence alignment of DNAJB13 and three paralogs from the type I DNAJ (HSP40) family shows the conservation of Met278 (red vertical arrow). Colors of residues correspond to the physicochemical properties of their side chain: red, small or hydrophobic (including aromatic, except tyrosine [Y]); blue, acidic; magenta, basic (except histidine [H]); and green, hydroxyl or sulfhydryl or amine group and glycine (G). Asterisks indicate identical residues in all sequences, and colons represent residues that share similar properties in all sequences.
Table 2.
Ciliary Investigations in Individuals DCP812, DCP813, and DCP856, in Whom DNAJB13 Mutations Were Identified
| Family | Individual | NO (nL/min)a | Abnormal Ciliab(TEMc) (%) | Beating Cilia (%) | CBF (Hz)d | Angle (°)d | Distance Traveled (μm/s)d |
|---|---|---|---|---|---|---|---|
| DC387 | DCP812 | 16.5 | 29 | 75 | 5 ± 1.5 | 72 ± 18 | 48.9 ± 13.7 |
| DCP813 | 17.3 | 37 | 60 | 4.3 ± 1.5 | 69 ± 12 | 39.8 ± 20.3 | |
| DC416 | DCP856 | lowe | inconclusive | 100 | 10.5 ± 0.5 | –f | –f |
| – | controlsg (mean ± SD) | >100 | – | 91 ± 13 | 8.9 ± 2 | 71.6 ± 6.6 | 66.7 ± 14.2 |
Abbreviations are as follows: CBF, ciliary beating frequency; NO, nitric oxide; and TEM, transmission electron microscopy.
Nasal NO was measured during apnea with the use of a chemoluminescence NO analyzer (NIOX Flex, Aerocrine, and Endono 8000, Seres). The mean value of the plateau was recorded. NO values above 100 nL/min were considered normal.
The abnormalities include mainly cilia with a “9 + 0” pattern and some rare cilia with a “9 + 1” or “8 + 1” pattern.
The method used is described in Tamalet et al.14
The CBF, angle, and distance traveled were evaluated on beating cilia.
The exact value is not available; the medical report mentions that the NO concentration is dramatically low, considering that a NO value below 100 nL/ml is low.
High-speed videomicroscopy was not performed.
According to Papon et al.5
Table 3.
Semen Characteristics of Individual DCP813, in Whom the DNAJB13 p.Met278Arg Substitution Was Identified
| pH | Volume of Ejaculate (mL) | Total Sperm Count (106/Ejaculate) | Total Motility: PR + NP (%) | Progressive Motility: PR (%) | Vitality (%) | Morphology: Typical Forms (%) | |
|---|---|---|---|---|---|---|---|
| 2009 | 7.9 | 2.8 | 1.4 | 12 | 0 | 25 | 17 |
| 2010 | 7.9 | 3.5 | 0.07 | 0 | 0 | – | 5 |
| 2015 | 7.9 | 1.5 | 0.6 | 0 | 0 | 10 | NA |
| Lower reference limits (WHO, 2010)16 | 7.2 | 1.5 | 39 | 40 | 32 | 58 | 18 |
According to the World Health Organization (WHO) standards, sperm motility is categorized into three types: progressive motility (PR), which corresponds to rapid and slow progressive spermatozoa (formerly grades a and b, respectively); non-progressive motility (NP), which corresponds to static spermatozoa (i.e., motile but non-progressive spermatozoa; formerly grade c); and immotile spermatozoa, for which no beating and no progression is observed (formerly grade d). Spermatozoa with progressive motility were always absent in semen from individual DCP813 (normal value > 32%). Static motility was observed for 12% of spermatozoa only in 2009. The subsequent evaluations that were performed displayed no motile spermatozoa at all.
With the aim of identifying the molecular basis of PCD in this family, we genotyped the genomic DNA of individuals DCP812 and DCP813 with the HumanCytoSNP-12Chip (Illumina) and analyzed the data with GenomeStudio and CNVPartition v.3.1.6 (Illumina). This study, which did not find any obvious gene deletion or duplication, identified a 16.4 Mb shared region of homozygosity on chromosome 11 (11q13.4), thereby revealing an unknown consanguinity (Figures S1 and S2). Whole-exome sequencing was subsequently performed in individual DCP813 with the Agilent SureSelect All Exon V5+UTRs 70-Mb Capture Kit. Data filtering was performed according to an autosomal-recessive mode of transmission and focused on gene variants located in the homozygous regions shared by DCP812 and DCP813 (Table S1) and present in sequence-variant databases at frequencies compatible with the incidence of PCD (because the higher estimate of the incidence of PCD is ∼1/15,000 individuals,17 the maximal theoretical frequency of a PCD allele is 0.008 [√1/15,000]). After this filtering process, variations were identified in the coding sequences of eight genes (Table S2). Among them, DNAJB13 (DnaJ heat shock protein family [Hsp40] member B13 [MIM: 610263]) retained our attention for the following reasons: in mice, DNAJB13, also known as TSARG6 (testis spermatogenesis apoptosis-related gene 6 protein), is detected in motile cilia, notably in the airways, the oviduct, and weakly in the brain,18, 19 as well as in sperm cells, where it localizes to the axoneme.20 Rsp16, which encodes a component of the RS multiprotein complex of flagella in the alga Chlamydomonas reinhardtii,21 was reported to be the ortholog of murine and human DNAJB13.18, 20, 21, 22 Indeed, sequence alignments indicate that human DNAJB13 shares 41% identity (and 57% similarity) with Rsp16, which is the highest score of identity among DNAJB proteins: the percentages of protein sequence identity between Rsp16 and each of the other three closest members of the DNAJB family (i.e., DNAJB1, DNAJB4, and DNAJB5) are lower (38%, 34%, and 35%, respectively). The importance of Rsp16 for flagellar beating and signal transduction from the CC to the dynein motors was suggested in Chlamydomonas reinhardtii: its silencing did not massively alter the structure of the RSs or the axoneme but delayed the initiation and propagation of flagellar bending; this resulted in twitching flagella and prevented the mutant cells from swimming.21 In addition, during sperm flagella assembly in the mouse testis, DNAJB13 was also shown to localize to the annulus, a ring-like structure found at the junction of the mid-piece and principal piece of the flagellum, and composed of Septin polymers.22 This localization at the annulus is transient given that, in mature epididymal sperm, DNAJB13 localizes solely to the RSs of the axoneme.22 Such transient localization, restricted to the annulus, suggests that DNAJB13 might have functions that are specific to sperm flagella, in addition to its functions in the RS multiprotein complexes.23
The sequence variation we identified in individual DCP813 is a homozygous transversion (c.833T>G) located in exon 8 of DNAJB13 (GenBank: NM_153614.3) (Figure 1B and Figure S1). This sequence variant has not been described in genomic-variant databases such as dbSNP, 1000 Genomes, the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) Exome Variant Server (EVS), and the Exome Aggregation Consortium (ExAC) Browser. At the structural level, DNAJB13 shares a conserved structure with type II DNAJ (HSP40) family members.24 The conserved region comprises the DNAJ domain, required for association with HSP70s, and the glycine and phenylalanine (G/F)-rich domain, a flexible spacer that is necessary for substrate binding and located downstream of the DNAJ domain25, 26 (Figure 1B). DNAJB13 has a C-terminal domain named DNAJ_C (Figure 1B), which is particularly conserved in three other members of the type II DNAJ (HSP40) family (Figure S3A), which is involved in substrate binding and presentation.24, 27 The c.833T>G variation identified in DCP813 is predicted to replace a neutral and non-polar methionine with a charged arginine. At the protein level, this missense variation (p.Met278Arg) is located in the DNAJ_C domain of DNAJB13 (Figure 1B) and involves a residue totally conserved in all species studied so far, as well as in three paralogs from the type I DNAJ (HSP40) family (Figure 1C). Such a high degree of conservation of Met278 therefore strongly suggests that amino acid substitutions at this position could be detrimental to protein function. Sanger sequencing confirmed the presence of the c.833T>G (p.Met278Arg) variation in individuals DCP813 and DCP812 (Figures S4). Parental DNA samples were not available; however, SNP genotyping data indicate that the c.833T>G transversion is indeed present in the homozygous state in both siblings (Figures S1 and S2).
All together, these data prompted us to search for a DNAJB13 defect in an additional consanguineous individual (DCP856 from family DC416) in whom whole-genome SNP analysis identified a homozygous region containing the DNAJB13 locus (Figure S5). This analysis was performed by whole-exome sequencing, and the data were subsequently analyzed through a filtering process similar to that followed for the first family (Tables S3 and S4). This led to the identification of a homozygous DNAJB13 mutation in individual DCP856, in whom no mutations were found in RSPH1, RSPH3, RSPH4A, or RSPH9. The DNAJB13 mutation (c.68+1G>C) involves the invariant dinucleotide of the splice-donor site following exon 1 (Figure 1B) and is therefore expected to abolish normal DNAJB13 splicing. Parental DNA samples were available for this family, and both parents were found to be heterozygous for the mutation (Figure S6). As detailed in Table 1, individual DCP856, a 14-year-old female, has a typical clinical picture of PCD; although ciliary investigations showed normal CBF values (10.5 Hz), the NO concentration was dramatically low, considering that a NO value below 100 nL/min is low (Table 2). TEM analyses, which were performed on three samples obtained from different nasal and bronchial biopsies, were inconclusive because of the very low number of cilia.
To provide further arguments supporting the implication of DNAJB13 in PCD, we studied its expression pattern in various human tissues. The detection of DNAJB13 in cilia and flagella and its localization within the RS complex have been reported in mice and Chlamydomonas reinhardtii, but so far information has been limited in humans. Data from the EMBL-EBI Expression Atlas (Riken Famtom5 Project) and the Human Protein Atlas indicate that DNAJB13 is highly expressed in human adult testis (and to a lesser extent in the lungs) and that the protein is present in germ cells at late stages of spermatogenesis. To further document the expression pattern of this gene in humans, we first performed qRT-PCR analysis on a tissue panel and found that DNAJB13 is mainly expressed in the trachea and testis; high amounts of transcripts were also detected in airway epithelial cells (AECs) obtained by nasal biopsy of healthy individuals (Figure S7A). Accordingly, DNAJB13 was readily detectable by western blot in human AECs and sperm (Figure S7B). In addition, studies by immunofluorescence microscopy revealed that DNAJB13 co-localizes with α-tubulin along the length of cilia and flagella (Figure S7C). Therefore, similar to what is found in mice and Chlamydomonas reinhardtii, DNAJB13 specifically localizes to the axoneme of cilia and flagella in humans.
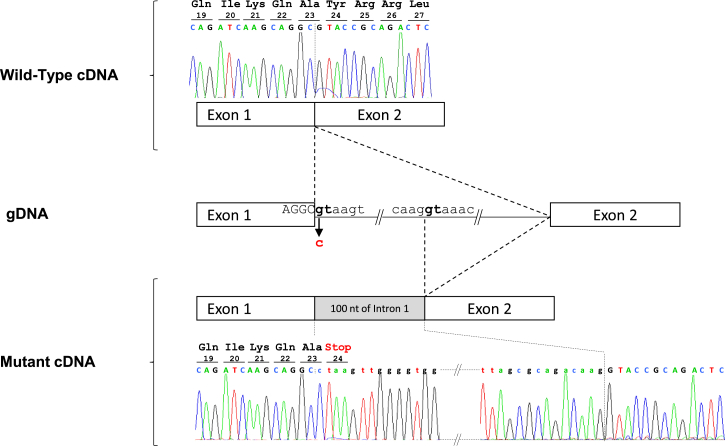
To assess the functional consequences of the c.68+1G>C mutation identified in individual DCP856 on the processing of DNAJB13 transcripts, we isolated total RNA from her airway cells and analyzed DNAJB13 transcripts after RT-PCR amplification followed by Sanger sequencing. As shown in Figure 2, the c.68+1G>C mutation, involving the splice-donor site of intron 1, results in the activation of a cryptic splice-donor site located within intron 1, thereby adding the first 100 nucleotides of intron 1 to exon 1. This abnormal transcript, which contains a premature stop codon in the very first nucleotides of the included intronic sequence (p.Tyr24∗) (Figure 2), would lead to a severely truncated DNAJB13 lacking the DNAJ, G/F-rich, and DNAJ_C domains and/or to the absence of protein production through activation of nonsense-mediated mRNA decay. The c.68+1G>C mutation found in the homozygous state in individual DCP856 is, therefore, a loss-of-function mutation.
Figure 2.
Functional Consequences of the DNAJB13 c.68+1G>C Mutation on RNA Splicing and Expected Consequences at the Protein Level
(Top) Nucleotide sequence of RT-PCR products spanning exons 1 and 2 and generated from total RNA obtained from nasal cells of a control individual.
(Middle) Schematic representation of a DNAJB13 genomic fragment spanning the intronic mutated site identified in individual DCP856 shows the cryptic splice-donor site that is activated in the presence of the c.68+1G>C mutation. The mutation (in red) involves the invariant dinucleotide (in bold) of the splice-donor site of intron 1.
(Bottom) Nucleotide sequence of RT-PCR products spanning exons 1 and 2 and generated from total RNA obtained from nasal cells of individual DCP856. The electrophoregram shows the retention of the first 100 nucleotides of intron 1 in DNAJB13 transcripts and the resulting premature stop codon.
With the aim of assessing the impact of the p.Met278Arg missense variant identified in individuals DCP812 and DCP813 on DNAJB13, we first performed in silico analyses with bioinformatics tools (Sift, Panther, PolyPhen-2, and PROVEAN), which all predicted a damaging effect of the variant on the protein (data not shown). As mentioned above, the p.Met278Arg substitution is located in the DNAJ_C domain of DNAJB13, which shares high homology with the DNAJ_C domains of DNAJB1, DNAJB4, and DNAJB5 (Figure S3A). Therefore, using the available human crystal structure of the DNAJB1 DNAJ_C domain (residues 164–340 from PDB: 3AGX) as a template, we modeled the structure of the DNAJB1 DNAJ_C domain (residues 140–316) in which the non-polar Leu302 residue, corresponding to Met278 in DNAJB13, was replaced by a methionine residue (Figure S3B). This representation shows that the environment of the methionine of interest is mainly non-polar. The replacement of that methionine by a polar and charged residue (arginine) is therefore expected to disrupt the predicted hydrophobic interactions in the wild-type (WT) structure, particularly those between that methionine and the conserved non-polar residues located in its close environment (i.e., Leu172, Val254, and Leu276). Given the high homology between the DNAJ_C domains of DNAJB1 and DNAJB13 (Figure S3A), similar consequences are expected for DNAJB13 carrying the p.Met278Arg substitution.
To experimentally investigate the functional consequences of the c.833T>G (p.Met278Arg) substitution found in individuals DCP812 and DCP813, we compared the biochemical properties of DNAJB13 carrying the p.Met278Arg substitution (DNAJB13_Met278Arg), generated by site-directed mutagenesis, to those of a WT protein (DNAJB13_WT). Although several DNAJ proteins have been shown to be able to dimerize through their C-terminal region,24, 28, 29, 30, 31 this property has never been documented for mammalian DNAJB13 proteins. We therefore tested whether DNABJ13_WT was able to form dimers and the impact of the p.Met278Arg substitution on dimerization. Co-immunoprecipitation experiments were performed in CHO-K1 cells transiently co-expressing GFP-tagged DNAJB13_WT and FLAG-tagged DNAJB13_WT. As shown in Figure S8A, GFP-DNAJB13_WT specifically co-precipitated with FLAG-DNAJB13_WT when an anti-FLAG antibody was used for immunoprecipitation, whereas it did not in a control experiment performed with an irrelevant FLAG-tagged protein (i.e., FLAG-Baf60). To confirm these results, we carried out reverse experiments by using an anti-GFP antibody, which also showed that the GFP-tagged DNAJB13 co-precipitated with the FLAG-tagged DNAJB13 (Figure S8B). This indicates that DNAJB13 can form homodimers. We subsequently performed similar experiments to assess the ability of DNAJB13_Met278Arg to form dimers by using constructs encoding FLAG- and GFP-tagged DNAJB13_Met278Arg, generated by site-directed mutagenesis. Mutant GFP-DNAJB13_Met278Arg co-precipitated with FLAG-DNAJB13_Met278Arg (Figure S8C), and these results were confirmed by dimerization assays on native polyacrylamide gels (data not shown). This indicates that the p.Met278Arg substitution does not preclude the formation of DNAJB13 homodimers.
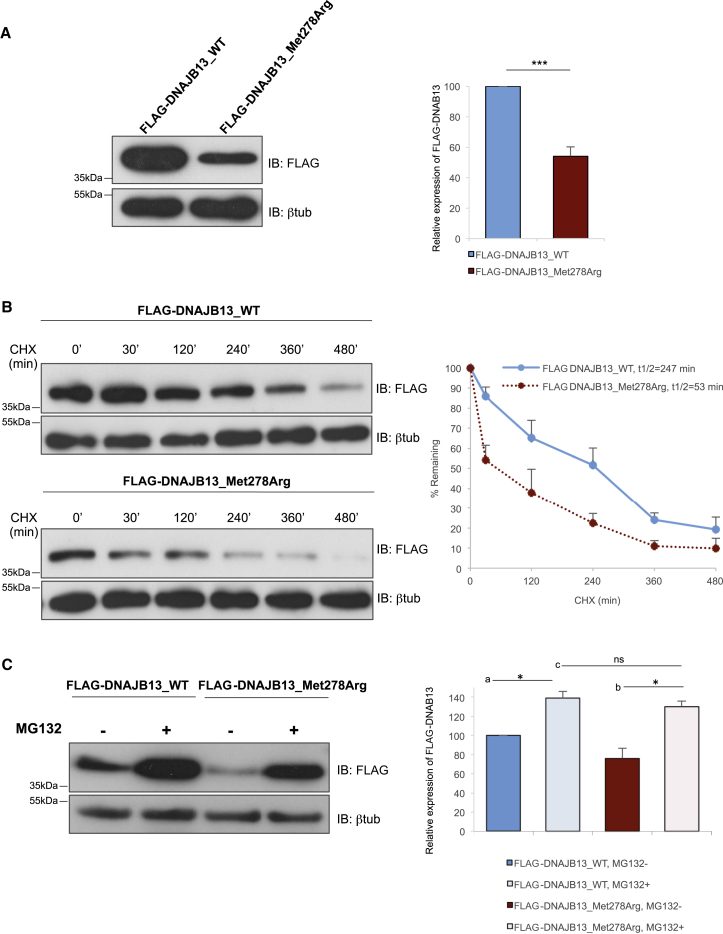
Most importantly, while conducting the co-immunoprecipitation experiments, we systematically observed less DNAJB13_Met278Arg than DNAJB13_WT, which we had to counteract by increasing the amount of the expression plasmid used in cell transfection. This observation prompted us to analyze the amount and stability of DNAJB13_Met278Arg. Total amounts of WT and mutant proteins produced in CHO-K1 cells transiently transfected with equal amounts of DNA were quantified with β-tubulin as an internal loading control. As shown in Figure 3A, the amount of mutant protein was only 54% of that of the WT protein. Similar results were obtained in a second cell type (germ cell 1 line [GC1]), thus excluding cell-type-related artifacts (Figure S9).
Figure 3.
Impact of the DNAJB13 p.Met278Arg Substitution on Protein Amounts, Protein Stability, and Proteasomal Degradation
(A) Left: CHO-K1 cells were transiently transfected with 0.5 μg of the plasmids encoding FLAG-DNAJB13_WT and FLAG-DNAJB13_Met278Arg. After 24 hr, the proteins were extracted, and equal amounts of protein extracts were subjected to SDS-PAGE on a 14% gel and immunoblotted with an anti-FLAG antibody. β-tubulin was used as a loading control. Right: the FLAG signal was quantified with ImageJ software and normalized to the amount of β-tubulin. The results are shown as the mean of ten independent experiments and are expressed as a percentage of the WT condition. Asterisks indicate that the mean is significantly different; t test p value = 0.0001.
(B) Left: CHO-K1 cells were transiently transfected with 0.5 μg of the plasmids encoding FLAG-DNAJB13_WT and FLAG-DNAJB13_Met278Arg. Cells were treated with 200 μg/mL of cycloheximide (CHX), and protein extracts were prepared at the indicated time points. Equal amounts of protein extracts were subjected to SDS-PAGE and immunoblotted with an anti-FLAG antibody. β-tubulin was used as a loading control. Right: the FLAG signal was quantified with ImageJ software and normalized to the amount of β-tubulin. The results are shown as the mean of three independent experiments and are expressed as a percentage of the WT, non-treated condition (p value = 0.008). Obtained values were plotted against time, and the half-lives of the DNAJB13 proteins were quantified.
(C) Left: CHO-K1 cells were transiently transfected with 0.5 μg of the plasmids encoding FLAG-DNAJB13_WT and FLAG-DNAJB13_Met278Arg. After 24 hr, the cells were treated with 20 μM of MG132 for 6 hr and then lysed for protein extraction in Laemmli sample buffer. Samples were subjected to SDS-PAGE on 14% gels and immunoblotted with an anti-FLAG antibody. β-tubulin was used as a loading control. Right: the FLAG signal was quantified with ImageJ software and normalized to the amount of β-tubulin. The results are shown as the mean of four independent experiments and are expressed as a percentage of the WT, non-treated condition. Asterisks indicate that the means are significantly different; t test p values = 0.04 (a) and 0.05 (b). “ns” indicates that the mean is not significantly different; t test p value = 0.25 (c).
To test whether the low amount of DNAJB13_Met278Arg resulted from protein instability, we next compared the half-life of each protein upon inhibition of protein synthesis with cycloheximide (CHX). To this end, DNAJB13_WT and DNAJB13_Met278Arg were transiently produced in CHO-K1 cells, which were then treated with 200 μg/mL of CHX for 8 hr. Quantification of protein amounts at different time points during CHX treatment showed that the half-life of DNAJB13_Met278Arg was 53 min, whereas that of DNAJB13_WT was 247 min (n = 3, p value = 0.02) (Figure 3B). This substantial reduction of the protein half-life shows that DNAJB13_Met278Arg is less stable than its WT counterpart.
We next performed transient transfection while simultaneously inhibiting proteasome-dependent degradation by using the peptide-aldehyde inhibitor MG132. Protein amounts from total cellular extracts were quantified. These cellular extracts consist of the detergent-soluble fraction; they also comprise the insoluble fractions that are expected to contain intracellular aggregates of misfolded proteins when protein clearance is compromised. As shown in Figure 3C, substantial accumulation of DNAJB13_Met278Arg was observed upon inhibition of the proteasome up to the same level as that of the WT protein. This strongly suggests that the difference in the amounts of WT and mutant proteins in non-treated cells results from enhanced proteasome-dependent degradation.
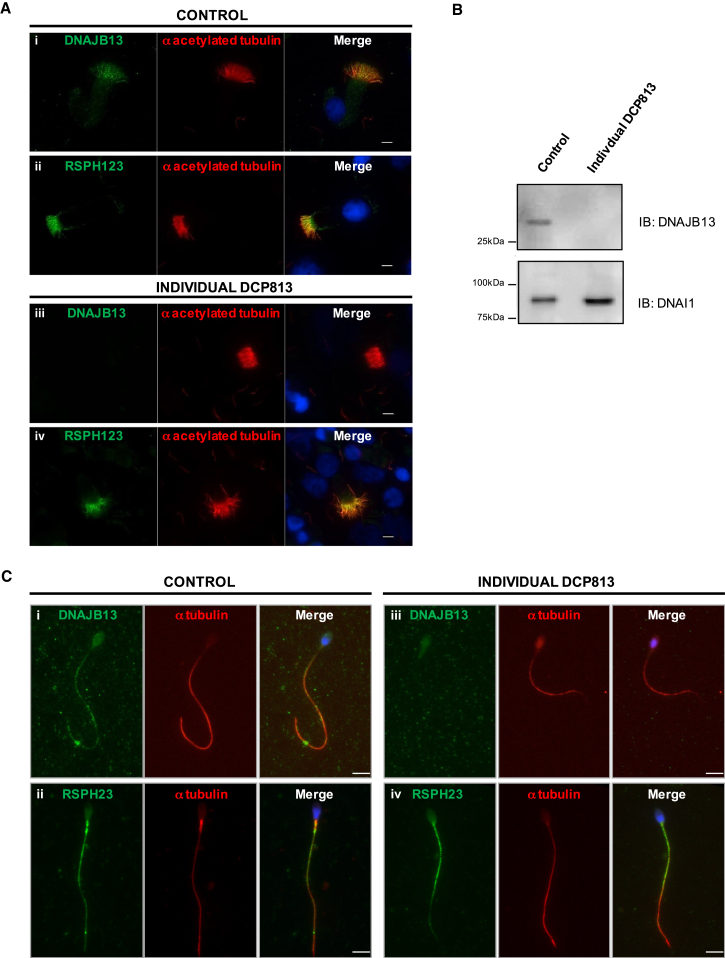
To further confirm the pathogenicity of the p.Met278Arg substitution, we analyzed the protein content of cilia and sperm from the male individual carrying this mutation (DCP813). We collected AECs by nasal biopsy and analyzed the localization of DNAJB13 and several axonemal components by means of immunofluorescence analysis. In contrast to what was observed in control cells, DNAJB13 was undetectable in cilia from individual DCP813, whereas other components of the RS multiprotein complex, such as RSPH9, RSPH11, and RSPH23, were readily detected (Figure 4A and Figure S10). Staining with an antibody that recognizes DYDC1 and DYDC2, two ciliary components displaying sequence similarity to Chlamydomonas reinhardtii RSP2, indicated that one or both of these proteins were also present along the cilia and at the basal region of cilia in individual DCP813 (Figure S10). The complete absence of DNAJB13 from this individual, who carries the p.Met278Arg substitution in the homozygous state, was confirmed by western blot analysis, whereas DNAI1, a component of the ODAs, was properly detected (Figure 4B). Semen samples of individual DCP813 were also analyzed by western blot; however, because of his severe oligozoospermia (Table 3), the sperm quantity was not sufficient to reach detectable levels of endogenous proteins. Nevertheless, we were able to analyze a few spermatozoa from the ejaculate by light microscopy and to perform immunofluorescence assays. Sperm cells had irregular caliber and length of their flagellum and abnormal head morphology (Figure S11). Most importantly, in spermatozoa from individual DCP813, DNAJB13 was undetectable, whereas RSPH23 displayed normal staining along the flagellum (Figure 4C).
Figure 4.
Amount and Localization of DNAJB13 and RSPH23 in AECs and Spermatozoa from a Healthy Control Individual and Individual DCP813
(A) DNAJB13 (i and iii) was present in the cilia of AECs from a healthy control but absent from the cilia of individual DCP813. RSPH23 (ii and iv) was detected within cilia of AECs from both the healthy control individual and individual DCP813. Scale bars represent 5 μm.
(B) Western blot analysis shows the absence of DNAJB13 in AECs from individual DCP813 (control cells are shown for comparison). DNAI1, a component of the ODAs, was used as a positive control and was detected in cells from both the healthy control individual and individual DCP813.
(C) In contrast to control sperm (i), the sperm flagellum from individual DCP813 (iii) did not display DNAJB13, whereas RSPH23 co-localized with tubulin in the sperm flagellum from both the control individual and individual DCP813 (ii and iv). Scale bars represent 5 μm.
All together, these data show that the p.Met278Arg substitution, which replaces a highly conserved hydrophobic methionine with a basic residue, considerably affects the structure and stability of DNAJB13 and results in the complete absence of DNAJB13 in vivo. The structural and functional defects observed in cilia and flagella from PCD individuals carrying the p.Met278Arg substitution undoubtedly result from the reduced availability of DNAJB13 in these organelles, a result in keeping with the loss of function associated with the DNAJB13 splice mutation identified in the second family (individual DCP856).
Overall, this study demonstrates the key role that DNAJB13 plays in upholding the integrity of the CC of motile cilia and flagella and its implication in PCD. As mentioned above, DNAJB13 is a member of the type II DNAJ (HSP40) protein family, whose members function as co-chaperones by regulating the ATPase activity of HSP70 proteins and stabilizing their interaction with client substrates.32 Several chaperones, including HSP70s, are present in cilia and flagella,33, 34, 35 but the co-chaperone activity of DNAJB13 toward HSP70 has so far not been properly established because (1) DNAJB13 does not contain the canonical His-Pro-Asp tripeptide motif, known to be indispensable for stimulating the ATPase function of HSP70,21, 32 and (2) Rsp16 was found to be devoid of co-chaperone properties toward HSP70 in vitro.21 Nevertheless, the co-chaperone activity of DNAJB13 cannot be excluded because DNAJB13 could function without activating ATP hydrolysis.21, 24 Alternatively, it could solely be a structural RS component acting independently of HSP70.
The structural organization of the axoneme relies on RSs, which are T-shaped multi-protein complexes that connect the peripheral doublets to the CC. Studies performed in Chlamydomonas reinhardtii have shown that the RS assembly process comprises two steps: a 12S RS precursor particle is initially assembled in the cell body and is then transported by the intraflagellar-transport complex to the flagellar tip for maturation into a 20S axonemal spoke, containing the 23 RS proteins identified to date.12 Rsp16, the ortholog of DNAJB13, is found in the RS neck as a homodimer and is present in the 20S particle but absent from the 12S precursor particle; it has been shown to be transported separately and to join the precursor particle in the flagella.13 Consistent with this finding, Yang et al. generated a Chlamydomonas Rsp16 RNAi strain, in which they observed the presence of RSs in the axoneme, but the spoke heads, which are normally in close proximity to the CC, were less defined, and tilted stalks were more frequent.21 In the current study, we have shown that loss-of-function mutations in DNAJB13 induce a PCD phenotype characterized by CC defects, more precisely the loss of the CP (“9 + 0” pattern). This observation, which is reminiscent of the situation reported for mutations in genes encoding other RS components (i.e., RSPH1, RSPH4A, RSPH9, and RSPH13),7, 8, 9, 10 strongly supports an important role of DNAJB13 in stabilizing the CCs. Our data also suggest that in humans, like in Chlamydomonas reinhardti, DNAJB13 might not be implicated in the cytoplasmic assembly of the RS pre-complex, nor in the anchoring of the RS pre-complex to the axoneme. DNAJB13 is, therefore, presumably one of the last RS components to be directly incorporated into the axoneme.
Previous published work has shown that in addition to being present in the RS multiprotein complex of mouse cilia and sperm flagella, DNAJB13 transiently localizes to the annulus during sperm cell differentiation.22 The transient localization of DNAJB13 to the annulus, concomitant to flagellar assembly, suggests that it has a specific function in this process. Indeed, DNAJB13 expression, together with that of 139 genes specific to flagella and/or spermiogenesis, is directly regulated by the transcription factor RFX2, whose deletion in mice prevents spermiogenesis and flagellar elongation.36 Given the severely reduced sperm count in individual DCP813, we did not have access to the ultrastructure of the sperm flagellum. Sperm analysis was not carried out for any of the male individuals carrying mutations in RSPH1, RSPH3, RSPH4A, or RSPH9, the RS genes implicated to date in PCD with CC defects.7, 8, 9, 10 HSV analysis showed that mutations in HYDIN induced asthenozoospermia and a reduced bending capacity of the sperm flagellum, but no TEM analysis of the sperm was available.11 Despite the absence of TEM evidence, it is very likely that the CP is also affected in the axoneme of the spermatozoa, thus leading to CC defects and asthenozoospermia. Additional defects of the flagella in spermatozoa lacking DNAJB13 might also occur as a result of (1) the existence of properties specific to sperm flagella rather than cilia and (2) the lack of DNAJB13 at the annulus during spermiogenesis.
In conclusion, this work shows the key role that DNAJB13 plays in preserving the integrity of the CC in humans. The function of DNAJB13, a protein that belongs to a large family of heat-shock proteins and whose loss-of-function substitutions are responsible for PCD and male infertility, is critical for both respiratory cilia and sperm flagella, given that none of its paralogs appears to be able to compensate for its deficiency.
Acknowledgments
We thank all the individuals and their family for their cooperation, as well as all the referring physicians. We thank Baptiste Rode for critical reading of the manuscript. This work was supported by INSERM, the Centre National de la Recherche Scientifique, the Université Paris Descartes, the Université Pierre et Marie Curie, grants from the Agence Nationale de la Recherche (MUCOFERTIL 12-BSV1-0011-01 and MASFLAGELLA 14-CE15-0002-03), grants from the Fondation pour la Recherche Médicale (DEQ20120323689), the Legs Poix from the Chancellerie des Universités, and the Milena Carvajal ProKartagener Foundation.
Published: August 4, 2016
Footnotes
Supplemental Data include 11 figures, 4 tables, and 6 movies and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.06.022.
Web Resources
Clustal Omega software, http://www.ebi.ac.uk/Tools/msa/clustalo/
EMBL EBI Expression Atlas, http://www.ebi.ac.uk/gxa/home
ExAC Browser, http://exac.broadinstitute.org/
Human Protein Atlas, http://www.proteinatlas.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
PROVEAN, http://provean.jcvi.org
Supplemental Data
References
- 1.Afzelius B.A. The immotile-cilia syndrome: a microtubule-associated defect. CRC Crit. Rev. Biochem. 1985;19:63–87. doi: 10.3109/10409238509086788. [DOI] [PubMed] [Google Scholar]
- 2.Satir P., Christensen S.T. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 3.Curry A.M., Rosenbaum J.L. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil. Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- 4.Smith E.F., Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papon J.F., Coste A., Roudot-Thoraval F., Boucherat M., Roger G., Tamalet A., Vojtek A.M., Amselem S., Escudier E. A 20-year experience of electron microscopy in the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2010;35:1057–1063. doi: 10.1183/09031936.00046209. [DOI] [PubMed] [Google Scholar]
- 6.Teves M.E., Nagarkatti-Gude D.R., Zhang Z., Strauss J.F., 3rd Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton (Hoboken) 2016;73:3–22. doi: 10.1002/cm.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kott E., Legendre M., Copin B., Papon J.F., Dastot-Le Moal F., Montantin G., Duquesnoy P., Piterboth W., Amram D., Bassinet L. Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet. 2013;93:561–570. doi: 10.1016/j.ajhg.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowles M.R., Ostrowski L.E., Leigh M.W., Sears P.R., Davis S.D., Wolf W.E., Hazucha M.J., Carson J.L., Olivier K.N., Sagel S.D. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care Med. 2014;189:707–717. doi: 10.1164/rccm.201311-2047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castleman V.H., Romio L., Chodhari R., Hirst R.A., de Castro S.C., Parker K.A., Ybot-Gonzalez P., Emes R.D., Wilson S.W., Wallis C. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeanson L., Copin B., Papon J.F., Dastot-Le Moal F., Duquesnoy P., Montantin G., Cadranel J., Corvol H., Coste A., Désir J. RSPH3 Mutations Cause Primary Ciliary Dyskinesia with Central-Complex Defects and a Near Absence of Radial Spokes. Am. J. Hum. Genet. 2015;97:153–162. doi: 10.1016/j.ajhg.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olbrich H., Schmidts M., Werner C., Onoufriadis A., Loges N.T., Raidt J., Banki N.F., Shoemark A., Burgoyne T., Al Turki S., UK10K Consortium Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am. J. Hum. Genet. 2012;91:672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C., Compton M.M., Yang P. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol. Biol. Cell. 2005;16:637–648. doi: 10.1091/mbc.E04-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamalet A., Clement A., Roudot-Thoraval F., Desmarquest P., Roger G., Boulé M., Millepied M.C., Baculard T.A., Escudier E. Abnormal central complex is a marker of severity in the presence of partial ciliary defect. Pediatrics. 2001;108:E86. doi: 10.1542/peds.108.5.e86. [DOI] [PubMed] [Google Scholar]
- 15.Papon J.F., Bassinet L., Cariou-Patron G., Zerah-Lancner F., Vojtek A.M., Blanchon S., Crestani B., Amselem S., Coste A., Housset B. Quantitative analysis of ciliary beating in primary ciliary dyskinesia: a pilot study. Orphanet J. Rare Dis. 2012;7:78. doi: 10.1186/1750-1172-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 17.Bush A., Chodhari R., Collins N., Copeland F., Hall P., Harcourt J., Hariri M., Hogg C., Lucas J., Mitchison H.M. Primary ciliary dyskinesia: current state of the art. Arch. Dis. Child. 2007;92:1136–1140. doi: 10.1136/adc.2006.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan J., Ekwurtzel E., Kvist U., Hultenby K., Yuan L. DNAJB13 is a radial spoke protein of mouse ‘9+2’ axoneme. Reprod. Domest. Anim. 2010;45:992–996. doi: 10.1111/j.1439-0531.2009.01473.x. [DOI] [PubMed] [Google Scholar]
- 19.Li W., Liu G. DNAJB13, a type II HSP40 family member, localizes to the spermatids and spermatozoa during mouse spermatogenesis. BMC Dev. Biol. 2014;14:38. doi: 10.1186/s12861-014-0038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan J., Yuan L. A heat-shock protein 40, DNAJB13, is an axoneme-associated component in mouse spermatozoa. Mol. Reprod. Dev. 2008;75:1379–1386. doi: 10.1002/mrd.20874. [DOI] [PubMed] [Google Scholar]
- 21.Yang C., Owen H.A., Yang P. Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9 + 2 flagella. J. Cell Biol. 2008;180:403–415. doi: 10.1083/jcb.200705069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan J., Kinoshita M., Yuan L. Spatiotemporal association of DNAJB13 with the annulus during mouse sperm flagellum development. BMC Dev. Biol. 2009;9:23. doi: 10.1186/1471-213X-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toure A., Rode B., Hunnicutt G.R., Escalier D., Gacon G. Septins at the annulus of mammalian sperm. Biol. Chem. 2011;392:799–803. doi: 10.1515/BC.2011.074. [DOI] [PubMed] [Google Scholar]
- 24.Cheetham M.E., Caplan A.J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wall D., Zylicz M., Georgopoulos C. The conserved G/F motif of the DnaJ chaperone is necessary for the activation of the substrate binding properties of the DnaK chaperone. J. Biol. Chem. 1995;270:2139–2144. doi: 10.1074/jbc.270.5.2139. [DOI] [PubMed] [Google Scholar]
- 26.Perales-Calvo J., Muga A., Moro F. Role of DnaJ G/F-rich domain in conformational recognition and binding of protein substrates. J. Biol. Chem. 2010;285:34231–34239. doi: 10.1074/jbc.M110.144642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G., Lu G.X., Xing X.W. Molecular cloning of TSARG6 gene related to apoptosis in human spermatogenic cells. Acta Biochim. Biophys. Sin. (Shanghai) 2004;36:93–98. doi: 10.1093/abbs/36.2.93. [DOI] [PubMed] [Google Scholar]
- 28.Yang H.M., Liu G., Nie Z.Y., Nie D.S., Deng Y., Lu G.X. Molecular cloning of a novel rat gene Tsarg1, a member of the DnaJ/HSP40 protein family. DNA Seq. 2005;16:166–172. doi: 10.1080/10425170500129736. [DOI] [PubMed] [Google Scholar]
- 29.Sha B., Lee S., Cyr D.M. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y.Y., Hong X.G., Wang C.C. The C-terminal (331-376) sequence of Escherichia coli DnaJ is essential for dimerization and chaperone activity: a small angle X-ray scattering study in solution. J. Biol. Chem. 2005;280:22761–22768. doi: 10.1074/jbc.M503643200. [DOI] [PubMed] [Google Scholar]
- 31.Borges J.C., Fischer H., Craievich A.F., Ramos C.H. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J. Biol. Chem. 2005;280:13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- 32.Qiu X.B., Shao Y.M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloch M.A., Johnson K.A. Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J. Cell Sci. 1995;108:3541–3545. doi: 10.1242/jcs.108.11.3541. [DOI] [PubMed] [Google Scholar]
- 34.Stephens R.E., Lemieux N.A. Molecular chaperones in cilia and flagella: implications for protein turnover. Cell Motil. Cytoskeleton. 1999;44:274–283. doi: 10.1002/(SICI)1097-0169(199912)44:4<274::AID-CM5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 35.Seixas C., Casalou C., Melo L.V., Nolasco S., Brogueira P., Soares H. Subunits of the chaperonin CCT are associated with Tetrahymena microtubule structures and are involved in cilia biogenesis. Exp. Cell Res. 2003;290:303–321. doi: 10.1016/s0014-4827(03)00325-2. [DOI] [PubMed] [Google Scholar]
- 36.Kistler W.S., Baas D., Lemeille S., Paschaki M., Seguin-Estevez Q., Barras E., Ma W., Duteyrat J.L., Morlé L., Durand B., Reith W. RFX2 Is a Major Transcriptional Regulator of Spermiogenesis. PLoS Genet. 2015;11:e1005368. doi: 10.1371/journal.pgen.1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.