Abstract
The physiological functions of natural killer (NK) cells in human immunity and reproduction depend upon diverse interactions between killer cell immunoglobulin-like receptors (KIRs) and their HLA class I ligands: HLA-A, HLA-B, and HLA-C. The genomic regions containing the KIR and HLA class I genes are unlinked, structurally complex, and highly polymorphic. They are also strongly associated with a wide spectrum of diseases, including infections, autoimmune disorders, cancers, and pregnancy disorders, as well as the efficacy of transplantation and other immunotherapies. To facilitate study of these extraordinary genes, we developed a method that captures, sequences, and analyzes the 13 KIR genes and HLA-A, HLA-B, and HLA-C from genomic DNA. We also devised a bioinformatics pipeline that attributes sequencing reads to specific KIR genes, determines copy number by read depth, and calls high-resolution genotypes for each KIR gene. We validated this method by using DNA from well-characterized cell lines, comparing it to established methods of HLA and KIR genotyping, and determining KIR genotypes from 1000 Genomes sequence data. This identified 116 previously uncharacterized KIR alleles, which were all demonstrated to be authentic by sequencing from source DNA via standard methods. Analysis of just two KIR genes showed that 22% of the 1000 Genomes individuals have a previously uncharacterized allele or a structural variant. The method we describe is suited to the large-scale analyses that are needed for characterizing human populations and defining the precise HLA and KIR factors associated with disease. The methods are applicable to other highly polymorphic genes.
Keywords: KIR, HLA class I, highly polymorphic, gene content diversity, immunity
Introduction
The human leukocyte antigen (HLA) complex of chromosome 6 is the most polymorphic region of the human genome.1 This variation is driven by pressure to resist diverse pathogens but also underlies susceptibility to autoimmunity and other inflammatory diseases of major importance to human health.2 HLA class I molecules are expressed on the surface of most tissue cells, where they interact with receptors on the surface of lymphocytes, effector cells of the immune system.3 Natural killer (NK) cells are innate and adaptive lymphocytes that destroy infected or tumor cells with aberrant expression of HLA class I; they also regulate trophoblast invasion during early pregnancy.4 NK cell activity is genetically modulated through differential expression of polymorphic killer cell immunoglobulin-like receptors (KIRs) that recognize HLA class I molecules.5 Only recently has the KIR genomic region been characterized to high resolution.6 Consequently, re-examination of diseases with long-established associations with specific HLA polymorphisms is revealing a strong and collective influence from KIR polymorphism.7, 8, 9, 10
The KIR locus in chromosomal region 19q13.4 is characterized by unusually high diversity in the numbers of both genes and their alleles.11 The region varies in size from 100 to 350 kb as a result of structurally diverse haplotypes with duplicated segments, large deletions, and gene fusions.12, 13 As a consequence of this plasticity, the 13 distinct KIR genes (KIR2DL1 [MIM: 604936], KIR2DL2 [MIM: 604937], KIR2DL4 [MIM: 604945], KIR3DL1 [MIM: 604946], KIR3DL2 [MIM: 604947], KIR2DS1 [MIM: 604952], KIR2DS2 [MIM: 604953], KIR2DS3 [MIM: 604954], KIR2DS4 [MIM: 604955], KIR2DS5 [MIM: 604956], KIR2DL5 [MIM: 605305], KIR3DL3 [MIM: 610095], and KIR3DP1 [MIM: 610604]) are combined in numerous ways. Haplotypes have between 4 and 20 KIR genes, and the most common KIR region haplotype has seven genes.14 To varying degrees, each KIR gene is polymorphic, and more than 600 KIR alleles are currently defined.15 KIR and HLA class I (HLA-A [MIM: 142800], HLA-B [MIM: 142830], and HLA-C [MIM: 142840]) polymorphism are actively co-evolving,16 suggesting that many more KIR alleles and haplotypes await discovery. During the last three decades, over 10,000 HLA class I alleles have been characterized in specialized clinical HLA laboratories,15 and similar intensive study will be needed for characterizing KIR diversity.
The function of an HLA class I molecule is to bind a peptide, usually a nonamer, inside a cell and take it to the cell surface, where the complex of peptide and HLA class I is engaged by KIRs and other lymphocyte receptors.17, 18 On healthy cells, the peptides bound by HLA class I molecules derive from normal human proteins and do not stimulate an immune response. On infected or transformed cell surfaces, pathogen-specific or tumor-specific peptides are bound to HLA class I, and gross changes in the surface level of HLA class I can be induced. All such differences activate lymphocytes and the immune response.4, 19 For the interactions between KIRs and HLA class I molecules to be effective, they have to respond to a wide diversity of tumors and pathogens, many of which are rapidly evolving.20 This has been achieved with a diversity of interactions within each individual and differences in those interactions from one individual to another. The latter provides barriers that can impede the spread of infection within families, communities, and populations.
Crucial features that distinguish KIR and HLA alleles from those of most other genes are the depth, breadth, and functional importance of their sequence divergence. Thus, alleles can differ by multiple nucleotide substitutions, and three or four alternative nucleotides are present at functionally critical positions. KIR and HLA alleles segregate as constituents of distinct lineages, which are further diversified by intra-genic and inter-genic recombination.13, 21 In turn, these lineages are maintained in all human populations, and both genomic regions exhibit clear evidence of the impact of balancing selection.22, 23 Moreover, the strong, highly reproducible signals of natural selection observed for the HLA class I and KIR regions suggest that their genomic variation is critical for human survival.24, 25
The development of methods for assessing the nature and extent of KIR genomic diversity has been limited by the complexity of the region. The widely used methods that exist for typing KIRs focus principally on gene content.12, 26, 27, 28 In contrast, the methods being used for determining allelic variation are costly, time consuming,6, 16, 29 and unsuitable for high-throughput studies. The results of the few allele-level population studies of KIRs,16, 29, 30, 31, 32 however, show that such investigation is likely to be informative. For example, some KIRs are restricted to population groups of specific geographic ancestry.30, 31 Other KIRs have lost expression but appear common and widely distributed.29, 32 To extend such studies to other populations, as well as disease cohorts, we have developed a sequencing and bioinformatics method that determines complete KIR and HLA class I genomic diversity.
Material and Methods
Overview
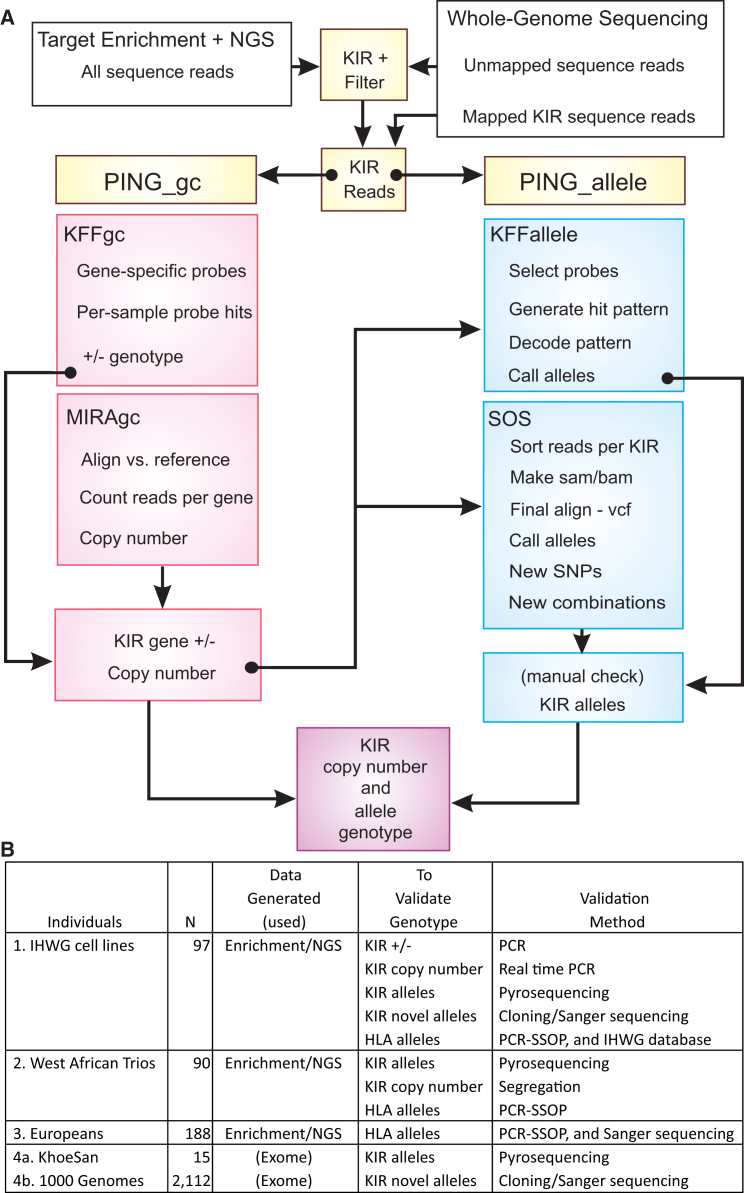
To target KIR and HLA class I genes for next-generation nucleotide sequencing (NGS), we designed sets of specific oligonucleotide probes to capture the KIR region (140–240 kb) and HLA-A, HLA-B, and HLA-C (each ∼3 kb) from libraries prepared from sheared genomic DNA. We then developed a bioinformatics pipeline (PING [Pushing Immunogenetics to the Next Generation]) specifically to convert sequence data obtained from the highly polymorphic KIR genes into high-resolution genotypes. A summary of the pipeline is shown in Figure 1A. PING first sorts the sequence reads to isolate those that represent fragments from the KIR genomic region from those that do not (a process termed filtering). PING then obtains the final KIR genotypes from these filtered reads by using a composite of two core modules that describe the gene and allele content for each individual and also return information on newly identified SNPs and recombinant alleles. The first module (PING_gc), which determines the KIR gene copy number, is used to inform the second module (PING_allele), which generates allele data (Figure 1A and Figure S1). Each module is split into two sub-modules. KIR Filter Fish (KFF), which is used in both main modules, probes the KIR sequence data with specific sequence search strings and determines which genes (KFFgc) or alleles (KFFallele) are present. The function served by KFF is equivalent to genotyping with sequence-specific oligonucleotide probes (SSOPs).33 To complement KFF, MIRAgc (based around the program MIRA)34 and Son of SAMtools (SOS; based around SAMtools)35 create alignments to reference sequences in order to determine the gene and allele content, respectively. The output is designed to comply with the genotype list (GL string) format that is used for reporting HLA and KIR data by clinical transplantation laboratories.36 We validated the typing obtained from the complete capture, NGS, and bioinformatics method (hereafter referred to as the capture/NGS method) by using standard molecular techniques, and we further tested the bioinformatics component by using existing datasets from whole-genome sequencing experiments. A summary of the data generated or otherwise obtained is shown in Figure 1B. KIR and HLA class I allele sequences used for probe design and as reference data were obtained from the Immuno Polymorphism Database (IPD; see Web Resources).15 Throughout this paper, any unique DNA sequence that spans a coding region (coding DNA sequence [CDS]) is considered a distinct allele. An explanation of KIR and HLA nomenclature is given in Appendix A.
Figure 1.
Pipeline for Analyzing Sequence Data from Highly Polymorphic and Structurally Divergent KIR Haplotypes
(A) The PING (Pushing Immunogenetics to the Next Generation) pipeline has two broad arms and two modules. The first module (PING_gc) determines KIR gene copy numbers, and the second module (PING_allele) determines their alleles. Within each module are two arms. The first arm (KFFgc and KFFallele) is an analysis independent of any alignment or assembly and uses virtual probes to mine the raw data. The second arm (MIRAgc and SOS) performs filtering and alignment of reads to reference sequences. Thus, copy-number and allele genotypes are each derived by two independent methods. The techniques are described fully in the Material and Methods.
(B) Data generated by the method described herein (1–3) and those obtained from other sources (4). The table shows the number of individuals or cell lines used for validation, the genotyping results to be validated, and the independent laboratory method used for this purpose.
Human Subjects and Data
Ethical approval for this study was obtained from the Stanford University Administrative Panels on Laboratory Care and Human Subjects in Medical Research and the Committee on Human Research at the University of California, San Francisco. Written informed consent was obtained from all individuals.
To develop and validate the capture/NGS method, we generated data from three sources of human genomic DNA:
-
1.
A Panel of IHWG Lymphoblastoid B Cell Lines. Genomic DNA was extracted from 97 International Histocompatibility Working Group (IHWG) cell lines. These cells have been used extensively in developing methods for genotyping polymorphic loci, including KIR and HLA.37, 38, 39, 40, 41 Most of the cell lines (93%) are homozygous for HLA-A, HLA-B, and HLA-C.37, 38, 39, 40, 41 A substantial majority of the IHWG cells (80%) are derived from donors of European origin and represent many of the common HLA alleles.42, 43 Also studied was genomic DNA from a chimpanzee B cell line, derived from Clint44 (Yerkes pedigree number C0471), a chimpanzee of the Pan troglodytes verus (western chimpanzee) subspecies and subject of the chimpanzee genome project.44
-
2.
West African Trios. Genomic DNA samples from 30 family trios (both of the parents and one child) from Mali in West Africa were analyzed.45
-
3.
European Control Samples. De-identified DNA samples from 188 unrelated healthy individuals of European origin, with no history of chronic disease, were studied. These samples were selected at random from a larger dataset (n = 500) developed as controls for genome-wide association studies (GWASs) of multiple sclerosis (MIM: 126200).46 These samples were used because their high-resolution HLA-A, HLA-B, and HLA-C genotypes had been determined by Sanger sequencing.46
To validate the PING pipeline, we used existing sequence-read data from an additional two sources, described below. To extract KIR-specific sequences from these datasets, we used SAMtools 0.1.18.35 to identify read pairs that mapped within the KIR region (hg19 coordinates: 19:55,228,188–55,383,188) or an unallocated chromosome 19 region (GenBank: GL000209.1) corresponding to an alternative KIR haplotype.
-
4.
15 KhoeSan individuals.47 These individuals had also been genotyped for KIR genes via standard lower-throughput methods of pyrosequencing and Sanger sequencing.16
-
5.
The 1000 Genomes Project data.48 All 2,532 of the whole-exome-sequenced individuals described in the May 2013 release were targeted.48 To ensure sufficient quantity of sequence reads for the analysis, we excluded samples if fewer than 25 reads mapped to exon 3 of KIR3DL2 or KIR3DL1/2v (see Appendix A). 420 of the 1000 Genomes samples were excluded on this basis, and the remaining 2,112 were genotyped. When previously uncharacterized KIR alleles were identified in the 1000 Genomes dataset, genomic DNA from the source samples was purchased from the Coriell Biorepository for confirmation of their sequence by standard molecular methods.
Laboratory Methods
Design of Capture Oligonucleotide Probes
To account for variation in the gene content of the KIR region, we targeted a panel of independently generated reference KIR haplotypes6, 14 that together represent all of the 13 recognized KIR genes. First, we designed probes against the two complete KIR haplotypes (GenBank: FP089703 and FP089704) that were generated from the PGF cell line, which was the source of the human reference sequence for the KIR region.6 We used end-to-end tiling with strand swapping to design non-overlapping 80-mer probes to match these reference sequences. We then designed a similar set of probes by using a further 27 complete KIR haplotype sequences6, 14 and all KIR sequences included in the January 2013 release of the IPD-KIR database.15 In this second stage, probes that differed by more than three nucleotides from the corresponding segment of the initial reference haplotypes were selected for use. We did not mask any repetitive elements in the target haplotypes. The KIR genomic region targeted by the probes is equivalent to that covered by chr19: 55,228,188–55,383,188 (UCSC Genome Browser hg19) and an unmapped chromosome 19 region (GenBank: GL000209.1), which are the two KIR haplotypes present in the hg19 reference genome. In a similar manner to the KIR probes, we designed probes against the alleles of the classic HLA class I genes present in the PGF cell line, which was also the source of the human reference sequence for the HLA region.1, 14 These probes were supplemented with probes designed against the 6,795 HLA class I sequences reported in the January 2013 release of the IPD-HLA database.15 A total of 10,456 capture probes were used.
Preparation of Biotinylated Capture Probes
The set of capture oligonucleotides, each one comprising a unique sequence flanked by the common sequences 5′-GGTGATTGCGTATCT-3′ (PTL3) and 5′-CATGTCGTGGGAATT-3′ (PTR3), was synthesized by CustomArray. This set of oligonucleotides was pooled and amplified in a single PCR using primers with sequences corresponding to PTL3 and PTR3. The PCR comprised 1× Titanium Taq buffer (Clontech), 1 μM each of biotin-PTL3 and -PTR3 primers (Integrated DNA Technologies), 0.2 μM dNTPs with 12.5% dUTP (Roche), 1 μL (1 unit) Uracil-DNA Glycosylase (UDG; New England Biolabs), 1 M betaine, 3 μL (15 units) AmpliTherm Polymerase (Epicenter), 0.2 ng of the pool of capture oligonucleotides, and H2O added to create a final volume of 100 μL. PCR cycling conditions were as follows: 37°C for 10 min, 95°C for 3 min, 95°C for 30 s, 55°C for 30 s, 72°C for 30 s (×28), 72°C for 10 min, and a hold at 10°C.
The biotinylated PCR product (100 μL aliquot) was then bound to streptavidin-coated magnetic beads (Illumina) that had been pre-washed with 100 μL 6× hybridization buffer (HB: 1 M NaCl, 0.5 M phosphate buffer, and 0.05% Tween-20) and suspended in 100 μL 12× HB. The incubation was carried out for 30 min at room temperature in HB and with agitation. The beads, now coated with biotinylated oligonucleotides, were then washed: once with 100 μL 6× HB, twice with 100 μL 0.2× HB, once with 100 μL 0.1 nM NaOH and 100 μL 10 mM EDTA, and lastly, once with 100 μL 0.2× HB. Biotinylated oligonucleotides were eluted from the beads with 0.1 mM EDTA and then concentrated via speed vacuum to a final concentration of 2.5 nM for each capture probe.
Library Preparation, Enrichment, and Sequencing
The protocol was based on the TruSeq Nano method for library preparation (Illumina). The DNA samples we used are described below. For each sample, 300 ng genomic DNA (as determined by Qubit instrument, Thermo Fisher Sceintific) was sheared into 800 bp fragments with a Covaris S220 instrument (Covaris). The library preparation was then performed according to the manufacturer’s instructions, whereby 96 unique “dual index” combinations were used individually to label the library obtained from each DNA sample, and the following modifications: (1) for cleaning and size selecting the samples after end repair, 70.2 μL sample purification beads plus 89.8 μL H20 were added to a 100 μL sample, and (2) in the final PCR, the 72°C extension time was changed from 30 to 90 s to account for the 800 bp fragment length.
Enrichment of HLA and KIR sequences was performed according to a modified version of the Nextera Rapid Capture Exome enrichment protocol (Illumina), a solution-based target-capture assay. The libraries of genomic DNA, indexed uniquely for each sample as described above, were pooled prior to their hybridization with the capture probes. Thus, each hybridization mix (100 μL) contained 96 uniquely indexed sequence libraries (62.5 ng for each library and 6,000 ng in total), 50 pM of each biotinylated capture probe, and HB (CT3 and all subsequent buffers from Illumina). The hybridization mix was incubated at 95°C for 10 min, gradually cooled by 2°C/min to 58°C, and then maintained at 58°C for 90 min. In this reaction, fragments of genomic DNA that contained targeted KIR and HLA sequences became specifically hybridized to biotinylated capture probes.
In the next reaction, 100 μL of streptavidin-coated magnetic beads were used to separate the specific hybridized genomic DNA away from the non-specific un-hybridized genomic DNA. The biotin present in hybrid DNA molecules was bound to streptavidin on the beads, leaving the non-specific DNA in solution. The DNA preparation enriched with the targeted KIR and HLA genes was then eluted from the beads. Binding of the hybridization product to the beads was achieved by 30 min incubation with agitation at 1,000 rpm on a plate shaker at room temperature. To clean the product, we removed the streptavidin beads from solution by using a magnetic separator, mixed them with 200 μL Enrichment Wash Solution (Illumina), and incubated them at 50°C for 30 min. This wash step was repeated. To elute the enriched DNA from the beads, we added 23 μL of elution mix (made from 1.5 μL 2M NaOH plus 28 μL Elution Buffer 1, Illumina), incubated for 5 min at room temperature and neutralized with 4 μL Elute Target Buffer 2 (Illumina). The eluted material was then subjected to a second round of enrichment from the hybridization step onward. After the gradual cooling step, the hybridization mix was maintained at 58°C for 14–18 hr.
An aliquot of 10 μL of the enriched DNA preparation was subject to PCR amplification in a 50 μL reaction mix containing 5 μL of a PCR primer cocktail, 15 μL of resuspension buffer, and 20 μL of Nextera Enrichment Amplification Mix. PCR cycling was performed as follows: 98°C for 30 s; 17 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s; and a final elongation step at 72°C for 5 min. Amplified material was purified with 40 μL of sample purification beads and eluted in 30 μL resuspension buffer (Illumina).
NGS Strategies
Set 1: IHWG Cell Lines. The enriched libraries were sequenced with a HiSeq 2000 instrument and sequencing chemistry (Illumina). Samples were clustered and paired-end sequencing was performed with the TruSeq SBSv3-HS Kit (Illumina). The sequencing read length was 2 × 101 bp.
Set 2: Trios and Chimpanzee. The enriched libraries obtained from these samples were sequenced with a HiSeq 2500 instrument and sequencing chemistry (Illumina). The sequencing read length was 2 × 250 bp. These samples were also genotyped for HLA-A, HLA-B, and HLA-C with SSOPs and for KIR3DL1 and KIR3DL2 by pyrosequencing23 (see Appendix A).
Set 3: European Control Samples. These samples were analyzed with a MiSeq instrument (Illumina) with V3 chemistry, and the sequencing read length was 2 × 300 bp.
Enrichment Efficiency
We estimated enrichment efficiency by mapping unprocessed sequence reads to the human reference sequence (hg19) with the Burrows-Wheeler Aligner (BWA-MEM49; k = 16) and counting the number of reads that mapped within the target coordinates of hg19.
Validating the Results from Capture/NGS via Established Methods
Previously described protocols were used for pyrosequencing16 and real-time PCR.12 Standard Sanger DNA sequencing reactions were performed in forward and reverse directions with the BigDye Terminator v.3.1 and were analyzed with a 3730 DNA Analyzer (Applied Biosystems). To verify the sequences of previously uncharacterized alleles, we cloned PCR products by using the pCR2.1-TOPO vector (Invitrogen) and sequenced them with M13 and internal primers. For each individual in whom an allele was identified, five or more clones corresponding to the allele were sequenced. HLA class I genotyping was performed with Luminex-bead-based SSOP hybridization, as described previously16 except where indicated otherwise.
Bioinformatics Methods: PING Pipeline
Harvesting KIR-Specific Reads
For sequence data obtained by the capture/NGS method, sequence reads specific to the KIR region were identified and harvested with Bowtie 2.50 We concatenated the 29 sequenced KIR haplotypes and all KIR alleles from the IPD-KIR database6, 14, 15 to create a single reference file for this purpose. The equivalent of 70,000 reads (at 2 × 300 bp) that passed this filter stage were taken per sample for KIR genotyping.
KIR-Gene-Content Module: PING_gc
This module is divided into two complementary components: the first (KFFgc) is based on string searches of raw data, and the second (MIRAgc) is based on read depth following alignment to reference sequences.
Determining KIR Gene Content with Virtual Probes: KFFgc. The sequence-read files specific to the KIR region were first processed with the “FASTQ Quality Trimmer” function of the FASTX-Toolkit v.0.0.13 (see Web Resources). From an alignment of all human KIR sequences (IPD-KIR release 2.6.1, February 17, 2015),15 all possible 32 bp sequences specific to just one of the KIR genes were generated, and those sequences covering polymorphic positions were removed. For each KIR gene, this process produced a set of gene-specific but not allele-specific probes. For each gene, ten such probes were selected at random and used for determining KIR gene content. We then counted the total number of exact matches to each probe sequence and its reverse complement in the forward and reverse (read1.fastq and read2.fastq, respectively) sequence-read files for each sample. Because every KIR haplotype has one copy of KIR3DL3,6, 14, 16 the presence or absence of other KIR genes was determined from the mean number of probe hits per target KIR divided by the mean number of KIR3DL3 probe hits on KIR3DL3. We used a ratio of 0.2 as the threshold, and values above this were considered positive.
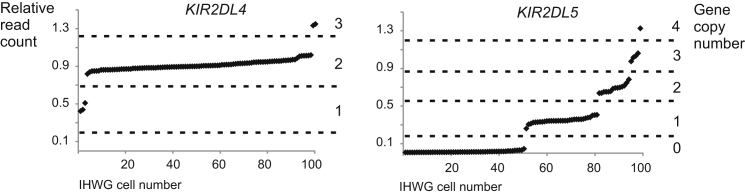
Determining KIR Gene Copy Number by Analyzing Read Depth: MIRAgc. As shown previously, the depth of reads aligned to a reference can be used for estimating structural variation on a genomic scale.51, 52 We incorporated this concept into the KIR genotyping pipeline. Using MIRA 4.0.2,34 we aligned the KIR-region-specific sequence-read pairs simultaneously against one reference sequence for each KIR gene. Using values extracted from the “contigstats” results table from the MIRA output, we divided the number of reads that aligned to each KIR gene by the number of reads aligning to KIR3DL3. This comparison produced a clustering of read ratios correlating with differences in KIR gene content (Figure 2 and Figure S2). For example, for KIR2DL4, which is absent from some haplotypes and duplicated on others,13, 53 we observed three clusters of read ratios, corresponding to one, two, or three copies of KIR2DL4 (Figure 2). For KIR2DL5, which can be present at centromeric and telomeric locations in the KIR haplotype,6, 54 we observed five clusters of read ratios, corresponding to individuals with zero, one, two, three, or four copies of KIR2DL5 (Figure 2). Representative examples of clustering results derived for all the KIR are given in Figure S2.
Figure 2.
KIR Gene Copy-Number Genotype Determined by Read Depth
The ratio of reads mapping to a specific KIR gene to those mapping to KIR3DL3 can be used for calculating KIR copy number. The results from 97 samples are shown and sorted by ratio. KIR2DL4 (left) was present in one, two, or three copies per individual in the sample set, and KIR2DL5 (right) was present in zero, one, two, three, or four copies.
In very rare cases, there can be copy-number variation of KIR3DL3. For example, an individual with duplicated KIR3DL3 on one haplotype was observed in a study of >3,500 subjects.12 Such individuals would then show an unusual copy number for all KIR genes and hence would be identified for manual inspection. The individual identified with a duplicated KIR3DL3 had a normal genotype of two KIR3DL2 copies,12 so in this case KIR3DL2 could be used as a substitute standard.
KIR-Allele-Genotyping Module: PING_allele
This module is also divided into two complementary components, the first (KFFallele) is based on string searches of raw data, and the second (SOS) is based on read depth following alignment to reference sequences. A summary of the KFFallele workflow is shown in Figure S1A, and SOS is shown in Figures S1B and S1C.
High-Resolution KIR Genotyping with Virtual Probes: KFFallele. For each KIR gene, an alignment of coding-region alleles was generated. From these, every possible unique 32-mer sequence that did not overlap an exon boundary was generated, and the resulting pool was screened for the removal of all 32-mer sequences present in another KIR gene. A “hit table” was generated with the original allele alignment and bespoke R scripting (allgenos_hit_table_2.R). The hit table was passed on to a random-forest algorithm in the “RandomForest” package of the R statistical software.55 This algorithm ranks the probes according to the amount of information they contribute to the final answer. The purpose of the random-forest step is to obtain the most efficient subset of probes, thereby increasing the computational speed of genotype assignment. The output consists of a series of probe subsets that have an increasing proportion of the total set (5%, 10%, 15%, etc.). Empirical testing found that the most efficient subset is usually the 40% most informative probes. We applied the final probe subset to the original allele alignment to produce a hit table of the expected probe pattern for all possible genotypes (again with allgenos_hit_table_2.R). The number of exact matches to each probe sequence or its reverse complement in the forward and reverse files (read1.fastq and read2.fastq, respectively) for each sample was then counted. Aggregate counts above a threshold of 10 were considered positive, and the results were compared with the genotype hit table with a bespoke R script (KFFsums.R) for assigning the KIR allele genotype.
KIR Sequence-Alignment Genotype by SOS. Data used for generating filters and reference sequences were obtained from the IPD-KIR database (release 2.6.1, 17 February 2015) and the set of 29 complete KIR haplotype sequences.6, 14, 15 For each given KIR gene, all available allele sequences were selected for use as a positive filter, and all allele sequences of all other KIR genes were used as a negative filter. We selected sequence reads specific to the given KIR by mapping them to the positive filter with Bowtie 2.50 We performed this mapping non-stringently (≥97% nucleotides matched) to allow for the detection of unknown SNPs. All reads that aligned to the positive filter were retained, and those that did not were excluded. The retained reads were mapped stringently (≥99% match) to the negative filter, and in this case, those that aligned were excluded. Finally, the selected reads were aligned to a single reference sequence that was chosen for each KIR gene (Figure S3), and the SNP variants were ascertained with SAMtools/BCFtools version 1.2.35, 50
We analyzed the resulting variant call files (VCFs) with a custom R script (jSOS) to generate genotypes based on the known combinations of SNPs (i.e., the unique KIR alleles available from IPD). A post-filtered and aligned read depth of 20 was used as the minimum for calling the genotype at any given nucleotide position. The jSOS algorithm determines all of the possible allele combinations on the basis of the genotypes of those SNPs that achieve the threshold read depth. Thus, if a specific SNP fails to reach the threshold, the ambiguity of the final allele call will increase, but an incorrect allele-level genotype will not be returned. When the combination of SNPs does not correspond to a known pair of alleles, jSOS identifies that a novel combination of known SNPs is present. In these situations, the known allele most likely present is identified in addition to the new combination (we view this as the most parsimonious genotype, and the least parsimonious could be two novel combinations present). The jSOS algorithm also identifies novel SNPs. When manual confirmation was sought, such as these cases of novel polymorphism, alignments for visual inspection were created with MIRA 4.0.2 and examined with Gap4 of the Staden Package.56
HLA Class I Genotypes
The HLA class I allele compositions were determined with NGSengine 1.7.0 (GenDX software), kindly provided by Wietse Mulder and Erik Rozemuller, with the “IMGT 3.18.0 combined” reference set. There was no pre-filtering for HLA genes, and the data were analyzed directly with the software. The one exception was the removal of reads mapping to HLA-Y, an HLA class I pseudogene present in a subset of HLA haplotypes.57 The presence or absence of HLA-Y was determined via sequence-specific string searches of the FASTQ data. For the IHWG cells, these assignments agreed with those previously determined by PCR.43 We further validated the assignment of HLA class I alleles by analyzing the sequence data with Assign MPS 1.0 (Conexio Genomics), kindly provided by Damian Goodridge. We resolved any discrepancies between the two methods, or with previously obtained results, by designing virtual probes that distinguish the two possibilities in question and searching the unprocessed sequence-read data (as described for KFF). Then, sequence reads specific to the locus under question were extracted and aligned to reference sequences (as described for SOS) and inspected manually. Here, the reference sequences used were chosen on the basis of the HLA class I genotype of the respective sample. The majority of HLA class I alleles are not fully characterized through all exons and introns (>95%).15 Thus, for some of the validations, the second or third fields of resolution were compared.
Haplotype Assignments
Haplotypes derived from homozygous cell lines were unambiguous. Haplotypes were assigned for the family trios by segregation. For all other individuals, centromeric and telomeric allele-level KIR haplotypes were assigned for each individual according to the expectation-maximization algorithm of haplo.stats implemented in the R programming language (see Web Resources).
Results
Validation of the KIR Capture Method
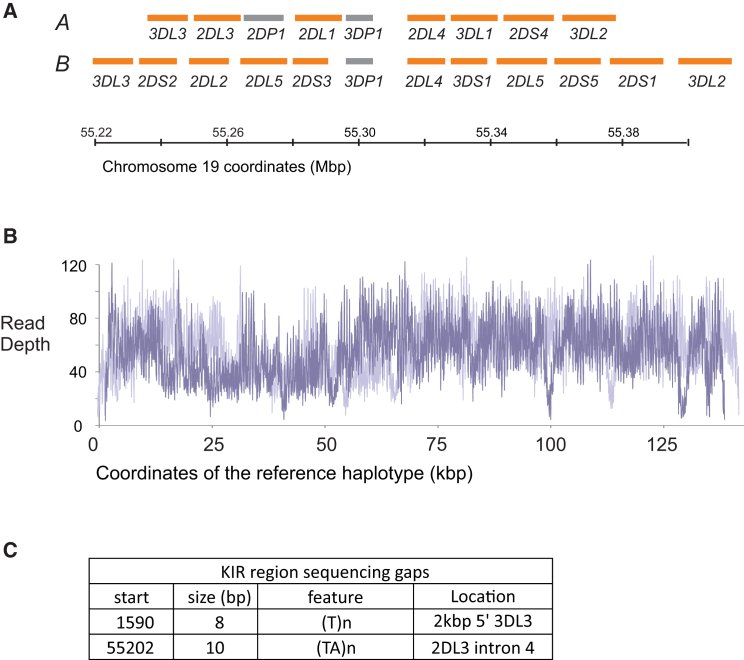
We applied our capture/NGS method to DNA extracted from 97 publically available cell lines (sample set 1, Material and Methods), originally collected by the IHWG to facilitate study of HLA genes.40 One of these cell lines is PGF, whose KIR haplotypes have been characterized previously by standard methods.6, 14 We therefore used the data we obtained from PGF cells to assess the quality of the KIR sequences produced. The PGF KIR sequence reads were mapped back onto the two conventionally determined haplotype sequences. For PGF KIR haplotype 1 (137,813 bp; GenBank: FP089703), our method gave 100% coverage, a mean read depth of 49.4×, and a read depth of >10× for 99.5% of the haplotype (Figures 3A and 3B). For PGF KIR haplotype 2 (142,732 bp; GenBank: FP089704), we obtained 99.99% coverage, a mean read depth of 49.8×, and a read depth >10× for 99.5% of the haplotype. Our haplotype 2 sequence contains two short gaps, comprising 18 nucleotides in total (Figure 3C). The missing sequences are not predicted to be of functional importance (Figure 3C). Thus, our method gives full coverage of the KIR region when it targets haplotypes that were included in the probe design. However, for population studies, it is important that our probe sets can cope with a wider (and unknown) range of diversity without any “allelic dropout.” As a test, we applied our method to genomic DNA from the subject of the chimpanzee genome project.44 We mapped the obtained reads to the two full KIR haplotype sequences (GenBank: BX842589 and AC155174) that were previously characterized from this individual.25, 58 These sequences represent both haplotypes of the KIR region and include 10 of the 14 chimpanzee KIR genes. We obtained 98.8% coverage for both of the haplotypes and mean read depths of 130× for haplotype 1 and 110× for haplotype 2. This success in capturing the chimpanzee KIR region strongly indicates that our method captures the full range of human KIR haplotypes.
Figure 3.
The KIR Region Is >99.99% Covered by Sequence Data
(A) Target KIR region on chromosome 19: the gene locations are shown in orange, and pseudogenes are shown in gray. The KIR region varies in gene content, and shown are examples of two frequent A and B haplotypes. The KIR prefix is omitted from the gene names for clarity (see Appendix A). The human reference build hg19 (UCSC Genome Browser) has a KIR A haplotype. Underneath is a KIR B haplotype shown to scale.
(B) Read depth after stringent alignment of sequence reads (no base pairs mismatched, and duplicates were removed) from the PGF cell line to the PGF reference KIR haplotypes 1 (light purple) and 2 (dark purple).
(C) Coordinates and features of two short gaps in PGF KIR haplotype 2. The location of the gaps is shown on the right.
Efficiency of the KIR Capture Method
To estimate the efficiency of the capture process, we mapped all sequence reads generated for each individual back to the human genome and counted those that fell within the target coordinates. According to this measure, the mean enrichment efficiency of the optimized 2 × 300 bp sequencing runs (sample set 3) was 87.01% (SD = 5.01). Because the target region represents <0.01% of the human genome, compared to whole-genome sequencing, this represents a significant (10,000×) reduction in the sequencing capacity required for analyzing the target.
Specificity of Sequence-Read Harvesting
To begin the bioinformatics analysis of KIR, we used a panel of reference haplotype sequences as filters to harvest any sequence reads that could map to the KIR region from the main pool of sequenced fragments (described in the Material and Methods). Because both the capture probes and the reference sequences for harvesting reads were designed with complete KIR haplotypes that did not have repetitive elements masked, we performed a further test for specificity on the harvested KIR reads. By analyzing 70,000 of these read pairs per individual, we showed that a mean of three read pairs (modal value = 0) could map outside the target region of human genome build hg19. Thus, the combination of our capture/NGS method and KIR sequence-read harvesting is highly specific. We also note that generating 2 × 100 bp sequence reads (instead of 2 × 300 bp) revealed that up to 12% of the harvested reads potentially originate from repetitive elements outside the KIR region. However, 100% of these reads map to a 1.8 kb LINE insertion that is located in intron 6 of KIR3DL213 (Figure S4) and does not overlap with any known control elements. Thus, these reads do not affect the subsequent analysis.
Measurement of KIR Gene Copy Number: PING_gc
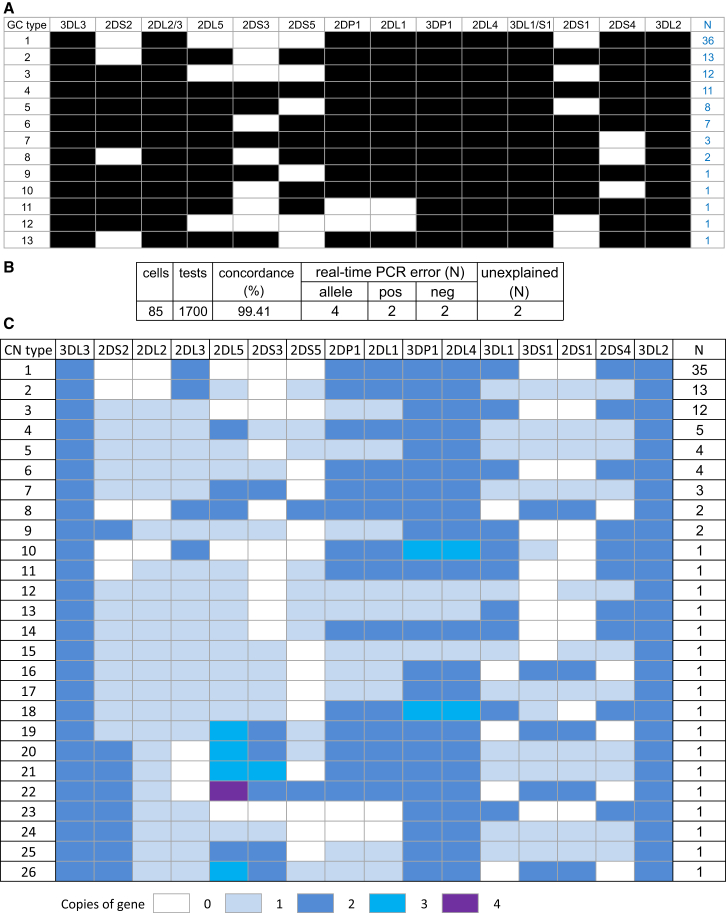
The PING_gc component of PING specifically determines gene copy number. KIR3DL3, a single-copy gene common to all KIR haplotypes,6, 14, 16 is used as the standard to which other KIR genes are compared (Material and Methods). To assess the correlation between read ratio and KIR gene content, we applied PING_gc to the sequence data generated from the 97 IHWG samples. The first PING_gc module (called KFFgc) produced 13 distinct KIR gene presence or absence genotypes (Figure 4A), identical to those obtained by established methods.26, 59, 60 We then applied the second PING module, MIRAgc, and used the observed clustering (Figure 2) to set threshold values for determining KIR gene copy numbers (Figure S3). To validate these results, we studied DNA samples from 85 of the same cell lines by using an established real-time PCR method for quantifying KIR genes.12 We observed 99.4% concordance between the results obtained by PING_gc and those obtained from real-time PCR (Figure 4B). Of ten discordant results from 1,700 determinations, four involved rare alleles that were not detected by the primers of the real-time PCR assay (KIR2DL2∗009 and KIR3DL2∗076, the latter of which was discovered during this study). Of the other six discordant results, two were due to false positives of the real-time PCR (as shown independently by a standard PCR method),27 two were just below the threshold values of the real-time PCR (but were clearly positive with PING_gc and standard PCR), and two remain unexplained (Figure 4B). Thus, the discrepancies were likely due to low-frequency errors in sensitivity or specificity of the real-time PCR method. We conclude that analysis of high-throughput sequencing data with the PING_gc module provides precise measurement of KIR gene content and copy number and gives almost 100% accuracy. Using PING_gc, we identified 26 distinct KIR gene copy-number genotypes in the 97 cell lines analyzed (Figure 4C). Two cells have duplicated KIR3DP1-KIR2DL4-KIR3DL1/S1 segments (copy-number genotypes 10 and 18; Figure 4C), and three cells have haplotypes lacking KIR2DL4 (genotypes 12, 13, and 15; Figure 4C). In summary, determination of copy number alone increases the resolution of KIR genotyping and is an important step toward understanding the role of KIR polymorphism in disease.61 We next sought to include the allele-calling components in validation of the PING pipeline in order to achieve full resolution of KIR genotypes.
Figure 4.
KIR Gene-Content and Copy-Number Genotypes
(A) Gene-content genotypes derived from all 97 cell lines by the PING pipeline. A black box indicates that a gene is present, and a clear box indicates the absence of a gene. One example from each observed gene-content genotype (GC type) is shown, and the number observed is shown on the right.
(B) Independent validation of the KIR copy-number genotypes by real-time PCR12 on 85 samples. There were four discrepancies due to alleles undetected by real-time PCR (allele). There were two false positives (pos) and two false negatives (neg) by real-time PCR. Two discrepancies remain unexplained.
(C) Gene copy-number genotypes derived from all 97 cell lines by PING_gc. A colored rectangle indicates the presence of a gene, and the shades represent the copy number as indicated in the key. One example from each observed gene copy-number genotype (CN type) is shown, and the number observed is shown on the right.
High-Resolution Genotypes of KIR Alleles: PING_allele
PING_allele determines KIR allele genotypes according to all known KIR coding-sequence alleles (Material and Methods). PING_allele was first validated with whole-exome data from a sample of 15 KhoeSan individuals.47 For these individuals, the KIR copy-number and allele data produced by PING matched the data obtained previously by the established methods of Sanger sequencing and pyrosequencing-based genotyping of KIR genes.16, 31 Because KIR3DL1, KIR3DS1, and KIR3DL2 of lineage II KIR (see Appendix A) exhibit high polymorphism and structural variation,13, 23, 62 they were chosen as a further test of PING_allele. Using the capture/NGS method, we applied the pipeline to data obtained from 30 family trios from Mali in West Africa (sample set 2, Material and Methods). In this highly heterozygous population, we identified 18 KIR3DL1/S1, 15 KIR3DL2, and 3 KIR3DL1/2v alleles (Table S1A). These alleles were authenticated by established pyrosequencing and Sanger sequencing methods (Material and Methods), as well as by their segregation in the trios. KIR3DL1/2v is a KIR3DL1-KIR3DL2 fusion gene that segregates with KIR3DL1/S1 and encodes a functional protein.13 Importantly, we correctly identified individuals with distinct combinations of KIR3DL1/S1 alleles, KIR3DL1/2v fusion genes, and KIR3DL1/S1-deleted haplotypes (Figure 5). To expand the analysis, we next analyzed 2,112 individuals from the 1000 Genomes dataset.48 From their exome sequences, we identified 50 KIR3DL1/S1, 46 KIR3DL2, and 5 KIR3DL1/2v alleles (Table S1A), as well as 14 KIR3DL1/S1 duplication and 13 KIR3DL1/S1 deletion haplotypes (Table S1B). Such duplicated and deleted KIR haplotypes were detected in all 26 populations represented in the 1000 Genomes dataset (Table S1B). These results demonstrate that the capture/NGS method coupled with the copy-number and allele components of the PING pipeline can correctly identify the extensive and complex variation of lineage II KIR molecules. The KIR3DL1/S1 and KIR3DL2 genotypes obtained for all individuals analyzed from the 1000 Genomes Project are shown in Table S1C.
Figure 5.
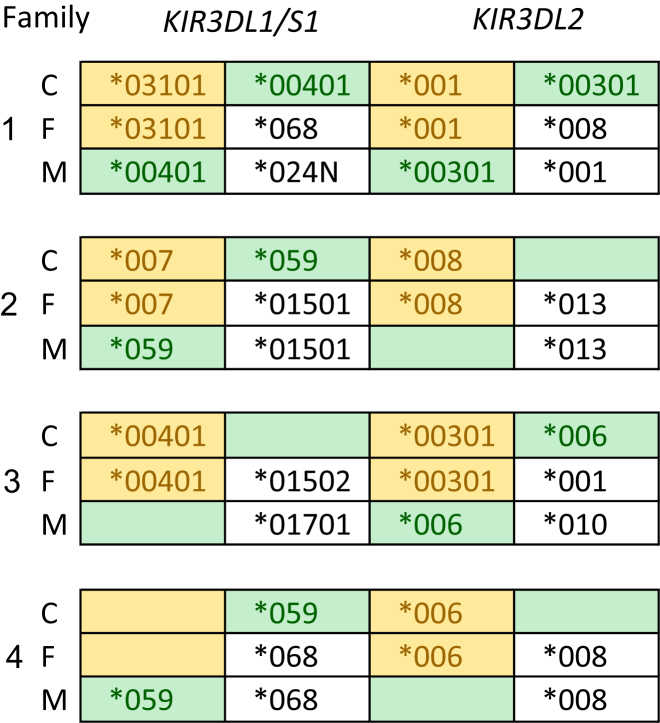
High-Resolution Allele-Level Genotyping of KIR Genes
Four examples of high-resolution allele and copy-number genotypes of lineage II KIR genes and their segregation in family trios: C, child; F, father; and M, mother. Colored boxes show the segregating alleles. All members of family 1 have two alleles each of KIR3DL1/S1 and KIR3DL2. Family 2 shows segregation of the KIR3DL1/2v fusion gene (the allele named KIR3DL1∗059), which consists of exons 1–6 from KIR3DL1 and 7–9 from KIR3DL2.13, 63 For clarity, KIR3DL1/2v is shown as an allele of KIR3DL1, so there is no allele of KIR3DL2 on this haplotype. Family 3 shows segregation of a haplotype that lacks KIR3DL1/S1 and is marked by the presence of KIR3DL2∗006. The gene copy numbers were determined by PING_gc, which indicated that one copy of KIR3DL1 is present in each of individuals 3C and 3M and two copies are present in 3F. Family 4 shows segregation of both the KIR3DL1/2v and the KIR3DL1 negative haplotypes to the child.
Identification of Novel Alleles by PING
Hereafter, we use “novel” to describe KIR variants that were previously undiscovered but identified and characterized by the methods described here. The PING pipeline identifies such novel alleles by the presence of either one or more novel SNPs or a novel combination of known SNPs (Material and Methods). To test this “new allele discovery” component of PING, we again used the lineage II KIR genes. In the course of analyzing the 1000 Genomes data, we identified 100 novel alleles: 33 KIR3DL1/S1, 65 KIR3DL2, and 2 KIR3DL1/2v alleles. Defining these alleles are 88 novel SNPs (39 in KIR3DL1/S1 and 49 in KIR3DL2; Tables S2A and S2B) and 17 novel combinations of known SNPs (Table S2C). Sequences of all the novel alleles were validated by standard methods: PCR amplification from genomic DNA of source material and subsequent cloning and/or Sanger sequencing (Material and Methods). Of the 2,112 individuals studied, 229 (10.8%) have at least one novel lineage II KIR allele (Table S1C). A total of 333 different KIR3DL1/S1-KIR3DL2 haplotypes were identified in this analysis (Table S1C).
High-Resolution KIR Allele and Copy-Number Genotypes
We applied PING_allele to the KIR sequence data obtained from the 97 IHWG cells. This analysis of 13 KIR genes identified 144 different KIR sequences: 128 corresponding to established alleles and 16 representing novel alleles. The latter were all shown to be authentic by the standard methods described above for KIR3DL1/S1 and KIR3DL2 (Table S2D). By considering all 144 KIR variants, we identified a minimum of 104 centromeric and 42 telomeric KIR haplotypes in the cell panel (Figure 6 and Table S3). Consistent with our results are KIR genotyping data obtained previously from the IHWG cells, which achieved a limited discrimination of alleles.6, 59 These analyses demonstrate that PING accurately processes high-throughput sequence data to give accurate high-resolution KIR genotypes. The high-resolution KIR genotypes of the IHWG cell panel have been compiled (Table S3), providing a resource for future investigation.
Figure 6.
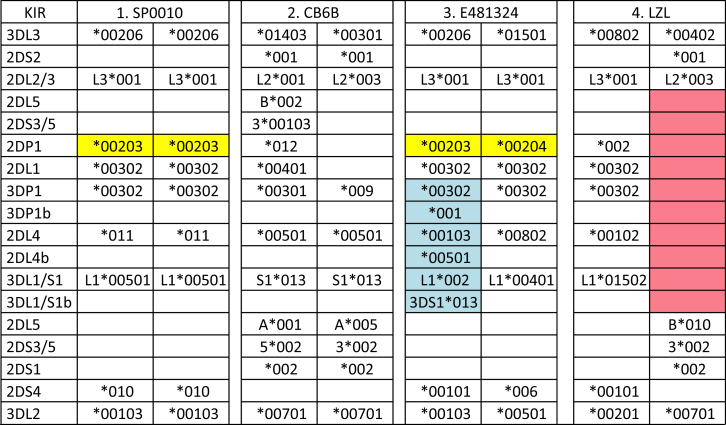
High-Resolution KIR Allele and Copy-Number Genotypes of 97 IHWG Cells
Four examples of high-resolution allele and copy-number genotypes of KIR. Individual 1 (SP0010) is homozygous for the KIR A haplotype. Individual 2 (CB6B) has two different B haplotypes. Individual 3 (E481324) has a duplication of three loci (in blue shading: denoted as 3DP1b, 2DL4b, and 3DL1/S1b). Individual 4 (LZL) has a deletion of the central segment of the KIR haplotype (red). Yellow shading denotes alleles that were identified in the current study. The full genotypes for each IHWG cell are given in Table S3.
Validation of the High-Resolution HLA Class I Capture Method
Because KIR and HLA class I glycoproteins are functionally interacting receptors and ligands, the capture/NGS method was designed to capture and analyze the two gene families in the same reaction (Material and Methods). To validate the HLA class I sequences obtained by this method, we first analyzed the panel of 97 IHWG cell lines. In previous studies,38, 39, 40 including high-density whole-genome SNP analysis,43 90 of the IHGW cells were judged to be homozygous for HLA-A, HLA-B, and HLA-C, and five of the other seven lines were found to be homozygous for two of the three HLA class I genes. Our high-throughput sequencing results are completely concordant with the genotypes previously determined by conventional methods15 (Table S4A). In the course of validation, we defined two novel HLA-C alleles that encode distinctive proteins (Table S4B). Because our method gives full-length genomic sequences, including introns and flanking regions, all HLA-A, HLA-B, and HLA-C alleles of the IHWG cells are now defined at a much higher resolution (“four field”; see Appendix A) than previously achieved.
The IHWG cells represent an unusual, and highly selected, sampling of the human population because they are HLA homozygous, and many of them derive from consanguineous individuals. To extend the study to heterozygous individuals, we applied the capture/NGS method to 30 West African family trios from Mali (sample set 2, Material and Methods) and 188 individuals selected at random from a panel of healthy Europeans (sample set 3). The capture/NGS method achieved full coverage of the HLA class I genes, and an example of the result obtained from one individual is shown in Figure 7A. In the Africans, 22 HLA-A, 30 HLA-B, and 15 HLA-C alleles were identified. These allele sequences agreed completely with HLA class I genotypes we determined by standard probe-based and Sanger sequencing methods. They were also consistent with the observed segregation of HLA-A, HLA-B, and HLA-C alleles within the family trios (Tables S4C–S4D). In total, 170 distinct HLA class I alleles were identified in the three validation sets (IHWG cell lines, African trios, and Europeans). These 170 alleles include 62 of the 69 fundamental allele types defined by the first two digits of the HLA nomenclature (Figure 7B) and thus cover most, if not all, of the breadth of HLA class I allelic diversity.21 Thus, it is likely that all HLA-A, HLA-B, and HLA-C alleles can be captured and sequenced by our method. In support of this thesis, the capture/NGS method robustly detects and sequences B∗73:01 (E.A., unpublished data), a unique, archaic allele that has by far the most divergent HLA class I gene sequence in the modern human population.64 Further demonstrating our method’s robust capacity to target divergent sequences, we successfully captured and sequenced all alleles of Patr-A, Patr-B, and Patr-C (Table S4E), the chimpanzee orthologs of HLA-A, HLA-B, and HLA-C, respectively, from the chimpanzee that was the subject of the chimpanzee genome project.44 In summation, the breadth and depth of our results give confidence that our method will be able to capture the full range of HLA class I alleles.
Figure 7.
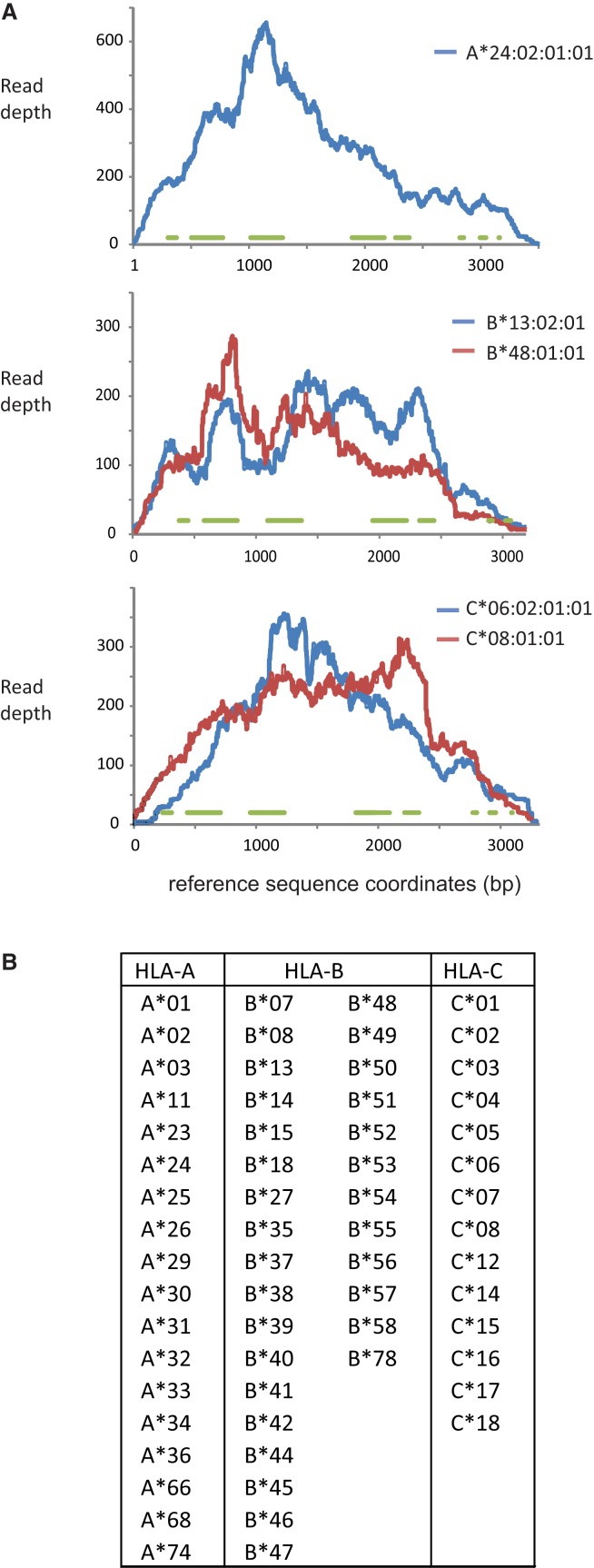
Capture of HLA Class I Genes for High-Resolution Allele Genotyping
(A) Shown is the read depth across each of the HLA class I genes from a representative sample (chosen by virtue of having a read number closest to the mean number of HLA-specific reads). Green lines indicate the coordinates of the exons that were covered. To generate this figure, we obtained full gene sequences (∼3 kb each) from IPD to represent all five HLA class I alleles known to be present in this sample (the sample is homozygous for a common allele of HLA-A). Sequence reads were filtered to be specific to HLA-A, HLA-B, and HLA-C and then aligned to these references with high stringency. The read depth was measured with SAMtools/BCFtools.
(B) The major HLA class I allele types detected in this study.
Discussion
We developed an integrated capture/NGS method to characterize completely the structure and sequence of the highly polymorphic KIR and HLA class I genes. The approach enables a focused and extensive definition of this physiologically important variation, which is not possible with any other single method. Our method is also well suited for genotyping the large cohorts required for insightful study of population genetics and disease association, as well as donor selection for clinical transplantation. All components of the method were validated with panels of DNA samples that represent the observed range of human variation for these complex genomic regions. Both the method and the information we have obtained during its development should prove valuable resources for future studies.
We used DNA from well-characterized immortalized human B cell lines as reference materials during the design and optimization of the laboratory and bioinformatics methods. These panels of IHWG and 1000 Genomes cell lines are generally available for other researchers (see Web Resources). For validation, we focused on lineage II KIR genes because they exhibit some of the most extreme and complex genomic variation within the human KIR locus. We identified and distinguished deletions and duplications of KIR3DL1/S1 and the presence of the KIR3DL1/2v fusion gene and also defined the alleles of these genes. This was achieved by a combination of quantitative assessment of read depth, virtual sequence probing, and reference alignment. Such independent verification is critical for characterizing structural KIR variants, which are not detected by methods that depend only on the alignment of sequence reads to reference haplotypes.65, 66 In summary, the validation experiments demonstrate our method to be robust and capable of detecting the full range of KIR genomic variation.
With few exceptions, studies of KIR in human populations and disease cohorts have analyzed KIR gene content, but not allelic diversity.67, 68 Such studies were seminal for showing how KIR genomic diversity can shape the immune response and provide resistance to disease.69 Studies of gene content also uncovered the influence of KIR diversity on the success of reproduction and bone marrow transplantation.9, 11 The few studies that have focused on specific KIR genes and their allelic diversity and copy number have refined these disease associations and implicated specific alleles.61, 70 In the course of validating our method, we identified and characterized 116 novel KIR alleles. This knowledge of KIR polymorphism makes substantial contributions to the KIR database.15 For example, the number of KIR3DL2 alleles was doubled and is now in excess of 100. We also show that 476 (22.5%) of the 1000 Genomes individuals have at least one example of a structural variant or novel allele of KIR3DL1/S1 or KIR3DL2 (Table S1C). All of these genomic variations have potential to influence NK cell function, but they are not visible to typing at the level of KIR gene content. A strong case can therefore be made that high-resolution knowledge of KIR diversity, in all its forms, will identify additional disease associations and improve the understanding of those already known.
The study of human populations and their evolutionary dynamics, ancestry, and disease has benefited from GWAS methods, which genotype numerous SNP markers in large cohorts of individuals. Such analysis of the KIR region has been impractical because its extraordinary structural diversity leaves few locations suitable for designing binary SNP markers, and many of the KIR genotyping results fail routine quality-control filters. Thus, the “immunochip,” which focuses on immune-system genes71 and has refined the role of HLA-associated diseases, includes relatively few informative SNPs in the KIR region. These SNPs are located in KIR3DL3, KIR2DL4, KIR3DL1/S1, and KIR3DL2, which were previously assumed to be present in one copy on every haplotype.14 Our study demonstrates that this is not the reality. In more than 10% of 1000 Genomes individuals, one of these four KIR genes is deleted, duplicated, or part of a fusion gene. We conclude that genotyping SNPs within the KIR locus by using standard binary measurement is of little practical value.
To compensate for the absence of suitable SNPs within the KIR genes, a recently described imputation method should accurately re-assess the diversity of KIR gene content for many of the reported GWASs.28 Imputation of HLA class I alleles from GWAS data has been informative for studies of immune-mediated diseases.72 Imputation of HLA alleles varies in its efficiency, particularly in non-European individuals, partly because >10,000 alleles are described but also because imputation relies on linkage disequilibrium, which can extend for shorter genomic tracts in non-Europeans than in Europeans.15, 72 Many of the KIR variants and polymorphisms identified by our method are not evenly distributed across human populations. For example, the KIR3DL1/2v fusion gene is restricted to Africans, who exhibit the lowest linkage disequilibrium worldwide. Thus, it is unlikely that imputation will be able to resolve all of the structural and allelic diversity of KIR.
We employed short-read technology because of its high fidelity. Pressing this point, all of the novel SNPs identified by our method were confirmed by independent and well-established sequencing methods. The capture method we used will probably soon be adapted to obtain longer fragment sizes and read lengths, which should become increasingly valuable as the sequencing error rates decrease.73 Because we are able to capture and sequence the chimpanzee KIR region, our method most likely captures the extent of human KIR diversity. Thus, there is limited allelic dropout. Alternative methods that do not suffer allelic dropout are whole-genome approaches. Because our method targets large numbers of individuals and has a low impact on sequencing instruments and reagent resources, our assay provides an economic and practically viable alternative to whole-genome experiments. We also note that our bioinformatics pipeline can obtain accurate KIR genotypes from any whole-genome sequencing experiments of sufficient mean depth. The pipeline is also designed for application to any highly polymorphic gene system. Our approach is designed to genotype very large numbers of individuals while having a low impact on computer resources. In these properties, it differs from the population-graph method of allele designation that has been applied to HLA.74 However, this method could be a valuable complement to our methods if it is also applied to KIR.
The first KIR cDNA sequences were reported in the late 1990s.75, 76 This led to research that revealed the unanticipated scope of genetic complexity and diversity of the human KIR gene family.27 The method we describe here will facilitate determination of a complete description of KIR variation in the human population, its interaction and co-evolution with HLA class I, and its influence on physiology, disease, and immunotherapy.
Conflicts of Interest
S.N. and M.R. are employees of Illumina.
Acknowledgments
This study was supported by US NIH grants U01 AI090905, R01 GM109030, R01 AI17892, and U19 AI119350.
Published: August 4, 2016
Footnotes
Supplemental Data include four figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.06.023.
Appendix A: KIR Nomenclature
Throughout this paper, any unique DNA sequence that spans a coding region (otherwise known as a CDS) is considered a distinct allele. Those alleles that encode a unique protein sequence define an allotype. KIR genes and alleles are named by the KIR Nomenclature Committee, formed from members of the WHO Nomenclature Committee for factors of the HLA system and members of the HUGO Genome Nomenclature Committee.15 The IPD-KIR database is part of the IPD, which is listed in the Web Resources section.
In the nomenclature for alleles, the digit (2 or 3) and letter (D) following the KIR prefix indicate whether two (2D) or three (3D) immunoglobulin (Ig)-like domains are present in the encoded protein. After the letter D is another letter: L, S, or P. The letters L and S refer to the relative length of the cytoplasmic tail: either short (S) or long (L). Long-tailed KIRs are inhibitory receptors, whereas short-tailed KIRs are activating receptors. P denotes a pseudogene. Next is a number that distinguishes KIR encoded by different genes but with the same domain number and signaling function. There are two instances where a pair of KIR gene names were combined to form one name after it became apparent that they occupied the same locus—these are KIR2DL2/3 and KIR3DL1/S1. Following the gene name, there are three fields of numbers that distinguish the various types of alleles. The first field, of three digits, distinguishes alleles that encode different allotypes. Thus, these alleles encode proteins with different amino acid sequences. The second field, of two digits, distinguishes alleles that encode the same allotype but differ by one or more synonymous substitutions in the coding region. The third field, also of two digits, distinguishes alleles with the same coding region sequence but with one or more substitutions in introns, flanking regions, or other parts of the gene. For example, KIR3DL2 is a receptor with three Ig-like domains and inhibitory signaling function. KIR3DL2∗001 and KIR3DL2∗002 are alleles that encode allotypes with different amino acid sequences, whereas KIR3DL2∗00101 and KIR3DL2∗00102 both encode the KIR3DL2∗001 allotype but differ by a synonymous nucleotide substitution in the coding sequence. KIR3DL2∗0010101 and KIR3DL2∗0010102 also encode the KIR3DL2∗001 allotype, but they differ by nucleotide substitutions in the introns.
There are four phylogenetically distinguished lineages of human KIR.77 Lineage I and III KIR molecules possess two Ig-like domains. Lineage I molecules (KIR2DL4 and KIR2DL5) interact with HLA ligands of low polymorphism, including HLA-G, and lineage III molecules (KIR2DL1– KIR2DL3 and KIR2DS1– KIR2DS5) interact with specific allotypes of HLA-B and HLA-C molecules. Lineage II (KIR3DL1/S1 and KIR3DL2) and V (KIR3DL3) molecules have three Ig-like domains. Lineage II molecules interact with allotypes of HLA-A or HLA-B,78 and the ligand for lineage V remains unknown. We focused on lineage II because it is characterized by extensive structural and sequence diversity.13, 23 Although most human KIR haplotypes have two lineage II genes, KIR3DL1/S1 and KIR3DL2, some haplotypes have deleted KIR3DL1/S1, some have duplicated KIR3DL1/S1, and some have an in-frame fusion of KIR3DL1 and KIR3DL2, termed KIR3DL1/2v.13 There are three divergent lineages of KIR3DL1/S1: the inhibitory KIR3DL1∗005 and KIR3DL1∗015, which are divergent and highly polymorphic, and activating KIR3DS1, which is less polymorphic.23
KIR haplotypes form two groups: A and B. KIR A haplotypes have a fixed content of seven genes and two pseudogenes, whereas KIR B haplotypes vary in gene content.6, 14, 27 There are around 20 common KIR B haplotypes with different gene content and numerous additional KIR B haplotypes that are rare.6, 12, 27
HLA class I alleles are named according to a hierarchical set of fields separated by colons, and each contains as many different digits as is needed to distinguish all alleles. The first field distinguishes the major alleles, which differ by multiple nucleotide and amino acid substitutions (e.g., HLA-A∗01 and HLA-A∗02). The second field distinguishes the subtypes of each major allele (e.g., HLA-A∗02:01 and HLA-A∗02:153), which encode allotypes that differ by one or more amino acid substitutions (e.g., HLA-A∗02:01 and HLA-A∗02:153). The third field distinguishes subtypes that encode proteins with identical amino acid sequences but differ by one or more synonymous substitutions within the protein-coding exons (e.g., HLA-A∗02:01:01 and HLA-A∗02:01:02). The fourth field distinguishes subtypes that have identical coding-region sequence but differ by one or more nucleotide substitutions within introns or the transcribed 3′ and 5′ flanking regions (e.g., HLA-A∗02:01:01:01 and HLA-A∗02:01:01:02). In addition, suffix letters are used to denote known expression variants (e.g., N denotes a null allele, for which the encoded protein is not expressed at the cell surface).
Accession Numbers
The KIR and HLA class I allele sequences reported in this article were submitted to GenBank and the Immuno Polymorphism Database (IPD).15 Their official (IPD-designated) names and GenBank accession numbers are given below and in Table S2 (KIR prefixes are excluded for brevity): 3DL1∗00103 (LN606766), 3DL1∗00104 (KP784298), 3DL1∗00105 (KP784297), 3DL1∗00404 (KP784290), 3DL1∗00504 (KP784291), 3DL1∗00505 (KP784294), 3DL1∗01506 (KP784293), 3DL1∗05902 (KP784300), 3DL1∗077 (LN606765), 3DL1∗088 (LN606767), 3DL1∗089 (LN606768), 3DL1∗090 (LN606769), 3DL1∗091 (LN606770), 3DL1∗092 (LN606771), 3DL1∗093 (LN606772), 3DL1∗094N (LN606773), 3DL1∗095 (KP784289), 3DL1∗096 (KP784285), 3DL1∗097 (KP784286), 3DL1∗098 (KP784301), 3DL1∗099 (KP784288), 3DL1∗100 (KP784299), 3DL1∗101 (KP784292), 3DL1∗102 (KP784295), 3DL1∗103 (KP784296), 3DL1∗109 (KP784287), 3DS1∗01304 (KP784279), 3DS1∗01305 (KP784278), 3DS1∗01306 (KP784276), 3DS1∗01307 (KP784284), 3DS1∗104 (KP784277), 3DS1∗105 (KP784280), 3DS1∗106 (KP784283), 3DS1∗107 (KP784281), 3DS1∗108 (KP784282), 3DL2∗00106 (KJ535483), 3DL2∗081 (KJ535484), 3DL2∗073 (KJ535485), 3DL2∗071 (KJ535486), 3DL2∗080 (KJ535487), 3DL2∗00303 (KJ535488), 3DL2∗00703 (KJ535489), 3DL2∗069 (KJ535490), 3DL2∗070 (KJ535491), 3DL2∗072 (KJ535492), 3DL2∗074 (KJ535493), 3DL2∗068 (KJ535494), 3DL2∗00304 (KJ535495), 3DL2∗079 (KJ535496), 3DL2∗077 (KJ535497), 3DL2∗01002 (KJ535498), 3DL2∗076 (KJ535500), 3DL2∗00203 (KJ535501), 3DL2∗082 (KJ535502), 3DL2∗078 (KJ535503), 3DL2∗101 (LN995832), 3DL2∗00708 (LN995833), 3DL2∗100 (KP784305), 3DL2∗102 (LN995834), 3DL2∗00705 (LN649139), 3DL2∗089 (LN649146), 3DL2∗00502 (LN649136), 3DL2∗095 (LN649155), 3DL2∗00204 (LN649156), 3DL2∗00706 (LN649150), 3DL2∗084 (LN649140), 3DL2∗093 (LN649152), 3DL2∗087 (LN649143), 3DL2∗00707 (LN649153), 3DL2∗098 (KP784302), 3DL2∗094 (LN649154), 3DL2∗01003 (LN649149), 3DL2∗00107 (LN649144), 3DL2∗085 (LN649141), 3DL2∗086 (LN649142), 3DL2∗088 (LN649145), 3DL2∗090 (LN649147), 3DL2∗097 (KP784303), 3DL2∗091 (LN649148), 3DL2∗096 (LN649157), 3DL2∗092 (LN649151), 3DL2∗00704 (LN606764), 3DL2∗01004 (KP784304), 3DP1∗01002 (KP893537), 3DP1∗015 (KP893538), 3DL2∗108 (LN999781), 3DL2∗107 (LN999782), 3DL2∗104 (LN999783), 3DL2∗109 (LN999784), 3DL2∗00602 (LN999785), 3DL2∗106 (LN999786), 3DL2∗04302 (LN999787), 3DL2∗00709 (LN999788), 3DL2∗01004 (LN999790), 3DL2∗103 (LN999791), 3DL2∗01902 (LN999792), 3DL2∗021 (LN999793), 3DL2∗07902 (LN999794), 3DL2∗06002 (LN999795), 3DL2∗105 (LN999796), 3DL2∗00109 (LN999797), 3DL2∗10002 (LN999798), 2DL1∗032N (KP893536), 2DL3∗034 (KP784272), 2DL5A∗021 (KP784273), 2DP1∗00203 (KP784307), 2DP1∗00204 (KP784309), 2DP1∗015 (KP784306), 2DP1∗016 (KP784275), 2DP1∗017 (KP784308), 2DP1∗018 (KP784274), 2DP1∗019 (KP784310), 2DS3∗008 (KP784269), 2DS5∗015 (KP784270), 2DS5∗016 (KP784271), HLA-C∗01:02:30 (KP893072), and HLA-C∗07:18 (KP893073).
Web Resources
1000 Genomes, http://www.1000genomes.org
Coriell Biorepository, https://catalog.coriell.org/
FASTX-Toolkit, http://hannonlab.cshl.edu/fastx_toolkit/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
GenDx NGSengine, http://www.gendx.com/products/ngsengine
Haplo Stats, http://www.mayo.edu/research/labs/statistical-genetics-genetic-epidemiology/software
Immuno Polymorphism Database (IPD), http://www.ebi.ac.uk/ipd/
International Histocompatibility Working Group (IHWG), http://www.ihwg.org/
OMIM, http://www.omim.org/
PING executable R files, https://github.com/wesleymarin/
PING scripts, https://web.stanford.edu/∼n0rmski/projectH/
R statistical software, http://www.r-project.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
(A) Lists the alleles of KIR3DL1/S1 and KIR3DL2 that have established IPD curated names (see Robinson et al.3 in Document S1) (i.e., “known alleles”) and were identified in the family trios (Trio) or 1000 Genomes (1000G) data sets, and the number of times each was observed. Orange shading indicates KIR3DL1/2v fusion gene alleles.
(B) Shows structural and allelic variation of KIR3DL1/S1. (i) Alleles that characterize haplotypes with duplicated KIR3DL1/S1. (ii) Alleles that characterize haplotypes with deleted KIR3DL1/S1. At the left is shown the alleles of KIR3DL1/S1 and KIR3DL2 present on each of the structural variant haplotypes identified in the 1000 Genomes data. The KIR2DL4 alleles and copy numbers were also determined in each case because the genes are usually duplicated or deleted in tandem, as observed here for all haplotypes. † indicates likely identical to KIR3DL1/S1 duplicated haplotype identified previously (see Gomez-Lozano et al.4 and Norman et al.5 in Document S1). At the center in blue text is shown the number of observations for each haplotype and at the right the populations in which each haplotype was observed are listed according to the standard 1000 Genomes three-letter codes (see Abecasis et al.6 in Document S1).
(C) Lists the KIR3DL1/S1 and KIR3DL2 genotypes obtained for each individual analyzed from the 1000 Genomes. Green (SNP) and yellow (recombination) shading indicate alleles identified for the first time in the current study. Blue shading indicates duplicated KIR3DL1/S1, red indicates deleted KIR3DL1/S1, and orange indicates KIR3DL1/2v.
(A) Novel alleles of 3DL1/S1 identified in 1000 Genomes individuals. Shown are the KIR3DL1/S1 alleles identified for the first time from PING bioinformatics analysis of the 1000 Genomes data. From left to right: the individual in whom the allele was first identified, their population (three digit 1000 Genomes code), GenBank accession numbers, official names (designated by IPD) and coding changes. † - KIR3DL1∗05902 and KIR3DL1∗098 are variants of KIR3DL1/2v. Codons are numbered according to the mature protein.
(B) Novel alleles of KIR3DL2 identified in 1000 Genomes individuals and identified by novel SNPs. Shown are the novel KIR3DL2 alleles defined by newly-identified SNPs using PING (SOS) to analyze the 1000 Genomes data. From left to right: the individual in whom the allele was first identified, their population (three digit 1000 Genomes code), GenBank accession numbers, official names (designated by IPD) and coding changes. Codons are numbered according to the mature protein. LP indicates leader peptide. (+ indicates allele is also characterized by recombination that generates a novel polypeptide sequence)
(C) Novel alleles of KIR3DL2 identified in 1000 Genomes individuals and identified by novel combinations of known SNPs. Shows the alleles of KIR3DL2 identified in the 1000 Genomes data that are characterized by novel combinations of known SNPs. The center column shows one closest allele and the distinguishing SNP. The right column indicates if the recombination generated a novel polypeptide sequence (Y/N).
(D) Novel KIR alleles identified in the IHWG cells. Shown are the novel KIR alleles identified in the IHWG cells. From left to right: cell line where the allele was first identified, accession numbers, official names and coding changes. Codons are numbered according to the mature protein.
Shown are the complete KIR genotypes from 97 IHWG cell lines. The colors blue, red, green and mauve denote likely haplotypes, divided into centromeric (KIR3DL3-KIR3DP1) and telomeric segments (KIR2DL4-KIR3DL2). Highlighted in yellow are the alleles discovered for the first time here (described in Figure 6). Cyan indicates haplotype with duplicated KIR3DP1-2DL4-3DL1/S1, pale yellow indicates deleted segment. Seven of the cell lines are homozygous for all of the KIR alleles (hom), including the four that are also homozygous through the KIR region according to whole genome SNP genotype (SP0010, LKT3, MT14B, SPL) and three others (BGE, PF04015, WT47).
(A) Shown are the HLA class I genotypes obtained from 97 IHWG cell lines using the enrichment and sequencing process. Homozygous genotypes are indicated with gray text for the second allele (two copies of the allele are assumed to be present). When analyzed at two-field resolution the alleles shown are consistent with those previously reported (shown at the right), with exception of one allele call indicated in red text; presence of A∗02:01 was confirmed in BGE using alternative methods (Material and Methods) and the sample shown heterozygous in the HLA-A region using high-density SNP analysis (see Norman et al.7 in Document S1). Highlighted in yellow are two alleles discovered for the first time here (described in panel B). Blue shading indicates presence of HLA-Y, confirmed in each case using PCR7.
(B) Shows the accession numbers, IPD designated names and characteristics of two HLA-C alleles discovered in the IHWG cell lines.
(C) Shown are the HLA class I genotypes obtained from 30 West African family trios using the enrichment and sequencing process. Colors indicate the segregating haplotypes. C - child, F - father, M - mother. HLA genotypes determined by SSOPs are shown at the right. Blue shading indicates presence of HLA-Y.
(D) HLA class I genotypes obtained from 188 randomly-selected donors of European origin. The genotypes obtained by Sanger sequencing are shown at the right. Gray boxes indicate discrepancies (n=7) between the two genotype calling methods, which were all resolved on visual inspection of the sequence alignments (the correct type is indicated in the gray box). Blue shading indicates presence of HLA-Y.
(E) MHC (equivalent to HLA) -A, -B and -C genotypes obtained from chimpanzee. At the left are shown the genotypes obtained using capture/NGS and the right using Sanger sequencing.
(F) Virtual probes used to determine presence of HLA-Y pseudogene from fastq read data.
References
- 1.Horton R., Wilming L., Rand V., Lovering R.C., Bruford E.A., Khodiyar V.K., Lush M.J., Povey S., Talbot C.C., Jr., Wright M.W. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 2.Trowsdale J., Knight J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinkernagel R.M., Doherty P.C. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- 4.Vivier E., Raulet D.H., Moretta A., Caligiuri M.A., Zitvogel L., Lanier L.L., Yokoyama W.M., Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell K.S., Purdy A.K. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132:315–325. doi: 10.1111/j.1365-2567.2010.03398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyo C.W., Guethlein L.A., Vu Q., Wang R., Abi-Rached L., Norman P.J., Marsh S.G., Miller J.S., Parham P., Geraghty D.E. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLaren P.J., Carrington M. The impact of host genetic variation on infection with HIV-1. Nat. Immunol. 2015;16:577–583. doi: 10.1038/ni.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn R.S., Moslehi H., Martin M.P., Abad-Santos M., Bowcock A.M., Carrington M., Liao W. Inhibitory KIR3DL1 alleles are associated with psoriasis. Br. J. Dermatol. 2016;174:449–451. doi: 10.1111/bjd.14081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancusi A., Ruggeri L., Urbani E., Pierini A., Massei M.S., Carotti A., Terenzi A., Falzetti F., Tosti A., Topini F. Haploidentical hematopoietic transplantation from KIR ligand-mismatched donors with activating KIRs reduces nonrelapse mortality. Blood. 2015;125:3173–3182. doi: 10.1182/blood-2014-09-599993. [DOI] [PubMed] [Google Scholar]
- 10.Hollenbach J.A., Pando M.J., Caillier S.J., Gourraud P.A., Oksenberg J.R. The killer immunoglobulin-like receptor KIR3DL1 in combination with HLA-Bw4 is protective against multiple sclerosis in African Americans. Genes Immun. 2016;17:199–202. doi: 10.1038/gene.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parham P., Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang W., Johnson C., Jayaraman J., Simecek N., Noble J., Moffatt M.F., Cookson W.O., Trowsdale J., Traherne J.A. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norman P.J., Abi-Rached L., Gendzekhadze K., Hammond J.A., Moesta A.K., Sharma D., Graef T., McQueen K.L., Guethlein L.A., Carrington C.V. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson M.J., Torkar M., Haude A., Milne S., Jones T., Sheer D., Beck S., Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson J., Halliwell J.A., Hayhurst J.D., Flicek P., Parham P., Marsh S.G. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norman P.J., Hollenbach J.A., Nemat-Gorgani N., Guethlein L.A., Hilton H.G., Pando M.J., Koram K.A., Riley E.M., Abi-Rached L., Parham P. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjorkman P.J., Saper M.A., Samraoui B., Bennett W.S., Strominger J.L., Wiley D.C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 18.Boyington J.C., Motyka S.A., Schuck P., Brooks A.G., Sun P.D. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 19.Zhang N., Bevan M.J. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das J., Khakoo S.I. NK cells: tuned by peptide? Immunol. Rev. 2015;267:214–227. doi: 10.1111/imr.12315. [DOI] [PubMed] [Google Scholar]
- 21.Parham P., Lomen C.E., Lawlor D.A., Ways J.P., Holmes N., Coppin H.L., Salter R.D., Wan A.M., Ennis P.D. Nature of polymorphism in HLA-A, -B, and -C molecules. Proc. Natl. Acad. Sci. USA. 1988;85:4005–4009. doi: 10.1073/pnas.85.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeGiorgio M., Lohmueller K.E., Nielsen R. A model-based approach for identifying signatures of ancient balancing selection in genetic data. PLoS Genet. 2014;10:e1004561. doi: 10.1371/journal.pgen.1004561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norman P.J., Abi-Rached L., Gendzekhadze K., Korbel D., Gleimer M., Rowley D., Bruno D., Carrington C.V., Chandanayingyong D., Chang Y.H. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 24.Hughes A.L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 25.Abi-Rached L., Moesta A.K., Rajalingam R., Guethlein L.A., Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010;6:e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gómez-Lozano N., Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens. 2002;59:184–193. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- 27.Uhrberg M., Valiante N.M., Shum B.P., Shilling H.G., Lienert-Weidenbach K., Corliss B., Tyan D., Lanier L.L., Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 28.Vukcevic D., Traherne J.A., Næss S., Ellinghaus E., Kamatani Y., Dilthey A., Lathrop M., Karlsen T.H., Franke A., Moffatt M. Imputation of KIR Types from SNP Variation Data. Am. J. Hum. Genet. 2015;97:593–607. doi: 10.1016/j.ajhg.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vierra-Green C., Roe D., Hou L., Hurley C.K., Rajalingam R., Reed E., Lebedeva T., Yu N., Stewart M., Noreen H. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS ONE. 2012;7:e47491. doi: 10.1371/journal.pone.0047491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gendzekhadze K., Norman P.J., Abi-Rached L., Graef T., Moesta A.K., Layrisse Z., Parham P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc. Natl. Acad. Sci. USA. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemat-Gorgani N., Edinur H.A., Hollenbach J.A., Traherne J.A., Dunn P.P., Chambers G.K., Parham P., Norman P.J. KIR diversity in Māori and Polynesians: populations in which HLA-B is not a significant KIR ligand. Immunogenetics. 2014;66:597–611. doi: 10.1007/s00251-014-0794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yawata M., Yawata N., Draghi M., Little A.M., Partheniou F., Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saiki R.K., Bugawan T.L., Horn G.T., Mullis K.B., Erlich H.A. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986;324:163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- 34.Chevreux B., Pfisterer T., Drescher B., Driesel A.J., Müller W.E., Wetter T., Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milius R.P., Mack S.J., Hollenbach J.A., Pollack J., Heuer M.L., Gragert L., Spellman S., Guethlein L.A., Trachtenberg E.A., Cooley S. Genotype List String: a grammar for describing HLA and KIR genotyping results in a text string. Tissue Antigens. 2013;82:106–112. doi: 10.1111/tan.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorak M.T., Shao W., Machulla H.K., Lobashevsky E.S., Tang J., Park M.H., Kaslow R.A. Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun. 2006;7:450–467. doi: 10.1038/sj.gene.6364315. [DOI] [PubMed] [Google Scholar]
- 38.Marsh S.G.E., Packer R., Heyes J.M., Bolton B., Fauchet R., Charron D., Bodmer J.G. The International Histocompatibility Workshop Cell Panel. In: Charron D., editor. Genetic Diversity of HLA: Functional and Medical Implications. EDK; 1996. pp. 26–28. [Google Scholar]
- 39.Mickelson, E., Hurley, C., Ng, J., Tilanus, M., Carrington, M., Marsh, S.G.E., Rozemuller, E., Pei, J., Rosielle, J., Voorter, C., et al. (2006). 13th IHWS Shared Resources Joint Report. IHWG Cell and Gene Bank and Reference Cell Panels. In Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference, J.A. Hanson, ed. (IHWG Press), pp. 523–553.
- 40.Yang S.Y., Milford E., Hammerling U., Dupont B. Description of the Reference Panel of B-Lymphoblastoid Cell Lines for Factors of the HLA system: The B-Cell Line Panel Designed for the Tenth International Histocompatibility Wprkshop. In: Dupont B., editor. Immunobiology of HLA Histocompatibility Testing. Springer-Verlag; 1987. pp. 11–19. [Google Scholar]
- 41.Yang Y., Chung E.K., Wu Y.L., Savelli S.L., Nagaraja H.N., Zhou B., Hebert M., Jones K.N., Shu Y., Kitzmiller K. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mack S.J., Cano P., Hollenbach J.A., He J., Hurley C.K., Middleton D., Moraes M.E., Pereira S.E., Kempenich J.H., Reed E.F. Common and well-documented HLA alleles: 2012 update to the CWD catalogue. Tissue Antigens. 2013;81:194–203. doi: 10.1111/tan.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norman P.J., Norberg S.J., Nemat-Gorgani N., Royce T., Hollenbach J.A., Shults Won M., Guethlein L.A., Gunderson K.L., Ronaghi M., Parham P. Very long haplotype tracts characterized at high resolution from HLA homozygous cell lines. Immunogenetics. 2015;67:479–485. doi: 10.1007/s00251-015-0857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 45.Ba A., Beley S., Chiaroni J., Bailly P., Silvy M. RH diversity in Mali: characterization of a new haplotype RHD∗DIVa/RHCE∗ceTI(D2) Transfusion. 2015;55:1423–1431. doi: 10.1111/trf.13109. [DOI] [PubMed] [Google Scholar]
- 46.Isobe N., Keshevan A., Gourraud P.A., Zhu A.H., Datta E., Schlaeger R., Caillier S.J., Santaniello A., Lizee A., Himmelstein D.S. Effects of HLA genetic risk burden on MRI disease phenotypes in multiple sclerosis. JAMA Neurol. 2016;73:795–802. doi: 10.1001/jamaneurol.2016.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kidd J.M., Sharpton T.J., Bobo D., Norman P.J., Martin A.R., Carpenter M.L., Sikora M., Gignoux C.R., Nemat-Gorgani N., Adams A. Exome capture from saliva produces high quality genomic and metagenomic data. BMC Genomics. 2014;15:262. doi: 10.1186/1471-2164-15-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon S., Xuan Z., Makarov V., Ye K., Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin M.P., Bashirova A., Traherne J., Trowsdale J., Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J. Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 54.Gómez-Lozano N., Gardiner C.M., Parham P., Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–319. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- 55.R Development Core Team . R Foundation for Statistical Computing; 2008. A language and environment for statistical computing. [Google Scholar]
- 56.Bonfield J.K., Whitwham A. Gap5--editing the billion fragment sequence assembly. Bioinformatics. 2010;26:1699–1703. doi: 10.1093/bioinformatics/btq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe Y., Tokunaga K., Geraghty D.E., Tadokoro K., Juji T. Large-scale comparative mapping of the MHC class I region of predominant haplotypes in Japanese. Immunogenetics. 1997;46:135–141. doi: 10.1007/s002510050252. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J.G., Bashirova A., Palmer S., Sims S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia, C.A., Robinson, J., Shilling, H.G., Hayhurst, J.D., Flicek, P., Parham, P., Madrigal, J.A., and Marsh, S.G. (2006). KIR Gene Characterisation of HLA Homozygous Cell Lines. In Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference, J.A. Hansen, ed. (IHWG Press), pp. 1203–1207.
- 60.Cook M.A., Norman P.J., Curran M.D., Maxwell L.D., Briggs D.C., Middleton D., Vaughan R.W. A multi-laboratory characterization of the KIR genotypes of 10th International Histocompatibility Workshop cell lines. Hum. Immunol. 2003;64:567–571. doi: 10.1016/s0198-8859(03)00042-9. [DOI] [PubMed] [Google Scholar]
- 61.Pelak K., Need A.C., Fellay J., Shianna K.V., Feng S., Urban T.J., Ge D., De Luca A., Martinez-Picado J., Wolinsky S.M., NIAID Center for HIV/AIDS Vaccine Immunology Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9:e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou L., Jiang B., Chen M., Ng J., Hurley C.K. The characteristics of allelic polymorphism in killer-immunoglobulin-like receptor framework genes in African Americans. Immunogenetics. 2011;63:549–559. doi: 10.1007/s00251-011-0536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shilling H.G., Guethlein L.A., Cheng N.W., Gardiner C.M., Rodriguez R., Tyan D., Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J. Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 64.Abi-Rached L., Jobin M.J., Kulkarni S., McWhinnie A., Dalva K., Gragert L., Babrzadeh F., Gharizadeh B., Luo M., Plummer F.A. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Church D.M., Schneider V.A., Steinberg K.M., Schatz M.C., Quinlan A.R., Chin C.S., Kitts P.A., Aken B., Marth G.T., Hoffman M.M. Extending reference assembly models. Genome Biol. 2015;16:13. doi: 10.1186/s13059-015-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gargis A.S., Kalman L., Bick D.P., da Silva C., Dimmock D.P., Funke B.H., Gowrisankar S., Hegde M.R., Kulkarni S., Mason C.E. Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat. Biotechnol. 2015;33:689–693. doi: 10.1038/nbt.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.González-Galarza F.F., Takeshita L.Y., Santos E.J., Kempson F., Maia M.H., da Silva A.L., Teles e Silva A.L., Ghattaoraya G.S., Alfirevic A., Jones A.R., Middleton D. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43:D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hollenbach J.A., Nocedal I., Ladner M.B., Single R.M., Trachtenberg E.A. Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics. 2012;64:719–737. doi: 10.1007/s00251-012-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bashirova A.A., Martin M.P., McVicar D.W., Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu. Rev. Genomics Hum. Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 70.Martin M.P., Qi Y., Gao X., Yamada E., Martin J.N., Pereyra F., Colombo S., Brown E.E., Shupert W.L., Phair J. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parkes M., Cortes A., van Heel D.A., Brown M.A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 72.de Bakker P.I., Raychaudhuri S. Interrogating the major histocompatibility complex with high-throughput genomics. Hum. Mol. Genet. 2012;21(R1):R29–R36. doi: 10.1093/hmg/dds384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodwin S., Gurtowski J., Ethe-Sayers S., Deshpande P., Schatz M.C., McCombie W.R. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015;25:1750–1756. doi: 10.1101/gr.191395.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dilthey A., Cox C., Iqbal Z., Nelson M.R., McVean G. Improved genome inference in the MHC using a population reference graph. Nat. Genet. 2015;47:682–688. doi: 10.1038/ng.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Colonna M., Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 76.Wagtmann N., Biassoni R., Cantoni C., Verdiani S., Malnati M.S., Vitale M., Bottino C., Moretta L., Moretta A., Long E.O. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 77.Rajalingam R., Parham P., Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J. Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 78.Vivian J.P., Duncan R.C., Berry R., O’Connor G.M., Reid H.H., Beddoe T., Gras S., Saunders P.M., Olshina M.A., Widjaja J.M. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Lists the alleles of KIR3DL1/S1 and KIR3DL2 that have established IPD curated names (see Robinson et al.3 in Document S1) (i.e., “known alleles”) and were identified in the family trios (Trio) or 1000 Genomes (1000G) data sets, and the number of times each was observed. Orange shading indicates KIR3DL1/2v fusion gene alleles.
(B) Shows structural and allelic variation of KIR3DL1/S1. (i) Alleles that characterize haplotypes with duplicated KIR3DL1/S1. (ii) Alleles that characterize haplotypes with deleted KIR3DL1/S1. At the left is shown the alleles of KIR3DL1/S1 and KIR3DL2 present on each of the structural variant haplotypes identified in the 1000 Genomes data. The KIR2DL4 alleles and copy numbers were also determined in each case because the genes are usually duplicated or deleted in tandem, as observed here for all haplotypes. † indicates likely identical to KIR3DL1/S1 duplicated haplotype identified previously (see Gomez-Lozano et al.4 and Norman et al.5 in Document S1). At the center in blue text is shown the number of observations for each haplotype and at the right the populations in which each haplotype was observed are listed according to the standard 1000 Genomes three-letter codes (see Abecasis et al.6 in Document S1).
(C) Lists the KIR3DL1/S1 and KIR3DL2 genotypes obtained for each individual analyzed from the 1000 Genomes. Green (SNP) and yellow (recombination) shading indicate alleles identified for the first time in the current study. Blue shading indicates duplicated KIR3DL1/S1, red indicates deleted KIR3DL1/S1, and orange indicates KIR3DL1/2v.
(A) Novel alleles of 3DL1/S1 identified in 1000 Genomes individuals. Shown are the KIR3DL1/S1 alleles identified for the first time from PING bioinformatics analysis of the 1000 Genomes data. From left to right: the individual in whom the allele was first identified, their population (three digit 1000 Genomes code), GenBank accession numbers, official names (designated by IPD) and coding changes. † - KIR3DL1∗05902 and KIR3DL1∗098 are variants of KIR3DL1/2v. Codons are numbered according to the mature protein.
(B) Novel alleles of KIR3DL2 identified in 1000 Genomes individuals and identified by novel SNPs. Shown are the novel KIR3DL2 alleles defined by newly-identified SNPs using PING (SOS) to analyze the 1000 Genomes data. From left to right: the individual in whom the allele was first identified, their population (three digit 1000 Genomes code), GenBank accession numbers, official names (designated by IPD) and coding changes. Codons are numbered according to the mature protein. LP indicates leader peptide. (+ indicates allele is also characterized by recombination that generates a novel polypeptide sequence)
(C) Novel alleles of KIR3DL2 identified in 1000 Genomes individuals and identified by novel combinations of known SNPs. Shows the alleles of KIR3DL2 identified in the 1000 Genomes data that are characterized by novel combinations of known SNPs. The center column shows one closest allele and the distinguishing SNP. The right column indicates if the recombination generated a novel polypeptide sequence (Y/N).
(D) Novel KIR alleles identified in the IHWG cells. Shown are the novel KIR alleles identified in the IHWG cells. From left to right: cell line where the allele was first identified, accession numbers, official names and coding changes. Codons are numbered according to the mature protein.
Shown are the complete KIR genotypes from 97 IHWG cell lines. The colors blue, red, green and mauve denote likely haplotypes, divided into centromeric (KIR3DL3-KIR3DP1) and telomeric segments (KIR2DL4-KIR3DL2). Highlighted in yellow are the alleles discovered for the first time here (described in Figure 6). Cyan indicates haplotype with duplicated KIR3DP1-2DL4-3DL1/S1, pale yellow indicates deleted segment. Seven of the cell lines are homozygous for all of the KIR alleles (hom), including the four that are also homozygous through the KIR region according to whole genome SNP genotype (SP0010, LKT3, MT14B, SPL) and three others (BGE, PF04015, WT47).
(A) Shown are the HLA class I genotypes obtained from 97 IHWG cell lines using the enrichment and sequencing process. Homozygous genotypes are indicated with gray text for the second allele (two copies of the allele are assumed to be present). When analyzed at two-field resolution the alleles shown are consistent with those previously reported (shown at the right), with exception of one allele call indicated in red text; presence of A∗02:01 was confirmed in BGE using alternative methods (Material and Methods) and the sample shown heterozygous in the HLA-A region using high-density SNP analysis (see Norman et al.7 in Document S1). Highlighted in yellow are two alleles discovered for the first time here (described in panel B). Blue shading indicates presence of HLA-Y, confirmed in each case using PCR7.
(B) Shows the accession numbers, IPD designated names and characteristics of two HLA-C alleles discovered in the IHWG cell lines.
(C) Shown are the HLA class I genotypes obtained from 30 West African family trios using the enrichment and sequencing process. Colors indicate the segregating haplotypes. C - child, F - father, M - mother. HLA genotypes determined by SSOPs are shown at the right. Blue shading indicates presence of HLA-Y.
(D) HLA class I genotypes obtained from 188 randomly-selected donors of European origin. The genotypes obtained by Sanger sequencing are shown at the right. Gray boxes indicate discrepancies (n=7) between the two genotype calling methods, which were all resolved on visual inspection of the sequence alignments (the correct type is indicated in the gray box). Blue shading indicates presence of HLA-Y.
(E) MHC (equivalent to HLA) -A, -B and -C genotypes obtained from chimpanzee. At the left are shown the genotypes obtained using capture/NGS and the right using Sanger sequencing.
(F) Virtual probes used to determine presence of HLA-Y pseudogene from fastq read data.