Abstract
Purpose of review
This article provides an overview of advances in the induced pluripotent stem cell field to model cardiomyopathies of inherited inborn errors of metabolism and acquired metabolic syndromes in vitro.
Recent findings
Several inborn errors of metabolism have been studied using “disease in a dish” model, including Pompe disease, Danon disease, Fabry disease, and Barth syndrome. Disease phenotypes of complex metabolic syndromes, such as diabetes mellitus and aldehyde dehydrogenase 2 deficiency have also been observed.
Summary
Differentiation of patient- and disease-specific induced pluripotent stem cell-derived cardiomyocytes has provided the capacity to model deleterious cardiometabolic diseases to understand molecular mechanisms, perform drug screens, and identify novel drug targets.
Keywords: induced pluripotent stem cells, cardiomyocytes, metabolic heart disease
Introduction
Metabolic diseases often disrupt normal cellular processes in the heart and result in cardiac dysfunction, life-threatening arrhythmias, and ultimately increased mortality (1,2). Frequently, metabolic diseases are categorized to inborn errors of metabolism (IEM) and acquired metabolic syndrome, the latter of which is defined as a group of risk factors that raise the risk of heart disease in adulthood. In vitro modeling of cardiometabolic diseases utilizing human cardiac tissue has posed a challenge due to morbidity associated with tissue biopsy and lack of tissue availability. Animal models have enabled critical advances in understanding the biology of heart diseases, but there remain questions of translatability to humans due to interspecies differences in physiology (3). Additionally, recapitulating accurate phenotypes of IEM in vivo has been challenging because transgenic animal models of these disorders are often nonviable with early mortality. Finally, metabolic syndrome is polygenic in nature and influenced by epigenetic or environmental factors, and therefore it has been difficult to model the disease using transgenic techniques.
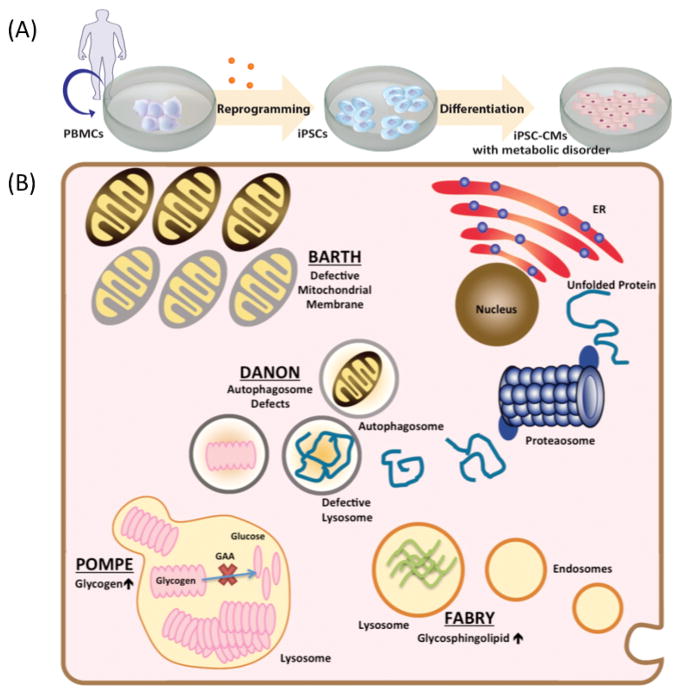
Since its advent in the mid-2000s, induced pluripotent stem cell (iPSC) technology has provided investigators a new methodology to model pathophysiology of heart diseases and thereby identify novel therapeutic targets (4,5). In recent years, researchers have studied various cardiomyopathies and cardiac channelopathies using patient-specific iPSC-derived cardiomyocytes (iPSC-CMs) (6), including arrhythmogenic right ventricular cardiomyopathy (7,8), familial dilated cardiomyopathy (9–11), familial hypertrophic cardiomyopathy (12), and long QT syndrome (13–15). Advances in replicating significant mutation-induced dysregulation of cells using iPSC-CM platforms have also allowed the study of deleterious metabolic diseases in a petri dish (Figure 1A). This review will encapsulate the current standing of iPSC-based disease modeling for IEM (Figure 1B) and common metabolic syndromes and how ultimately, the iPSC field can attain the goals of personalized medicine, to understand and treat disease at a patient-specific level (16).
Figure 1. Current iPSC-CM models of inherited errors in metabolism.
(A) A schematic view of the process to recapitulate metabolic heart disease in a dish, with steps to reprogram human peripheral blood mononuclear cells to iPSCs and then differentiate to cardiomyocytes. (B) Cellular metabolic processes in inborn errors of metabolism successfully recapitulated in iPSC-CMs with correlated in vitro phenotypes. Pompe disease (glycogen storage disorder Type II) showed glycogen accumulation in lysosomes. Danon disease (glycogen storage disorder Type IIb) showed defects in early autophagosome vacuoles, which lead to cellular hypertrophy. Barth Syndrome (mitochondrial disorder) showed impaired cardiolipin remodeling resulting in reduced mitochondrial supercomplex formation. Fabry disease (lysosomal storage disorder) showed increased glycosphingolipid accumulation in lysosomes.
Cardiomyopathy with inborn errors of metabolism
IEM are heritable monogenic disorders that often present congenitally with multi-system manifestations affecting various organs including the heart, nervous system, and skeletal muscle. Mutations in IEM commonly disrupt energy metabolism, such as glycogen and lysosomal storage, lipid metabolism, fatty acid oxidation and transport, and respiratory chain function. Studies have shown that approximately 5% of IEM have associated cardiac morbidity and mortality (1). Cardiac involvement in IEM often presents as dilated (DCM) or hypertrophic cardiomyopathy (HCM), as well as life-threatening arrhythmias (Table 1).
Table 1.
Induced pluripotent stem cell models of cardiomyopathies associated with inborn errors of metabolism
| Disease | Year (Ref) | Specific mutation | iPSC-CM in vitro phenotype | Cardiac muscle clinical phenotype |
|---|---|---|---|---|
| Glycogen Storage disorder | ||||
| Pompe Disease (Type II) | 2011 (17) | Unspecified GAA gene mutations | Lysosomal glycogen accumulation; increased autophagosome structures; mitochondrial dysfunction | Hypertrophy, hyperdynamic systolic function with outflow obstruction |
| 2014 (18) | Compound heterozygote mutations: [del ex18] exon 18 deletion and [del525T] single nucleotide exon 2 deletion in GAA gene Compound heterozygote mutation: (IVS1-13T>G) and deletion of exon 18 in GAA gene |
Elevated lysosomal glycogen; larger glycogen granules observed in infantile compared to late onset PD iPSC-CMs | ||
| 2015 (19) | Homozygous exon 18 deletion in GAA Heterozygous exon 18 deletion in GAA |
Lysosomal glycogen accumulation; autophagic dysfunction - autophagosome accumulation and higher electrophoretic mobility of lysosome-associated membrane proteins | ||
| 2015 (20) | Heterozygous mutation c.796C>T and c.1316T>A in GAA | Lysosomal glycogen accumulation | ||
| Danon Disease (Type IIb) | 2015 (21) | 2 base pair insertion in exon 2 of LAMP-2 gene (129-130 insAT) Single point mutation in intron 1 of LAMP-2 gene (IVS-1 c.64+1 G>A) |
Increased early autophagic vacuoles, lack of mature autophagic vacuoles; Heart failure feature: cellular hypertrophy; Oxidative stress features: fragmented, short mitochondria, poor mitochondrial network | Hypertrophy, reduced ejection fraction, and altered calcium handling consistent with heart failure |
| Mitochondrial disease | ||||
| Barth Syndrome | 2011 (22) | Missense mutation in TAZ1 gene (590G>T) Splice site mutation in TAZ1 gene (110-1AG>AC) Missense mutation in TAZ1 (170G>T) |
Impaired cardiolipin remodeling: absent mature CL and accumulation of precursor, MLCL; decreased respiratory chain supercomplex formation; altered mitochondrial oxygen consumption; increased ROS CM differentiation unsuccessful due to cell death |
Dilated cardiomyopathy |
| 2014 (23) | Frameshift mutation in TAZ gene (c.517delgG) Missense mutation in TAZ gene (c.328T>C) |
Impaired cardiolipin remodeling; lower ATP levels; impaired sarcomerogenesis; defective contractility | ||
| Lysosomal storage disease | ||||
| Fabry disease | 2013 (24) | Hemizygous mutation in exon 3 of GLA gene c.485G>A | Membranous cytoplasmic bodies in lysosomes CM differentiation unsuccessful due to cell death |
Hypertrophic cardiomyopathy, left ventricular hypertrophy, systolic and diastolic dysfunction, arrhythmias |
| 2014 (25) | Hemizygous mutation in exon 3 of GLA gene c.485G>A Hemizygous mutation in exon 5 of GLA gene c.658C>T |
Lysosomal globotriaosylceramide accumulation; electron dense storage inclusions (zebra bodies); disorganized contractile fibers with peripheral localization | ||
Carbohydrate Metabolism
Glycogen Storage Disease
Pompe disease
Pompe disease (PD) is an autosomal recessive, type II Glycogen storage disease resulting from complete loss of acid alpha-glucosidase (GAA), which hydrolyzes glycogen to glucose (26,27). Significant cellular and ultrastructural pathology observed with electron microscopy is caused by abnormal accumulation of lysosomal glycogen primarily in cardiac and skeletal muscle, causing DCM or HCM characteristics. Enzyme replacement therapy (ERT) using recombinant human GAA (rhGAA) has been a mainstay therapy for PD patients (28). However, ERT is not curative, with varying degree of response, and has adverse effects such as immunologic reactions and life-threatening arrhythmias, warranting efforts to further understand the pathophysiology of PD with iPSC modeling.
Huang et al. first modeled PD in iPSCs by conditionally over-expressing GAA at the time of reprogramming to evade initial metabolic defects of the GAA mutation and allow successful generation of iPSCs from two patients with infantile-onset PD (17). Mutant PD iPSCs exhibited ultrastructural abnormalities and mitochondrial dysfunction. Upon differentiation into cardiomyocyte-like cells, the cells reconstituted cardinal features of PD, showing a 2-fold increase in glycogen content, deteriorating mitochondria, and formation of autophagosome-like structures. When treated with rhGAA, PD iPSC-CMs demonstrated positive functional response with significant reduction in glycogen. Additionally, several molecular markers were found to be up- or down-regulated with rhGAA therapy, which could offer options for monitoring therapeutic response in PD patients. Raval et al. then used iPSC-CMs derived from two patients with infantile-onset PD to further investigate its underlying molecular mechanisms (19). In normal physiological states, the lysosome degrades intracellular pathogens, damaged organelles and proteins via the autophagy pathway. Defects were identified in LAMP Golgi-based glycosylation secondary to lysosomal glycogen accumulation with higher protein mobility and subsequent alkalization. These investigators linked this to subsequent hypertrophic remodeling, which provided novel pathophysiologic insight into the development of cardiomyopathy associated with PD.
Danon disease
Danon disease is an X-linked, type IIb glycogen storage disorder associated with skeletal dysfunction and a myriad of heart disease, including HCM, DCM, and increased arrhythmogenicity (29). Deficient mutations in the LAMP-2 gene prevent complete fusion of lysosomes and autophagosomes resulting in impaired autophagy. Two patient-specific iPSC-CMs with LAMP-2 gene mutations were generated and compared to a rescued cell line with overexpression of a common Danon disease isoform, LAMP-2B (LAMP 2B OE) (21). The Danon iPSC-CMs exhibited key features of heart failure including cellular hypertrophy, delayed calcium handling, and increased oxidative stress, which were then absolved in the LAMP-2B OE cells. Additionally, significant decline in mature autophagocytic vacuoles in Danon iPSC-CMs as compared to LAMP-2B OE was observed, suggestive of impaired autophagy due to LAMP-2 deficiency. Treatment of Danon iPSC-CMs with antioxidant therapy resulted in significantly reduced apoptotic and stress response events. This study unraveled a mechanism of LAMP-2 in cardiac pathology of Danon disease and discerned a prospective therapy. In the future, functional correction of deficient LAMP-2 with genome editing techniques could be conducted in similar models to further study diseased and rescued characteristics.
Mitochondrial Disease
Barth syndrome
Barth syndrome (BTHS) is an X-linked mitochondrial disorder with mutations in tafazzin (TAZ1), a phospholipid acyltransferase that impairs remodeling of cardiolipin (CL) to its mature form. As CL is a primary constituent of the mitochondrial membrane, TAZ1 mutations result in a characteristic triad of disease: cardiomyopathy, skeletal myopathy and neutropenia (30). The first BTHS patient-specific iPSCs were derived from three patients with TAZ1 mutations (22). Reprogrammed BTHS iPSCs displayed impaired CL remodeling, with decreased levels of mature CL leading to scarce formation of mitochondrial supercomplexes and subsequent incomplete energy transformation. Wang et al. then investigated whether restoration of TAZ levels via transfection with modified RNA (modRNA) would rescue the phenotype (23). Cellular contractility and sarcomerogenesis were studied using three cell lines: [1] “diseased” BTHS iPSC-CMs derived from two patients harboring either a missense or frameshift TAZ mutation, [2] “rescued” BTHS iPSC-CMs by transfection with synthetic modRNA to restore TAZ levels, and [3] “induced” BTHS iPSC-CMs by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas-9 mediated genome editing to create a pathogenic mutant TAZ. While irregular alignment of sarcomeres was observed in both “diseased” and “induced” BTHS iPSC-CMs, restoration of intact sarcomerogenesis was seen in the “rescued” cell lines. Intriguingly, there was variability in sarcomere organization with respect to mutation types, with the TAZ frameshift mutation showing more pronounced disorganization than the missense mutation. This finding suggests a wide spectrum of disease phenotypes based on different mutations and highlights the feasibility of iPSC technology to correlate specific mutant types with in vitro phenotypes to further predict disease severity as well as response to therapy.
Similar results were observed on thin tissue constructs generated from iPSC-CMs, called “heart-on-a-chip,” which allows investigation of CM contractile properties including twitch stress, and diastolic and peak systolic stresses in vitro (23). Using this method, the authors found significant decline in contractility with lower twitch and peak systolic stress in both “diseased” and “induced” BTHS iPSC-CM, compared to “rescued” cells. When these cells were treated with precursors of CL, such as linoleic acid, or with the anti-oxidant mitoTEMPO, there was profound improvement in sarcomeric organization and contractile dysfunction, suggesting potential therapeutic opportunities.
Lysosomal Storage Disorder
Fabry disease
Fabry Disease (FD) is an X-linked lysosomal storage disorder caused by deficiency in alpha-galactosidase A with consequent accumulation of globotriaosylceramide (GL-3) in the heart, brain, and kidneys, leading to damage of these organs (31). Recently, Itier et al. derived FD iPSC-CMs from two patient with GLA mutations to understand the role of GL-3 accumulation in the pathogenesis of FD-associated cardiomyopathy (25). The FD iPSC-CMs demonstrated progressive lysosomal accumulation of GL-3, increased lysosomal storage inclusions (zebra bodies), and disorganized contractile fibers. Presently, ERT with alpha-galactosidase A is the main treatment strategy available for FD by degrading GL-3 and thereby reducing its tissue accumulation; however, long-term reduction of GL-3 was not observed. Investigators sought to identify a novel sustained treatment of GL-3 reduction utilizing the FD iPSC-CM model. Substrate reduction therapy (SRT) inhibits glucosylceramide synthase, a rate-limiting step in glycosphingolipid synthesis, and was postulated to be a potential alternative therapy to ERT with encouraging results in animal models. When tested in iPSC-CMs, SRT had effectively prevented and reduced GL-3 deposits in FD cardiomyocytes compared to wild type. These findings confirmed the therapeutic potential of SRT and identified an alternative pathway for GL-3 clearance using a small molecule inhibitor.
Cardiomyopathy associated with acquired metabolic disorder/syndrome
Metabolic syndrome is a constellation of at least 3 of the following conditions: central obesity, elevated triglycerides, hyperglycemia, hypertension, and reduced high-density lipoprotein. These factors predispose individuals to developing diabetes mellitus and cardiovascular disease-associated mortality (2). The recent rise in obesity and insulin resistance with subsequent development of metabolic syndrome necessitates the ability to recapitulate this polygenic disease to better understand its pathogenesis and identify specific treatment options (Table 2) (34).
Table 2.
Induced pluripotent stem cell models of cardiomyopathies in acquired metabolic syndromes
| Year (Ref) | Specific mutation | iPSC-CM in vitro phenotype | Cardiac muscle clinical phenotype | |
|---|---|---|---|---|
| Type II Diabetes | 2014 (32) | Two diabetic extreme phenotypes: Fast progression with CVD within 5 years Slow progression without CVD for 15 years Specific mutations not identified |
iPSC-CMs exposed to diabetic milieu: Disorganized hypertrophy, intracellular lipid accumulation, altered calcium transients, cellular hypertrophy, oxidative stress Fast progression patient iPSC-CMs: Sarcomeric disarray, lipid-induced toxicity, reduced calcium transient frequency Slow progression patient iPSC-CMs: Less sarcomeric disarray, no lipid-induced toxicity or reduction in frequency of calcium transients |
Systolic dysfunction and left ventricular hypertrophy, with subsequent risk of heart failure |
| Ischemia with ALDH2 deficiency | 2014 (33) | Common heterozygous mutation in ALDH2 gene (ALDH2*2/1) | Increased oxidative stress, increased production of toxic aldehydes, decreased cell growth, decreased viability, increased cell apoptosis | Cardiac dysfunction and increased infarct size after myocardial infarction resulting from oxidative stress and nitroglycerin intolerance |
Diabetes-induced cardiomyopathy
Diabetes mellitus (DM) is a leading cause of cardiac morbidity and mortality with a projected increased prevalence of 300 million individuals by the year 2025 (35). Individuals with DM have a 2- to 5-fold increased risk for developing heart failure, independent of other known risk factors, such as hypertension or coronary artery disease (36). In a diabetic state, body metabolism is abnormally driven by fatty acid oxidation rather than glucose and lactate metabolism, leading to lipid accumulation with resultant toxicity to cardiomyocytes. Diabetic cardiomyopathy presents with diastolic or systolic dysfunction, and ultimately advanced heart failure (35). Causes of diabetic cardiomyopathy are thought to be rather multifactorial, including both epigenetic/environmental factors and genetic predisposition. Given this complex nature of disease, there has been lack of adequate animal models.
Recently, Drawnel et al. modeled a diabetic environment in iPSC-CMs by inducing fatty acid metabolism dependence with initial insulin treatment in the absence of glucose, followed by the addition of diabetic mediators, high glucose, endothelin 1, and cortisol (32). DM iPSC-CMs accurately produced features of diabetic cardiomyopathy in vitro, including cellular hypertrophy, abnormal sarcomeric organization, lipid accumulation, decreased calcium transients, and evidence of oxidative stress. Gene expression studies showed up-regulation of cellular metabolism genes and down-regulation of genes controlling protein synthesis. To study the underlying genetic predisposition of diabetic cardiomyopathy, investigators then generated iPSC-CMs from a patient with fast progression of cardiovascular disease (CVD) within 5 years of Type II DM versus slow progression of disease (no CVD after 15 years of Type II DM). These iPSC-CMs exhibited features of DM phenotypes but with varying extent of in vitro phenotypic severity. The fast progressive iPSC-CMs showed evidence of sarcomeric disarray, abnormal calcium handling, and lipid-induced toxicity, whereas the slow progressive type displayed only mild sarcomeric disorganization. Gene expression studies comparing fast versus slow progression iPSC-CMs identified up-regulation in glucocorticoid signaling and calcineurin-NFAT signaling, which are key signaling pathways in hypertrophic remodeling. The DM iPSC-CM model helped to demonstrate feasibility of iPSC-CM platforms to model polygenic disease, which could open many doors for therapeutic opportunities including rapid drug screening (32).
ALDH polymorphism and CM
Deficiency in the aldehyde dehydrogenase 2 (ALDH2) enzyme is found in about 8% of the population, with a predominance in East Asians, and is most commonly associated with alcohol intolerance, including facial flushing, nausea, and tachycardia (37). However, deficiency in ALDH2 has also been associated with poor cardiovascular outcomes (33). This is because during ischemia, oxidative stress ensues with a rise in reactive oxygen species (ROS), lipid peroxidation, and 4-hydroxynonenal (4HNE), a toxic aldehyde. Normally, ALDH2 functions to detoxify 4HNE and induce vasodilation with nitroglycerin (38). A defect in ALDH2 function may result in cardiac dysfunction and impaired recovery following an ischemic event. Recently, Ebert et al. evaluated effects of this polymorphism in patient fibroblasts with ALDH2 deficiency versus control fibroblasts (33). Elevated ROS and 4HNE were observed in the enzyme deficient cells. When these cells were treated with 4HNE, a toxic aldehyde, there was a decline in cell growth and viability, whereas treatment with Alda-1, a small molecular activator of ALDH2, restored cell growth and viability, suggesting potential relationship between oxidative stress and cell cycle arrest in ALDH2 deficient fibroblasts. To investigate the role of ALDH2 deficiency in cardiomyocytes, patient specific iPSC-CMs were then generated. Similar to the fibroblast study, when ALDH2 deficient iPSC-CMs were subjected to ischemia, elevation in ROS and reduced cell viability were observed, while treatments with Alda-1 and an ROS scavenger reversed these findings. Through transcriptome analysis, the authors then identified activation of JNK with subsequent activation of c-Jun-related apoptotic signaling following ischemic stress. The restoration of cell viability with the administration of JNK inhibitor validated the proposed pathway and demonstrated unique insight into the ischemic consequences in patients with an ALDH2 polymorphism using iPSC model.
Discussion
Important advances in the iPSC field have provided new research capacity to study metabolic disorders and associated heart disease in ways previously unavailable in animal or alternative in vitro models. Human iPSC-CM platforms provide a powerful tool that allows drug screening specific to individual patients with variable disease expression. However, these advances have not arisen without additional questions.
Despite the increased potential to recapitulate disease, there remain significant challenges in reconstitution of metabolic mitochondrial diseases (39). Phenotypes of Leigh and mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke (MELAS) syndrome were successfully studied in iPSC-derived skeletal and neuronal cells demonstrating significant mitochondrial dysfunction (40). Carnitine palmitoyltransferase II (CPTII), an enzyme essential for fatty acid transport, was also reconstituted and treated in iPSC-derived skeletal muscle cells (41). However, methodologies to produce cardiomyocytes in these diseases have been hindered by the overwhelming cell death occurring during differentiation to cardiomyocytes. Further efforts are warranted to optimize differentiation conditions to generate iPSC-CM platforms that will successfully recapitulate cardiac disease phenotypes associated with mitochondrial diseases, which include, but are not limited to Sengers, Kearns-Sayre, Leigh, and MELAS syndromes and CPT II deficiency (42–44).
The immature-fetal like characteristic of iPSC-CMs has also been a cause of concern for investigators and its direct application to adult heart disease (6). Efforts to develop mature CMs have been successful with prolonged culture protocols and alterations in extracellular matrix dynamics (45). However, fetal characteristics may pose an advantage for studying IEM where diseases commonly present early in life. Interestingly, it has also been postulated that iPSC-CMs allow characterization of metabolic profiles earlier on during differentiation and are not maturation dependent. This is evident by several studies presented in this review, where metabolic markers were commonly analyzed rather than cardiac sarcomere markers.
A progressing area in the cardiac stem cell field is the application of genome editing technologies, including zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) (46). Genome editing provides the opportunity to introduce and repair mutations, thereby establishing and rescuing a disease phenotype (47,48). This approach eliminates a need to recruit patients with specific mutations and also bypasses the step required to reprogram adult somatic stem cells to iPSCs. Additionally, genome editing can directly establish causation between a mutation and its disease by creating genome-edited isogenic cells with wide type protein (15). The culmination of these features provides the potential to pharmaceutically target specific disease-inducing mutations. A recent study showed significant reduction in LDL cholesterol with in vivo genome editing of loss of function mutations in the PCSK9 gene (49). This exciting result demonstrates the tremendous applications of both genome editing and iPSC models for drug discovery.
Conclusion
Undoubtedly, both iPSCs and iPSC-CMs have allowed recapitulation of previously challenging phenotypes to model in vivo and in vitro. To date, several inborn errors of metabolism and metabolic syndromes have been recapitulated in a patient-specific manner and have provided insight into disease mechanisms as well as identification of novel drug targets. Advances in genome editing technologies have provided further capacity to induce and rescue a disease phenotype in vitro with potential for in vivo application. These innovations provide optimism for further exploration of additional metabolic disorders, such as mitochondrial disorders, that remain challenging to model in iPSC-CMs.
Key points.
This article provides an overview of current metabolic heart disease iPSC models.
Inborn errors of metabolism with significant cardiomyopathy have been modeled using iPSC-CMs.
Acquired metabolic syndromes, such as diabetes mellitus cardiomyopathy and ischemic cardiac changes associated with aldehyde dehydrogenase 2 polymorphism have been modeled using iPSC-CMs.
Acknowledgments
We thank Dr. Elena Matsa and Blake Wu for critical reading of the manuscript.
Financial Support and sponsorship
We are grateful for funding support from American Heart Association 13EIA14420025, Burroughs Wellcome Foundation, National Institutes of Health (NIH) R01 HL126527, NIH R01 HL128170, NIH R01 HL130020 (JCW), and the Stanford Medical Scholars Research Program (AMC).
Footnotes
Conflicts of Interest
There are no conflicts of interest.
References
- 1.Byers SL, Ficicioglu C. Infant with cardiomyopathy: When to suspect inborn errors of metabolism? World J Cardiol. 2014;6(11):1149–55. doi: 10.4330/wjc.v6.i11.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik S, Wong ND, Franklin SS, Kamath TV, Gilbert JL, Pio JR, et al. Impact of the metabolic syndrome on mortality from coronary heart disease, cardiovascular disease, and all causes in United States adults. Circulation. 2004;110(10):1245–50. doi: 10.1161/01.CIR.0000140677.20606.0E. [DOI] [PubMed] [Google Scholar]
- 3.Houser SR, Margulies KB, Murphy AM, Spinale FG, Francis GS, Prabhu SD, et al. Animal models of heart failure: A scientific statement from the American Heart Association. Circ Res. 2012;111(1):131–50. doi: 10.1161/RES.0b013e3182582523. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Wilson K, Wu JC. Induced pluripotent stem cells. J Am Med Assoc. 2015;313(16):1613–4. doi: 10.1001/jama.2015.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Matsa E, Burridge PW, Wu JC. Human stem cells for modeling heart disease and for drug discovery. Sci Transl Med. 2014;6(239):239ps6. doi: 10.1126/scitranslmed.3008921. This review summarizes recent advances in the use of iPSC technology to model heart disease, screen for new drugs, and test candidate drugs for cardiotoxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, et al. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet. 2013;6(6):557–68. doi: 10.1161/CIRCGENETICS.113.000188. [DOI] [PubMed] [Google Scholar]
- 8.Wen J-Y, Wei C-Y, Shah K, Wong J, Wang C, Chen H-SV. Maturation-based model of arrhythmogenic right ventricular dysplasia using patient-specific induced pluripotent stem cells. Circ J. 2015;79:1402–8. doi: 10.1253/circj.CJ-15-0363. [DOI] [PubMed] [Google Scholar]
- 9.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4(130):130ra47–130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, et al. Epigenetic regulation of phosphodiesterases 2A and 3A underlies compromised β-adrenergic signaling in an iPSC model of dilated cardiomyopathy. Cell Stem Cell. 2015;17(1):89–100. doi: 10.1016/j.stem.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349(6251):982–6. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12(1):101–13. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363(15):1397–409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 14.Matsa E, Dixon JE, Medway C, Georgiou O, Patel MJ, Morgan K, et al. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur Heart J. 2014;35(16):1078–87. doi: 10.1093/eurheartj/eht067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014;64(5):451–9. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins FHV. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang HP, Chen PH, Hwu WL, Chuang CY, Chien YH, Stone L, et al. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum Mol Genet. 2011;20(24):4851–64. doi: 10.1093/hmg/ddr424. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi T, Kawagoe S, Otsu M, Shimada Y, Kobayashi H, Hirayama R, et al. The generation of induced pluripotent stem cells (iPSCs) from patients with infantile and late-onset types of Pompe disease and the effects of treatment with acid-α-glucosidase in Pompe’s iPSCs. Mol Genet Metab. 2014;112(1):44–8. doi: 10.1016/j.ymgme.2014.02.012. [DOI] [PubMed] [Google Scholar]
- *19.Raval KK, Tao R, White BE, De Lange WJ, Koonce CH, Yu J, et al. Pompe disease results in a golgi-based glycosylation deficit in human induced pluripotent stem cell-derived cardiomyocytes. J Biol Chem. 2015;290(5):3121–36. doi: 10.1074/jbc.M114.628628. This is the first study to identify the mechanism of impaired autophagy and resultant cardiac hypertropy in Pompe disease iPSC-CMs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato Y, Kobayashi H, Higuchi T, Shimada Y, Era T, Kimura S, et al. Disease modeling and lentiviral gene transfer in patient-specific induced pluripotent stem cells from late-onset Pompe disease patient. Mol Ther — Methods Clin Dev. 2015;2(May):15023. doi: 10.1038/mtm.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashem SI, Perry CN, Bauer M, Han S, Clegg SD, Ouyang K, et al. Brief report: oxidative stress mediates cardiomyocyte apoptosis in a human model of Danon disease and heart failure. Stem Cells. 2015;(33):2342–50. doi: 10.1002/stem.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudek J, Cheng IF, Balleininger M, Vaz FM, Streckfuss-Bömeke K, Hübscher D, et al. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. 2013;11(2):806–19. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- **23.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20(6):616–23. doi: 10.1038/nm.3545. This study combined novel iPSC and tissue engineering approaches to elucidate the mechanism underlying cardiomyopathy in Barth syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawagoe S, Higuchi T, Otaka M, Shimada Y, Kobayashi H, Ida H, et al. Morphological features of iPS cells generated from Fabry disease skin fibroblasts using Sendai virus vector (SeVdp) Mol Genet Metab. 2013;109(4):386–9. doi: 10.1016/j.ymgme.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Itier JM, Ret G, Viale S, Sweet L, Bangari D, Caron A, et al. Effective clearance of GL-3 in a human iPSC-derived cardiomyocyte model of Fabry disease. J Inherit Metab Dis. 2014:1013–22. doi: 10.1007/s10545-014-9724-5. [DOI] [PubMed] [Google Scholar]
- 26.Hers HG. alpha-Glucosidase deficiency in generalized glycogenstorage disease (Pompe’s disease) Biochem J. 1963;86(1959):11–6. doi: 10.1042/bj0860011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Ploeg AT, Reuser AJ. Pompe’s disease. Lancet. 2008;372(9646):1342–53. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 28.Chen LR, Chen CA, Chiu SN, Chien YH, Lee NC, Lin MT, et al. Reversal of cardiac dysfunction after enzyme replacement in patients with infantile-onset Pompe disease. J Pediatr. 2009;155(2):271–5. e2. doi: 10.1016/j.jpeds.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Boucek D, Jirikowic J, Taylor M. Natural history of Danon disease. Genet Med. 2011;13(6):563–8. doi: 10.1097/GIM.0b013e31820ad795. [DOI] [PubMed] [Google Scholar]
- 30.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580(23):5450–5. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 31.Zarate YA, Hopkin RJ. Fabry’s disease. Lancet. 2008;372(9647):1427–35. doi: 10.1016/S0140-6736(08)61589-5. [DOI] [PubMed] [Google Scholar]
- *32.Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M, et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014:810–20. doi: 10.1016/j.celrep.2014.09.055. This study modeled a diabetic environment in iPSC-CMs to successfully recapitulate features of diabetic cardiomyopathy in vitro. [DOI] [PubMed] [Google Scholar]
- **33.Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, et al. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Transl Med. 2014;6(255) doi: 10.1126/scitranslmed.3009027. This study used an iPSC-CM platform to investigate the role of ALDH2 polymorphism in cardiac complications after ischemic stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015:e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 35.Boudina S, Abel ED. Diabetic cardiomyopathy revisited. Circulation. 2007;115(25):3213–23. doi: 10.1161/CIRCULATIONAHA.106.679597. [DOI] [PubMed] [Google Scholar]
- 36.Kannel WB, Daniel L. Diabetes and cardiovascular disease. The Framingham study. 1979 doi: 10.1001/jama.241.19.2035. [DOI] [PubMed] [Google Scholar]
- 37.Larson HN. Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “Asian” variant. J Biol Chem. 2005;280(34):30550–6. doi: 10.1074/jbc.M502345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C-H, Budas GR, Churchill EN, Disatnik M-H, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–5. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finsterer J, Kothari S. Cardiac manifestations of mitochondrial disorders. Int J Cardiol. 2014;177:754–63. doi: 10.1016/j.ijcard.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Ma H, Folmes CDL, Wu J, Morey R, Mora-Castilla S, Ocampo A, et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature. 2015 doi: 10.1038/nature14546. [DOI] [PubMed] [Google Scholar]
- 41.Yasuno T, Osafune K, Sakurai H, Asaka I, Tanaka A, Yamaguchi S, et al. Functional analysis of iPSC-derived myocytes from a patient with carnitine palmitoyltransferase II deficiency. Biochem Biophys Res Commun. 2014;448(2):175–81. doi: 10.1016/j.bbrc.2014.04.084. [DOI] [PubMed] [Google Scholar]
- 42.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–60. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebert AD, Diecke S, Chen IY, Wu JC. Reprogramming and transdifferentiation for cardiovascular development and regenerative medicine: where do we stand? 2015:1–14. doi: 10.15252/emmm.201504395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmgren D. Cardiomyopathy in children with mitochondrial disease clinical course and cardiological findings. Eur Heart J. 2003;24(3):280–8. doi: 10.1016/s0195-668x(02)00387-1. [DOI] [PubMed] [Google Scholar]
- 45.Tzatzalos E, Abilez OJ, Shukla P, Wu JC. Engineered heart tissues and induced pluripotent stem cells: Macro- and microstructures for disease modeling, drug screening, and translational studies. Adv Drug Deliv Rev. 2015 doi: 10.1016/j.addr.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hotta A, Yamanaka S. From Genomics to Gene Therapy: Induced pluripotent stem cells meet genome editing. Annu Rev Genet. 2015;49:47–70. doi: 10.1146/annurev-genet-112414-054926. [DOI] [PubMed] [Google Scholar]
- 47.Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karakikes I, Ameen M, Termglinchan V, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes: insights into molecular, cellular, and functional phenotypes. Circ Res. 2015;117(1):80–8. doi: 10.1161/CIRCRESAHA.117.305365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Q, Strong A, Patel KM, Ng S-L, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–92. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]