Abstract
Purpose
Piwi-interacting RNAs (piRNAs) are a broad group of noncoding small RNAs that have important biological functions in germline cells and can maintain genome integrity via silencing of retrotransposons. In this study, we aimed to explore the associations between genetic variants of important genes involved in piRNA biogenesis and male infertility with spermatogenic impairment.
Methods
To this end, five single-nucleotide polymorphisms (SNPs) in the ASZ1, PIWIL1, TDRD1, and TDRD9 genes were genotyped by TaqMan allelic discrimination assays in 342 cases of nonobstructive azoospermia (NOA) and 493 controls.
Results
The SNP rs77559927 in TDRD1 was associated with a reduced risk of spermatogenic impairment. The genotypes TC and TC + CC showed odds ratios and 95 % confidence intervals of 0.73 (0.55–0.98, P = 0.034) and 0.73 (0.56–0.97, P = 0.030), respectively, in patients with NOA compared with those in the controls.
Conclusion
Thus, our results provided the first epidemiological evidence supporting the involvement of TDRD1 genetic polymorphisms in piRNA processing genes in determining the risk of spermatogenic impairment in a Han Chinese population.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0738-9) contains supplementary material, which is available to authorized users.
Keywords: TDRD1, Piwi-interacting RNA, Polymorphism, Spermatogenesis impairment, Nonobstructive azoospermia
Introduction
Infertility has been reported to affect 10–15 % of couples; approximately half of these cases are related to male infertility problems [1, 2]. Spermatogenic impairment is the most common form of male infertility, and many genetic factors have been shown to contribute to this disorder [3–6]. Spermatogenesis is a unique and complex developmental process regulated by many genes [7]. Thus, mutations or other alterations may cause spermatogenic impairment and male infertility [8].
Piwi-interacting RNAs (piRNAs) are a broad group of noncoding small RNAs with a length of 26–31 nucleotides. These RNAs are abundantly expressed in animal gonads [9] and exhibit repeat-derived sequences. piRNAs were first discovered in Drosophila and mapped to serve as an endogenous defense mechanism to suppress retrotransposon activity by silencing gene expression [10]. To date, however, the molecular mechanisms that mediate piRNA-induced DNA methylation remain unclear. Previous reviews [11, 12] have indicated that piRNAs play an essential role in maintaining DNA integrity in germline cells.
Numerous genes are involved in the generation of piRNAs. In particular, proteins encoded by Piwi genes are essential for the biogenesis and function of piRNAs [13, 14]. In addition, genes such as piwi and tdrd have been implicated in the biogenesis of piRNAs [15–18]. Tudor domain-containing proteins or Tudor domain-related proteins (TDRDs) have been extensively studied in the context of Piwi proteins [19], playing a key role in piRNA biogenesis. TDRD9 is expressed in mitotic spermatogonia, meiotic spermatocytes, and haploid spermatids in the mouse testis [20]. In mice, the Piwi clade consists of Miwi2, Mili, and Miwi. Miwi has been shown to be expressed in meiotic pachytene germ cells, Miwi2 has been reported to be localized only in embryonic stages, and Mili has been found at all stages but is enriched in spermatocytes [15, 21].
Piwi genes and piRNA biogenesis-associated genes have been shown to be expressed at various stages of germ cell development in the testis [22]. Studies in animal models with deletions of these genes have consistently shown impairment of gametogenesis. Aravin and Bourc’his reported that mutations in the Mili and Miwi2 genes lead to elimination of DNA methylation around transposable elements and sterility in male mice [23]. Consistent with studies in flies, mutations in the Miwi2 gene have been shown to result in the accumulation of DNA damage [24]. Houwing et al. studied the Piwi pathway proteins Ziwi and Zili in zebrafish. In their study, eggs from female zebrafish with mutations in the zili gene were shown to exhibit defects in the meiotic process as both Piwi-piRNA complexes and piRNA biogenesis-associated proteins are localized in the nucleus in Drosophila [25]. These results reflect the potential role of piRNA proteins in regulating DNA transcription and cell cycle progression.
Considering the essential role of piRNA biogenesis pathway genes in spermatogenesis, we hypothesized that genetic variants in these genes would potentially affect spermatogenesis. Therefore, to test our hypothesis, we performed genotyping analyses for five SNPs in these piRNA pathway genes and carried out a case-control study on the association between these SNPs and susceptibility to spermatogenic impairment.
Materials and methods
Patient recruitment and sample collection
Three hundred forty-two (342) patients with nonobstructive azoospermia (NOA) were strictly screened by clinicians as cases from two hospitals in China. In order to confirm either azoospermia, all patients underwent at least two semen analyses, and no sperms were found after centrifugation of the ejaculate at 3000×g for 10 min, according to World Health Organization (WHO) guidelines [26]. For a diagnosis of NOA, all patients underwent standard clinical examinations. Patients with a history of Y chromosome microdeletions, karyotype anomalies, genetic abnormalities, cryptorchidism, and orchitis were excluded [27]. Finally, only NOA patients without known pathogenicity factors were included in this study. Additionally, patients having unique occupational exposure that was thought to affect semen quality were excluded. Each patient donated 5 mL blood for genomic DNA extraction. Four hundred ninety-three (493) controls with normozoospermia were randomly recruited from the Shanghai Human Sperm Bank (Shanghai Province, China). The criteria for the controls included in this study were as follows: (1) sperm concentration more than 15 × 106/mL, (2) progressive motility over 32 %, and (3) round cells <1 × 106/mL, according to the WHO criteria. Both the control and case groups were from Han Chinese populations. This study was conducted following the approval of the Ethics Committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine, and all patients and controls provided written informed consent for participation in this genetic study.
SNP selection and genotype analyses
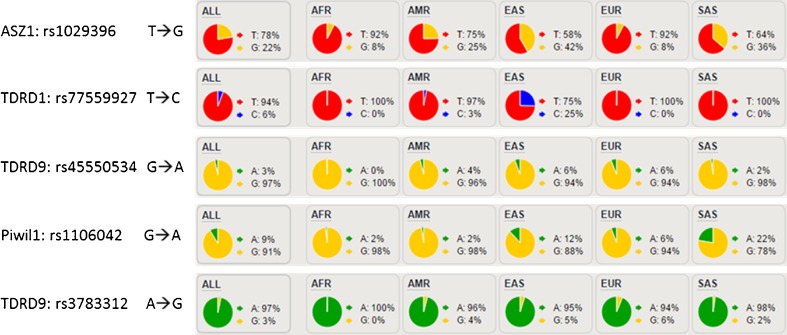
We first selected some genes associated with the piRNA biogenesis pathway as target genes to prepare RNA probes and performed target capture sequencing for 59 NOA patients. Some SNPs were found with higher frequencies in patients with NOA than in Han Chinese in Beijing (CHB) and Han Chinese South (CHS) populations in China. Based on the 1000 Human Genomes Project, SNPs in Han Chinese with higher frequencies compared with those in other populations were selected as the target sites. Finally, a total of five potential functional SNPs in four genes were selected randomly, including rs1029396 in ASZ1, rs1106042 in PIWIL1, rs77559927 in TDRD1, and rs45550534 and rs3783312 in TDRD9 (Fig. 1, Table 1, Supplementary Tables 1 and 2). In the case of multiple SNPs in the same haplotype block (linkage coefficient r2 < 0.8), only one was selected. The allele frequencies of these SNPs in CHB and CHS were significantly higher than the 1000G global frequency, particularly for rs77559927 in TDRD1. Therefore, we speculated that these SNPs might have special functions in the Chinese population.
Fig. 1.
Five potential functional SNPs in four genes selected randomly
Table 1.
piRNA biogenesis-associated genes and polymorphisms evaluated in this study
| SNP | Chr | Gene | Alleles | SNP property | Sense change |
|---|---|---|---|---|---|
| rs45550534 | 14 | TDRD9 | A/G | 3′UTR_exon35 | ND |
| rs77559927 | 10 | TDRD1 | T/C | 5′UTR_exon1 | ND |
| rs1029396 | 7 | ASZ1 | T/G | nonsynon_exon6 | p.Lys216Thr |
| rs1106042 | 12 | PIWIL1 | G/A | nonsynon_exon13 | p.Arg527Lys |
| rs3783312 | 14 | TDRD9 | A/G | 3′UTR_exon35 | ND |
ND not determined
Genomic DNA from the patients was isolated from peripheral blood leukocytes. The SNP genotyping work was performed using an improved multiplex ligation detection reaction (iMLDR) technique developed by GENESKY Biotechnologies Inc. (Shanghai, China). Two types of negative controls were used for each plate, i.e., (1) double-distilled water as a template and (2) DNA sample without primers. Duplicate tests were designed, and the results were consistent. A random sample accounting for ~5 % (n = 20) of all DNA samples was directly sequenced using Big Dye Terminator version 3.1 and an ABI3730XL automated sequencer (Applied Biosystems, Foster City, CA, USA) to confirm the results of iMLDR.
Statistical analyses
Statistical analyses were performed with the Statistical Analysis System (version 9.1.3; SAS Institute, Cary, NC, USA). The Hardy-Weinberg equilibrium was tested using Hardy-Weinberg equilibrium calculator [28]. The risk of spermatogenic failure was estimated as the odds ratios (ORs) and 95 % confidence intervals (95 % CIs) using unconditional univariate logistic regression. All P values were two-sided, with P = 0.05 considered the threshold of significance.
Results
Characteristics of the study population
The clinical characteristics of the 342 cases and 493 controls are shown in Table 2. We found that the testis volume in the cases was significantly smaller than that in the controls (P < 0.01). No significant differences in age or body mass index (BMI) were detected between the two groups (P > 0.05).
Table 2.
Patient and control characteristics
| Characteristic | Controls (n = 493) | Patients with NOA (n = 342) | All participants (n = 845) |
|---|---|---|---|
| Age (years) | 26.95 ± 3.40 | 27.30 ± 4.05* | 27.13 ± 4.42 |
| Body mass index | 22.51 ± 2.27 | 23.28 ± 3.24* | 22.85 ± 2.81 |
| Right testicle volume (mL) | 15.48 ± 1.76 | 5.81 ± 1.66** | 11.53 ± 5.06 |
| Left testicle volume (mL) | 15.43 ± 1.87 | 5.87 ± 1.77** | 11.531 ± 5.04 |
*P > 0.05, **P < 0.01
Association of spermatogenic failure with the TDRD1 gene
All SNP frequencies were consistent with Hardy-Weinberg equilibrium (HWE) in controls, which had a p value of less than 0.0001 for deviation from HWE. The associations between SNPs in piRNA biogenesis-related genes and the risks of male infertility are shown in Table 3 and Supplementary Table 3. The genotype frequency of rs77559927 in the TDRD1 gene was significantly different between patients with NOA and controls for the χ2 trend test (P = 0.0467). Significantly decreased risk of spermatogenic impairment was found for carriers of the rs77559927 TC genotype of TDRD1 when compared with homozygous carriers of the T allele in patients with NOA (OR 0.73, 95 % CI 0.55–0.98; P = 0.034). The homozygous carriers of the C allele may also have a lower risk of spermatogenic impairment (OR 0.75, 95 % CI 0.41–1.39). However, because the number of individuals with this genotype was too small, our study did not have sufficient statistical power to identify any differences. Instead, when we combined the rs77559927 TC and CC genotypes, assuming a dominant model effect, the combined rs77559927 TC + CC variant genotypes were associated with a 26.6 % reduced risk of spermatogenic impairment (OR 0.73, 95 % CI 0.56–0.97). This OR value suggested that the variant rs77559927T > C may be a protective factor against the risk of spermatogenic impairment. The SNP rs77559927 is a T to C change in the 5′ untranslated region (UTR) of exon 1 of the TDRD1 gene, which may lead to increased or decreased expression of certain transcript isoforms. The altered expression of TDRD1 may affect the roles of piRNAs and Piwi proteins during spermatogenesis, resulting in alterations in spermatogenesis. With regard to the other SNPs, no significant differences in distribution frequencies were identified among patients with NOA and controls (Supplementary Table 4).
Table 3.
Association of rs77559927 with NOA risk
| SNP | Genotype | Controls (n = 493) | Patients (n = 342) | OR (95 % CI) | P corrected | P value | ||
|---|---|---|---|---|---|---|---|---|
| rs77559927 | N | % | N | % | ||||
| TT | 255 | 52.0 | 203 | 59.4 | 1.00 | |||
| TC | 208 | 42.0 | 121 | 35.4 | 0.73 (0.55–0.98) | 0.17 | 0.034 | |
| CC | 30 | 6.1 | 18 | 5.3 | 0.75 (0.41–1.39) | 1.00 | 0.366 | |
| Dominant model | TT vs. TC + CC | 0.73 (0.56–0.97) | 0.15 | 0.030 | ||||
| Recessive model | CC vs. TT + TC | 0.86 (0.47–1.56) | 1.00 | 0.616 | ||||
| P trend | 0.047 | |||||||
| P for HWE | 0.144 | |||||||
Italic values indicate significant findings (P < 0.05)
OR odds ratio, CI confidence interval, P trend P value for X 2 trend test, P corrected P value after correction
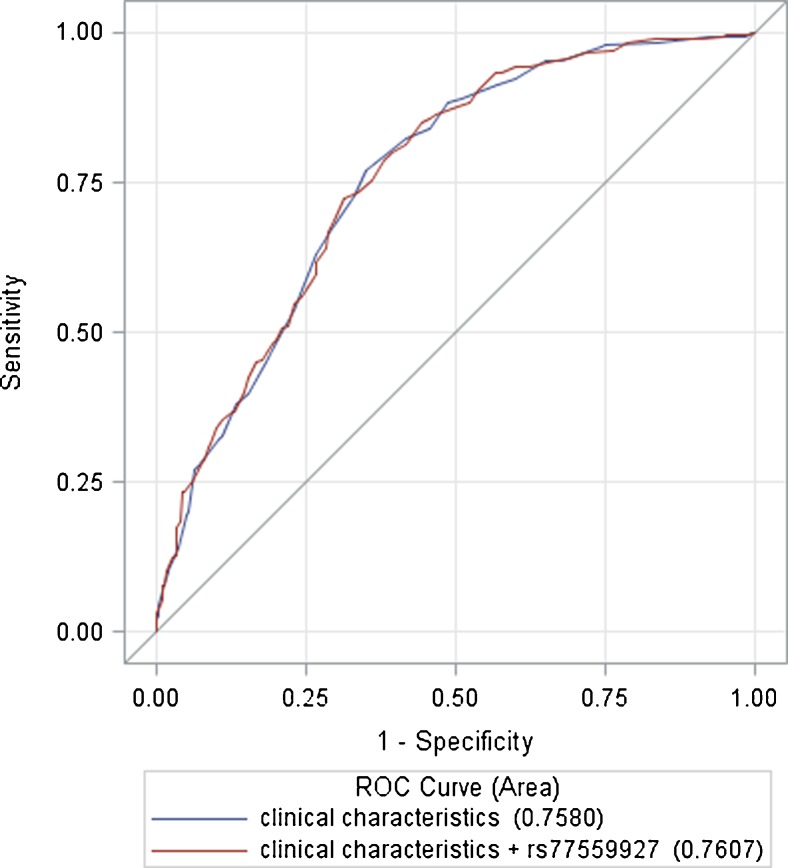
We also assessed the discriminative accuracy of the prediction model with and without addition of the identified SNP to the significant demographic and clinical features (age, BMI) by comparing the area under the receiver operating characteristic curve (AUC; Fig. 2). When adding the dominant model of rs77559927 in the prediction model of oligospermia, the AUC increased to 0.76 (95 % CI 0.73–0.79) compared with the model that only included age and BMI (AUC 0.758; 95 % CI 0.726–0.790; P = 0.336).
Fig. 2.
Area under the receiver operating characteristic curve
Discussion
Spermatogenesis defects may have various causes; for example, some conditions, such as cryptorchidism and orchitis, can cause NOA, and other known genetic factors, such as Y chromosome microdeletions and Klinefelter’s syndrome, are directly associated with spermatogenesis failure [29–31]. In this study, we aimed to identify unknown genetic factors that may be associated with spermatogenesis defects in patients with unexplained NOA. Therefore, we enrolled patients according to strict inclusion and exclusion criteria.
piRNAs have a wide range of functionalities, which are not always easy to identify. Numerous studies have found associations between piRNA biogenesis-related genes and fertility [11, 12]. However, the exact functional mechanism of Piwi/piRNA complexes remains unclear. Based on the findings of all reviewed studies, piRNA biogenesis-associated genes have been shown to be vital for spermatogenesis [32].
Studies on spermatogenesis impairment have focused on genetic variants or polymorphisms in candidate genes [33, 34]. However, available information regarding how genetic variants influence gene expression levels in spermatogenesis is limited. If a genetic polymorphism is located in the 3′UTR, it may influence messenger RNA (mRNA) stability and change the ability of mRNAs to bind microRNAs; this may result in decreased gene expression owing to mRNA cleavage or translational repression [35]. Alternatively, if a gene polymorphism is located in one of the binding motifs for splicing, pre-RNA splicing activity may be decreased, resulting in downregulation of genes involved in spermatogenesis [36, 37].
As mentioned previously, some studies in humans have also reported an association between piRNA biogenesis-associated gene polymorphisms and infertility [38]. Thus, variations in piRNA pathway genes may play a role in spermatogenic impairment. Accordingly, in this study, we evaluated the possible associations between common SNPs in piRNA pathway genes and male infertility with spermatogenesis impairment in Chinese patients with NOA and fertile controls to test whether polymorphisms in piRNA pathway genes were involved in human spermatogenic impairment. Because of phenotypic multiplicity in male infertility, only patients with NOA were recruited to avoid interference from other etiological factors in our study. However, we found no significant differences in the frequencies of SNP genotypes in the TDRD9, PIWIL1, and ASZ1 genes between patients with NOA and controls. Because our sample size was limited, the results indicated that these SNPs had relatively weak or no associations with spermatogenic impairment.
Notably, we detected significant differences in the frequencies of genotypes between patients with NOA and controls in the SNP rs77559927. The frequencies of genotypes TC and CC were significantly lower in patients with NOA than those in controls, suggesting that the variant rs77559927 T > C was associated with a lower risk of spermatogenic impairment (OR = 0.73), indicating that the genotypes TC and CC may have some protective effects on spermatogenic impairment. However, the mechanisms through which this SNP affects spermatogenic impairment are not yet clear. The SNP rs77559927 is a T to C change at the 5′-UTR of exon 1 of the TDRD1 gene, which may lead to increased or decreased expression of certain transcript isoforms. Altered expression of TDRD1 may affect the roles of piRNAs and Piwi proteins during spermatogenesis, subsequently altering spermatogenesis; thus, this SNP may alter the susceptibility to male infertility with oligospermia. Further studies are required to examine this hypothesis.
In conclusion, in this study, we investigated the relationships between polymorphisms in piRNA pathway genes and male infertility with spermatogenesis impairment in humans. The results of this study revealed that the SNP rs77559927 in TDRD1 was negatively associated with male infertility with oligospermia and may have some protective effects against spermatogenic impairment in the Chinese population. Furthermore, additional independent validation studies and functional experiments are needed to determine the specific role of the SNP rs77559927 in the TDRD1 gene. Because the sample size in this study was limited and restricted to the Chinese population, the findings of this study need to be validated in further studies with larger samples and other ethnic populations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 31 kb)
(DOCX 13 kb)
(XLS 525 kb)
(XLS 23 kb)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Funding
This study was supported by the Shanghai Municipal Commission of Health and Family Planning (No. 2013GY08), the Shanghai Hospital Development Center (Grant No: SHDC12014236), and the National High-Tech Research and Development Program (863) of China (2015AA020404).
Footnotes
Capsule Thus, our results provided the first epidemiological evidence supporting the involvement of TDRD1 genetic polymorphisms in piRNA processing genes in determining the risk of spermatogenic impairment in a Han Chinese population.
Xiao-Bin Zhu and Jian-Qi Lu contributed equally to this work.
References
- 1.Dohle GR, Colpi GM, Hargreave TB, Papp GK, Jungwirth A, Weidner W, et al. EAU guidelines on male infertility. Eur Urol. 2005;48(5):703–11. doi: 10.1016/j.eururo.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Okabe M, Ikawa M, Ashkenas J. Male infertility and the genetics of spermatogenesis. Am J Hum Genet. 1998;62(6):1274–81. doi: 10.1086/301895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online. 2007;14(6):734–45. doi: 10.1016/S1472-6483(10)60677-3. [DOI] [PubMed] [Google Scholar]
- 4.Krausz C, Escamilla AR, Chianese C. Genetics of male infertility: from research to clinic. Reproduction. 2015;150(5):R159–74. doi: 10.1530/REP-15-0261. [DOI] [PubMed] [Google Scholar]
- 5.Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian J Androl. 2012;14(1):40–8. doi: 10.1038/aja.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toshimori K, Ito C, Maekawa M, Toyama Y, Suzuki-Toyota F, Saxena DK. Impairment of spermatogenesis leading to infertility. Anat Sci Int. 2004;79(3):101–11. doi: 10.1111/j.1447-073x.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 7.Stouffs K, Seneca S, Lissens W. Genetic causes of male infertility. Ann Endocrinol (Paris) 2014;75(2):109–11. doi: 10.1016/j.ando.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Stouffs K, Tournaye H, Liebaers I, Lissens W. Male infertility and the involvement of the X chromosome. Hum Reprod Update. 2009;15(6):623–37. doi: 10.1093/humupd/dmp023. [DOI] [PubMed] [Google Scholar]
- 9.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 10.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during drosophila melanogaster development. Dev Cell. 2003;5(2):337–50. doi: 10.1016/S1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 11.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135(1):3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 12.Yadav RP, Kotaja N. Small RNAs in spermatogenesis. Mol Cell Endocrinol. 2014;382(1):498–508. doi: 10.1016/j.mce.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29(19):3301–17. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24(22):2493–8. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. [DOI] [PubMed]
- 16.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2(6):819–30. doi: 10.1016/S1534-5807(02)00165-X. [DOI] [PubMed] [Google Scholar]
- 17.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24(7):636–46. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu X., Zhi E., and Li Z., MOV10L1 in piRNA processing and gene silencing of retrotransposons during spermatogenesis. Reproduction, 2015. 149(5). [DOI] [PubMed]
- 19.Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci U S A. 2009;106(48):20336–41. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17(6):775–87. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–17. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Dev Growth Differ. 2012;54(1):78–92. doi: 10.1111/j.1440-169X.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- 23.Aravin AA, Bourc’his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22(8):970–5. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 26.World HO. Laboratory manual of the WHO for the examination of human semen and sperm-cervical mucus interaction. Ann Ist Super Sanita. 2001;37(1):I–XII. [PubMed] [Google Scholar]
- 27.Li LB, Xia YK, Li XS, Lu J, Ma MF, Song L, et al. An analysis on chromosome X, Y and 18 in the spermatozoa of asthenospermia patients by triple-color fluorescence in situ hybridization. Zhonghua Nan Ke Xue. 2008;14(3):211–4. [PubMed] [Google Scholar]
- 28.Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169(4):505–14. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadziselimovic F. Cryptorchidism, its impact on male fertility. Eur Urol. 2002;41(2):121–3. doi: 10.1016/S0302-2838(01)00040-9. [DOI] [PubMed] [Google Scholar]
- 30.Krausz C, Hoefsloot L, Simoni M, Tuttelmann F, European Academy of A., and European Molecular Genetics Quality N EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2(1):5–19. doi: 10.1111/j.2047-2927.2013.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernaeve V, Staessen CG, Van SA, Devroey P, Tournaye H. Can biological or clinical parameters predict testicular sperm recovery in 47, XXY Klinefelter’s syndrome patients? Hum Reprod. 2004;19(5):1135–9. doi: 10.1093/humrep/deh253. [DOI] [PubMed] [Google Scholar]
- 32.Pandey RR, Tokuzawa Y, Yang Z, Hayashi E, Ichisaka T, Kajita S, et al. Tudor domain containing 12 (TDRD12) is essential for secondary PIWI interacting RNA biogenesis in mice. Proc Natl Acad Sci U S A. 2013;110(41):16492–7. doi: 10.1073/pnas.1316316110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francesca N, Csilla K. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online. 2008;16(4):504–13. doi: 10.1016/S1472-6483(10)60457-9. [DOI] [PubMed] [Google Scholar]
- 34.Nishimune Y, Tanaka H. Infertility caused by polymorphisms or mutations in spermatogenesis-specific genes. J Androl. 2006;27(3):326–34. doi: 10.2164/jandrol.05162. [DOI] [PubMed] [Google Scholar]
- 35.Qin Y, Xia Y, Wei W, Han X, Lu C, Ji G, et al. Genetic variants in microRNA biogenesis pathway genes are associated with semen quality in a Han-Chinese population. Reprod Biomed Online. 2012;24(4):454–61. doi: 10.1016/j.rbmo.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Sanford JR, Ellis J, Cáceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem Soc Trans. 2005;33(Pt 3):443–6. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- 37.Zhoucun A, Sizhong Z, Yuan Y, Yiongxin M, Li L, Wei Z. Single nucleotide polymorphisms of the gonadotrophin-regulated testicular helicase (GRTH) gene may be associated with the human spermatogenesis impairment. Hum Reprod. 2006;21(3):755–9. doi: 10.1093/humrep/dei388. [DOI] [PubMed] [Google Scholar]
- 38.Sarkardeh H, Totonchi M, Asadpour O, Sadighi Gilani MA, Zamani EM, Almadani N, et al. Association of MOV10L1 gene polymorphisms and male infertility in azoospermic men with complete maturation arrest. J Assist Reprod Genet. 2014;31(7):865–71. doi: 10.1007/s10815-014-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 31 kb)
(DOCX 13 kb)
(XLS 525 kb)
(XLS 23 kb)