Abstract
Fungi are generally benign members of the human mucosal flora or live as saprophytes in the environment. However, they can become pathogenic, leading to invasive and life threatening infections in vulnerable patients. These invasive fungal infections are regarded as a major public health problem on a similar scale to tuberculosis or malaria. Current treatment for these infections is based on only four available drug classes. This limited therapeutic arsenal and the emergence of drug-resistant strains are a matter of concern due to the growing number of patients to be treated, and new therapeutic strategies are urgently needed. Adaptation of fungi to drug pressure involves transcriptional regulation, in which chromatin dynamics and histone modifications play a major role. Histone deacetylases (HDACs) remove acetyl groups from histones and actively participate in controlling stress responses. HDAC inhibition has been shown to limit fungal development, virulence, biofilm formation, and dissemination in the infected host, while also improving the efficacy of existing antifungal drugs toward Candida spp. In this article, we review the functional roles of HDACs and the biological effects of HDAC inhibitors on Candida spp., highlighting the correlations between their pathogenic effects in vitro and in vivo. We focus on how HDAC inhibitors could be used to treat invasive candidiasis while also reviewing recent developments in their clinical evaluation.
Keywords: HDAC, chromatin, acetylation, Candida, HDAC inhibitors
Invasive fungal infections have become a major public health problem, with up to two million cases worldwide each year (Brown et al., 2012). In developed countries, disseminated candidiasis, mostly caused by the yeasts Candida albicans, C. glabrata, and C. parapsilosis, remains the predominant threat, with more than 400,000 cases per year (Brown et al., 2012). Antifungal treatments are currently based on only four classes of drugs: polyenes, principally represented by amphotericin B; triazoles; echinocandins; and pyrimidines (Denning and Bromley, 2015). The emergence of strains resistant to this limited arsenal makes the need for novel therapeutic agents urgent (Denning and Bromley, 2015).
Candida albicans is the predominant cause of invasive candidiasis, and is also the most extensively studied Candida species. Its great success as a pathogen is linked to its capacity to survive in the bloodstream, to invade tissues and to effectively adapt to a range of host niches. One of its key virulence traits is its morphological plasticity; its ability to shift from a yeast form to a hyphal form has been clearly linked to virulence (Sudbery, 2011; Gow et al., 2012). Hyphal forms adhere better to mucosal niches, making it easier to maintain their colonization. This colonization can lead to epithelial rupture, dissemination of the pathogen in the bloodstream and ultimately invasion of deep-seated tissues. Other virulence factors, such as the white-to-opaque switch, the GUT (Gastrointestinally IndUced Transition) or gray phenotypes, cell wall plasticity, adherence, and biofilm formation favor development in the host (Polke et al., 2015).
Candida albicans’ capacity to adapt to various environmental conditions, including drug pressure, is linked to a complex interplay of stress-signaling responses (Fuchs and Mylonakis, 2009; Shor and Perlin, 2015). This signaling alters the transcription program to adapt the production of proteins, causing the emergence of the biological state that will be the most beneficial for yeast survival and development. Transcriptional regulation requires transcription factors to bind their DNA template and subsequently recruit dedicated machinery for transcription repression or activation. Classical histone modifications, such as acetylation, methylation, and phosphorylation have been shown to play a role in regulating stress responses, antifungal tolerance and virulence in C. albicans and C. glabrata (Liu et al., 2005; Rai et al., 2012; Stevenson and Liu, 2013; Kim et al., 2015; Tscherner et al., 2015). In particular, reversible acetylation by various histone acetyltransferases (HATs) and histone deacetylases (HDACs, also known as lysine deacetylases or KDACs) is crucial to chromatin-mediated transcriptional regulation. Recent studies have suggested that inhibiting fungal HDACs may have beneficial and synergistic effects, reducing the virulence and growth of Candida spp., while also decreasing their tolerance and resistance to existing antifungal drugs (Al-Dhaheri and Douglas, 2010; Wurtele et al., 2010; Stevenson and Liu, 2011; Hnisz et al., 2012; Lu et al., 2012; Nobile et al., 2014; Rajasekharan et al., 2015; Li et al., 2015; Pfaller et al., 2015; Zhang and Xu, 2015).
In this review, we briefly summarize the advances made in the characterization of HDACs in Candida spp. We have also correlated the functional roles of HDACs and the in vitro biological properties of HDAC inhibitors on Candida spp., mostly C. albicans, with their in vivo effects and discussed the potential for development of new antifungal compounds.
HDACs In Candida Species
To date, a total of 11 HDACs have been identified in C. albicans and C. glabrata (Figure 1; Tables 1 and 2). These two species represent the major proportion of the fungal infections clinically observed and regroup most of the molecular information accumulated on fungal HDACs. Three main classes of HDACs have been originally described in C. albicans based on the key S. cerevisiae enzymes (Trojer et al., 2003; Kim et al., 2015). The key enzyme for Class I HDACs is Rpd3, for Class II it is Hda1, and for Class III it is Sir2, a sirtuin. This family of enzymes uses a specific enzymatic chemistry based on the cofactor nicotinamide adenine dinucleotide (NAD).
FIGURE 1.
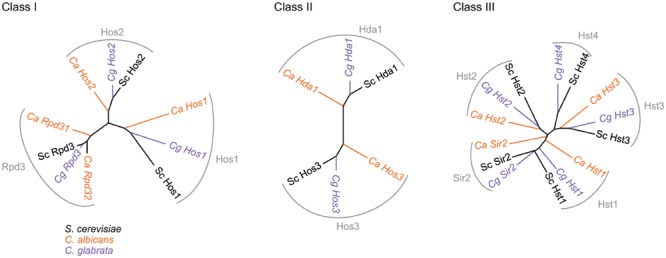
Phylogenetic trees representing HDACs in S. cerevisiae, C. albicans, and C. glabrata. Percent identities matrices between these HDACs are presented in Supplementary Table S1.
Table 1.
Gene and protein accession numbers for HDACs expressed in C. albicans.
| Classes | Name | Gene name | Protein ID | Reference | |
|---|---|---|---|---|---|
| I | Rpd3 type | Rpd31 | CR_02760C | Q5A209 | Srikantha et al., 2001 |
| Rpd32 | C3_07000W | Q5ADP0 | Hnisz et al., 2009 | ||
| Hos1 | C4_06010C | Q59Q78 | Srikantha et al., 2001 | ||
| Hos2 | C3_00780W | Q5A839 Q5A7T9 | Srikantha et al., 2001 | ||
| II | Hda1 type | Hda1 | CR_02050C | Q5A960 | Klar et al., 2001; Srikantha et al., 2001; Hnisz et al., 2009; Zacchi et al., 2010 |
| Hos3 | C4_02300W | Q5AF34 | Srikantha et al., 2001 | ||
| III | Sirtuin | Sir2 | C2_01330C | O59923 | Klar et al., 2001; Maglott et al., 2007; Fu et al., 2008; Nobile et al., 2012 |
| Hst1 | C1_09050W | Q5AQ47 | Maglott et al., 2007; Hnisz et al., 2009 | ||
| Hst2 | CR_01800C | Q5A985 | Maglott et al., 2007; Hnisz et al., 2009; Nobile et al., 2012 | ||
| Hst3 | C5_01340W | Q5A1W9 | Enjalbert et al., 2006; Maglott et al., 2007; Wurtele et al., 2010; Singh et al., 2011; Stevenson and Liu, 2011 | ||
| Fungi only | Set3 | C1_14140C_A | Q59ZX1 | Uhl et al., 2003; Maglott et al., 2007; Hnisz et al., 2009, 2010 | |
Table 2.
Gene and protein accession numbers for HDACs expressed in C. glabrata.
| Classes | Name | Gene name | Protein ID | Reference | |
|---|---|---|---|---|---|
| I | Rpd3 type | Rpd3 | CAGL0B01441g | Q6FXA7 | This study |
| Hos1 | CAGL0D01430g | Q6FWB7 | Dujon et al., 2004 | ||
| Hos2 | CAGL0A03322g | Q6FY81 | This study | ||
| II | Hda1 type | Hda1 | CAGL0J03454g | Q6FPJ0 | This study |
| Hos3 | CAGL0J06974g | Q6FP35 | Dujon et al., 2004 | ||
| III | Sirtuin | Sir2 | CAGL0C05357g | Q6FWI7 | Dujon et al., 2004 |
| Hst1 | CAGL0K01463g | Q6FNA6 | This study | ||
| Hst2 | CAGL0L08668g | Q6FKU1 | Dujon et al., 2004; Domergue et al., 2005 | ||
| Hst3 | CAGL0H08239g | Q6FRI7 | Dujon et al., 2004 | ||
| Hst4 | CAGL0F05621g | Q6FU79 | This study | ||
| Fungi only | Set3 | CAGL0G04499g | Q6FT89 | This study | |
Class I HDACs: Rpd31, Rpd32, Hos1, and Hos2
Interestingly, C. albicans possesses two genes which are potential orthologs of S. cerevisiae’s RPD3. These genes are now designated as RPD31 and RPD32. The current annotation of these genes has given rise to some confusion, with discrepancies between the original publication (Srikantha et al., 2001) and the current annotation in the Candida Genome Database (CGD; Assembly 22, version s06-m01-r01). Table 1 presents updated information on the gene and protein accession numbers. Hos1 was first identified and cloned 15 years ago in C. albicans, but its functional role has remained elusive (Srikantha et al., 2001). Hos2 was initially described in the CGD as a Class III enzyme with sirtuin activity (Karthikeyan et al., 2013), but it is now presented as a member of the class I family (Kim et al., 2015). In vitro analysis of the enzymatic activity of recombinant Hos2 showed it to be inactive on acetylated histones but capable of deacetylating acetylated tubulin (Karthikeyan et al., 2013). These findings remain to be confirmed in vivo.
Class II HDACs: Hda1 and Hos3
Hda1 was identified in 2001 and was shown to play an important role in hyphal development (see below). Hos3 was also described in 2001, but its function has yet to be studied in detail in Candida spp. (Srikantha et al., 2001).
Class III HDACs: Sir2 and Hst Proteins
The sirtuin family, a group of NAD+-dependent HDACs, is conserved between some yeasts and humans. Sirtuins were first characterized in Candida spp. in 1999, with the identification and cloning of the SIR2 gene in C. albicans (Pérez-Martín et al., 1999). Sir2 deacetylates histones, specifically lysine 16 on histone H4, it is also important for silencing at telomeres and ribosomal genomic regions (Freire-Benéitez et al., 2016). Interestingly, SIR2 is not present in all Candida spp. Thus, for example in C. lusitaniae, no Sir functionality for heterochromatic silencing in subtelomeric and pericentric regions has been detected, while in other species such as C. albicans or C. glabrata, an ancestral gene was duplicated to generate HST1 and SIR2 (Froyd et al., 2013; Kapoor et al., 2015).
Hst are also members of the sirtuin family; Hst1 is a component of the Set3 HDAC complex, while Hst3 is involved in nucleosome assembly. With the HAT Rtt109, Hst3 dynamically controls the level of lysine 56 acetylation on histone H3 (Rundlett et al., 1996; Wurtele et al., 2010).
Other HDACs: Set3
Set3 is an NAD+-dependent HDAC, which, in S. cerevisiae, forms a 7-subunit complex (Set3C) containing HDAC and non-HDAC proteins in C. albicans (Hnisz et al., 2010, p. 2). Four of these proteins, Set3, Hos2, Snt1 and Sif2, constitute the core complex and are essential for Set3C assembly, while three others (Hos4, Hst1, and Cpr1) are peripheral. Set3, Hos2, and Hst1 have HDAC activity. In addition, the PhD finger domain of Set3 binds methylated H3K4 and recruits the Set3C complex to chromatin in S. cerevisiae (Kim et al., 2012). This complex is conserved in C. albicans, where it is important for morphogenesis (Nobile et al., 2014).
Functional Roles of HDACs in Candida albicans
HDACs and Yeast-to-Hyphae Transition
Candida albicans exists in various morphological forms: an ovoid-shaped yeast phase is commonly found on mucosal and skin surfaces, where it is well tolerated by the immune system; hyphal forms possess a long tube-like extension to provide increased potential invasiveness. Both forms contribute to disseminated infections, but the ability to reversibly switch from one form to the other has been directly linked to virulence. The yeast-to-hyphae transition is controlled by various pathways which were recently reviewed (Sudbery, 2011).
The functional role of many HDACs has been linked to the yeast-to-hyphae transition (Figure 2A). First, Hda1 was reported to be important for a specific chromatin state during hyphal elongation and maintenance (Lu et al., 2011); C. albicans strains deleted for the HDA1 gene are unable to maintain hyphal development. Hda1 is recruited by the transcription factor Brg1 and establishes a chromatin state which is not permissive to Nrg1 repressor binding. Thus, Nrg1 is unable to bind the promoter regions of hypha-specific genes and prevent their expression (Lu et al., 2011, 2012). And in the absence of Nrg1, Hda1 also maintains a nucleosomal structure compatible with the expression of the hyphal genes.
FIGURE 2.
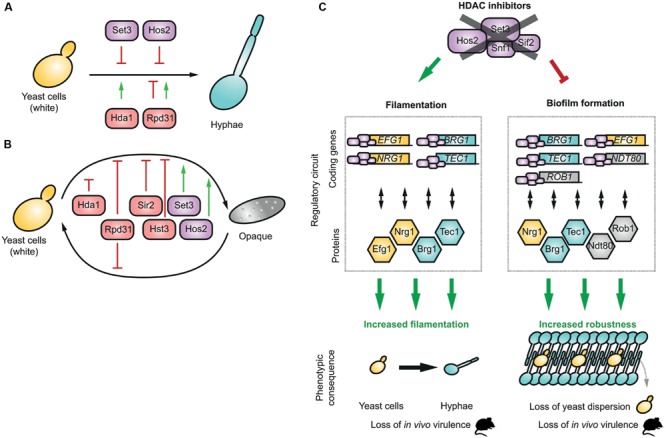
HDACs and morphogenesis in C. albicans. Phenotypic effect of different HDACs during filamentation (A) and the white to opaque transition (B), (C) HDACs regulate the expression of key transcription factors of regulatory circuits, which regulate the gene expression program during filamentation and the formation of biofilms. HDAC inhibition deregulates the transcription regulatory circuit, generating a hyperfilamentation phenotype and a loss of virulence in vivo. Similarly, upon treatment with HDAC inhibitors, biofilms are more robust but have a decreased yeast dispersion and a loss of virulence in vivo.
The yeast-to-hyphae transition is also controlled by Rpd31, which acts both as a repressor and an activator (Figure 2A). In yeast cells, Rpd31 is repressing the expression of hyphal-specific genes such as HWP1 and ECE1 and under non-hyphae-inducing conditions, these genes are activated when RPD31 or SSN6 are deleted (Lee et al., 2015). However, under filament-inducing conditions, the Rpd31-Ssn6 complex promotes filamentous elongation by triggering the expression of the master regulator UME6, a key factor in hyphal differentiation (Lee et al., 2015). Finally, the Set3C complex has been identified as a repressor of the yeast-to-hyphae transition (Figure 2A). In hyphae-inducing conditions in vitro, set3-, and hos2-null mutant strains had hyperfilamentous phenotypes (Hnisz et al., 2010, p. 3). The same authors later showed that the set3 mutant induces transient upregulation of EFG1 and NRG1, and downregulation of other hyphal associated genes, such as BRG1 and TEC1 (Hnisz et al., 2012).
HDACs and the White-to-Opaque Transition
Candida albicans colonies are typically white and smooth, but under some specific conditions, such as genetic conversion at the mating-type locus, a morphological white-to-opaque switch can occur. White cells have been showed to be more virulent in murine models (Kvaal et al., 1997). In human systemic infections, white cells are more likely to be isolated, whereas opaque cells may be better adapted to colonization (Morschhäuser, 2010).
The white-to-opaque transition involves a set of transcription factors responsible for the control of genes specific to white and opaque cells (Hernday et al., 2013). These factors are naturally linked to the transcription and chromatin machinery. Strains deleted for either HDA1 or RPD31 showed an enhanced ability to switch from the white to the opaque state, while only the rpd31Δ mutant displayed increased reverse opaque-to-white switch (Figure 2B). Thus, Hda1 selectively represses the white-to-opaque switch and Rpd31 suppresses the transition in both directions (Klar et al., 2001; Srikantha et al., 2001).
The sirtuins Hst3 and Sir2 were identified as switch repressors, whereas Set3C HDACs (Set3 and Hos2) were recognized as key activators of the white-to-opaque switch (Figure 1) (Pérez-Martín et al., 1999; Hnisz et al., 2009; Stevenson and Liu, 2011).
HDACs and Biofilm Formation
Candida albicans can form biofilms – multicellular structures of mixed communities of microorganisms containing yeast and hyphal forms surrounded by a self-produced extracellular matrix – which commonly develop on implanted medical devices, such as intravascular catheters or prostheses, as well as on mucosal surfaces. Biofilms create secondary infectious foci in hematogenous disseminated candidiasis through the release of yeast cells into the bloodstream. They are also an important source of antifungal resistance because the extracellular matrix hinders drug diffusion (Taff et al., 2013; Perlin et al., 2015).
Set3C HDACs have been shown to be important for the development of biofilms (Nobile et al., 2014). Thus, deletion of SET3 and HOS2 decreases biofilm formation and biomass, and these mutants seem to be more resistant to mechanical shearing and yeast dispersion in vivo (Nobile et al., 2014). The Set3C complex binds to five of the six biofilm master regulators, namely NRG1, BRG1, TEC1, NDT80, and ROB1. Notably, Nrg1, which is transiently repressed by Set3C during filamentation, is involved in the regulation of cellular dispersion (Uppuluri et al., 2010).
Role of HDACs in Virulence in vivo
Several studies have investigated the role of HDACs in C. albicans virulence through in vivo experiments, mostly assessing survival rates after systemic injection of wild-type and mutant strains.
The RPD31 deletion induced filamentation defects and attenuated virulence when injected into mice (Srikantha et al., 2001; Lee et al., 2015). These results are consistent with the hyphae-inducing conditions found in animal models. Likewise, the set3 mutant displayed a hyperfilamentous phenotype in vitro. This phenotype was confirmed in vivo in mouse kidneys, but, surprisingly, it was associated with attenuated virulence (Hnisz et al., 2010) (Figure 2C). This attenuated virulence could be linked to the Set3C-mediated transcription regulation which includes transient downregulation of EFG1 and NRG1 and induction of BRG1 and TEC1 (Hnisz et al., 2012). In addition, Hst3 deletion leads to increased H3K56 acetylation, decreased cell viability with abnormal filamentous growth and genomic instability. In vivo, this deletion attenuates the virulence of C. albicans in mice models (Wurtele et al., 2010).
HDACs and Antifungal Resistance
Histone acetylation dynamics and HDACs have been shown to be involved in the development of resistance to antifungal drugs. Notably, Li et al. (2015) showed that the expression of HDA1 and RPD3 was increased during acquisition of azole resistance, but decreased once resistance had been established. Hda1 and Rpd3 control the acetylation of Hsp90, a protein involved in the development of drug resistance in various fungi (Cowen and Lindquist, 2005; Robbins et al., 2012). Inactivation of HDACs in C. albicans phenocopies the genetic and pharmacological inhibition of Hsp90, restoring azole susceptibility by blocking the Hsp90-dependent response involved in azole resistance (Cowen and Lindquist, 2005).
Some authors initially hypothesized that this effect was achieved because HDACs directly influenced the expression of efflux transporter genes involved in azole resistance. However, recent studies suggest that the deletion of HDACs or the use of HDAC inhibitors could decrease the expression of efflux transporters as a part of a general decrease of histone acetylation and its consequence on transcription regulation (Li et al., 2015).
Functional Roles of HDACs in Candida glabrata
In the species distribution of invasive candidiasis, C. glabrata ranks second after C. albicans (Brown et al., 2012). Chromatin remodeling and sirtuin-family HDACs are required for C. glabrata to adapt to stressful conditions such as survival inside phagocytes, adhesion and maintenance of colonization, or multidrug resistance (Rai et al., 2012; Orta-Zavalza et al., 2013; De Las Peñas et al., 2015).
Sir2 is important for the regulation of cell adhesion; its absence reduces silencing and many subtelomeric adhesin-encoding EPA genes are derepressed (De Las Peñas et al., 2015). Hst1 in C. glabrata is recruited by the transcription factor Sum1 and contributes to the repression of PDR1 and CDR1, which regulate the expression of efflux pumps. Thus, when Hst1 is deleted, these genes are upregulated and azole resistance is enhanced (Orta-Zavalza et al., 2013).
Effects of HDAC Inhibitors on Candida spp. in vitro
Inhibitors of mammalian HDAC enzymes were first developed nearly 35 years ago. As soon as the first molecules were identified, several pioneer studies analyzed their effects on yeast HDACs.
Non-selective Inhibitors of Class I and II HDACs
Trichostatin A (TSA) is a well-known HDAC inhibitor. It was first isolated from a culture broth of Streptomyces platensis and was initially presented as a fungistatic drug inhibiting growth of Trichophyton and Aspergillus (Tsuji et al., 1976). Rapidly, however, it was shown to act on the differentiation of mammalian cells and to inhibit their HDACs (Yoshida et al., 1987, 1990). Ten years later, TSA was tested on pathogenic yeasts, including C. albicans where it induced a dramatic increase in white-to-opaque transition (Klar et al., 2001). This phenotype is fully compatible with Hda1 and/or Rpd31 inhibition (see above, Klar et al., 2001; Srikantha et al., 2001). TSA was also shown to trigger the yeast-to-hyphae conversion of C. albicans through inhibition of Set3C HDACs (Hnisz et al., 2010). Finally, the deletion of Hos2, a Set3C subunit, but none of the other HDACs, phenocopies the TSA induced yeast-to-hyphae transition (Hnisz et al., 2010). No in vitro assessment of TSA on Candida HDACs has been yet reported but TSA is active on purified Rpd3, Hda1 and Hos3 in S. cerevisiae (Carmen et al., 1999). Therefore, the phenotypes observed in Candida spp. are likely to be mediated by a direct inhibition of the Rpd3, Hda1, and a Hos2 enzyme as a TSA treatment phenocopies the deletion of these HDACs.
Sodium butyrate is another well-known HDAC inhibitor. In 1978, this fatty acid was shown to inhibit mammalian HDACs. In 2002, it was tested in C. albicans with other HDAC inhibitors (Candido et al., 1978; Smith and Edlind, 2002). Sodium butyrate alone was shown to have minimal effects on growth, heat sensitivity, and germ tube formation in C. albicans (Smith and Edlind, 2002), although some reports suggested that it inhibited growth and biofilm formation in C. albicans, C. parapsilosis, and Cryptococcus neoformans, while also enhancing the functions of macrophages in vitro (Candido et al., 1978). Whether its effect on HDAC enzymes is direct or not remain to be determined.
Some uracil-based compounds have been identified, among which suberoylanilide hydroxamic acid (SAHA), also known as vorinostat. This compound is currently licensed for clinical use for the treatment of cancer. When tested against C. albicans strains, it displayed relatively low antifungal activity (Mai et al., 2007), although another study showed that the same concentration could reduce the pathogenicity of C. albicans by decreasing its adherence to cultured human cells by 90%, and significantly inhibiting serum-induced germination (Simonetti et al., 2007). Finally, apicidin, a cyclic tetrapeptide, displays limited direct antifungal activity against C. albicans (Smith and Edlind, 2002).
Sirtuin Inhibitors
Nicotinamide is a vitamin and precursor of NAD+ and a well-described inhibitor of Class III HDACs, including Sir2 (Landry et al., 2000; Bitterman et al., 2002; Sanders et al., 2007). It was shown to have broad antifungal activity against several pathogenic Candida and Aspergillus species. In particular, the addition of nicotinamide to wild-type C. albicans cells led to morphological alterations and strong growth inhibition in vitro, these effects are thought to be mediated through inhibition of H3K56 deacetylation (Wurtele et al., 2010).
Thus, the in vitro effects of these non-selective HDAC inhibitors used alone were only studied in C. albicans and results were somewhat conflicting. Further studies including C. glabrata will be needed.
Selective HDAC Inhibitors
In addition to the pan-HDACs inhibitors, such as TSA and SAHA, a fungal-specific Hos2 inhibitor, MGCD290, has been developed (Pfaller et al., 2009). No enzymatic data currently evaluates its effect on purified Hos2, but in vitro, this compound alone showed a modest activity against Candida spp., with minimum inhibitory concentrations (MICs) ranging from 0.5 to 16 μg/mL, depending on the species. However, in combination with azoles, MGCD290 was active against a broad range of fungi, including molds such as Aspergillus spp., and was promoted as the way forward for the development of a new class of clinical drugs.
More generally, the HDAC enzymatic activity is dependent of key residues which have been highly conserved through evolution (Lombardi et al., 2011 and Supplementary Figure S1). Sequence alignments of class I and class II HDACs from C. albicans and human reveal that Set3 is the most divergent enzyme, with <20% of identity with human or other C. albicans HDACs (Supplementary Figure S1; Supplementary Table S2). The functional study of this enzyme has demonstrated its importance for the biology of C. albicans and its virulence in vivo (see section Functional Roles of HDACs in Candida albicans). Altogether, Set3 appears to be an exceptional candidate for the development fungal specific HDAC inhibitors. Alternatively, Hos1, Hos3, and Hst3 could also constitute new potential targets (Supplementary Table S2).
A new generation of inhibitors has currently been developed and targets selectively human HDAC isoforms, such as HDAC1/2, HDAC3, or HDAC8 (for review, Falkenberg and Johnstone, 2014). This illustrates that high levels of selectivity can be reached among human HDACs. Similarly, it is very likely that specific compounds could probably target specifically fungal HDACs. Structural studies showed that HDAC8 active site is very malleable and adapts its conformation when different inhibitors are bound (Somoza et al., 2004). Future work will hopefully generate more structural information on fungal HDACs and provide additional insights to the quest for specific fungal inhibitors.
The Clinical Potential of HDAC Inhibition in Candida Infections
HDAC Inhibitors Bolster Existing Antifungal Drugs and Limit the Emergence of Resistance
When used alone, HDAC inhibitors seem to display only a modest anti-Candida activity, however, their potential increases exponentially when they are used in combination with existing antifungal agents. This activity is observed not only with planktonic cells but also with biofilms.
Thus, HDAC inhibitors can significantly enhance azole activity in vitro (Smith and Edlind, 2002; Mai et al., 2007; Li et al., 2015). Combination of TSA with fluconazole, itraconazole, or voriconazole significantly reduced trailing growth (the phenotypic expression of drug-tolerance) and/or the azole MICs, in C. albicans, C. parapsilosis, and C. tropicalis (Smith and Edlind, 2002; Mai et al., 2007). Similar effects were observed in C. albicans when SAHA or other hydroxamate-based inhibitors were used in combination with fluconazole. These observations are consistent with the increased azole susceptibility of the HDA1 or RPD3 mutants of C. albicans (Mai et al., 2007; Zhang and Xu, 2015). MGCD290 also potentiates the activity of triazoles against Candida spp. in vitro (Pfaller et al., 2009, 2015). This synergy between HDAC inhibitors and antifungals is not limited to azoles, and TSA was shown to enhance the activity of other antifungal agents acting on membrane synthesis, including terbinafine, although it had no effect on the activities of amphotericin B and 5-fluorocytosine (Smith and Edlind, 2002). MGCD290 was also found to potentiate the echinocandins, although this synergistic effect was less pronounced than the effect with azoles (Pfaller et al., 2015).
Apart from its activity against Candida spp., MGCD290 was shown to have a synergistic activity with azoles against Aspergillus, Rhizopus, Mucor, Fusarium, Scedosporium, Rhodotorula, and Trichosporon genus (Pfaller et al., 2009).
One of the growing threats in the treatment of invasive candidiasis is the emergence of multidrug resistance, including echinocandin resistance, especially in C. glabrata (Maubon et al., 2014). Synergy between HDAC inhibitors, azoles (especially fluconazole) and echinocandins was also demonstrated for the treatment of several resistant strains of C. albicans, C. glabrata, or C. krusei (Pfaller et al., 2009; Li et al., 2015, p. 3). In vitro, MGCD290 also decreased the echinocandin-resistance of C. glabrata, C. albicans, and C. krusei isolates. Moreover, in most azole- or echinocandin-resistant strains, combination with a HDAC inhibitor led to a shift from resistance to greater susceptibility (Pfaller et al., 2015). Similarly, several echinocandin- or azole-resistant C. albicans isolates were as sensitive to nicotinamide as susceptible strains upon combined treatment with a HDAC inhibitor (Wurtele et al., 2010).
Through similar studies, several HDAC inhibitors were shown to enhance the action of antifungal drugs against fungi present in biofilms. Thus, Al-Dhaheri and Douglas (2010) showed that, in the presence of amphotericin B, TSA or apicidin, sodium butyrate significantly reduced viability of Candida spp. in biofilms. Similarly, sodium valproate, an organic compound used as an anticonvulsive agent which has been shown to be a HDAC inhibitor, used in combination with amphotericin B showed synergistic antifungal activity on biofilms produced by C. albicans, C. krusei, and C. parapsilosis (Göttlicher et al., 2001; Phiel et al., 2001; Al-Dhaheri and Douglas, 2010). Valproate was the most effective agent against C. krusei, while butyrate had the greatest impact on C. albicans. Using a biofilm formation assay, butyrate alone showed antifungal activity against C. albicans, C. parapsilosis, and C. neoformans (Hnisz et al., 2012). A combination therapy based on flavonoids and butyrate also significantly reduced a C. tropicalis biofilm (Rajasekharan et al., 2015).
Even though they are preliminary, these results with HDAC inhibitors on biofilms are encouraging. Despite their lack of specificity, HDAC inhibitors may be used at high concentrations in the particular context of lock therapy, which involves the direct application of very high local doses of active drugs to contaminated catheters. In addition, combinatorial strategies against biofilms have recently gained interest for the treatment of Candida infections associated with biofilms on devices (Liu et al., 2005). Indeed, the disruption of the membrane or the cell wall by antifungal agents may help promote the uptake of compounds that are active inside cells.
Use of HDAC Inhibitors in Animal Models and Clinical Studies
The animal experiments described above confirmed that genetic inhibition of HDACs produced strains with attenuated virulence (Figure 2). These experiments are important for functional studies, but genetic knock-out models are not entirely predictive of the ability of HDAC inhibitors to cure Candida infection in vivo.
Indeed, until now, only three HDAC inhibitors have been tested as therapeutic agents in animal models: the sirtuin inhibitor nicotinamide (intraperitoneal injection), valproic acid (intraperitoneal injection) and the Hos2 inhibitor MGCD290 (oral route). Wurtele et al. (2010) demonstrated that nicotinamide, mimics the in vitro effects of Hst3 repression, leading to a loss of virulence in mice. This antifungal effect requires the presence of the acetyltransferase RTT109 which acetylates H3K56, suggesting that nicotinamide exerts its therapeutic effect through inhibition of Hst3p-mediated H3K56 deacetylation. Paradoxically, the intra-peritoneal injection of high doses of valproic acid in a disseminated mice model of candidiasis was associated with accelerated (mean time to death: 21.5 days vs. >40 days) and increased mortality (44% vs. 75%, P = 0.02; Roger et al., 2011). Similarly, MGCD290 was tested in a murine model of invasive candidiasis in combination with fluconazole (Besterman J., presented at Interscience Conference on Antimicrobial Agents and Chemotherapy IAAC in 2012 in San Francisco, CA, USA). The results of this study indicated that kidney fungal loads in animals receiving both MGCD290 and fluconazole were significantly lower than fungal loads in animals treated with fluconazole alone. These preliminary results on the use of HDAC inhibitors in murine models of candidiasis are conflicting, but there are also major differences between these in vivo models. In the future, the relevancy of such in vivo experiment will probably need to be attested to provide clear information on the therapeutic potential of the tested compounds. Also, HDAC inhibitors still need to be rigorously tested under a range of experimental conditions to examine both their toxicity and antifungal efficacy. Indeed, as several regulatory and signaling pathways/mechanisms are highly conserved between fungal and human eukaryotic cells, the use of a non-selective HDAC inhibitor (acting upstream these pathways) exposes to the risk of unwanted adverse effects. However, HDAC inhibitors toxicity does not seem to be a major issue yet, but the commercially available HDAC is today were only recently approved. The most common serious adverse events reported with HDAC inhibitors in cancerology were cytopenia (thrombocytopenia, anemia, neutropenia, or leukopenia), pyrexia, infection, sepsis, or cardiac toxicity. Other frequent adverse reactions are fatigue, nausea and diarrhea (Mottamal et al., 2015). In a recent phase 2 clinical study, the combination of oral MGCD290 and fluconazole in patients with moderate or severe vulvo-vaginal candidiasis, although well tolerated, did not significantly improve outcome compared with fluconazole alone (Augenbraun et al., 2013). This result does not support the therapeutic use of this HDAC inhibitor in this particular clinical context. Also, there is currently no available data suggesting that patients treated with HDACs inhibitors are less susceptible to Candida infection, and more specifically designed studies, among onco-hematological patients, are needed to answer this issue. Thus, for now, the clinical utility of HDAC inhibitors remains to be validated, and further research is more necessary than ever. The recent and expanding use of the Galleria mellonella larvae model, which inter alia allows high-throughput screening of chemical libraries for the discovery of new antifungal compounds will probably facilitate the discovery of more selective and efficient HDAC inhibitors (Lionakis, 2011).
Conclusion
The activity of HDAC proteins is essential for the functionality of chromatin in all eukaryotic cells. In Candida species, most HDACs contribute to life cycle regulation, morphogenic plasticity, and biofilm formation; they are also involved in azole- and echinocandin-resistance. Therefore, their genetic or chemical inhibition can affect yeast virulence and its capacity to form biofilms while also enhancing the efficacy of existing antifungal drugs, even toward resistant strains. Three HDAC inhibitors now have FDA-approval for clinical use and a dozen compounds are included in clinical trials for cancer therapy. Obviously, the development of fungal-specific drugs would have a high clinical impact as they would avoid inhibition of endogenous host HDACs, therefore probably reducing side effects. These molecules already represent a great potential to create new antifungal treatments, especially given their behavior and how well they are tolerated in patients. Along with other compounds targeting innovative machineries (i.e., mitochondrial function, glycosylphosphatidylinositol biosynthesis, vesicle transport) which are currently in preclinical development, they may be incorporated in the antifungal pipeline. However, in vivo tests should be expanded to include more diverse animal models, including invertebrates.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MCh is supported by a fellowship from the Région Rhône Alpes (France). MCo and JG’s groups received funding from the ANR trough the FungiBET ANR-14-CE16-0027-02 program. JG’s group has also received support from French National Agency for Research (ANR) through the EpiGam ANR-11-PDOC-0011 and the FP7 Marie Curie Action “Career Integration Grant” EpiGam2 304003.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01238
References
- Al-Dhaheri R. S., Douglas L. J. (2010). Apoptosis in Candida biofilms exposed to amphotericin B. J. Med. Microbiol. 59 149–157. 10.1099/jmm.0.015784-0 [DOI] [PubMed] [Google Scholar]
- Augenbraun M., Livingston J., Parker R., Lederman S., Chavoustie S., Morgan F., et al. (2013). “Fluconazole and MGCD290 in vulvo vaginal candidiasis (VVC): results from a randomized phase II study,” in Poster 1330 in IDWeek 2013 San Francisco, CA. [Google Scholar]
- Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A. (2002). Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277 45099–45107. 10.1074/jbc.M205670200 [DOI] [PubMed] [Google Scholar]
- Brown G. D., Denning D. W., Gow N. A. R., Levitz S. M., Netea M. G., White T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4 165rv13 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- Candido E. P., Reeves R., Davie J. R. (1978). Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14 105–113. 10.1016/0092-8674(78)90305-7 [DOI] [PubMed] [Google Scholar]
- Carmen A. A., Griffin P. R., Calaycay J. R., Rundlett S. E., Suka Y., Grunstein M. (1999). Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. U.S.A. 96 12356–12361. 10.1073/pnas.96.22.12356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen L. E., Lindquist S. (2005). Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309 2185–2189. 10.1126/science.1118370 [DOI] [PubMed] [Google Scholar]
- De Las Peñas A., Juárez-Cepeda J., López-Fuentes E., Briones-Martín-Del-Campo M., Gutiérrez-Escobedo G., Castaño I. (2015). Local and regional chromatin silencing in Candida glabrata: consequences for adhesion and the response to stress. FEMS Yeast Res. 15 fov056. 10.1093/femsyr/fov056 [DOI] [PubMed] [Google Scholar]
- Denning D. W., Bromley M. J. (2015). Infectious disease. How to bolster the antifungal pipeline. Science 347 1414–1416. 10.1126/science.aaa6097 [DOI] [PubMed] [Google Scholar]
- Domergue R., Castaño I., De Las Peñas A., Zupancic M., Lockatell V., Hebel J. R., et al. (2005). Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308 866–870. 10.1126/science.1108640 [DOI] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., et al. (2004). Genome evolution in yeasts. Nature 430 35–44. 10.1038/nature02579 [DOI] [PubMed] [Google Scholar]
- Enjalbert B., Smith D. A., Cornell M. J., Alam I., Nicholls S., Brown A. J. P., et al. (2006). Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17 1018–1032. 10.1091/mbc.E05-06-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg K. J., Johnstone R. W. (2014). Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 13 673–691. 10.1038/nrd4360 [DOI] [PubMed] [Google Scholar]
- Freire-Benéitez V., Price R. J., Tarrant D., Berman J., Buscaino A. (2016). Candida albicans repetitive elements display epigenetic diversity and plasticity. Sci. Rep. 6 22989 10.1038/srep22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyd C. A., Kapoor S., Dietrich F., Rusche L. N. (2013). The deacetylase Sir2 from the yeast Clavispora lusitaniae lacks the evolutionarily conserved capacity to generate subtelomeric heterochromatin. PLoS Genet. 9:e1003935 10.1371/journal.pgen.1003935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.-H., Meng F.-L., Hu Y., Zhou J.-Q. (2008). Candida albicans, a distinctive fungal model for cellular aging study. Aging Cell 7 746–757. 10.1111/j.1474-9726.2008.00424.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. B., Mylonakis E. (2009). Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8 1616–1625. 10.1128/EC.00193-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlicher M., Minucci S., Zhu P., Krämer O. H., Schimpf A., Giavara S., et al. (2001). Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20 6969–6978. 10.1093/emboj/20.24.6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N. A. R., van de Veerdonk F. L., Brown A. J. P., Netea M. G. (2012). Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10 112–122. 10.1038/nrmicro2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernday A. D., Lohse M. B., Fordyce P. M., Nobile C. J., DeRisi J. L., Johnson A. D. (2013). Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol. Microbiol. 90 22–35. 10.1111/mmi.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Bardet A. F., Nobile C. J., Petryshyn A., Glaser W., Schöck U., et al. (2012). A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 8:e1003118 10.1371/journal.pgen.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Majer O., Frohner I. E., Komnenovic V., Kuchler K. (2010). The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 6:e1000889 10.1371/journal.ppat.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Schwarzmüller T., Kuchler K. (2009). Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol. Microbiol. 74 1–15. 10.1111/j.1365-2958.2009.06772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S., Zhu L., Froyd C., Liu T., Rusche L. N. (2015). Regional centromeres in the yeast Candida lusitaniae lack pericentromeric heterochromatin. Proc. Natl. Acad. Sci. U.S.A. 112 12139–12144. 10.1073/pnas.1508749112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan G., Paul-Satyaseela M., Dhatchana Moorthy N., Gopalaswamy R., Narayanan S. (2013). Functional characterization of Candida albicans Hos2 histone deacetylase. F1000Res. 2 238 10.12688/f1000research.2-238.v3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J.-E., Lee J.-S. (2015). Histone deacetylase-mediated morphological transition in Candida albicans. J. Microbiol. 53 805–811. 10.1007/s12275-015-5488-3 [DOI] [PubMed] [Google Scholar]
- Kim T., Xu Z., Clauder-Münster S., Steinmetz L. M., Buratowski S. (2012). Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150 1158–1169. 10.1016/j.cell.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Srikantha T., Soll D. R. (2001). A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics 158 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal C. A., Srikantha T., Soll D. R. (1997). Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect. Immun. 65 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., et al. (2000). The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97 5807–5811. 10.1073/pnas.110148297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-E., Oh J.-H., Ku M., Kim J., Lee J.-S., Kang S.-O. (2015). Ssn6 has dual roles in Candida albicans filament development through the interaction with Rpd31. FEBS Lett. 589 513–520. 10.1016/j.febslet.2015.01.011 [DOI] [PubMed] [Google Scholar]
- Li X., Cai Q., Mei H., Zhou X., Shen Y., Li D., et al. (2015). The Rpd3/Hda1 family of histone deacetylases regulates azole resistance in Candida albicans. J. Antimicrob. Chemother. 70 1993–2003. 10.1093/jac/dkv070 [DOI] [PubMed] [Google Scholar]
- Lionakis M. S. (2011). Drosophila and Galleria insect model hosts. Virulence 2 521–527. 10.4161/viru.2.6.18520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. T., Lee R. E. B., Barker K. S., Lee R. E., Wei L., Homayouni R., et al. (2005). Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49 2226–2236. 10.1128/AAC.49.6.2226-2236.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi P. M., Cole K. E., Dowling D. P., Christianson D. W. (2011). Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 21 735–743. 10.1016/j.sbi.2011.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Su C., Liu H. (2012). A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog. 8:e1002663 10.1371/journal.ppat.1002663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Su C., Wang A., Liu H. (2011). Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 9:e1001105 10.1371/journal.pbio.1001105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D., Ostell J., Pruitt K. D., Tatusova T. (2007). Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 35 D26–D31. 10.1093/nar/gkl993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai A., Rotili D., Massa S., Brosch G., Simonetti G., Passariello C., et al. (2007). Discovery of uracil-based histone deacetylase inhibitors able to reduce acquired antifungal resistance and trailing growth in Candida albicans. Bioorg. Med. Chem. Lett. 17 1221–1225. 10.1016/j.bmcl.2006.12.028 [DOI] [PubMed] [Google Scholar]
- Maubon D., Garnaud C., Calandra T., Sanglard D., Cornet M. (2014). Resistance of Candida spp. to antifungal drugs in the ICU: where are we now? Intensive Care Med. 40 1241–1255. 10.1007/s00134-014-3404-7 [DOI] [PubMed] [Google Scholar]
- Morschhäuser J. (2010). Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47 94–106. 10.1016/j.fgb.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Mottamal M., Zheng S., Huang T. L., Wang G. (2015). Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Mol. Basel Switz. 20 3898–3941. 10.3390/molecules20033898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Fox E. P., Hartooni N., Mitchell K. F., Hnisz D., Andes D. R., et al. (2014). A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio 5 e01201–e1214. 10.1128/mBio.01201-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Fox E. P., Nett J. E., Sorrells T. R., Mitrovich Q. M., Hernday A. D., et al. (2012). A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148 126–138. 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orta-Zavalza E., Guerrero-Serrano G., Gutiérrez-Escobedo G., Cañas-Villamar I., Juárez-Cepeda J., Castaño I., et al. (2013). Local silencing controls the oxidative stress response and the multidrug resistance in Candida glabrata. Mol. Microbiol. 88 1135–1148. 10.1111/mmi.12247 [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J., Uría J. A., Johnson A. D. (1999). Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 18 2580–2592. 10.1093/emboj/18.9.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin D. S., Shor E., Zhao Y. (2015). Update on antifungal drug resistance. Curr. Clin. Microbiol. Rep. 2 84–95. 10.1007/s40588-015-0015-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Messer S. A., Georgopapadakou N., Martell L. A., Besterman J. M., Diekema D. J. (2009). Activity of MGCD290, a Hos2 histone deacetylase inhibitor, in combination with azole antifungals against opportunistic fungal pathogens. J. Clin. Microbiol. 47 3797–3804. 10.1128/JCM.00618-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Rhomberg P. R., Messer S. A., Castanheira M. (2015). In vitro activity of a Hos2 deacetylase inhibitor, MGCD290, in combination with echinocandins against echinocandin-resistant Candida species. Diagn. Microbiol. Infect. Dis. 81 259–263. 10.1016/j.diagmicrobio.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Phiel C. J., Zhang F., Huang E. Y., Guenther M. G., Lazar M. A., Klein P. S. (2001). Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 276 36734–36741. 10.1074/jbc.M101287200 [DOI] [PubMed] [Google Scholar]
- Polke M., Hube B., Jacobsen I. D. (2015). Candida survival strategies. Adv. Appl. Microbiol. 91 139–235. 10.1016/bs.aambs.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Rai M. N., Balusu S., Gorityala N., Dandu L., Kaur R. (2012). Functional genomic analysis of Candida glabrata-macrophage interaction: role of chromatin remodeling in virulence. PLoS Pathog. 8:e1002863 10.1371/journal.ppat.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekharan S. K., Ramesh S., Bakkiyaraj D. (2015). Synergy of flavonoids with HDAC inhibitor: new approach to target Candida tropicalis biofilms. J. Chemother. 27 246–249. 10.1179/1973947814Y.0000000186 [DOI] [PubMed] [Google Scholar]
- Robbins N., Leach M. D., Cowen L. E. (2012). Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep. 2 878–888. 10.1016/j.celrep.2012.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger T., Lugrin J., Le Roy D., Goy G., Mombelli M., Koessler T., et al. (2011). Histone deacetylase inhibitors impair innate immune responses to toll-like receptor agonists and to infection. Blood 117 1205–1217. 10.1182/blood-2010-05-284711 [DOI] [PubMed] [Google Scholar]
- Rundlett S. E., Carmen A. A., Kobayashi R., Bavykin S., Turner B. M., Grunstein M. (1996). HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. U.S.A. 93 14503–14508. 10.1073/pnas.93.25.14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders B. D., Zhao K., Slama J. T., Marmorstein R. (2007). Structural basis for nicotinamide inhibition and base exchange in Sir2 enzymes. Mol. Cell 25 463–472. 10.1016/j.molcel.2006.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Perlin D. S. (2015). Coping with stress and the emergence of multidrug resistance in fungi. PLoS Pathog. 11:e1004668 10.1371/journal.ppat.1004668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti G., Passariello C., Rotili D., Mai A., Garaci E., Palamara A. T. (2007). Histone deacetylase inhibitors may reduce pathogenicity and virulence in Candida albicans. FEMS Yeast Res. 7 1371–1380. 10.1111/j.1567-1364.2007.00276.x [DOI] [PubMed] [Google Scholar]
- Singh R. P., Prasad H. K., Sinha I., Agarwal N., Natarajan K. (2011). Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J. Biol. Chem. 286 25154–25170. 10.1074/jbc.M111.233569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. L., Edlind T. D. (2002). Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upregulation. Antimicrob. Agents Chemother. 46 3532–3539. 10.1128/AAC.46.11.3532-3539.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza J. R., Skene R. J., Katz B. A., Mol C., Ho J. D., Jennings A. J., et al. (2004). Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12 1325–1334. 10.1016/j.str.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Srikantha T., Tsai L., Daniels K., Klar A. J., Soll D. R. (2001). The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183 4614–4625. 10.1128/JB.183.15.4614-4625.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J. S., Liu H. (2011). Regulation of white and opaque cell-type formation in Candida albicans by Rtt109 and Hst3. Mol. Microbiol. 81 1078–1091. 10.1111/j.1365-2958.2011.07754.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J. S., Liu H. (2013). Nucleosome assembly factors CAF-1 and HIR modulate epigenetic switching frequencies in an H3K56 acetylation-associated manner in Candida albicans. Eukaryot. Cell 12 591–603. 10.1128/EC.00334-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9 737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- Taff H. T., Mitchell K. F., Edward J. A., Andes D. R. (2013). Mechanisms of Candida biofilm drug resistance. Future Microbiol. 8 1325–1337. 10.2217/fmb.13.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P., Brandtner E. M., Brosch G., Loidl P., Galehr J., Linzmaier R., et al. (2003). Histone deacetylases in fungi: novel members, new facts. Nucleic Acids Res. 31 3971–3981. 10.1093/nar/gkg473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherner M., Zwolanek F., Jenull S., Sedlazeck F. J., Petryshyn A., Frohner I. E., et al. (2015). The Candida albicans histone acetyltransferase Hat1 regulates stress resistance and virulence via distinct chromatin assembly pathways. PLoS Pathog. 11:e1005218 10.1371/journal.ppat.1005218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji N., Kobayashi M., Nagashima K., Wakisaka Y., Koizumi K. (1976). A new antifungal antibiotic, trichostatin. J. Antibiot. 29 1–6. 10.7164/antibiotics.29.1 [DOI] [PubMed] [Google Scholar]
- Uhl M. A., Biery M., Craig N., Johnson A. D. (2003). Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C.albicans. EMBO J. 22 2668–2678. 10.1093/emboj/cdg256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P., Pierce C. G., Thomas D. P., Bubeck S. S., Saville S. P., Lopez-Ribot J. L. (2010). The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot. Cell 9 1531–1537. 10.1128/EC.00111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele H., Tsao S., Lépine G., Mullick A., Tremblay J., Drogaris P., et al. (2010). Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat. Med. 16 774–780. 10.1038/nm.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Kijima M., Akita M., Beppu T. (1990). Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265 17174–17179. [PubMed] [Google Scholar]
- Yoshida M., Nomura S., Beppu T. (1987). Effects of trichostatins on differentiation of murine erythroleukemia cells. Cancer Res. 47 3688–3691. [PubMed] [Google Scholar]
- Zacchi L. F., Schulz W. L., Davis D. A. (2010). HOS2 and HDA1 encode histone deacetylases with opposing roles in Candida albicans morphogenesis. PLoS ONE 5:e12171 10.1371/journal.pone.0012171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xu W. (2015). Histone deacetylase inhibitors for enhancing activity of antifungal agent: a patent evaluation of WO2014041424(A1). Expert Opin. Ther. Pat. 25 237–240. 10.1517/13543776.2014.981256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


