Abstract
The mitotic spindle is required for chromosome congression and subsequent equal segregation of sister chromatids. These processes involve a complex network of signaling molecules located at the spindle. The endocytic protein, clathrin, has a “moonlighting” role during mitosis, whereby it stabilizes the mitotic spindle. The signaling pathways that clathrin participates in to achieve mitotic spindle stability are unknown. Here, we assessed the mitotic spindle proteome and phosphoproteome in clathrin-depleted cells using quantitative MS/MS (data are available via ProteomeXchange with identifier PXD001603). We report a spindle proteome that consists of 3046 proteins and a spindle phosphoproteome consisting of 5157 phosphosites in 1641 phosphoproteins. Of these, 2908 (95.4%) proteins and 1636 (99.7%) phosphoproteins are known or predicted spindle-associated proteins. Clathrin-depletion from spindles resulted in dysregulation of 121 proteins and perturbed signaling to 47 phosphosites. The majority of these proteins increased in mitotic spindle abundance and six of these were validated by immunofluorescence microscopy. Functional pathway analysis confirmed the reported role of clathrin in mitotic spindle stabilization for chromosome alignment and highlighted possible new mechanisms of clathrin action. The data also revealed a novel second mitotic role for clathrin in bipolar spindle formation.
During mitosis, a mitotic spindle is established to pull sister chromatids toward opposite poles of the dividing cell. This ensures that upon mitotic exit, each newly formed daughter cell receives one copy of each chromosome. Premature chromosome segregation results in aneuploidy—a hallmark of many human cancers. This adverse situation is avoided by activation of the spindle assembly checkpoint (SAC).1 This signaling pathway consists of a number of protein complexes that monitor mitotic spindle assembly, delaying anaphase onset until all chromosomes are stably attached to kinetochores (KTs) (1).
In addition to components of the SAC (e.g. Bub1, Bub3, BubR1, Mad1, and Mad2), a large number of proteins associate with the mitotic spindle and regulate its assembly, stability, and function. Targeted approaches as well as proteomics have identified nearly one thousand experimentally validated spindle protein components (2, 3). These include structural proteins (e.g. tubulin, NUMA, and TPX2), microtubule nucleating and stabilizing (e.g. TPX2, MAP2, TACC3, and ch-TOG), destabilizing proteins (e.g. MCAK and Katanin) (4), motor proteins (e.g. kinesin family members and cytoplasmic dynein) (5, 6), and regulatory proteins such as protein kinases (e.g. Aurora B and Plk1) and phosphatases (e.g. PP2A) (7). Disruption of many of these components by siRNA or small molecule inhibitors leads to a plethora of mitotic phenotypes consistent with mitotic spindle dysfunction such as monopolar spindles, a reduced number of spindle microtubules, misaligned chromosomes, and loss of tension of kinetochore-fibers (K-fibers). These disruptions often lead to the activation of the SAC, however, if this fails, progression through metaphase may occur, leading to mis-segregation and aneuploidy. Despite the identification and characterization of a large number of mitotic spindle proteins, our understanding of the cross-talk between pathways that regulate the mitotic spindle remains poor.
Clathrin is a protein complex of three identical 190 kDa clathrin heavy chains (CHCs) arranged in a trimer (called a triskelion) of three “legs” connected by their C termini at a central vertex (8, 9). A globular N-terminal domain (TD) is found at the end of each leg. During endocytosis, clathrin can interact with multiple adapter proteins such as amphiphysin via its TD. Clathrin is best known for its role in clathrin-mediated endocytosis, where it cycles between the cytoplasmic triskelion and a polymerized coat on vesicles or membranes. However, during mitosis, a pool of clathrin localizes to the spindle (10, 11), where it is involved in organizing and stabilizing spindle microtubules, instead of endocytosis (10, 12). Clathrin spindle function is dependent on its trimerisation and its interaction with phosphorylated TACC3 and ch-TOG (11, 12). ch-TOG is important for microtubule outgrowth from spindle poles, whereas TACC3 binds microtubules (13, 14) and loads ch-TOG onto the spindle (15, 16). Aurora A mediates TACC3 phosphorylation (17, 18). The phospho-TACC3/clathrin/ch-TOG complex is then recruited to the spindle microtubules by a mechanism that is currently under debate (11, 12, 19). It is believed that at the spindle, this complex forms a structural bridge connecting two microtubules within a K-fiber to aid chromosome congression (10), with TACC3 directly interacting with microtubules (15, 16).
Both SAC activation and inhibition are targets for new classes of potential chemotherapeutic agents, many of which are currently under investigation in human cancer clinical trials. siRNA-mediated depletion of clathrin or inhibition by small molecules causes defective chromosome congression to the metaphase plate and persistent SAC activation (10, 20–23). This is analogous to the effects of Aurora A inhibitors, which are also SAC activators (24–28) and block clathrin recruitment to the spindle by blocking TACC3 recruitment (11). It is possible that SAC activation and the anticancer properties of Aurora A inhibitors are partly caused by the blocking of clathrin function at the spindle.
A thorough characterization and understanding of the molecular pathways that clathrin is involved in at the mitotic spindle is essential for addressing the fundamental issues of the molecular mechanisms of mitosis as well as for developing more potent antimitotic compounds as potential new therapeutics for cancer treatment. In this study, we carried out quantitative proteomics and phosphoproteomics of purified human mitotic spindles from clathrin-depleted metaphase cells. We identified a significant accumulation of mitotic spindle proteins and phosphorylation site occupancy in clathrin-depleted mitotic spindles. The results reveal that clathrin has two mitotic roles (1) in bipolar spindle formation and (2) in spindle stabilization and chromosome alignment. These findings provide insight into the cross-talk between signaling pathways that regulate these processes, specifically those that are regulated by clathrin.
EXPERIMENTAL PROCEDURES
siRNA Transfection and Mitotic Spindle Isolation
HeLa S3 cells were grown in RPMI media (Invitrogen, Mulgrave, Australia) containing 10% fetal bovine serum (FBS) (Invitrogen) to a density of 7.5 e6 cells per ml for each treatment (total of three treatments in one experiment). Two of these pools of cells were transfected with 250 μm siRNA that was directed either toward CHC [sense: 5′-GCAATGAGCTGTTTGAAGA-3′; antisense: 5′-TCTTCAAACAGCTCATTGC-3′] or Luc [sense: 5′-CGUACGCGGAAUACUUCGATT-3′; antisense: 5′-UCGAAGUAUUCCGCGUACGTT-3′] using the Neon™ Transfection System 100 μl kit (Invitrogen). One pool of HeLa S3 cells was left untransfected. The cells were then transferred to six multiflasks (BD Falcon, Noble Park, Australia), with five growth layers in each, per treatment for large-scale production of transfected or untransfected HeLa cells and grown for 72 h post-transfection at 37 °C. Penicillin-streptomycin (100 IU/ml and 100 μg/ml, respectively) (Invitrogen) was added 24 h post-transfection to all flasks. Cells were synchronized to pro-metaphase with 40 ng/ml Nocodazole (Sigma, Castle Hill, Australia) for 14 h before collection by mitotic shake-off. Mitotic spindle protein isolation was carried out as described by (69). Briefly, the cells were collected in PBS by mitotic shake-off and resuspended in 20 ml of RPMI medium. A small aliquot (750 μl to 1 ml) of the transfected HeLa lysate in RPMI was set aside before cell lysis and resuspended in lysis buffer containing 5 m Tris (pH 7.4), 1% Triton-X 100, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulphonyl fluoridie (PMSF), 40 μg/ml Leupeptin, a complete EDTA-free protease inhibitor (one tablet per 5 ml; Roche, Sydney, Australia) and phosphatase inhibitor mixture set II (1:100 diluted; Merck, Millipore [Calbiochem], Bayswater, Australia) at 4 °C. The lysate was sonicated and used to confirm the absence of clathrin by Western blots. The cells were allowed to progress through mitosis for ∼40 min at 37 °C until they reached metaphase. At ∼35 min, the mitotic spindles were stabilized by the addition of Taxol (5 mg/ml, Sigma). The cells were washed in a solution containing 5 μg/ml Taxol, 2 μg/ml Latrunculin B and 1 mm PMSF. The cells were centrifuged at 300 g and the pellet was resuspended in spindle lysis buffer (100 mm Pipes at pH 6.9, 1 mm MgSO4, 2 mm EGTA, 0.005% Nonidet P-40, 5 μg/ml Taxol, 2 μg/ml Latrunculin B, 200 μg/ml DNase I, 10 μg/ml RNase A, 20 U/ml Bensonase HC, 1 μg/ml Pepstatin, 1 μg/ml Leupeptin, 1 μg/ml Aprotinin, and 1 mm PMSF) for 20 min at 37 °C. The pellet was harvested (1000 g, 3 min) and incubated again in spindle lysis buffer for 5 min. The lysate was centrifuged again and washed twice (incubated for 20 min at 37 °C and spun at 2000 g for 5 min) with a low ionic strength buffer containing 1 mm Pipes pH 6.9 and 5 μg/ml Taxol. An aliquot of the isolated mitotic spindles (20 μl) was resuspended in 4% paraformaldehyde for confirmation of mitotic spindle isolation by microscopy. The spindle lysate was finally resuspended in 0.1 m Glycine, pH 2.8 and sonicated to disaggregate the spindles for proteomic analysis.
Immunoblotting
HeLa cell lysates were collected by centrifugation, washed with PBS, then resuspended in ice-cold lysis buffer for sonication [20 mm Tris-HCl pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.1 mm PMSF, 1% Triton X-100, 1 μg/ml Leupeptin, and EDTA-free Complete protease inhibitor mixture (Roche)] followed by incubation on ice for 30 min. The supernatant was collected following centrifugation at 13,000 rpm for 30 min at 4 °C. Cell lysates and purified isolated mitotic spindles were fractionated by SDS-PAGE for immunoblot analysis with the following antibodies: anti-EG5 (ab137535, Abcam, Melbourne, Australia), diluted 1:500; anti-CHC (610500, BD Biosciences, San Jose, USA), diluted 1:1000; anti-γ-Tubulin (T5326, Sigma), diluted 1:50,000; and anti-INCENP (ab86088, Abcam), diluted 1:500. Antibody bound to the indicated protein was detected by incubation with a horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc., Waterford, Australia). Blotted proteins were visualized using the SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific) or SuperSignal® West Dura Extended Duration Substrate (Thermo Scientific).
Immunofluorescence Microscopy
HeLa cells were grown on glass coverslips in RPMI and 10% FBS under 5% CO2. 90 nm Luc or CHC siRNA duplexes (see above) were transfected using Lipofectamine 2000 (Invitrogen). Control and treated cells were fixed in −20 °C methanol for 4 min 68–72 h after transfection. After fixation, samples were typically blocked for 40 min at RT in 3% BSA/PBS. Cells were then incubated with primary antibodies for 60–90 min, washed four times with PBS, stained with fluorescently labeled secondary antibodies, washed four times with PBS, and mounted with ProLong Gold (Life Technologies, Mulgrave, Australia). The following primary antibodies were used: anti-EG5 (ab137535, Abcam), diluted 1:100; anti-INCENP (ab3645, Abcam), diluted 1:100; anti-MCAK/KIF2C (ab42676, Abcam), diluted 1:200; anti-NuMA (#3888S, Cell Signaling Technology, Arundel, Australia), diluted 1:50; anti-RIF1 (ab13422, Abcam), diluted 1:50; anti-TPX2 (NB500–179, Novus), diluted 1:1000; anti-CHC (610500, BD Biosciences), diluted 1:200; anti-CHC (ab21679, Abcam), diluted 1:200; anti-CHC (SC-6579, SantaCruz Biotechnologies, Mulgrave, Australia), diluted 1:200; anti-TACC3 (SC-22773, SantaCruz Biotechnologies), diluted 1:200; anti-TACC3 (ab56595, Abcam), diluted 1:200; anti-TACC3-S558 (#5645S, Cell Signaling Technology), diluted 1:200; anti-α-Tubulin (T9026, Sigma), diluted 1:500; and Alexa Fluor 488-conjugated anti-α-Tubulin (322588, Life Technologies), diluted 1:1000. Secondary antibodies labeled with Alexa Fluor 488, Alexa Fluor 568 or Alexa Fluor 647 from Life Technologies (diluted 1:500) were used along with 1 μg/ml DAPI. Cells were imaged using an 100×, 1.4 NA U-Plan S-Apo objective on a microscope (IX70, Olympus) equipped for optical sectioning microscopy (DeltaVision, Applied Precision, Parramatta, Australia) using standard filters (DAPI: 390/18, 435/48; FITC: 475/28, 523/36; TRITC: 543/27, 594/45; Cy5: 632/22, 676/34) and a CCD camera (CoolSnapHQ2, Roper Scientific, Tucson, USA). Each z series (0.3 μm intervals) was acquired, deconvolved and projected using SoftWoRx (Applied Precision). All images were scaled using linear transformations in Adobe Photoshop CS or ImageJ. Photoshop CS and Illustrator CS were used to construct final figures.
The staining intensity of the validated proteins in untreated and knock-down cells was quantified in z projections by measuring the total intensity (IntDen) using the Measure plugin in ImageJ. For EG5, TPX2, and TACC3 proteins, 20 circular selections, each 20 pixels in diameter, were drawn at different positions in a mitotic spindle. All circles were drawn at similar positions and were separated by a similar distance in all analyzed cells. For NuMA, a selection of 100 pixels wide and 42 pixels high was drawn around each spindle pole. For p-TACC3-S558, a circle of 30 pixels in diameter was drawn around each spindle pole. For INCENP, three regions were used: a selection (84 pixels wide and 62 pixels high) was drawn around each spindle pole, and the third selection (305 pixels wide and 159 pixels high) was drawn around the metaphase plate. For KIF2C, a selection (377 pixels wide and 338 pixels high) covering the entire spindle was used. The IntDen values for each selection in a spindle were then summed to give a spindle total intensity value per analyzed cell. Each total intensity of a spindle was normalized by dividing it by the mean spindle total intensity of untreated cells from all experiments per studied condition.
Tryptic Digestion and Dimethylation Labeling
The spindle protein lysates were precipitated with TCA-acetone and dissolved in 6 m Guanidine-HCl. Equal protein amounts were used for the three treatments within each biological replicate. The Guanidine-HCl in the protein lysate solution was diluted with 0.1 m TEAB and samples were incubated in a reducing buffer containing 0.05% SDS and 0.005 m tris(2-carboxyethyl) phosphine (TCEP) (Sigma Aldrich, Australia) in 0.1 m TEAB at 85 °C for 5 min, then at 60 °C for 30 min. Samples were incubated at room temperature for 10 min with 5 mm S-methyl methane thiosulfonate (MMTS) (Sigma Aldrich, Australia). Trypsin (Promega, Alexandria, Australia) was added (1:20 trypsin: protein by weight) for 16 h at 37 °C. The tryptic peptides from the spindle lysates were each loaded onto Oasis HLB sorbent cartridges (Waters, Rydalmere, Australia) in 0.1% TFA. The peptides were eluted with 70% acetonitrile (ACN) in 0.1% formic acid and dried.
The peptides were then labeled with light, medium and heavy isotope dimethyl labels (70, 71), using (1) 5% CH2O and 500 mm NaBH3CN (light), (2) 5% C[2H]2O and 500 mm NaBH3CN (medium), and (2) 5% 13C[2H]2O and 500 mm NaB[2H]3CN (heavy) for untransfected, Luc siRNA-transfected and CHC siRNA-transfected spindle sample pools, respectively. Lipids were precipitated by the addition of 1% formic acid with vortexing for 10 min at 1500 rpm followed by a centrifugation at 13000 rpm for 5 min. The supernatant was dried down and dissolved in 0.1% formic acid. The combined sample was dried and enriched for phosphopeptides.
TiO2 Phosphopeptide Enrichment and Strong Cation Exchange Separation of Nonphosphopeptides
TiO2 [from a disassembled Titansphere column (GL Sciences, Tokyo, Japan)] was packed into micro-columns for chromatography as described previously (29) using a C8 plug (3 m Empore disk, Thermo Fisher Scientific, Scoresby, Vic, Australia). The dimethylated peptides were loaded into the micro-column slowly in a loading solution (60 μl) of 50% ACN and 5% TFA. The column was washed three times with the loading solution and the phosphopeptides were eluted using (1) a solution of 2% NH4OH (29.1% NH3 in water) and 20% ACN and then (2) a solution of 20% NH4OH and 20% ACN. These were pooled together for high pH-reverse phase and loaded in 10 mm ammonium formate (pH 10). They were eluted stepwise with 5%, 10%, 15%, 20%, 25%, 30%, and 80% ACN in 10 mm ammonium formate and each fraction was dried, resuspended in 10 μl of 0.1% formic acid, and analyzed by LC-MS/MS.
The unbound/nonphosphopeptide solution was separated by SCX chromatography on micro-columns. A GeLoader tip (Bio-Rad Laboratories Pty., Ltd, Gladesville, Australia) was packed with Strong Cation Exchanger (SCX) beads (material obtained from a dismantled SCX column within an iTRAQ reagent methods development kit, AB SCIEX, Scoresby, Victoria, Australia) in a manner similar to that described for TiO2 enrichment (29). The sample was loaded onto the SCX micro-column in a loading solution containing 10 mm KH2PO4 in 25% ACN at pH 3. The micro-column was washed twice with loading buffer and the nonphosphopeptides were eluted stepwise with 50, 100, 200, 300, 400, and 500 mm KCl at pH 3. The eluted samples were de-salted using a POROS R3 (Life Technologies, Mulgrave, Victoria, Australia) micro-column. The R3 flow-through was dried, resuspended in 10 μl of 0.1% formic acid and analyzed by LC-MS/MS.
Label-free Proteomics
HeLa S3 cells were grown in RPMI media (Invitrogen, Mulgrave, Australia) containing 10% FBS (Invitrogen) to a density of 7.5 e6 cells per ml. Cells were synchronized to prometaphase with 40 ng/ml Nocodazole (Sigma) for 14 h before collection by mitotic shake-off. Mitotic spindle protein isolation was carried out as previously described by Sillje et al., 2005. The proteins were reduced (0.05% SDS and 0.005 m TCEP) and alkylated (5 mm MMTS) as before. Trypsin was added (1:20 trypsin: protein by weight) for 16 h at 37 °C. TiO2 phosphopeptide enrichment was carried out as described above without fractionating the eluted peptides by high pH-reverse phase. The unbound/nonphosphopeptide solution was separated by SCX chromatography as described above.
ESI-LC MS/MS for the Dimethylation Labeling Approach
Samples were injected via the autosampler in 5 μl aliquots and the separation of peptides was done using a Dionex 3000 HPLC (Thermo Fisher Scientific, Mulgrave, Australia). Each sample was loaded onto to an in-house (75 μm inside diameter (i.d.) capillary column) ReproSil-Pur 120 C18-AQ (3 μm beads, Dr Maisch, Germany) packed 18 cm column. Mobile phase buffer A consisted of 0.1% formic acid in water and phase B consisted of 90% acetonitrile, 0.1% formic acid, and 9.9% water. The peptides were eluted using a 165 min gradient from 0% to 35% buffer B (90% ACN and 0.1% formic acid) at a rate of 250 nL min−1 and introduced into an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) via a 10 μm i.d. nanoelectrospray coated SilicaTip (New Objective, Woburn, MA). Following a single round of analysis for all fractions (both phospho and nonphosphopeptides), the respective phosphopeptide fractions within each replicate were pooled and loaded onto a longer in-house (75 μm i.d. capillary column) ReproSil-Pur 120 C18-AQ (3 μm beads, Dr Maisch, Germany) packed 35 cm column. The peptides were eluted using the same gradient and buffer conditions as above. Technical replicates of these were run until the samples lasted for maximum coverage. The same technique was applied to the respective SCX fractions of nonphosphopeptides within each replicate.
The LTQ-Orbitrap Velos mass spectrometer was operated using the following parameters: capillary temperature, 275 °C; nanospray source voltage 2.2 kV; Orbitrap automatic gain control target, 300000; S-lens RF level 69; MS ion time, 50 ms; and AGC target set to 1 × 106. A full MS scan in the mass area of 300–1800 Da was performed in the Orbitrap with a resolution of 30,000 FWHM and a target value of 1 × 106 ions. The ten most intense ions (default charge state +2) were selected from each full scan event for higher energy collision dissociation (HCD) and detected at a resolution of 7500 FWHM. The following parameters were used for HCD mode: minimum signal required, 10,000; isolation width, 2; normalized collision energy, 45; and activation time, 0.1s. A normalized collision energy of 40 was used to collect data from all the nonphosphopeptide fractions.
Data Processing
Raw data files from each sample generated on the LTQ-Orbitrap Velos mass spectrometer were processed using MaxQuant version 1.3.0.5 (Max Planck Institute of Biochemistry, Germany). Peak lists were searched against the UniProt reference proteome for Homo sapiens (downloaded 16th September 2013). Deamidation (N/Q), oxidation (M) and phosphorylation (S/T/Y) were specified as variable modifications. The digestion enzyme was set to trypsin, including cleavage before proline. The first search and main search were set to 20 and 6 ppm, respectively, allowing up to three missed cleavages and a maximum of seven modifications per peptide. MS/MS search parameters were set as follows: the MS/MS tolerance for FTMS and ITMS were set to 20 ppm and 0.5 Da, respectively (default setting). “Methylthio” (C) was used as a fixed modification, the protein and peptide false discovery rate (FDR) was set to 0.05 for the analysis of phosphopeptides, whereas the site FDR was set to 0.01. Only phosphosites with a localization probability >0.75 were reported. For the nonphosphopeptide analysis, an FDR of 0.01 was used for peptide, protein and site FDR. The minimum peptide length was set to 5. Minimum peptides for quantification was set to 2 using “unique and razor peptides.” A reversed database was searched to determine the FDR. All other parameters were the default settings.
Data Analysis
The phospho and nonphosphopeptide raw files were processed separately and so were files from Analysis 1 and Analysis 2 approaches as depicted in supplemental Fig. S5. The proteinGroups.txt output file from MaxQuant (nonphosphopeptide data set) was imported into Microsoft Excel for further analysis. The proteinsGroups.txt file thus processed separately from Analysis 1 and Analysis 2 was then combined manually. Reversed database identification and contaminants were deleted. The average ratios from the same protein found from both analyses was used. Protein abundance ratios (CHC/Luc) from each biological replicate were averaged and only those ratios determined from three or more replicates for each protein are reported. Significance tests (t test) were carried out on log2-transformed ratios and proteins with a p value < 0.05 were considered significant. It should be noted that because of the strict processing thresholds applied for calculation of protein abundance (minimum of two peptides per protein) in MaxQuant, single “unmodified” peptides are eliminated. Such peptides were found in the “Evidence.txt” file. Although the protein abundance of clathrin and TACC3 is high in HeLa cells, they may have been present at a very low abundance at the spindle compared with whole cell lysates. These were therefore not found in the actual files (proteinGroups.txt) used for processing. The average of all peptides found for clathrin from the separately processed phosphopeptide and nonphosphopeptide data sets found in the respective Evidence.txt files were therefore extracted.
The Phospho(STY).txt output file from MaxQuant was imported into Microsoft Excel. The Phospho(STY).txt file thus processed separately from Analysis 1 and Analysis 2 was then combined manually. Phosphosites with p > 0.75 were classified as Class I peptides. Peptides with the same phosphosite position were grouped and averaged (including Class II peptides with p < 0.75 within the same group). If, however, no Class I peptides were found within the grouped peptides, the phosphosites were eliminated from the analysis.
Each phosphopeptide ratio (CHC/Luc) was divided by the respective protein abundance ratio generated above for each respective biological replicate to correct for differences in protein abundance. There were instances where the protein ratio was not available to apply a correction (i.e. either nonphosphorylated peptides were not detected for that protein or the respective protein abundance ratio was not detected within a biological replicate). Such ratios/instances were not considered for phosphorylation abundance analysis. After correction, the average ratio for each phosphopeptide was calculated across the six biological replicates. Only phosphosites that were detected in three or more replicates are reported. Significance tests were carried out on log2-transformed ratios and phosphosites with p values < 0.05 were considered significant. The ratio intensities for phosphosites in the PhosphoSTY.txt file are reported for peptides that passed a certain internal threshold set by the algorithm MaxQuant uses. All peptides that passed or did not pass this threshold are present in the Evidence.txt file. As a result, phosphopeptides for the protein TACC3 were eliminated from further analysis using the PhosphoSTY files because a) not all peptides passed the internal algorithm criteria and b) those that did pass were finally found in <3 biological replicates. TACC3 is an important protein with regards to this study; therefore, we extracted information for data analysis of this protein from the Evidence.txt file. Further information is provided in the Results section.
Bioinformatics
The online tool JVENN was used to identify protein,phosphopeptide and kinase consensus motif overlaps between the lists compared (http://bioinfo.genotoul.fr/jvenn/example.html) (30). NetworkAnalyst (http://www.networkanalyst.ca) was used for protein interaction network analysis. Spindle protein prediction was performed using Predster (http://predster.cathdb.info/). Consensus kinase motifs in phosphopeptides were searched for using the ScanProsite tool (Prosite; ExPASy Bioinformatics resource Portal). Search Tool for the Retrieval of Interacting Genes/Proteins (STRING v9.1) (31) was also used for protein interaction assessment and GO enrichment.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (32) with the data set identifier PXD001603.
Experimental Design and Statistical Rationale
A human spindle proteome reference library was created to identify and classify as many proteins at the metaphase mitotic spindle. These were compared with various published literature sources and databases in an attempt to extend the list of spindle-associated proteins. The effect of clathrin-depletion on the metaphase mitotic spindle was then studied using the methods described above. All proteomic experiments were conducted with six biological replicates and at least three technical replicates (where indicated). For proteomic data analysis, a pair-wise t test was used to compare clathrin-knock-down spindle data with luciferase siRNA treated spindle data for the purposes of direct comparison of the respective effects. Instances that displayed p values < 0.05 following a t test were considered to be a significant change.
RESULTS
A Reference Mitotic Spindle Proteome
To investigate the effects of depletion of clathrin on the human mitotic spindle proteome and phosphoproteome, we first established a reference library of the human mitotic spindle. Mitotic spindles (containing KTs and centrosomes) from metaphase-synchronized HeLa cells were purified essentially as previously described (33). Enrichment of mitotic spindles was confirmed by microscopy, revealing that the samples contained predominantly spindle poles with associated microtubules (supplemental Fig. S1A), as previously described (33). Enrichment of mitotic spindles was also confirmed by immunoblotting selected key spindle proteins (supplemental Fig. S1B). We used label-based and label-free proteomics approaches to ensure comprehensive coverage of the spindle proteome and phosphoproteome, because these two approaches are known to result in identification of different subsets of the proteome/phosphoproteome (34–36). A label-free approach was used only for identification (supplemental Fig. S1C). Multiplex peptide dimethyl stable isotope labeling was used for both identification and quantification of spindle peptides and proteins (Fig. 1A). From six independent experimental replicates, we identified a combined total of 3046 proteins using the two approaches (supplemental Table S1). This number included 1345 and 2701 from the label-free (supplemental Table S2) and dimethyl labeling (supplemental Table S3 and S4) approaches, respectively, with 1000 proteins in common (supplemental Table S1).
Fig. 1.
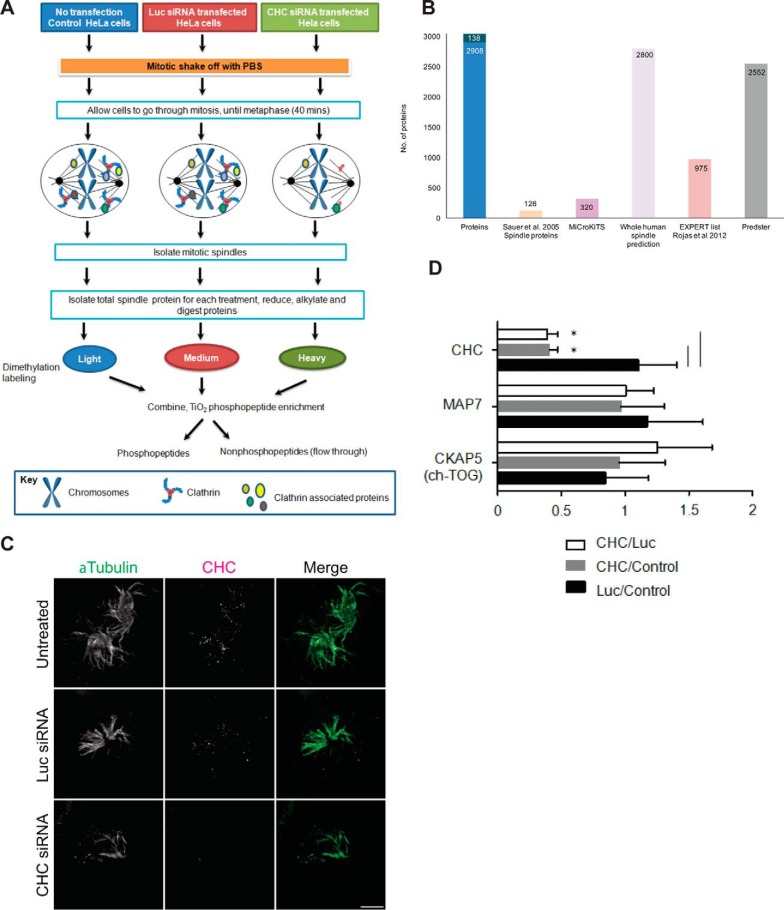
Mitotic spindle proteomics. A, Untransfected HeLa S3 cells and cells transfected with Luc-targeting siRNA or CHC-targeting siRNA were synchronized to pro-metaphase with Nocodazole and collected by mitotic shake-off. The workflow describes isolation of mitotic spindle proteins, tryptic digestion, dimethylation labeling of tryptic peptides, followed by preparation of each sample for analysis of phosphopeptides and nonphosphopeptides on the Orbitrap Velos mass spectrometer. B, Bar graph depicting the number of proteins that overlap between proteins identified in our study (Proteins) and proteins classified as localizing to the spindle by Sauer et al., 2005 (Sauer spindle list) (3), MiCroKiTS, predicted spindle proteins in the whole human proteome (SPIP), the Expert list (37) and the online tool Predster. C, Representative wide-field microscopy images of isolated mitotic spindles. Spindle structures of untreated cells and cells treated with siRNA against Luc or CHC stained for α-Tubulin (green) and CHC (magenta). Scale bar: 5 μm. D, Graph shows the relative change in abundance (mean ± S.E.) of clathrin, the microtubule- associated protein Map7 and ch-TOG by quantitative mass spectrometry. n≥3. *, p < 0.05.
We next assessed the breadth and specificity of the proteins that we identified as being spindle-associated by comparing our findings to the reported (3) and predicted (37) spindle proteomes (Fig. 1B; supplemental Table S1, supplemental Table S5). Sauer et al. (3) reported 156 spindle proteins. We identified 126 of these proteins (supplemental Table S1). We also identified an additional 219 proteins that have been classified as spindle and/or spindle-associated according to MiCroKiTS v4 (supplemental Table S1), an on-line database containing protein localization information from multiple organisms (2). We also compared our spindle proteome to the predicted spindle proteomes—Spindle Predictions Integrated Platform (SPIP) as well as the online tool Predster (SPINDLE mode) (37). Both platforms assign individual proteins with a Fischer score to indicate their likelihood of being a spindle protein, whereby the higher the score, the more likely it is to be a spindle protein. All, but 138 proteins (4.4% had no score indicating they are unlikely to be spindle proteins), had a Fisher score indicating that our spindle preparation has a high level of purity.
We next took a candidate approach and assessed the localization of RIF1. RIF1 had been assigned the highest Fischer score of 13.62, among the down-regulated proteins on clathrin depletion (supplemental Table S1, supplemental Table S3), indicating that it is highly likely to be spindle-associated. Immunofluorescence microscopy revealed that it localized along the spindle fibers (supplemental Fig. S2). Collectively, we report a spindle proteome consisting of 2908 proteins. Thus, we generated a spindle proteome fraction of high purity with 95.4% of proteins having been either experimentally validated or predicted to be spindle proteins.
The Clathrin-depleted Spindle Proteome
To assess the effect of depleting clathrin on the spindle proteome and phosphoproteome, we carried out quantitative LC-MS/MS of isolated mitotic spindles from untreated metaphase HeLa cells and metaphase HeLa cells transfected with CHC siRNA or control Luciferase (Luc) siRNA (Fig. 1A). Depletion of clathrin by its target siRNA was confirmed in total cell lysates (prior to mitotic spindle isolation) by Western blotting (supplemental Fig. S3), as well as in purified mitotic spindles by immunofluorescence microscopy (Fig. 1C). The microtubule arrays were similar in all three experimental conditions, suggesting that the cells were synchronized at a similar mitotic stage. Consistent with the role of clathrin in stabilizing K-fibers (11), we observed a reduction in the numbers of spindle fibers associated with each spindle pole following CHC siRNA treatment (Fig. 1C). Nevertheless, an equivalent amount of spindle proteins of similar purity were isolated in all experimental samples. To quantitate the changes in protein and phosphopeptide abundance by LC-MS/MS, mitotic spindles isolated from untreated, Luc-depleted and CHC-depleted cells were labeled with light, medium and heavy dimethyl isotope variants, respectively. An equal amount of each labeled sample was combined. Phosphopeptides were enriched for and separated for analysis. The nonphosphopeptide flow-through was fractionated by strong cation exchange to obtain comprehensive coverage (supplemental Fig. S1B). To obtain quantifiable changes because of clathrin depletion with a high level of confidence, we only assessed those proteins that were identified in at least three of the six independent biological replicates. Thus, a total of 835 proteins were analyzed (supplemental Table S3).
Analysis by tandem MS of the protein levels of CHC (supplemental Table S6), the microtubule-binding partner MAP7, and ch-TOG provided an independent assessment of the depletion efficiency as well as the specificity of the CHC targeted siRNA (Fig. 1D). No significant change in MAP7 abundance was detectable, demonstrating the specificity of the CHC siRNA. Consistent with previous reports (19), recruitment of ch-TOG to the mitotic spindle was not dependent on clathrin because we observed no reduction in its abundance in clathrin-depleted spindles (Fig. 1D). Nonphosphopeptides of TACC3 could not be identified to assess the effect of clathrin depletion on its spindle localization. However, the lack of detection of nonphosphorylated TACC3 in isolated spindle is consistent with the idea that the spindle localization of TACC3 is dependent on its phosphorylation (17, 18). Depletion of CHC caused a significant change in the abundance (up-regulated or down-regulated) of 121 out of 835 proteins (14%) compared with Luc siRNA-treated spindles (Fig. 2A and supplemental Table S3). Similar results were observed in CHC siRNA treated cells compared with untreated cells (supplemental Table S7 and supplemental Fig. S4). Consistent with these data, Luc siRNA treatment did not result in a statistically significant change compared with the untreated control (supplemental Table S8), indicating that Luc siRNA had no apparent effect on protein levels and/or spindle integrity. Thus, all subsequent analysis is discussed in the context of CHC siRNA versus Luc siRNA.
Fig. 2.
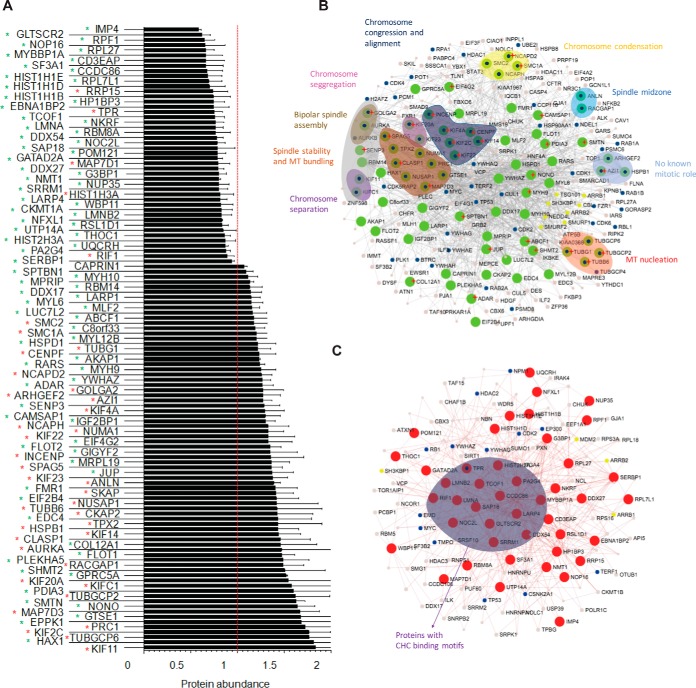
Depletion of clathrin causes a change in protein abundance of mitotic spindle proteins. A, Bar graph depicting total protein abundance of significantly altered proteins upon CHC knockdown. Data is presented as the ratio of CHC/Luc. Error bars represent S.E. n≥3, p < 0.05 (t test). Red asterisk, known spindle or spindle-associated proteins. Green asterisk, predicted spindle proteins. B, Network of significantly increased proteins upon CHC knock-down. Green nodes represent up-regulated proteins. Blue dots represent proteins involved in the cell cycle, yellow dots represent endocytic proteins. Groups of proteins involved in common mitotic functions are represented in the indicated colored balloon with matching colored text labels. Red start indicates those proteins with a putative clathrin binding motif. C, Network of significantly decreased proteins upon CHC knock-down. Red nodes represent down-regulated proteins. Blue dots represent proteins involved in the cell cycle, yellow dots represent endocytic proteins. Proteins with putative clathrin binding motifs are grouped into the purple balloon.
We next examined the protein interaction network of the 121 proteins that significantly changed in abundance following clathrin depletion (Fig. 2B and 2C) using NetworkAnalyst (38, 39). A significant number of up-regulated proteins (except for RRP15, nucleoprotein TPR, MAP7D1, and HIST1H3A) are known spindle proteins with key roles in processes that drive mitotic progression (Fig. 2B). Similar results were obtained by using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING v9.1; supplemental Table S9). In contrast, most of the down-regulated proteins are predicted spindle proteins with no known spindle role (Fig. 2C). The results show that depletion of clathrin causes an accumulation of proteins on the mitotic spindle that are associated with chromosome alignment and spindle assembly.
To gain insight into how clathrin might function with any of the proteins that were up- or down-regulated, we scanned the sequence of each protein for the presence of a clathrin-binding motif (L[IVLMF]X[IVLMF][DE]) using the Eukaryotic Linear Motif (ELM) resource (40). The results revealed that 43% (32 of 75 proteins) of up-regulated proteins and 24% (11 of 46 proteins) of down-regulated proteins contained at least one clathrin-binding motif (Table I). To test for specificity of the motif prediction, 1000 proteins from the HeLa proteome (41) were analyzed by ELM for the presence of a clathrin-binding motif. Of these, 27% of the 1000 proteins were found to contain at least one predicted clathrin-binding motif, illustrating that a significant proportion of the proteins that were up-regulated, but not down-regulated, are potential clathrin-binding proteins.
Table I. Proteins with clathrin-binding motifs as predicted by ELM.
| Protein name | Gene name | Protein | Change upon CHC depletion | No. of motifs | Amino acid position of clathrin-binding motif(s) in protein |
|---|---|---|---|---|---|
| Glioma tumor suppressor candidate region gene 2 protein | GLTSCR2 | Q9NZM5 | 0.62 | 1 | 86–90 |
| Coiled-coil domain-containing protein 86 | CCDC86 | Q9H6F5 | 0.67 | 1 | 32–36 |
| Nucleoprotein TPR | TPR | P12270 | 0.75 | 3 | 1008–1012; 328–332; 504–508 |
| Treacle protein | TCOF1 | E9PHK9 | 0.75 | 1 | 996–1000 |
| Prelamin-A/C;Lamin-A/C | LMNA | P02545 | 0.76 | 1 | 226–230 |
| ATP-dependent RNA helicase DDX54 | DDX54 | Q8TDD1 | 0.78 | 1 | 298–302 |
| Nucleolar complex protein 2 homolog | NOC2L | Q9Y3T9 | 0.78 | 1 | 365–369 |
| Transcriptional repressor p66-alpha | GATAD2A | B5MC40 | 0.79 | 1 | 387–391 |
| La-related protein 4 | LARP4 | Q71RC2 | 0.82 | 1 | 2–6 |
| Lamin-B2 | LMNB2 | J9JID7 | 0.83 | 2 | 203–207; 241–245 |
| Proliferation-associated protein 2G4 | PA2G4 | Q9UQ80 | 0.87 | 1 | 105–109 |
| Telomere-associated protein RIF1 | RIF1 | Q5UIP0 | 0.89 | 4 | 274–278; 429–433; 549–553; 750–754 |
| Spectrin beta chain, brain 1 | SPTBN1 | Q01082 | 1.09 | 3 | 1005–1009; 219–223; 560–564 |
| ATP-binding cassette sub-family F member 1 | ABCF1 | Q8NE71 | 1.17 | 1 | 472–476 |
| Structural maintenance of chromosomes protein 1A | SMC1A | Q14683 | 1.2 | 2 | 6–10; 978–982 |
| Tubulin gamma-1 chain;Tubulin gamma-2 chain | TUBG1;TUBG2 | P23258 | 1.22 | 1 | 66–70 |
| Centromere protein F | CENPF | P49454 | 1.23 | 5 | 1210–1214; 1751–1755; 1853–1857; 2268–2272; 784–788 |
| Myosin-9 | MYH9 | P35579 | 1.25 | 2 | 1426–1430; 1848–1852 |
| Condensin complex subunit 1 | NCAPD2 | Q15021 | 1.25 | 4 | 140–144; 176–180; 722–726; 938–942 |
| Double-stranded RNA-specific adenosine deaminase | ADAR | E7ENU4 | 1.27 | 1 | 377–381 |
| Golgin subfamily A member 2 | GOLGA2 | Q08379 | 1.27 | 1 | 504–508 |
| 5-azacytidine-induced protein 1 | AZI1 | Q9UPN4 | 1.27 | 1 | 99–103 |
| Sentrin-specific protease 3 | SENP3 | Q9H4L4 | 1.28 | 1 | 468–472 |
| Chromosome-associated kinesin KIF4A | KIF4A | O95239 | 1.29 | 3 | 182–186; 686–690; 920–924 |
| Calmodulin-regulated spectrin-associated protein 1 | CAMSAP1 | Q5T5Y3 | 1.3 | 3 | 26–30; 1233–1237; 1250–1254 |
| Kinesin-like protein KIF22 | KIF22 | Q14807 | 1.33 | 2 | 251–255; 443–447 |
| Eukaryotic translation initiation factor 4 gamma 2 | EIF4G2 | D3DQV9 | 1.33 | 2 | 116–120; 730–734 |
| Inner centromere protein | INCENP | Q9NQS7 | 1.36 | 1 | 31–35 |
| Sperm-associated antigen 5 | SPAG5 | Q96R06 | 1.36 | 3 | 312–316; 442–446; 522–526 |
| Junction plakoglobin | JUP | F5GWP8 | 1.37 | 1 | 315–319 |
| Tubulin beta-6 chain | TUBB6 | Q9BUF5 | 1.42 | 1 | 65–69 |
| Kinesin-like protein KIF14 | KIF14 | Q15058 | 1.44 | 1 | 1579–1583 |
| CLIP-associating protein 1 | CLASP1 | B7ZLX3 | 1.46 | 2 | 68–72; 1349–1353 |
| Collagen alpha-1(XII) chain | COL12A1 | Q99715 | 1.46 | 2 | 340–344; 2510–2514 |
| Serine hydroxymethyltransferase, mitochondrial | SHMT2 | P34897 | 1.51 | 1 | 381–385 |
| Kinesin-like protein KIF20A | KIF20A | O95235 | 1.55 | 1 | 798–802 |
| Protein disulfide-isomerase A3 | PDIA3 | P30101 | 1.6 | 1 | 47–51 |
| Gamma-tubulin complex component 2 | TUBGCP2 | Q9BSJ2 | 1.62 | 1 | 225–229 |
| Smoothelin | SMTN | P53814 | 1.62 | 2 | 583–587; 876–880 |
| Non-POU domain-containing octamer-binding protein | NONO | Q15233 | 1.64 | 1 | 274–278 |
| MAP7 domain-containing protein 3 | MAP7D3 | Q8IWC1 | 1.67 | 1 | 788–792 |
| Epiplakin | EPPK1 | P58107 | 1.68 | 9 | 103–107; 641–645; 916–920; 1602–1606; 2260–2264; 2794–2798; 3328–3332; 3862–3866; 4930–4934 |
| Kinesin-like protein KIF2C | KIF2C | Q99661 | 1.76 | 1 | 599–603 |
The Spindle Phosphoproteome Following Depletion of Clathrin
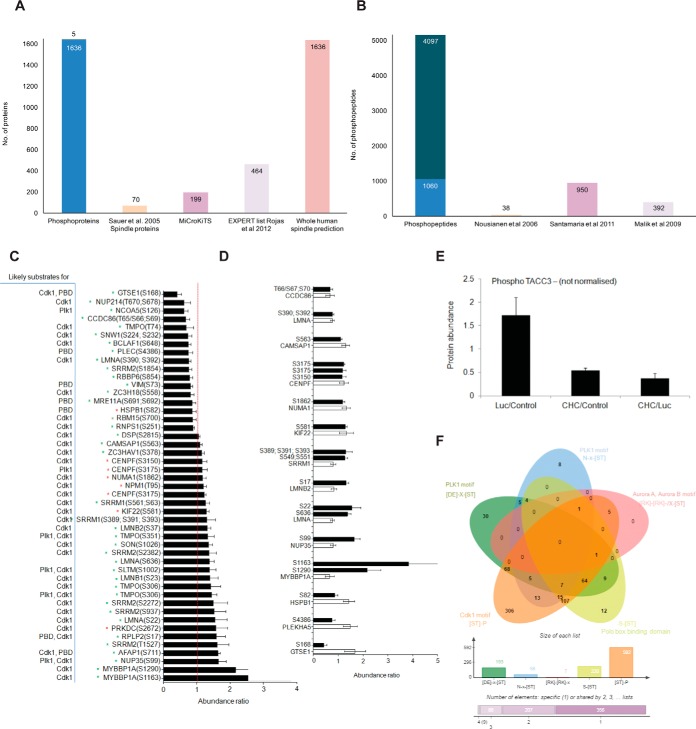
From the label-free (supplemental Table S10) and dimethyl labeling (supplemental Table S11) experiments described above, we identified 758 and 4784 phosphosites, respectively, following enrichment using TiO2 and fractionation by a high pH-reverse phase micro-column method. 385 were phosphosites identified in common between the two experimental methods, revealing a total of 5157 unique phosphosites identified. These collectively belonged to 1641 phosphoproteins, of which 70 are known spindle proteins (3), 199 are spindle/spindle-associated proteins according to MiCroKiTS, 464 are categorized to the “Expert” list and 1636 are classified as spindle-associated proteins using SPIP (37) (Fig. 3A; supplemental Table S12). We also manually compared all phosphopeptides (that contained the exact same residues within the tryptic peptide) to three other reported spindle phosphoproteomes (Fig. 3B) (42–44). We identified 4097 phosphopeptides that were unique to our spindle phosphoproteome. The other 1060 were found in previous studies. We quantified the change in abundance of 1259 phosphosites (identified in n>3 experimental replicates) in clathrin-depleted mitotic spindles (supplemental Table S11). The presence and abundance of at least two nonphosphopeptides within the same protein of each phosphopeptide were used to correct for the underlying change in protein abundance associated with each phosphopeptide. Phosphopeptides lacking corresponding protein abundance data were excluded from further analysis (supplemental Table S13). From a final list of 803 phosphosites that could be analyzed, 47 phosphosites had significantly altered site occupancy (p < 0.05; t test) in clathrin-depleted spindles, which included increased occupancy of 30 phosphosites and decreased occupancy of 17 phosphosites (Fig. 3C). The change in phosphorylation was then compared with those corresponding proteins whose abundance was changed significantly by clathrin depletion (Fig. 3D) to determine if the increase/decrease in phosphorylation was because of a corresponding significant change in protein abundance or to a phosphorylation event. Of the 47 phosphosites that had altered site occupancy, 27 had a corresponding change in protein abundance. The increase in phosphosite occupancy in the proteins SRRM1, LMNB2, LMNA, NUP35, and MYBBP1A, and decrease in the proteins HSPB1, PLEKHA5, and GTSE1, did not correlate with a significant change in the abundance of these proteins, indicating that these phosphorylation events are regulated by kinases/phosphatases. Although we were not able to detect any TACC3 nonphosphopeptides, we were able to detect TACC3 phosphosites. Despite not being able to normalize TACC3 phosphorylation based on protein abundance, we noted that the phosphosites quantified were significantly decreased in clathrin-depleted spindles compared with control spindles (Fig. 3E; supplemental Table S14), as previously reported (19).
Fig. 3.
Phospho-regulation upon CHC knock-down. A, Bar graph depicting the number of phosphoproteins that overlap between our study (Phosphoproteins) and proteins classified as localizing to the spindle by Sauer et al., 2005 (Sauer spindle list) (3), MiCroKiTS, the Expert list and predicted spindle proteins in the whole human proteome (37). B, Bar graph depicting the number of phosphosites that were identified in our study (Phosphosites) and those that were also identified in studies by (42, 43, 44). C, Bar graph depicting the normalized abundance of significantly altered phosphosites upon CHC knockdown. Data is presented as the ratio of CHC/Luc. Error bars represent S.E. n≥3, p < 0.05 (t test). Red asterisk, known spindle or spindle-associated proteins; Green asterisk, predicted spindle proteins. D, Bar graph depicting the normalized abundance of significantly altered phosphosites (white bars) among significantly altered proteins (black bars) upon CHC knockdown. Data is presented as the ratio of CHC/Luc. Error bars represent S.E. n≥3, p < 0.05 (t test). E, Total phosphoprotein abundance of TACC3 (nonnormalized), as determined by quantitative mass spectrometry. Error bars represent S.E. n≥3. w.r.t value of 1). F, Venn diagram depicting the overlap of consensus motifs for Plk1 (D/E-X-S/T), Aurora A/B kinases (R/K-R/K-X-S/T), Polo box-binding domain (S-S/T) and Cdk1 kinase (S/T-P) within the quantified phosphopeptides (803 normalized phosphopeptides). The size of each list is shown in the bar graph below.
We then analyzed our quantified data set of 803 phosphosites for the presence of a consensus site(s) for the mitotic kinases, Plk1, Aurora kinases and Cdk1 (supplemental Table S15). We used the ScanProsite tool (Prosite; ExPASy Bioinformatics resource Portal) to scan for the following motifs within the phosphopeptides: (1) D/E-X-S/T (Plk1), (2) R/K-R/K-X-S/T (Aurora A and B kinases), (3) S-S/T (Polo box-binding domain), and (4) S/T-P (Cdk1). A total of 616 unique phosphopeptides contained at least one consensus motif for Plk1, Cdk1 or Aurora kinases (Fig. 3F and supplemental Table S15). The majority of phosphosites contained at least one consensus motif for Cdk1. In line with this trend, 29 of 31 phosphosites that were up-regulated in CHC-depleted spindles were Cdk1 predicted substrates. Among the 17 down-regulated phosphosites, 10 were predicted Cdk1 substrates and five contained a polo box-binding domain or Plk1 motif including the phosphorylated residue. Collectively, these results indicate that clathrin function at the mitotic spindle is associated primarily with a small subset of Cdk1-dependent substrates.
Proteomic Validation and effect of Clathrin Depletion on Spindle Localization of Selected Proteins
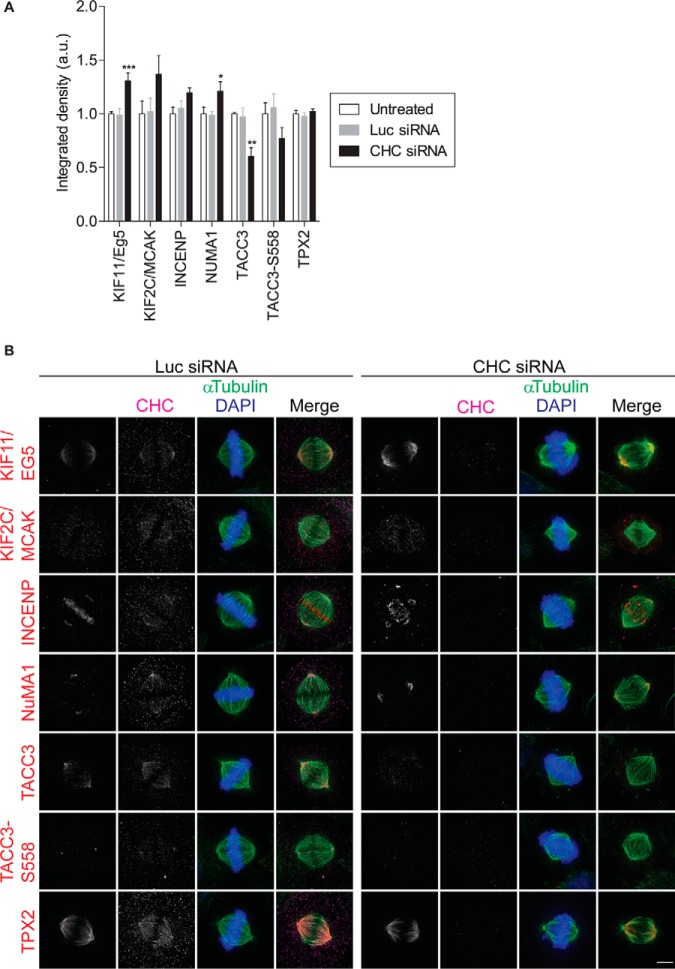
To experimentally verify our proteomics data set, we performed immunofluorescence microscopy, whereby we calculated the fluorescence intensity at the mitotic spindle of selected proteins whose abundance increased in clathrin-depleted spindles. This method was chosen as immunoblotting is not sufficiently sensitive and reliable to detect small but significant changes in protein abundance (45). We selected five critical spindle proteins for assessment by immunofluorescence microscopy: KIF11/Eg5, KIF2C/MCAK, INCENP, NUMA1, and TPX2. All five proteins assessed displayed their previously reported mitotic localization: spindle pole (Kif11/Eg5, INCENP, TPX2 and NUMA1), spindle microtubules (Kif11/Eg5, Kif2C/MCAK, TACC3, and TPX2) or both (Kif11/Eg5 and TPX2) in untreated and Luc siRNA-treated cells (Fig. 4). This provides additional validation of our spindle proteome. In line with our MS results, the fluorescence intensity of KIF11/Eg5, KIF2C/MCAK, INCENP, and NUMA1 increased at the mitotic spindle in clathrin-depleted metaphase cells compared with control cells (Fig. 4). We also assessed the enrichment of TACC3 and phosphorylated TACC3-S558 at the spindle as well as its spindle localization. Both TACC3 and TACC3-S558 decorated the spindle poles as well as the spindle fibers, as previously reported (19) and again, consistent with the lack of detection of these proteins in isolated spindles by MS, the spindle localization of both was reduced in clathrin-depleted cells (Fig. 4). We were unable to detect a significant increase in TPX2 (Fig. 4). Overall, the proteomics and immunofluorescence microscopy data was similar.
Fig. 4.
Validation of protein localization by immunofluorescence. A, Staining quantification of the selected proteins at the mitotic spindle. The graph (mean ± S.E.) shows the relative integrated fluorescence density of the indicated proteins. In total, n≥33 cells assessed per protein from three independent experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.0005 (One-way ANOVA). B, Representative immunofluorescence images showing the localization and abundance of KIF11/Eg5, KIF2C/MCAK, INCENP, NUMA1, TPX2, TACC3 and pTACC3-S558 in CHC siRNA- and control Luc siRNA-treated metaphase cells. Untreated HeLa cells and those treated with siRNA against Luc or CHC were fixed and labeled with the indicated antibodies. Maximum projections of deconvolved z series are shown. Scale bar: 5 μm.
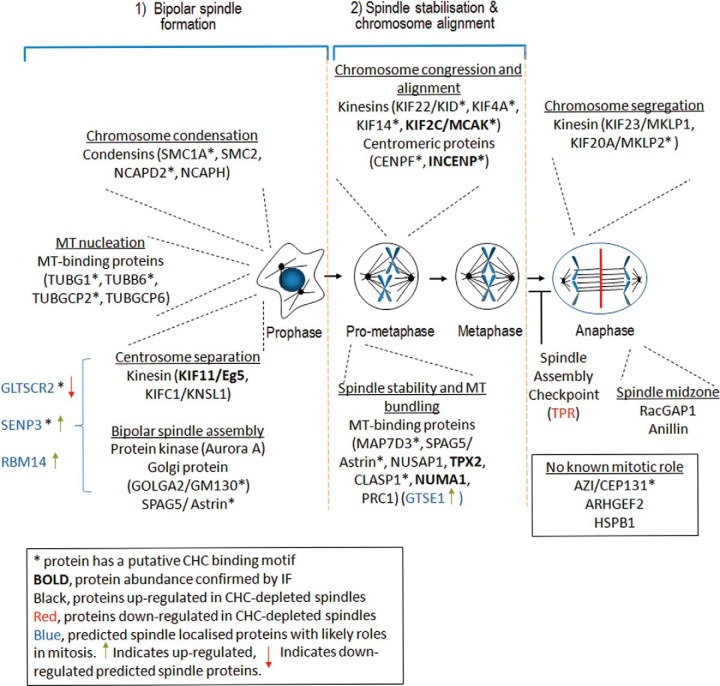
To investigate the link between these proteins and clathrin, we also determined if the specific spindle localization of all proteins assessed (KIF11/Eg5, KIF2C/MCAK, TACC3, TACC3-S558, INCENP, NUMA1, and TPX2) was perturbed in clathrin-depleted mitotic spindles (Fig. 4). Only the distribution of INCENP at the metaphase plate was slightly impaired, consistent with disruption of chromosome alignment. Among this subset of spindle proteins, clathrin only contributes to the spindle recruitment of TACC3. Instead, this data suggests that clathrin cooperates with most spindle proteins once they have arrived at the spindle to participate in multiple spindle functions including bipolar spindle formation, spindle stability, and chromosome alignment (Fig. 5).
Fig. 5.
Processes regulated by clathrin during mitosis. Clathrin has a moonlighting role during mitosis, whereby it stabilizes the mitotic spindle. Depicted is a global overview of the mitotic processes regulated by proteins whose abundance at the mitotic spindle is affected by the depletion of clathrin. The absence of clathrin disrupts bipolar spindle formation, spindle stabilization, chromosome alignment and chromosome segregation. We hypothesize that clathrin may also have a role in regulating the abundance of spindle proteins by transporting them as cargo between the spindle and cytoplasm.
DISCUSSION
In this study, we reveal the proteome and phosphoproteome of the mitotic spindle in the presence and absence of clathrin. Functional pathway analysis of the proteins disrupted by clathrin depletion confirmed the reported role of clathrin in stabilizing the mitotic spindle but also revealed a novel second role for clathrin in bipolar spindle formation. The findings provide insights into the molecular pathways that clathrin is associated with during these processes.
We report a comprehensive coverage of the spindle proteome and spindle phosphoproteome. A complete identification of the spindle proteome/phosphoproteome is challenging for the systems biology field because its assembly requires many distinct structural and regulatory proteins and once established it is a highly dynamic structure with many proteins only associating transiently in a spatiotemporal manner as the cell proceeds through mitosis. Nevertheless, large-scale proteomics experimental approaches have contributed substantially to the characterization of many spindle components, with the first spindle proteome reporting 795 proteins (containing 126 known spindle proteins (3)) and a spindle phosphoproteome of 736 phosphosites (containing 312 phosphosites from known spindle proteins) (43). We report a spindle proteome that consists of 3046 proteins (351 previously identified) and a spindle phosphoproteome of 5157 phosphosites (269 from known spindle proteins). Specifically, we identified 4097 new phosphosites in known and predicted spindle proteins. Of the 2695 proteins and 4888 phosphorylation sites that have not been previously reported to be spindle-associated, most are likely bona fide spindle components because >95% are predicted spindle proteins. In support of this idea, by using a candidate-based approach, we identified the telomere-associated protein, RIF1, as a component of the spindle, in which it localizes along microtubules. In agreement with a role for clathrin in stabilizing the mitotic spindle, the abundance of spindle components involved in this process was disrupted by clathrin depletion, including 9.2% of proteins and 2.2% of phosphorylation sites. This small global effect is expected given that clathrin acts to stabilize a subset of microtubules within the kinetochore fibers (10). To date, this role is known to involve its association with phosphorylated TACC3 and ch-TOG (11, 12). Our quantitative mass spectrometry findings vastly extend the link between clathrin and spindle stabilization because the abundance of the following groups of proteins involved in spindle stability was increased in clathrin-depleted spindles: spindle stability and microtubule bundling (MAP7D3, SPAG5/astrin, NUSAP1, TPX2, CLASP1, NUMA1, NUMA1 phosphorylated at S1862, PRC1, RBM14, GTSE1, and HAX1), chromosome congression and alignment (KIF22/KID, KIF22 phosphorylated at S581, KIF4A, KIF14, KIF2C/MCAK, CENPF, CENPF phosphorylated at S3150 and S3175, INCENP, and HAX1), as well as chromosome segregation (KIF23/MKLP1, KIF20A/MKLP2, GLTSCR2, SENP3, and RBM14) (Fig. 5). Overexpression of many of these such as NUMA1 (46), TPX2 (47), and SPAG5/astrin (48) leads to mitotic phenotypes consistent with clathrin depletion (10, 12) and include an increase in metaphase plate width, loss of spindle fibers, chromosome misalignment and mis-segregation. In contrast, we observed a reduction in TACC3 and phospho-TACC3 spindle localization in clathrin-depleted spindles, suggesting that clathrin contributes to spindle stabilization via additional pathways that do not involve TACC3. Another mitotic regulator that stabilizes kinetochore fibers is HURP/DLGAP5 (49). HURP/DLGAP5 was detected in our study but was not reported in the final quantitative results because it was detected in only two biological replicates. Nevertheless, clathrin depletion resulted in a 1.6 ± 0.47 (mean ± S.D.) fold increase in HURP levels. The overall trend in increased protein levels at the mitotic spindle suggests compensation for the lack of clathrin to aid in spindle stabilization.
One of the major findings of our study is that clathrin depletion increased the abundance of spindle proteins associated with a role in bipolar spindle formation, which occurs at the onset of mitosis. These proteins include microtubule-binding proteins, kinesin motor proteins, condensin components, and centromeric proteins. Functional pathway analysis revealed that they play key roles in centrosome separation (KIF11/Eg5 and KIFC2/KNSL1), microtubule nucleation (TUBG1, TUBB6, TUBGCP2, and TUBGCP6), chromosome condensation (SMC1A, SMC2, NCAPD2, and, NCAPH), and bipolar spindle assembly (Aurora A and GM130). Condensin I and II aid the displacement of excess INCENP from the arms of mitotic chromosomes for subsequent mitotic stages (37, 50), and thus the observed increase in the condensin subunits (SMC2, NCAPD2, and NCAPH) as well as in SMC1A might be because of the increase in INCENP. The centrosome protein AZI1/CEP131, has been classified as a spindle component but its mitotic role is unknown. Its spindle abundance also increased following clathrin depletion. The centrosome localization of AZI1/CEP131 suggests it may play a role in bipolar spindle formation and thus we hypothesize that this may involve co-operation with clathrin.
The depletion of clathrin also had a significant effect on the abundance of an additional 89 proteins (44 increased and 45 decreased) and 47 phosphorylation sites (27 increased and 20 decreased) in proteins that have not previously been classified as locating to the spindle. However, all of these are predicted spindle proteins. Four proteins can be linked to mitosis: RBM14, GTSE1, SENP3, and GLTSCR2 (Fig. 5). In an analogous manner to known spindle proteins, the spindle levels of all proteins, except GLTSCR2, increased following CHC depletion. RBM14 interacts with the centriole assembly factor STIL to prevent assembly of centriolar protein complexes, thus ensuring proper spindle assembly (51). GTSE1 is a microtubule plus-end tracking protein (+TIP) that binds directly to the microtubules. Its depletion results in disorganized microtubules (52). SENP3 is a SUMO isopeptidase that during mitosis interacts with Borealin, a chromosomal passenger complex protein involved in regulating chromosome congression, the SAC, and cytokinesis (53). This interaction results in desumoylation of Borealin. Similarly, SENP3 interacts with and desumoylates nucleophosmin (NPM1) (54), which locates to the mitotic centrosome and participates in centrosome and spindle formation as well as chromosome congression (55). GLTSCR2 is also an upstream negative regulator of NPM1, whereby it induces its nucleoplasmic translocation, leading to increased proteasomal-mediated degradation of NPM1 (56). The level of GLTSCR2 decreased in CHC-depleted spindles, and consistent with the role of GLTSCR2, NPM1 abundance was unaffected. GLTSCR2 may play a role in regulating NPM1 centrosome levels and therefore contribute to regulating bipolar spindle formation. Both GLTSCR2 and SENP3 contain putative clathrin-binding motifs and thus their localization/function at the spindle may be regulated by clathrin directly.
Our analysis did not show a significant effect of clathrin depletion on the abundance of SAC components or its regulators. This is surprising given that clathrin depletion activates the SAC (20). SAC activation would have resulted in an increase in the spindle abundance of the kinetochore components CENP-E and dynein, their target components ZW10 and ROD, and spindle checkpoint proteins such as Mad1, Mad2, BubR1 and Cdc20 (1). There are several reasons why we most likely were unable to observe a response in SAC proteins: (1) only MAD1 and ZW10 were detectable, however, they were not quantifiable because they were not detected in both control and clathrin-depleted samples or were detected in <3 biological replications; and (2) clathrin depletion does not cause a complete block in chromosome segregation, but rather a delay, and thus the time point taken for analysis may have prevented detection of SAC components. This incomplete block is likely because of redundancy, whereby alternate spindle assembly and stability pathways are up-regulated. For example, the spindle abundance of the microtubule-binding protein TPX2 is involved in spindle assembly. Its abundance is increased following CHC depletion and excess TPX2 leads to spontaneous microtubule assembly (57, 58). Future experiments that deplete CHC in combination with proteins that are up-regulated will determine if they function in the same or parallel pathways.
Several kinases and phosphatases play an important role in regulating protein phosphorylation throughout mitosis to ensure key events occur in a spatially and temporally regulated manner. Specifically, Aurora A kinase is involved in bipolar spindle assembly and subsequently in spindle stability (59, 60). Two key substrates are TPX2 and TACC3. Aurora A spindle abundance increased significantly following CHC depletion. However, this did not appear to result in an increase in phosphorylation of Aurora A-mediated substrates. No phosphorylation sites that changed in spindle abundance following CHC depletion were predicted Aurora A kinase substrates. Thus, we hypothesize that clathrin functions downstream of Aurora A and does not regulate its activity. However, the abundance of two Aurora A-binding partners, SPAG5/astrin and TPX2, increased on CHC-depleted spindles. Both are required for directing Aurora A to the mitotic spindle (61, 62). Consistent with this idea, Aurora A spindle levels increased following CHC-depletion. Thus, our data suggests that clathrin does not appear to be important for regulating Aurora A kinase activity but instead may contribute to regulating the levels of Aurora A at the spindle via its binding partners, SPAG5 and TPX2, thereby, contributing to spindle assembly and stability. This may be by direct interaction, at least with SPAG5, which contains three putative clathrin-binding motifs.
It was not surprising that the majority of phosphorylation sites that changed in abundance at the spindle following CHC depletion were predicted to be a substrate of Cdk1 and a few contained a putative motif for Plk1 or for PBD, which is a phospho-peptide-binding motif for Plk1. The availability of data resources that cater specifically to phosphatases is limited and thus we were unable to scan the phosphorylation sites that changed in spindle abundance for putative phosphatase motifs. However, given that the majority are SP/TP motifs, potential phosphatases with known roles in mitosis include Cdc14A, Cdc14B, PP1, PP2A, and Ptpcd-1 (63). These could be candidate phosphatases for dephosphorylating those proteins whose phosphorylation decreased in CHC-depleted spindles.
The only reported clathrin-interacting partner during metaphase is TACC3, and together they form a complex with either ch-TOG or GTSE1 (11, 19, 64, 65). This study identified 43 potential novel clathrin-binding partners (during metaphase based on the observations that their spindle abundance is dependent on clathrin and they contain at least one clathrin-binding motif (L[IVLMF]X[IVLMF][DE]), suggesting that clathrin participates in key mitotic functions via direct interactions. We reasoned that proteins that decreased in abundance in clathrin-depleted spindles represent interacting partners that are dependent on clathrin for their spindle localization. In contrast, the up-regulated proteins may associate with clathrin once located at the spindle to regulate specific functions. Twelve of the down-regulated proteins contained at least one clathrin-binding motif. Of these, only TPR is a known spindle protein, although the others are predicted spindle proteins. TPR is a component of the nuclear pore complex and binds to the SAC proteins MAD1 and MAD2. Depletion of TPR decreases the level of Mad1 at KTs, correlating with an inability to activate Mad2, which is required for inhibiting APC (Cdc20) and thus satisfying the SAC (66). Although we did not observe an effect on SAC proteins the decrease in TPR suggests a potential mechanism for how clathrin depletion activates the SAC. TPR is predicted to contain three clathrin-binding motifs and thus clathrin may directly associate with and tether TPR at the spindle. In contrast, 31 proteins that increased in spindle abundance following clathrin depletion contained at least one putative clathrin-binding motif and 15 of these were spindle-associated proteins. Analysis of the localization of the known clathrin-binding partner, TACC3, as well as KIF2C/MCAK and INCENP, which contain putative clathrin-binding motifs, revealed that the spindle localization of TACC3 and INCENP is dependent on clathrin. In contrast, the localization of KIF11/Eg5, NUMA1, and TPX2, which do not have a clathrin-binding motif, was unaffected. Clathrin is not exclusively localized to the spindle in metaphase cells. In the cytoplasm, it can form “classical” clathrin cages that sequester a pool of mitotic proteins such as TACC3, ch-TOG and GTSE1 (67). Thus, it is possible that clathrin also participates in a number of mitotic functions by binding spindle proteins such as cargo on clathrin-coated vesicles to traffic proteins toward the spindle (down-regulated in CHC-depleted spindles) or from the spindle (up-regulated in CHC-depleted spindles). This suggests that in addition to its role at the spindle, clathrin also contributes to regulating the spatial and temporal accumulation of spindle proteins for efficient mitotic progression.
The findings clearly demonstrate that we have moved well beyond generating a spindle components list. The Plk-dependent spindle proteome and spindle phosphoproteome have been identified and provide a solid platform to investigate and improve our understanding of the signaling pathways that Plk1 is associated with (44). Here, we further exemplify the power of using quantitative mass spectrometry by characterizing the clathrin-dependent spindle proteome and phosphoproteome. The findings provide a solid foundation for potential new mitotic roles for clathrin that have not previously been identified using standard molecular biology techniques, such as siRNA-mediated depletion and ectopic expression of mutants. Specifically, we show that the mitotic role of clathrin is not only in spindle stabilization but is also linked to a large number of mitotic processes spanning mitotic entry through to chromosome segregation. The data also provides substantial insight into clathrin-associated signaling pathways during metaphase.
Supplementary Material
Acknowledgments
We thank Dr. Mark Graham for advice and discussions on data analysis and processing as well as critical reading of the manuscript. We thank Dr. Kasper-Engholm Keller for help and advice with technical aspects of mass spectrometry and related trouble-shooting. Usage of the DeltaVision microscope was carried out within the Cell Imaging Facility at Westmead Millennium Institute/Westmead Research Hub.
Footnotes
Author Contribution: Cell culture, scale-up, sample preparation, proteomics, mass spectrometry, associated data analysis and manuscript writing was carried out by S.R.R. S.L.P. established the spindle isolation protocol in the laboratory. Immunofluroscence, microscopy and related image processing was carried out by N.F.R. MC contributed to experimental design, co-ordination of the project, data analysis and manuscript writing. P.J.R. contributed to experimental design and manuscript writing. All authors read and approved the manuscript.
* This work was supported by grants from the National Health and Medical Research Council (NH&MRC, APP1022218) of Australia (MC) and NH&MRC Career Development Award Fellowships (MC, 477104 & APP1069264) and for equipment from the Australian Cancer Research Foundation, the Ramaciotti Foundation and the Cancer Institute NSW.
 This article contains supplemental material.
This article contains supplemental material.
The authors declare no conflict of interest.
1 The abbreviations used are:
- SAC
- Spindle Assembly Checkpoint
- CHC
- Clathrin Heavy Chain
- Luc
- Luciferase
- TD
- Terminal Domain
- NUMA
- Nuclear mitotic apparatus protein 1
- TPX2
- Targeting protein for Xklp2
- TACC3
- Transforming acidic coiled-coil-containing protein 3
- ch- TOG
- Colonic and hepatic tumor overexpressed gene protein (CKAP5-Cytoskeleton-associated protein 5)
- CLASP1
- CLIP-associating protein 1
- TiO2
- Titanium dioxide
- KIF11
- Kinesin-like protein KIF11
- KIF2C
- Kinesin-like protein KIF2C
- INCENP
- Inner centromere protein
- GTSE1
- G2 and S phase-expressed protein 1.
REFERENCES
- 1. Musacchio A., and Salmon E. D. (2007) The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 2. Huang Z., Ma L., Wang Y., Pan Z., Ren J., Liu Z., and Xue Y. (2015) MiCroKiTS 4.0: a database of midbody, centrosome, kinetochore, telomere and spindle. Nucleic Acids Res. 43, D328–D334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sauer G., Korner R., Hanisch A., Ries A., Nigg E. A., and Sillje H. H. (2005) Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics 4, 35–43 [DOI] [PubMed] [Google Scholar]
- 4. Compton D. A. (2000) Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95–114 [DOI] [PubMed] [Google Scholar]
- 5. Sharp D. J., Rogers G. C., and Scholey J. M. (2000) Microtubule motors in mitosis. Nature 407, 41–47 [DOI] [PubMed] [Google Scholar]
- 6. Wittmann T., Hyman A., and Desai A. (2001) The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 3, E28–E34 [DOI] [PubMed] [Google Scholar]
- 7. Nigg E. A. (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32 [DOI] [PubMed] [Google Scholar]
- 8. Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S. C., Kirchhausen T., and Walz T. (2004) Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432, 573–579 [DOI] [PubMed] [Google Scholar]
- 9. Royle S. J. (2006) The cellular functions of clathrin. Cell Mol. Life Sci. 63, 1823–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Royle S. J., Bright N. A., and Lagnado L. (2005) Clathrin is required for the function of the mitotic spindle. Nature 434, 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Booth D. G., Hood F. E., Prior I. A., and Royle S. J. (2011) A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30, 906–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin C. H., Hu C. K., and Shih H. M. (2010) Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J. Cell Biol. 189, 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerald N. J., Damer C. K., O'Halloran T. J., and De Lozanne A. (2001) Cytokinesis failure in clathrin-minus cells is caused by cleavage furrow instability. Cell Motil. Cytoskeleton 48, 213–223 [DOI] [PubMed] [Google Scholar]
- 14. Niswonger M. L., and O'Halloran T. J. (1997) A novel role for clathrin in cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 94, 8575–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gergely F., Draviam V. M., and Raff J. W. (2003) The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17, 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kinoshita K., Noetzel T. L., Pelletier L., Mechtler K., Drechsel D. N., Schwager A., Lee M., Raff J. W., and Hyman A. A. (2005) Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 170, 1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LeRoy P. J., Hunter J. J., Hoar K. M., Burke K. E., Shinde V., Ruan J., Bowman D., Galvin K., and Ecsedy J. A. (2007) Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res. 67, 5362–5370 [DOI] [PubMed] [Google Scholar]
- 18. Giet R., McLean D., Descamps S., Lee M. J., Raff J. W., Prigent C., and Glover D. M. (2002) Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156, 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hood F. E., Williams S. J., Burgess S. G., Richards M. W., Roth D., Straube A., Pfuhl M., Bayliss R., and Royle S. J. (2013) Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J. Cell Biol. 202, 463–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith C. M., Haucke V., McCluskey A., Robinson P. J., and Chircop M. (2013) Inhibition of clathrin by pitstop 2 activates the spindle assembly checkpoint and induces cell death in dividing HeLa cancer cells. Mol. Cancer 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hinrichsen L., Harborth J., Andrees L., Weber K., and Ungewickell E. J. (2003) Effect of clathrin heavy chain- and alpha-adaptin-specific small inhibitory RNAs on endocytic accessory proteins and receptor trafficking in HeLa cells. J. Biol. Chem. 278, 45160–45170 [DOI] [PubMed] [Google Scholar]
- 22. Motley A., Bright N. A., Seaman M. N., and Robinson M. S. (2003) Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162, 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Royle S. J., and Lagnado L. (2006) Trimerisation is important for the function of clathrin at the mitotic spindle. J. Cell Sci. 119, 4071–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Glover D. M., Leibowitz M. H., McLean D. A., and Parry H. (1995) Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81, 95–105 [DOI] [PubMed] [Google Scholar]
- 25. Hoar K., Chakravarty A., Rabino C., Wysong D., Bowman D., Roy N., and Ecsedy J. A. (2007) MLN8054, a small-molecule inhibitor of Aurora A, causes spindle pole and chromosome congression defects leading to aneuploidy. Mol. Cell. Biol. 27, 4513–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katayama H., Zhou H., Li Q., Tatsuka M., and Sen S. (2001) Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J. Biol. Chem. 276, 46219–46224 [DOI] [PubMed] [Google Scholar]
- 27. Marumoto T., Honda S., Hara T., Nitta M., Hirota T., Kohmura E., and Saya H. (2003) Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J. Biol. Chem. 278, 51786–51795 [DOI] [PubMed] [Google Scholar]
- 28. Sasai K., Parant J. M., Brandt M. E., Carter J., Adams H. P., Stass S. A., Killary A. M., Katayama H., and Sen S. (2008) Targeted disruption of Aurora A causes abnormal mitotic spindle assembly, chromosome misalignment and embryonic lethality. Oncogene 27, 4122–4127 [DOI] [PubMed] [Google Scholar]
- 29. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., and Jorgensen T. J. D. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 30. Bardou P., Mariette J., Escudie F., Djemiel C., and Klopp C. (2014) jvenn: an interactive Venn diagram viewer. BMC. Bioinformatics. 15, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sillje H. H., and Nigg E. A. (2006) Purification of mitotic spindles from cultured human cells. Methods 38, 25–28 [DOI] [PubMed] [Google Scholar]
- 34. Megger D. A., Pott L. L., Ahrens M., Padden J., Bracht T., Kuhlmann K., Eisenacher M., Meyer H. E., and Sitek B. (2014) Comparison of label-free and label-based strategies for proteome analysis of hepatoma cell lines. Biochim. Biophys. Acta 1844, 967–976 [DOI] [PubMed] [Google Scholar]
- 35. Patel V. J., Thalassinos K., Slade S. E., Connolly J. B., Crombie A., Murrell J. C., and Scrivens J. H. (2009) A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome. Res. 8, 3752–3759 [DOI] [PubMed] [Google Scholar]
- 36. Pichler P., Kocher T., Holzmann J., Mazanek M., Taus T., Ammerer G., and Mechtler K. (2010) Peptide labeling with isobaric tags yields higher identification rates using iTRAQ 4-plex compared to TMT 6-plex and iTRAQ 8-plex on LTQ Orbitrap. Anal. Chem. 82, 6549–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rojas A. M., Santamaria A., Malik R., Jensen T. S., Korner R., Morilla I., de, Juan D., Krallinger M., Hansen D. A., Hoffmann R., Lees J., Reid A., Yeats C., Wehner A., Elowe S., Clegg A. B., Brunak S., Nigg E. A., Orengo C., Valencia A., and Ranea J. A. (2012) Uncovering the molecular machinery of the human spindle–an integration of wet and dry systems biology. PLoS. One. 7, e31813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xia J., Benner M. J., and Hancock R. E. (2014) NetworkAnalyst–integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 42, W167–W174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xia J., Gill E. E., and Hancock R. E. (2015) NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 10, 823–844 [DOI] [PubMed] [Google Scholar]
- 40. Dinkel H., Van Roey K., Michael S., Davey N. E., Weatheritt R. J., Born D., Speck T., Kruger D., Grebnev G., Kuban M., Strumillo M., Uyar B., Budd A., Altenberg B., Seiler M., Chemes L. B., Glavina J., Sanchez I. E., Diella F., and Gibson T. J. (2014) The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 42, D259–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo X., Trudgian D. C., Lemoff A., Yadavalli S., and Mirzaei H. (2014) Confetti: a multiprotease map of the HeLa proteome for comprehensive proteomics. Mol. Cell. Proteomics 13, 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Malik R., Lenobel R., Santamaria A., Ries A., Nigg E. A., and Korner R. (2009) Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome. Res. 8, 4553–4563 [DOI] [PubMed] [Google Scholar]
- 43. Nousiainen M., Sillje H. H., Sauer G., Nigg E. A., and Korner R. (2006) Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U.S.A. 103, 5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santamaria A., Wang B., Elowe S., Malik R., Zhang F., Bauer M., Schmidt A., Sillje H. H., Korner R., and Nigg E. A. (2011) The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics 10, M110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aebersold R., Burlingame A. L., and Bradshaw R. A. (2013) Western blots versus selected reaction monitoring assays: time to turn the tables? Mol. Cell. Proteomics 12, 2381–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bruning-Richardson A., Bond J., Alsiary R., Richardson J., Cairns D. A., McCormac L., Hutson R., Burns P. A., Wilkinson N., Hall G. D., Morrison E. E., and Bell S. M. (2012) NuMA overexpression in epithelial ovarian cancer. PLoS. One. 7, e38945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iyer J., and Tsai M. Y. (2012) A novel role for TPX2 as a scaffold and co-activator protein of the Chromosomal Passenger Complex. Cell Signal. 24, 1677–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yuan J., Li M., Wei L., Yin S., Xiong B., Li S., Lin S. L., Schatten H., and Sun Q. Y. (2009) Astrin regulates meiotic spindle organization, spindle pole tethering and cell cycle progression in mouse oocytes. Cell Cycle 8, 3384–3395 [DOI] [PubMed] [Google Scholar]
- 49. Sillje H. H., Nagel S., Korner R., and Nigg E. A. (2006) HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16, 731–742 [DOI] [PubMed] [Google Scholar]
- 50. Bakhrebah M., Zhang T., Mann J. R., Kalitsis P., and Hudson D. F. (2015) Disruption of a conserved CAP-D3 threonine alters condensin loading on mitotic chromosomes leading to chromosome hypercondensation. J. Biol. Chem. 290, 6156–6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shiratsuchi G., Takaoka K., Ashikawa T., Hamada H., and Kitagawa D. (2015) RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. EMBO J. 34, 97–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scolz M., Widlund P. O., Piazza S., Bublik D. R., Reber S., Peche L. Y., Ciani Y., Hubner N., Isokane M., Monte M., Ellenberg J., Hyman A. A., Schneider C., and Bird A. W. (2012) GTSE1 is a microtubule plus-end tracking protein that regulates EB1-dependent cell migration. PLoS. One. 7, e51259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Klein U. R., Haindl M., Nigg E. A., and Muller S. (2009) RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol. Biol. Cell 20, 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Haindl M., Harasim T., Eick D., and Muller S. (2008) The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Amin M. A., Matsunaga S., Uchiyama S., and Fukui K. (2008) Nucleophosmin is required for chromosome congression, proper mitotic spindle formation, and kinetochore-microtubule attachment in HeLa cells. FEBS Lett. 582, 3839–3844 [DOI] [PubMed] [Google Scholar]
- 56. Kim J. Y., Cho Y. E., An Y. M., Kim S. H., Lee Y. G., Park J. H., and Lee S. (2015) GLTSCR2 is an upstream negative regulator of nucleophosmin in cervical cancer. J. Cell Mol. Med. 19, 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le, Bot N., Vernos I., Karsenti E., and Mattaj I. W. (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83–93 [DOI] [PubMed] [Google Scholar]
- 58. Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., and Zheng Y. (2003) A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5, 242–248 [DOI] [PubMed] [Google Scholar]
- 59. Hochegger H., Hegarat N., and Pereira-Leal J. B. (2013) Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open. Biol. 3, 120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barr A. R., and Gergely F. (2007) Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120, 2987–2996 [DOI] [PubMed] [Google Scholar]
- 61. Du J., Jablonski S., Yen T. J., and Hannon G. J. (2008) Astrin regulates Aurora-A localization. Biochem. Biophys. Res. Commun. 370, 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kufer T. A., Sillje H. H., Korner R., Gruss O. J., Meraldi P., and Nigg E. A. (2002) Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qian J., Winkler C., and Bollen M. (2013) 4D-networking by mitotic phosphatases. Curr. Opin. Cell Biol. 25, 697–703 [DOI] [PubMed] [Google Scholar]
- 64. Cheeseman L. P., Booth D. G., Hood F. E., Prior I. A., and Royle S. J. (2011) Aurora A kinase activity is required for localization of TACC3/ch-TOG/clathrin inter-microtubule bridges. Commun. Integr. Biol. 4, 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., and Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee S. H., Sterling H., Burlingame A., and McCormick F. (2008) Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22, 2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Borner G. H., Antrobus R., Hirst J., Bhumbra G. S., Kozik P., Jackson L. P., Sahlender D. A., and Robinson M. S. (2012) Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197, 141–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jiang K., Toedt G., Montenegro, Gouveia S., Davey N. E., Hua S., van, der, Vaart B., Grigoriev I., Larsen J., Pedersen L. B., Bezstarosti K., Lince-Faria M., Demmers J., Steinmetz M. O., Gibson T. J., and Akhmanova A. (2012) A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800–1807 [DOI] [PubMed] [Google Scholar]
- 69. Sillje H. H., and Nigg E. A. (2006) Purification of mitotic spindles from cultured human cells. Methods 38, 25–28 [DOI] [PubMed] [Google Scholar]
- 70. Boersema P. J., Aye T. T., van Veen T. A., Heck A. J., and Mohammed S. (2008) Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics. 8, 4624–4632 [DOI] [PubMed] [Google Scholar]
- 71. Boersema P. J., Taouatas N., Altelaar A. F., Gouw J. W., Ross P. L., Pappin D. J., Heck A. J., and Mohammed S. (2009) Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol. Cell Proteomics. 8, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.