Abstract
Despite 40 years of control efforts, onchocerciasis (river blindness) remains one of the most important neglected tropical diseases, with 17 million people affected. The etiological agent, Onchocerca volvulus, is a filarial nematode with a complex lifecycle involving several distinct stages in the definitive host and blackfly vector. The challenges of obtaining sufficient material have prevented high-throughput studies and the development of novel strategies for disease control and diagnosis. Here, we utilize the closest relative of O. volvulus, the bovine parasite Onchocerca ochengi, to compare stage-specific proteomes and host-parasite interactions within the secretome. We identified a total of 4260 unique O. ochengi proteins from adult males and females, infective larvae, intrauterine microfilariae, and fluid from intradermal nodules. In addition, 135 proteins were detected from the obligate Wolbachia symbiont. Observed protein families that were enriched in all whole body extracts relative to the complete search database included immunoglobulin-domain proteins, whereas redox and detoxification enzymes and proteins involved in intracellular transport displayed stage-specific overrepresentation. Unexpectedly, the larval stages exhibited enrichment for several mitochondrial-related protein families, including members of peptidase family M16 and proteins which mediate mitochondrial fission and fusion. Quantification of proteins across the lifecycle using the Hi-3 approach supported these qualitative analyses. In nodule fluid, we identified 94 O. ochengi secreted proteins, including homologs of transforming growth factor-β and a second member of a novel 6-ShK toxin domain family, which was originally described from a model filarial nematode (Litomosoides sigmodontis). Strikingly, the 498 bovine proteins identified in nodule fluid were strongly dominated by antimicrobial proteins, especially cathelicidins. This first high-throughput analysis of an Onchocerca spp. proteome across the lifecycle highlights its profound complexity and emphasizes the extremely close relationship between O. ochengi and O. volvulus. The insights presented here provide new candidates for vaccine development, drug targeting and diagnostic biomarkers.
Despite 40 years of vector control and mass drug administration programs, onchocerciasis (river blindness) remains one of the most important of the neglected tropical diseases, infecting nearly 17 million people in sub-Saharan Africa (1). The disease is caused by a filarial nematode, Onchocerca volvulus, which resides within subcutaneous nodules and achieves a reproductive lifespan of >10 years (2). An adult female worm releases ∼1000 first-stage larvae (microfilariae, Mf)1 per day (3), which migrate to the skin and eyes, ultimately causing onchodermatitis and ocular pathology. Recent estimates of disease burden indicate that 4.2 million people are affected by severe itching, 746,000 experience poor vision and 265,000 individuals are rendered blind, resulting in a total global morbidity of almost 1.2 million years lived with disability (1, 4).
Historically, vector control was implemented on a massive scale, but the mainstay of current control efforts is a single anthelminthic, ivermectin, which suppresses microfilaridermia for several months following a single annual dose but does not kill the adult worms (5). However, resistance to ivermectin may be emerging in West Africa (6, 7), and this drug is contraindicated in individuals heavily co-infected with Loa loa (a filarial parasite coendemic in Central Africa) because of the risk of severe adverse events, such as potentially fatal encephalopathy (8). Two potential adulticidal treatments are under evaluation to accelerate elimination efforts: flubendazole, another anthelminthic used primarily for veterinary indications (9); and antibiotics such as tetracycline derivatives, which target the obligate Wolbachia endobacteria present in all stages of O. volvulus (10). These drugs will have to overcome bioavailability and safety issues (in the case of flubendazole (11)) or undergo a significant contraction in the duration of the regimen (in the case of doxycycline (10)) before they could be implemented on a wide scale. Vaccine development against onchocerciasis has a long history (12), but despite some recent breakthroughs with antigens such as a mutated form of cysteine proteinase inhibitor (13, 14), a vaccine candidate is yet to reach preclinical development.
An equally pressing challenge for onchocerciasis is rapid, sensitive and specific diagnosis of the disease in a format appropriate for rural Africa. The classical method, which is to examine skin snips for Mf, is far from ideal because of its insensitivity, capacity to cause significant discomfort, and the logistics associated with the biosafety of the biopsy punch (15). Immunoassays for O. volvulus antibodies are an important tool for monitoring the potential re-emergence of infection following regional elimination (16), but they can only be used in young children because of the longevity of the humoral response. Other diagnostic approaches include the diethylcarbamazine patch test (based on a hypersensitivity reaction in infected individuals (15)) or measuring transmission at the level of the vector by PCR of pooled blackflies (17). However, the desirability of a simple, noninvasive test to determine if an individual harbors one or more viable adult nematodes has spurred the hunt for onchocerciasis biomarkers in body fluids such as urine (18).
Our understanding of filarial genomics and molecular biology has been shaped largely by the publication of the Brugia malayi genome (19) and follow-up studies of its transcriptome (20–22), secretome (23–25), and structural proteome (26). This species is geographically restricted cause of lymphatic filariasis in humans, but is also popular as a laboratory model, as it will complete its lifecycle in jirds and will undergo limited development in mice (27). Furthermore, its availability from a central facility in Athens, Georgia, has greatly facilitated genomic and post-genomic studies on this parasite (28). However, B. malayi differs greatly from O. volvulus in its much shorter lifespan, location of Mf (which circulate in peripheral blood rather than migrating through the skin), and the lifestyle of the adult worms, which are located in the lymphatic vessels rather than nodules (27) and do not become heavily accreted with host material, unlike O. volvulus (29). The number of available filarial genomes has expanded recently with the publication of draft assemblies for Loa (30), the canine heartworm, Dirofilaria immitis (31), and the release of an unpublished O. volvulus genome assembly by the Wellcome Trust Sanger Institute (http://parasite.wormbase.org/Onchocerca_volvulus_prjeb513/Info/Index). Nevertheless, RNA and protein expression in Onchocerca spp. have yet to be explored in a high-throughput manner.
Here, we utilize the closest relative of O. volvulus, the bovine parasite O. ochengi (32, 33), to perform the first global expression study of an Onchocerca spp. across the major stages of the lifecycle (Fig. 1). For the past two decades, O. ochengi has been exploited in its natural host as an advanced screen for drug (34) and vaccine development (35), and has also revealed fundamental insights into the symbiosis between filariae and Wolbachia (36, 37). We show that the proteome of O. ochengi exhibits both qualitative and quantitative dynamic changes during development and that the protein families undergoing regulation have almost identical orthologs in O. volvulus. Furthermore, we quantify several hundred host and parasite proteins in fluid derived from O. ochengi nodules (Fig. 1), revealing the presence of a novel vaccine candidate (a 6-ShK-domain protein), homologs of transforming growth factor (TGF)-β, and bovine antimicrobial proteins that probably derive from neutrophils. A key unexpected finding is that many of the protein families exhibiting stage-specific enrichment across the O. ochengi lifecycle are associated with mitochondria, suggesting regulation of energy metabolism during development. These data provide a substantial new resource for the development of new vaccine, drug and diagnostic candidates for this chronically neglected disease.
Fig. 1.
The lifecycle of Onchocerca ochengi, highlighting the sampling strategy of this study. Simulium damnosum blackflies were blood-fed on naturally infected Cameroonian cattle harboring Mf in their skin. The blackflies were reared in the laboratory for 7 days and vL3 (1) were obtained on dissection of the head. To obtain NF, freshly excised intradermal nodules containing adult O. ochengi were pricked with a hypodermic needle (2), and the expressed liquid was collected. For the harvesting of adults, ethanol-fixed nodules were digested with collagenase to ensure minimal host contamination on AF (3), whereas AM (4) were obtained directly from freshly dissected nodules. Finally, iuMf (5) were collected from viable AF by puncture of the uteri and harvesting of culture supernatants.
EXPERIMENTAL PROCEDURES
Parasite Material
Intradermal nodules were excised from the skin of cattle slaughtered at Ngaoundéré abattoir, Adamawa Region, Cameroon, and dissected immediately in PBS to obtain adult males (AM) and adult females (AF; Fig. 1); or were fixed in 50% ethanol. The AM were frozen at −80 °C, while an incision was made in the body wall of viable, gravid AF and the uteri were carefully exposed. The uteri were punctured and the AF were transferred into RPMI medium (supplemented with 200 U/ml penicillin and 200 μg/ml streptomycin) and incubated at 37 °C. Periodically, supernatant fractions were removed from the AF cultures, spun at 300 g for 10 min, and intrauterine microfilariae (iuMf) were counted and assessed for viability (rapid motility). Individual batches displaying >10% sluggish or immotile iuMf were discarded. Following a single wash in PBS, iuMf were frozen as dry pellets at −80 °C. Skin Mf were not collected because of the high prevalence of coinfection with other Onchocerca spp. in cattle from this region (38) and the difficulty of reliably separating these species in large numbers. For the collection of vector-derived third-stage larvae (vL3), naturally infected cattle from the Adamawa Region were identified at local markets and transported to the Research Foundation for Tropical Diseases and Environment in Buea. Competent vectors (Simulium damnosum sensu lato) were fed on the bait animals, and vL3 were obtained from the blackflies after 7 days (Fig. 1) as previously described (39), except that the vL3 were purified on 20% Percoll gradients and stored at −80 °C in serum-free Grace's Insect Medium (Sigma-Aldrich) without full cryopreservation. Nodule fluid (NF) was harvested from abattoir-derived material as reported previously (40). The AM, iuMf, vL3 and NF were shipped to the UK on dry ice, whereas fixed nodules were transported chilled.
To obtain AF largely free of host tissue, we applied the enzymatic digestion method of Schulz-Key & Karam (41) to ethanol-fixed nodules, which were incubated at 35 °C with orbital shaking at 150 rpm in 0.05% type I bovine collagenase (Sigma-Aldrich). To remove collagenase and host debris, AF were washed in PBS for 2–3 h before any AM were located and discarded. Only young AF that could be liberated from all host tissue after 24 h of digestion were used for proteomic analysis. This reduced the proportion of bovine protein identifications from 70% in AF rinsed in PBS without collagenase digestion, to ∼30% after digestion (data not shown).
Protein Extraction
Soluble nematode whole body extracts (WBE) were prepared by adding pooled material for each O. ochengi life stage to fresh lysis buffer (40 mm Tris, 6 m urea, 1.5 m thiourea, 66 mm dithiothreitol (DTT, Sigma, St. Louis, MO), complete protease inhibitor mixture (Roche, Penzberg, Germany) and a 1:1 mixture of 1 mm glass and 0.1 mm zirconia-silica beads. Sample were homogenized using a Mini-Beadbeater (Biospec, Bartlesville, OK) for four, 1-min cycles at top speed (with 2-min rest periods on ice between cycles). Samples were centrifuged at 12,000 × g for 10 min (4 °C), and both pellet and the supernatant were retained. Protein concentrations were determined using the Pierce Coomassie Plus (Bradford) Protein Assay (Thermo Fisher Scientific, Waltham, MA).
Experimental Design and Statistical Rationale
To minimize potential bias and to increase coverage, extracted proteins from each life stage were analyzed using four methodologies. The soluble fraction was subjected to (a) direct tryptic digestion without fractionation, (b) anion exchange fractionation (AEX) of peptide mixtures, and (c) geLC-MS of proteins prior to tryptic digestion. Subsequently, the remaining insoluble pellet was exposed to trypsin to release additional peptides. Furthermore, three analytical replicates of the global (unfractionated) soluble WBE were performed for each life stage. For NF, five biological replicates were analyzed without fractionation in either protein or peptide space. Only protein identifications supported by ≥2 unique peptides present in ≥3 replicate analyses per stage were used for protein abundance comparisons in all cases.
Enrichment of protein domains was assessed using Pfam (EBI, v. 27.0 (42)) as previously described (43) using the gathering threshold as a cut-off. Briefly, a hypergeometric test for enrichment of Pfam domains in the observed proteome (for identifications supported by ≥2 unique peptides only) relative to the complete search database was performed using R (phyper). The Benjamini & Hochberg step-up FDR-controlling procedure was applied to the calculated p values (44), and enrichment was considered statistically significant where p < 0.01.
GeLC-MS
Proteins were fractionated using a NuPAGE® (Thermo Fisher Scientific) precast 4–12% Tris-Bis gradient gel. Each gel lane was cut into 20 slices and in-gel tryptic digestions were performed as described previously by Darby et al. (37).
Tryptic digestion
Soluble proteins (100 μg) were precipitated by addition of an equal volume of ice-cold 30% (w/v) TCA in acetone with incubation at −20 °C for 2 h. Samples were centrifuged at 12,000 × g for 10 min (4 °C) to pellet proteins. Pellets were washed three times with ice-cold acetone and allowed to air dry. Protein pellets were re-suspended in 25 mm ammonium bicarbonate, 0.1% (w/v) RapiGest SF (Waters, Milford, MA). Insoluble material from the initial worm homogenization step was washed three times with 25 mm ammonium bicarbonate and then suspended in 25 mm ammonium bicarbonate, 0.1% (w/v) RapiGest SF. The NF samples were centrifuged at 12,000 × g for 10 min (4 °C) and the supernatant retained. The NF protein concentration was determined as described above, and the sample was diluted in 25 mm ammonium bicarbonate, 0.1% (w/v) RapiGest SF. All protein samples were digested as described previously by Armstrong et al. (43).
Anion Exchange Fractionation
Peptides were fractionated by strong AEX chromatography into four fractions using a method described previously (45). Peptide samples were neutralized with absolute ammonium hydroxide and diluted fourfold in binding buffer (20 mm Britton-Robinson (BR) buffer, pH 11). The peptide mixture was applied to a stack of six layers of Empore anion exchange discs (Agilent) assembled inside a 200 μl pipette tip and centrifuged at 1000 × g for 5 min. The filter was washed with BR buffer and the wash was pooled with the flow-through. Peptides were successively eluted from the anion exchange membrane in 20 mm BR buffer at pH 8, pH 5, and pH 3. Each eluate was desalted and dried as above, before being re-suspended in a 0.1% (v/v) TFA, 3% (v/v) ACN solution for analysis by MS.
NanoLC MS ESI MS/MS Analysis
Peptide solutions were analyzed by on-line nanoflow LC using the nanoACQUITY-nLC system (Waters) coupled to an LTQ-Orbitrap Velos (Thermo Fisher Scientific) MS as reported previously (43). The gradient consisted of 3–40% ACN, 0.1% formic acid for 45 min (in-gel digests) or 150 min (nonfractionated soluble and insoluble digests), then a ramp of 40–85% ACN, 0.1% formic acid for 3 min in positive ionization mode.
Peptide samples separated by AEX were analyzed by on-line nanoflow LC using the Thermo EASY-nLC 1000 LC system (Thermo Fisher Scientific) coupled with Q-Exactive mass spectrometer (Thermo Fisher Scientific). Samples were loaded on a 50 cm Easy-Spray column with an internal diameter of 75 μm, packed with 2 μm C18 particles, fused to a silica nano-electrospray emitter (Thermo Fisher Scientific). The column was operated at a constant temperature of 35 °C. Chromatography was performed with a buffer system consisting of 0.1% formic acid (buffer A) and 80% ACN in 0.1% formic acid (buffer B). The peptides were separated by a linear gradient of 5–50% buffer B over 240 min at a flow rate of 300 nl/min. The Q-Exactive was operated in data-dependent mode with survey scans acquired at a resolution of 70,000. Up to the top 10 most abundant isotope patterns with charge states +2, +3, and/or +4 from the survey scan were selected with an isolation window of 2.0 Th and fragmented by higher energy collisional dissociation with normalized collision energies of 30. The maximum ion injection times for the survey scan and the MS/MS scans were 250 and 100 ms, respectively, and the ion target value was set to 1e6 for survey scans and 1e4 for the MS/MS scans. Repetitive sequencing of peptides was minimized through dynamic exclusion of the sequenced peptides for 20 s.
Protein Identification and Quantification
Spectra were imported into Progenesis QI (v. 2, Nonlinear Dynamics) and processed as described previously (43). Tandem MS data were searched against 13,991 partially revised Onchocerca gene models (see Proteogenomics below) and those from the Wolbachia symbiont, wOo (37) (UniProt release 2014_03; 647 protein sequences); together with predicted proteomes for the bovine host (Bos taurus, UniProt release 2014_02; 24,233 protein sequences) and a general contaminant database (GPMDB, cRAP version 2012.01.01; 115 protein sequences). The search parameters were a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.8 (LTQ-Orbitrap Velos) or 0.01 Da (Q-Exactive). Two missed tryptic cleavages were permitted. Carbamidomethylation (cysteine) was set as a fixed modification and oxidation (methionine) set as a variable modification. Mascot search results were further validated using the machine learning algorithm Percolator embedded within Mascot. The Mascot decoy database function was utilized and the false discovery rate was <1%, whereas individual percolator ion scores >13 indicated identity or extensive homology (p < 0.05). Mascot search results were imported into Progenesis QI as XML files. Relative protein abundance was calculated by the Hi-3 default method in Progenesis (46), in which the abundance of each peptide is calculated from all of its constituent peptide ions and the average abundance of the three top-ranked peptides is used to calculate the protein signal. Protein abundance was normalized across samples by average intensity utilizing the Normalyzer software package (47). Dirofilaria immitis excretory-secretory product (ESP) spectral data (MGF files obtained from (48)) were searched against the D. immitis theoretical proteome (http://parasite.wormbase.org/Dirofilaria_immitis_prjeb1797/Info/Index; 12,857 protein sequences) using the Mascot (version 2.3.02, Matrix Science) search engine with settings as described above. Data were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (49) with the data set identifier PXD002889 and DOI 10.6019/PXD002889.
Proteogenomics
An unpublished draft O. ochengi genome, generated by Gaganjot Kaur and Georgios Koutsovoulos of the Blaxter Laboratory, University of Edinburgh, was downloaded from WormBase ParaSite (http://parasite.wormbase.org/Onchocerca_ochengi_prjeb1809/Info/Index, version nOo.2.0; 13,990 predicted protein-coding genes). The genome assembly was obtained from pooled individuals, and has a Core Eukaryotic Genes Mapping Approach (50) score of 94.4%. However, it is relatively fragmented, comprising 1818 contigs with a N50 length of 12.7 kb. The Wellcome Trust Sanger Trust has released a highly contiguous reference genome for O. volvulus (http://parasite.wormbase.org/Onchocerca_volvulus_prjeb513/Info/Index, O_volvulus_Cameroon_v3; 12,143 predicted protein-coding genes; 703 scaffolds with an N50 length of 25.5 Mb) based on sequencing of a single individual, optical mapping, and extensive manual curation (Matthew Berriman and colleagues, unpublished). As split gene models (i.e. a single gene incorrectly annotated as two or more genes because of fragmentation between contigs and/or frameshift errors) affect the accuracy of protein identification and quantification, the ProteoAnnotator pipeline (51) was used to objectively assess the strength of evidence for peptide-spectrum matches against O. ochengi versus O. volvulus gene models.
The O. ochengi gene models from release nOo.2.0 were considered “official” models, whereas O. volvulus protein annotations from release WBPS2 were designated as an “alternative” set. In ProteoAnnotator, 642 protein ambiguity groups (PAGs) were identified in which the lead protein was derived from O. volvulus rather than O. ochengi; thus, these O. volvulus gene models were provisionally considered better matches to the MS data and were divided into four groups. In group 1 (n = 271), the PAGs contained only O. volvulus gene models, presumably because of unassembled regions of the O. ochengi draft genome or errors in assembly and prediction. For group 2 (n = 273), the lead O. volvulus gene model exceeded the length of all O. ochengi models in its PAG by ≥10%, which suggests that the O. ochengi models were a fragmented subset of the full-length O. volvulus model. In group 3 (n = 30), the PAGs contained multiple O. volvulus and O. ochengi gene models and the retained model (from either species) was selected by manual review of the position of unique peptides following alignment of all sequences in the PAG. Finally, in group 4 (n = 68), the lead protein was an O. volvulus model that was <10% longer than O. ochengi models in the same PAG. The search database was revised by adding O. volvulus sequences from group 1, replacing existing O. ochengi sequences with O. volvulus models for group 2, and amending O. ochengi models or replacing with O. volvulus sequences as appropriate for group 3. For group 4, we took a conservative approach and retained the unmodified gene models from the cognate genome. This amended database contained a total of 13,991 gene models, of which 572 were derived from O. volvulus.
Bioinformatics
Venn diagrams were created using the Venn diagrams freeware (http://bioinformatics.psb.ugent.be/webtools/Venn), whereas heat-maps were created using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E) freeware. Functional annotation by Clusters of Orthologous Groups was obtained via the WebMGA server (52). The conserved domain structure of selected proteins was also interrogated in InterProScan 5 (53); sequence alignment was performed using UniProt (Clustal Omega), and sequences were annotated using jalview2 (54). Protein-protein interactions were determined using STRING version 9.1 (55). Prediction of classical N-terminal signal peptides, non-classical secretion signatures, transmembrane domains, propeptide cleavage sites, N-glycosylation sites and mitochondrial targeting signals was performed using the SignalP 4.0 server (56), the SecretomeP 2.0 server (57), the TMHMM 2.0 Server (58), the ProP 1.0 server (59), the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc) and MitoProt (60), respectively. Orthologs to O. ochengi proteins in the B. malayi (Uniprot release 2013_08; 11,338 protein sequences), L. sigmodontis (http://parasite.wormbase.org/Litomosoides_sigmodontis_prjeb3075/Info/Index; 10,246 protein sequences), D. immitis (http://parasite.wormbase.org/Dirofilaria_immitis_prjeb1797/Info/Index; 12,857 protein sequences) and O. volvulus (http://parasite.wormbase.org/Onchocerca_volvulus_prjeb513/Info/Index; 12,143 protein sequences) theoretical proteomes were determined using reciprocal BLAST (61) with a bit score cut-off of 50.
For phylogenetic analysis of glutathione-domain proteins supported by peptide evidence, O. ochengi sequences containing the canonical glutathione transferase (GST) C-terminal and N-terminal domains (Pfam identifiers PF02798 and PF00043, respectively) were obtained from predicted protein-coding sequences available from the WormBase ParaSite. Protein sequences for GSTs from seven species-independent cytosolic GST classes (alpha, mu, omega, pi, sigma, theta, and zeta) and to glutathione-domain containing proteins belonging to the MAPEG (membrane associated proteins in eicosanoid and glutathione metabolism), mPGES2 (membrane-associated prostaglandin E synthase-2), CLIC (chloride intracellular channel protein), EF (eukaryotic elongation factors 1-gamma) and metaxin families were retrieved via BLAST analysis (61, 62) of the NCBI database at http://www.ncbi.nlm.nih.gov (non-redundant GenBank CDS translations, PDB, SwissProt, PIR and PRF, excluding those in env_nr; posted date - Aug 13, 2015 2:33 am). Arthropod and vertebrate glutathione-domain protein sequences were included in the analysis for comparative purposes as representatives of the intermediate and definitive hosts for O. ochengi and other filariae. The GST domain-containing protein homologs were subjected to multiple sequence alignment using MUSCLE (63). Phylogenetic bootstrap neighbor-joining trees (subjected to 1000 bootstrap replicates) were produced as PHYLIP output files according to the neighbor-joining method of Saitou & Nei (64) within ClustalX Version 2.1 (65, 66). Default settings for alignments were accepted using the GONNET protein weight matrices with PHYLIP tree format files viewed within the TREEVIEW program (67).
RESULTS
Overview of protein identifications and comparison with B. malayi
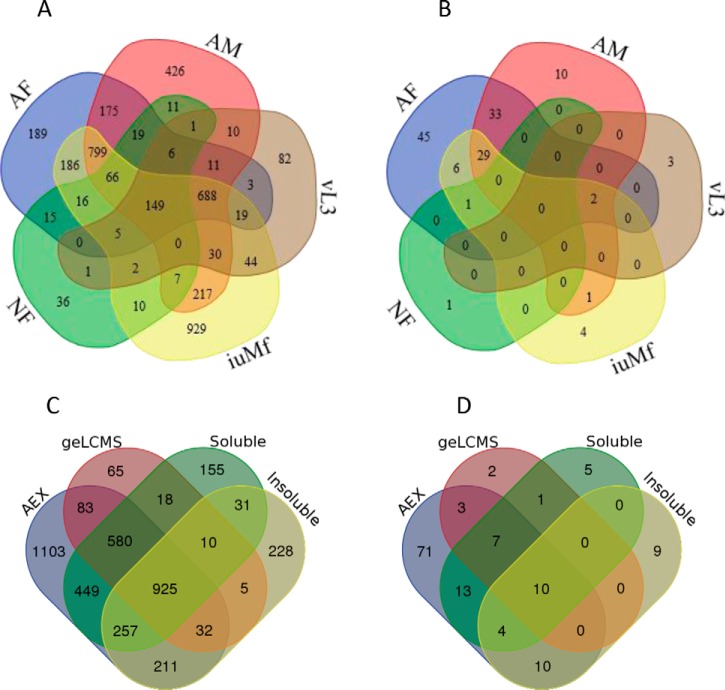
In this study, we took advantage of the remarkably close relationship between O. ochengi and O. volvulus to refine our MS search database using a proteogenomic approach based on gene predictions from both species. We identified 4260 unique O. ochengi proteins across NF and the four WBE examined, together with 135 proteins from the Wolbachia symbiont (wOo), representing 30.4% and 20.8% of the theoretical proteomes, respectively (Fig. 2A and 2B; supplemental Table S1). Both the greatest total number and largest stage-specific set of protein identifications were derived from iuMf, whereas 16.2% of proteins were shared between all WBE (Fig. 2A; supplemental Table S2). The most effective methodology for protein identification, by a wide margin, was fractionation by anion exchange (85.4% of nematode data set), whereas digestion of insoluble pellets uniquely contributed 5.4% of the total (Fig. 2C and 2D). However, the greatest proportion of predicted transmembrane proteins identified by a single approach (32.9%) was observed by digestion of insoluble pellets (supplemental Table S1). For wOo, 85.9% of all identifications were derived from a single stage (AF) (Fig. 2B; supplemental Table S2), which reflects the previously reported dynamics of Wolbachia in filarial nematodes and in particular, the large number of bacteria that accumulate in gravid AF during embryogenesis (68, 69). Classification of the detected wOo proteins by Clusters of Orthologous Groups, although purely qualitative (supplemental Fig. S1), clearly reflected the functional allocation of resources in wOo toward translation, post-translational processing and energy conversion, with little representation of coenzyme and secondary metabolism.
Fig. 2.
Venn sets of protein identifications by lifecycle stage and proteomic methodology. The total number of proteins identified in O. ochengi (A) and wOo (B) in each stage, and the total number of proteins identified in O. ochengi (C) and wOo (D) by methodology. Protein identifications contained in each set are provided in supplemental Table S2.
The only large-scale study of a filarial structural proteome published previously involved analysis of five lifecycle stages of B. malayi (the same WBE as the current study, plus mature Mf) by very extensive (∼100 fractions) separation of peptides by strong cation-exchange LC (26). Restricting the comparison to the stages common to both studies, this resulted in the identification of 6697 unique proteins (Fig. 3A; supplemental Table S3). Notably, the two studies accorded in attaining the greatest number of protein identifications from iuMf, and the least from vL3 (Fig. 3D and 3E; supplemental Table S3). Comparisons of orthologous groups revealed that 16.9% of identified B. malayi proteins had no ortholog in the O. ochengi genome, which was more than double the reciprocal figure (i.e. 8.1% of identified O. ochengi proteins had no ortholog in the B. malayi genome) (Fig. 3A). At the genome level, the number of non-orthologous genes in B. malayi is only ∼1.4 times greater than the non-orthologous complement in O. ochengi, suggesting that Bennuru et al. (26) achieved deeper sampling of low-abundance proteins.
Fig. 3.
Venn sets of proteins identified in O. ochengi compared with published data for B. malayi. The number of orthologous (blue circles, B. malayi; yellow circles, O. ochengi) and non-orthologous (lilac circles, B. malayi; beige circles, O. ochengi) proteins is shown for (A) all stages combined, (B) AM, (C) AF, (D) iuMf and (E) vL3. Note that the number of proteins per stage is lower than in Fig. 2 because of the exclusion of paralogous gene families. Protein identifications contained in each set are provided in supplemental Table S3.
To determine whether the protein identifications in each of the shared and unique sets exhibited significant differences, we performed a Pfam domain enrichment analysis between detected proteins and the complete theoretical proteome of B. malayi (or O. ochengi in the case of nonorthologous proteins from this species). A single domain, “regulators of G protein signaling” (RGS), was overrepresented in the set of B. malayi-only orthologs (supplemental Fig. S2). This was because of the detection by Bennuru et al. (26) of all six RGS proteins encoded by the B. malayi genome, whereas only two of 14 proteins containing RGS domains in the O. ochengi genome were identified. Interestingly, collagens were highly enriched in the set of non-orthologous O. ochengi proteins, especially those detected in iuMf only, suggesting a gene expansion in this species or loss of some collagen genes from B. malayi. However, most enriched domains (“GTP_EFTU_D2”, “pro_isomerase” and proteasome) were shared between the two parasites (supplemental Fig. S2). We conclude that there is no evidence of significant systematic bias between the two studies, although our vL3 data set is probably restricted to the most abundant proteins in this stage.
Protein Family Domain Enrichment Analysis
To identify the protein families that dominated the proteome of each lifecycle stage, we performed an enrichment analysis by comparing Pfam domain counts in observed proteins against those present in all sequences contained in the Onchocerca search database. A total of 64 Pfam domains were enriched in one or more lifecycle stages; although only two, actin and I-set domains, were significantly overrepresented in all WBE (p < 0.01; Fig. 4). The I-set domain represents the intermediate module from a larger family of immunoglobulin (Ig) domains, which included additional members that were significantly enriched in AF and vL3 only (Fig. 4). In total, 28 proteins were identified with I-set and/or Ig domains, of which 13 were detected in all WBE (supplemental Fig. S3).
Fig. 4.

Pfam enrichment analysis across the lifecycle of O. ochengi. Hierarchically clustered heat-map of protein families that were significantly overrepresented (p < 0.01) in the observed proteome (≥2 unique peptides) relative to the complete search database. The intensity of shading is proportional to fold-enrichment.
We found several members of the pleiotropic β-galactoside-binding protein family (“Gal-bind_lectin” or simply galectin; Fig. 4, supplemental Fig. S4) to be enriched in AF, iuMf and vL3, including almost identical orthologs (nOo_08682 and nOo_10718) of Ov-GBP-1 and Ov-GBP-2, respectively, which are previously described galectins from O. volvulus (70, 71). A more novel finding was the strong representation of Pfam domains associated with intracellular trafficking, including Arf, “Adaptin_N”, Snf7 and WD40, in AF and/or iuMf (Fig. 4, supplemental Fig. S5). However, in a clear example of how the Pfam enrichment analysis accorded with classical biochemical studies of filarial nematodes, vL3 was the only stage displaying significant overrepresentation of enzymes containing “glyco_hydro_18” and “peptidase_C1” domains (Fig. 4). The former group contained chitinases (supplemental Fig. S6A), whereas the peptidase domains were found in a group of seven cathepsins (supplemental Fig. S6B). Both of these enzyme families are known to be strongly associated with the L3 stage (72–75). A third enzymatic domain specifically enriched in vL3 was the Ras superfamily of small GTPases (Fig. 4, supplemental Fig. S6C), which comprises a very large family of regulators and signaling molecules with 56 members in Caenorhabditis elegans (76). To the best of our knowledge, Ras proteins have not been reported to be overrepresented in the L3 stage of filarial nematodes previously.
Redox and Detoxification Enzymes
All stages displayed significant Pfam enrichment for one or more enzyme families with roles in redox reactions and/or detoxification, comprising proteins with thioredoxin, GST or peroxiredoxin (AhpC-TSA/1-cysPrx_C) domains (Fig. 4). Although GST domains were only significantly overrepresented in AF and iuMf, two members of this family were widely distributed across all WBE (Fig. 5A). These belonged to two different cytosolic GST subgroups: nOo_00341 within the pi class and nOo_09064 from the sigma class (Fig. 5B).
Fig. 5.
Detection and phylogeny of GST-domain proteins observed across the O. ochengi lifecycle. A, Distribution of GST-domain proteins in each lifecycle stage (black, detected with ≥2 unique peptides; gray, detected with one unique peptide only; white, not detected). B, Phylogenetic neighbor-joining tree of GST-domain proteins identified from O. ochengi (in bold type) displaying classes present in C. elegans, Drosophila melanogaster, Homo sapiens, Mus musculus, and selected parasitic nematodes and platyhelminths. Numbers shown alongside branches are bootstrap values of 1,000 replications. Complete accession numbers for all GST-domain sequences are provided in supplemental Table S4. MAPEG, membrane associated proteins in eicosanoid and glutathione metabolism; mPGES2, membrane-associated prostaglandin E synthase 2; CLIC, chloride intracellular channel protein; EF, eukaryotic elongation factor 1-γ.
The pi-class GST nOo_00341 is orthologous to OvGST2 that is reportedly the dominant cytosolic GST of O. volvulus, abundant in all tissues and life stages (77). In contrast to this single pi-class member, we identified several distinct sigma-class GSTs (Fig. 5B), although only nOo_09064 was detected across the entire lifecycle. This GST is homologous to OvGST1 from O. volvulus, which has been reported to function as a glutathione-dependent prostaglandin D synthase and is novel among the GST superfamily in containing glycosylated residues (78). The third subgroup of GSTs that have been reported from O. volvulus is the omega class, represented by OvGST3, which contains a signal sequence and undergoes alternative splicing (79). This GST is the only member of the superfamily that is significantly up-regulated during exposure of O. volvulus AF to oxidative stressors (80). Moreover, OvGST3 exhibits highly restrictive expression in O. volvulus adult worms, in that it localizes exclusively to the eggshell of developing embryos from the morula stage onwards (79). Interestingly, although we detected several distinct omega-class GSTs (Fig. 5B), they were only robustly identified in AF, iuMf, and (in the case of nOo_09038) NF (Fig. 5A). This suggests that these GSTs may be expressed during embryogenesis, although further studies would be required to determine if they are found only in the eggshell.
Two canonical GST superfamily domains typically identify GSTs and reside in the N-terminal and C-terminal regions (PF02798 and PF00043, respectively). In addition to the cytosolic GST forms such as the pi and sigma classes, class-specific C- and N-terminal domains may also occur in other functionally distinct and unrelated proteins that may lack classical glutathione-conjugating activity, including the mPGES2 (81), CLIC (82), EF (83), and metaxin families (84). The single EF family member identified in the current study, eukaryotic elongation factor 1-γ, is essential for delivery of aminoacyl tRNAs to the ribosome and was present in all WBE as expected (Fig. 5A). However, we also identified seven proteins that contained a specific GST-like C-terminal domain (PF14497), including five from the metaxin family that were expressed in adult worms and iuMf (Fig. 5A). These represent components of the preprotein import complex of the mitochondrial outer membrane; although their function remains obscure (84, 85). The other proteins containing domain PF14497 were prostaglandin E synthase-2 from iuMf only and a CLIC family member, chloride intracellular channel EXC-4, detected in all WBE except vL3 (Fig. 5A). The former is a microsome-associated enzyme with a broad specificity for thiol cofactors (81), whereas the latter is essential for the formation and maintenance of correct tubular architecture in the excretory canal of C. elegans (86).
Peroxiredoxins (Prx; also known as thioredoxin peroxidases, TPX) are cysteine-dependent antioxidant enzymes that play a major role in the scavenging of reactive oxygen and nitrogen species (87). In O. volvulus, Ov-TPX-2 is a member of the Prx1 family of typical 2-Cys Prxs, and is expressed from the late L1 stage in the vector through to adult worms, where it is associated with the body wall, intestine and uterus (88). Embryos and Mf of O. volvulus do not express Ov-TPX-2, but are “bathed” in this protein in utero, and it can still be detected on the Mf surface after birth (88). The O. ochengi Prxs, nOo_08778 and nOo_10285, are almost identical orthologs of Ov-TPX-2, whereas nOo_02155 represents a closely related Prx1 isoform with an N-terminal extension of ∼40 amino acids. In accordance with the O. volvulus data, we observed these Prx1 proteins in all WBE, and two isoforms were also significantly enriched in NF (Fig. 4; see Secreted Onchocerca proteins in nodule fluid below). A fourth Prx1 family member, OVOC4328, was only identified from AM extracts and in common with Bm-TPX-1, contained a predicted N-terminal mitochondrion-targeting domain (89). Indeed, the C. elegans ortholog of OVOC4328, PRDX-3, has a key role in the detoxification of mitochondrial hydrogen peroxide generated by the electron transport chain (90). We also identified a single member of the Prx6 family (composed predominantly of 1-Cys members), which is orthologous to OvPXN-2 from O. volvulus (91). As reported for O. volvulus, this was detected consistently throughout the lifecycle (91).
Significant enrichment for thioredoxin domains was apparent in AF, iuMf and vL3 (Fig. 4), although most of the 14 proteins detected that contained these domains were also identified in AM. Orthologs for two of these proteins (OVOC4952 and nOo_05700) included a protein disulfide isomerase from O. volvulus (92) and DPY-11 from C. elegans (93), respectively. Both of these proteins are localized to the hypodermal syncytium (as well as iuMf in O. volvulus) and are thought to have a role in the catalysis of disulfide bond formation in cuticular collagens (92, 93). Furthermore, an ortholog of OVOC4952 was identified on the surface of AF in L. sigmodontis, although not in ESP (43). Two additional proteins detected in all WBE (nOo_01735 and OVOC82) were orthologs of a bi-functional protein disulfide isomerase/transglutaminase from Dirofilaria immitis that has been characterized experimentally (94). The transglutaminase activity may be required to form the covalent ε-(γ-glutamyl) lysine isopeptide bonds that are also present in nematode cuticles (94).
Mitochondrial Proteins
Mitochondria are dynamic organelles that constantly undergo fission and fusion depending on the energy requirements of the cell and levels of oxidative stress. We identified a number of proteins involved in mitochondrial fission and fusion among the “dynamin_N” Pfam group that was significantly enriched only in iuMf (Fig. 4). These included FZO-1, a “mitofusin” that is required for the formation of tubular mitochondrial structures in the body wall muscle cells of C. elegans (95); and EAT-3, a second mitofusin that is essential for optimal ATP production and resistance to damage caused by free radicals (96). Recently, experiments utilizing C. elegans mutants have demonstrated a major role for FZO-1 in the rate of growth of L1 and in energy metabolism as measured by oxygen consumption (97). A third dynamin-domain protein identified in iuMf, dynamin-related protein-1, is involved in mitochondrial fission and control of apoptosis (98).
The mitochondrial tRNAs of chromadorean nematodes are unusual in that all lack the typical cloverleaf tRNA structure, with 20 classified as “T-armless” and the remaining two as “D-armless” (99). These tRNAs require specialized elongation factor-Tu proteins, termed TUFM-1 and TUFM-2, to recognize the T-armless and D-armless molecules, respectively. Several elongation factors, eukaryotic translation initiation factors and peptide chain release factors contributed to the significant enrichment of “GTP_EFTU” domains in AF (Fig. 4), most of which were cytosolic and detected in all WBE except vL3. However, a TUFM-1 homolog was also identified in all WBE, whereas a TUFM-2 homolog was restricted to AF and iuMf. The latter binds solely to two D-armless tRNASer molecules in chromadorean nematodes (100), and we only detected the mitochondrial form of seryl-tRNA synthetase in iuMf, which contains the Pfam domain “tRNA-synt_2b” (significantly enriched in iuMf; Fig. 4).
Several proteins with pyridine nucleotide-disulfide oxidoreductase (Pyr_redox_2) domains were significantly overrepresented in vL3 only (Fig. 4). These included apoptosis-inducing factor-1 and dihydrolipoamide dehydrogenase, which in healthy cells have roles in redox homeostasis of the respiratory chain and the mitochondrial matrix, respectively (101, 102). A third Pyr_redox_2 domain protein with a mitochondrial-targeting signal peptide was homologous to C. elegans thioredoxin reductase (TrxR)-1, a cytosolic enzyme (103). However, the mitochondrial targeting sequence suggests that it is functionally more closely related to C .elegans TrxR-2, which is the mitochondrial isoform in this species.
Both vL3 and iuMf exhibited enrichment for proteins containing peptidase family M16 domains (Fig. 4), with five and seven members detected with confidence (≥2 peptides), respectively. Four of these (nOo_00369, nOo_00741, nOo_04056, and nOo_04374) resembled core proteins of the cytochrome bc1 complex or mitochondrial processing peptidase (MPP) (which are closely related (104)). The role of MPPs is to cleave the N-terminal import signal from nuclear-encoded mitochondrial proteins, and the core proteins of the bc1 complex may function as specialized MPP proteins that target the signal peptide of the Rieske iron-sulfur protein (105); although such peptidase activity has not been demonstrated for C. elegans core proteins (104). An additional peptidase family M16 domain protein (nOo_01854), homologous to the mitochondrial presequence protease Cym1, was robustly detected in iuMf only. The role of Cym1 is to degrade the cleaved presequences generated by MPPs in the mitochondrial matrix and release the fragments to the cytosol (106). Because MPPs and Cym1 are also encoded by the nucleus, they should feature an N-terminal mitochondrial targeting sequence. We identified this signal in the O. ochengi MPP homologs (Fig. 6), but not in the Cym1 homolog nor in nOo_04749, a further peptidase family M16 domain protein identified in all WBE and annotated as “insulin-degrading enzyme.” However, manual curation of the nOo_04749 gene model with reference to the O. volvulus ortholog, OVOC6041, revealed the correct start site and a mitochondrial targeting peptide. This suggests that nOo_04749 is also a MPP, although a highly divergent member of the family (Fig. 6).
Fig. 6.
Domain structure and lifecycle distribution of peptidase family M16 domain proteins. Schematic representation of O. ochengi peptidase family M16 domain proteins and their orthologs in O. volvulus. Locus tags containing the suffix “m” refer to modified O. ochengi gene models that were manually corrected using O. volvulus gene models as a template. Text in blue refers to the lifecycle stages in which the proteins were detected (≥2 unique peptides). Protein signature identifiers from InterProScan 5 (53) are provided for the N- and C-terminal peptidase M16 domains.
Quantitative Differences Between Stages
To further explore potential differences in protein expression across the O. ochengi lifecycle, we compared protein abundance by a label-free method (Hi-3) for 1313 proteins for which we had detected ≥2 unique peptides across two MS platforms and three sample preparation methodologies (supplemental Table S5). Each lifecycle stage showed a cluster of protein expression containing a variable number of unique proteins (supplemental Fig. S7A) that displayed some overlap with the qualitative Pfam enrichment analysis, but was not dependent on tallies of conserved domains. As most proteins derived from wOo were of low abundance, only 13 endosymbiont products fulfilled our criteria for quantification by Hi-3. The pattern of protein expression was clearly related to the known dynamics of Wolbachia replication as determined for B. malayi, in which Mf and L3 have the lowest bacterial numbers, AM harbor an intermediate level, and gravid AF contain the largest bacterial population (68) (supplemental Fig. S7B).
Secreted Onchocerca proteins in nodule fluid
In NF, we identified 94 O. ochengi and 498 bovine proteins with ≥2 unique peptides in ≥3 biological replicates. As expected, the most abundant filarial proteins in NF all featured classical or non-classical secretion signatures (Fig. 7A). The ESP repertoire from NF displayed remarkable parallels with that of L. sigmodontis gravid AF, with shared orthologs for abundant transthyretin-like and von Willebrand factor type-d domain proteins (43), cysteine proteinase inhibitors (107), and poorly characterized filarial antigens (Av33 (108), Ov16 (109), and RAL-2 (110)). In addition, an uncharacterized NF protein (nOo_00893) had a ML (lipid-binding) domain and was orthologous to an abundant secreted L. sigmodontis protein sharing this same motif (43). However, the most important similarity between the NF and L. sigmodontis secretomes was the presence of a ShK domain protein (43), which is related to the metridin-like cnidarian toxins that can block Kv1.3 potassium channels in memory T-cells (111) (Fig. 7B). Recently, a synthetic C-terminal ShK domain peptide from a B. malayi astacin was demonstrated to exhibit Kv1.3-channel-blocking activity for human T-cells, albeit with lower potency than cnidarian ShK peptides (112). In L. sigmodontis, the most abundant secreted ShK-domain protein (nLs_04059) is not an astacin and is unusual in containing six ShK domains. We detected several peptides that matched to an O. volvulus ortholog (OVOC2486) of nLs_04059, and this sequence enabled us to reconstruct the O. ochengi ortholog from two split gene models (nOo_12220 and nOo_06172) covering the central portion along with the N- and C-terminal domains located on other contigs. With the exception of a longer N-terminal region immediately downstream of the signal peptide, the combined “nOo_12220/nOo_06172” gene model is identical to OVOC2486 (Fig. 7B). The proteins from both Onchocerca spp. exhibit a single lysyltyrosine dyad in the sixth ShK domain (as opposed to two in nLs_04059), which is essential for Kv1.3 channel-blocking activity (111). It is unclear whether cleavage of the sixth ShK domain (the only one containing a LysTyr dyad) from the remainder of the molecule is required for activity in vivo, because although nLs_04059 has a propeptide cleavage site upstream of the sixth domain, the only predicted cleavage site within the Onchocerca proteins is located in the middle of the molecule (Fig. 7B).
Fig. 7.
Abundant O. ochengi proteins in nodule fluid and structure of a ShK-domain protein. A, Proteins in NF were quantified by the Hi-3 method and ranked by abundance; the top 30 (from a total of 94 robustly identified) are shown. An asterisk indicates prediction of a classical signal peptide, whereas a plus sign demarks proteins predicted to be secreted by a non-classical pathway. B, Reconstruction of a full-length, six ShK-domain protein in O. ochengi using the O. volvulus gene model OVOC2486 as a template, and alignment against the L. sigmodontis ortholog nLs_04059. Highlighted features comprise predicted signal peptides (orange boxes), ShK toxin-like domains (black boxes), conserved cysteine residues (yellow shading), predicted propeptide cleavage sites (red boxes), and lysyltyrosine dyads (bold type). Residues in different colors represent translations from separate contigs.
The high level of concordance between the NF and L. sigmodontis secretomes was maintained when the complete NF dataset was compared with those from other published filarial secretome studies (supplemental Fig. S8). Remarkably, only three proteins were observed in every study, including triosephosphate isomerase, which has recently been shown to have an essential role in Mf production in B. malayi (113). As noted by us previously (43), the three B. malayi secretome studies display a limited degree of overlap, highlighting the potential impact of methodology on proteomic data; although we observed a strong correlation between our NF dataset and that of Moreno & Geary (24) for B. malayi (supplemental Fig. S8). Seven proteins were shared uniquely between the NF, L. sigmodontis and D. immitis secretomes, including orthologs of nematode secreted protein 22U (114), a cyclophilin (23), and a single transthyretin-like protein.
A striking feature of NF, which has not been observed previously in high-throughput secretome analyses of filarial nematodes, was the presence of two TGF-β homologs (Fig. 8). The TGF-β superfamily has members present in all multicellular organisms, and is involved in development, regulation of the mammalian immune response and immunomodulation by parasites. The prototypic TGF-β family member, human TGF-β1, has an N-terminal moiety composed of a signal peptide, a proregion and a protease cleavage site (RXXR), targeting of which releases the C-terminal active domain capable of binding to the receptor. The active domain of TGF-β1 contains nine cysteines necessary for correct folding and dimerization (115).
Fig. 8.
TGF-β homologs detected in O. ochengi nodule fluid. Alignment of O. ochengi TGF-β homologs from NF against orthologs from O. volvulus and B. malayi. Highlighted features comprise predicted signal peptides (red boxes), start methionines (red type), tetrabasic protease cleavage site (cyan shading), conserved cysteines (yellow type and black shading), potential N-glycosylation motifs (blue boxes), and peptides detected by MS (orange type). The green box represents the corrected C terminus of nOo_02612.
The first TGF-β family member we identified, nOo_02612, has an identical ortholog from O. volvulus, OVOC7660, and is closely related to a B. malayi TGF-β family member, Bm-TGH-2. Although the predicted sequence of nOo_02612 showed a C-terminal extended region, lacking the CXC motif characteristic of TGF-β family members, closer inspection revealed that this was because of miscalling of an intron-exon boundary (Fig. 8). The second TGF-β family member detected in NF, nOo_06906, also has an identical O. volvulus ortholog, OVOC7290, and displays significant homology to another B. malayi protein, Bm9118 (Fig. 8). However, Bm9118 lacks the protease cleavage site and first three cysteines of the TGF-β family active domain, suggesting that the gene model may be incomplete. All peptides detected in NF from nOo_02612 were contained in the proregion of the protein, whereas all peptides identified from nOo_06906 were located in the active C-terminal domain (Fig. 8), indicating that nOo_06906 at least is present in secretions in its active form.
As reported above (see Redox and detoxification enzymes), the repertoire of Prxs in NF (one Prx6 and two Prx1 isoforms) overlapped with those detected in all WBE, although a single Prx1 observed in all of the structural proteomes (nOo_02155) was not found in NF. This was orthologous to a molecule identified on the surface of L. sigmodontis (TPX-1, nLs_01344) that was also not secreted (43). Unexpectedly, nodule fluid also contained six filarial immunoglobulin-domain proteins (supplemental Fig. S3) that are known to localize to extracellular matrices in C. elegans. One of these proteins (OVOC10067) was homologous to the perlecan-like proteoglycan UNC-52; a major component of the basement membrane of contractile tissues, including the pharynx and anus, in developing embryos and subsequent stages of C. elegans (116). In addition to its structural role, UNC-52 is required for control of directionality during axon outgrowth in C. elegans (117); whereas another immunoglobulin-domain protein detected in NF, hemicentin, is involved in the anchoring of mechanosensory neuron axons (118). Furthermore, a peroxidasin homolog related to C. elegans PXN-2 was present in iuMf and NF, and this was orthologous to a L. sigmodontis peroxidasin that was observed in the secretome of AF and immature Mf (43). In C. elegans, PXN-2 is located in the extracellular matrix and is required for late embryonic elongation, muscle attachment, and motoneuron axon guidance choice (119). Finally, a surprising component of NF was a giant (predicted molecular weight, ∼690 kDa) immunoglobulin-domain protein homologous to C. elegans DIG-1 (supplemental Fig. S9), the largest secreted protein from any organism described to date (120).
Bovine Proteins in Nodule Fluid
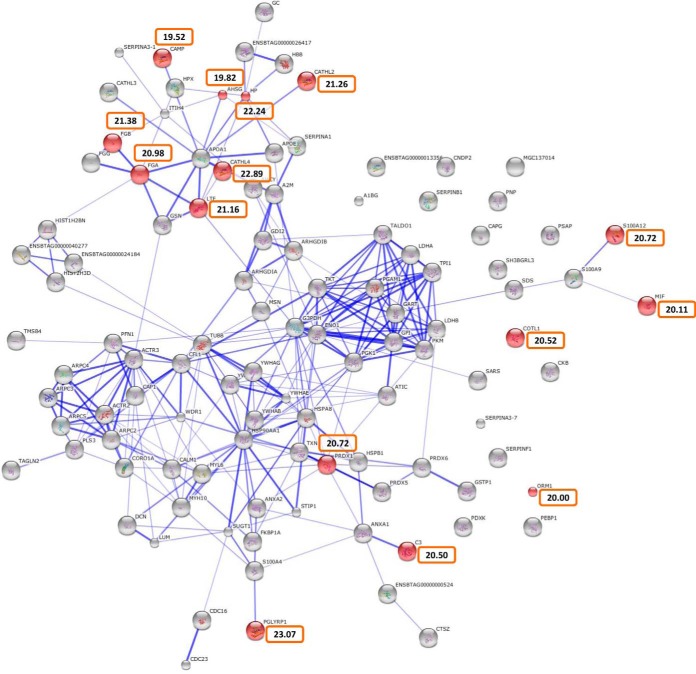
In addition to serum components, the bovine proteins quantified in NF displayed a high abundance of molecules involved in innate immunity. The predominant 100 bovine proteins were analyzed for interaction networks and GO term enrichment (Fig. 9). The most numerous host cell type in both O. ochengi and O. volvulus nodules is neutrophils, which form a dense layer around the adult nematodes without causing any apparent damage to the parasites (36, 121, 122). Accordingly, several proteins known to be abundant in bovine neutrophils were significantly enriched within the GO term “defense response” (FDR-corrected p < 0.01), including cathelicidin-2 (bactenecin-5), cathelicidin-4 (indolicidin), cathelicidin-7 (bovine myeloid antimicrobial peptide-34) (123, 124), haptoglobin (125),PGLYRP1 (peptidoglycan recognition protein (126)), and S100A12 (calgranulin C) (127) (Fig. 9). All of these proteins are major constituents of neutrophil granules except for calgranulin C, which is cytosolic. This has been purified from extracts of collagenase-treated O. volvulus (128), suggesting a tight association with the parasite surface, and has some antifilarial activity, especially against Mf (129).
Fig. 9.
Protein interaction network of the most abundant bovine proteins in nodule fluid. The 100 most abundant bovine proteins in nodule fluid, as determined by the Hi-3 method, were subjected to network analysis. The significantly enriched GO term “defense response” (FDR-corrected p < 0.01) is highlighted by the red nodes, and orange boxes contain normalized (log2) abundance values. CAMP, cathelicidin-7; CATHL2, cathelicidin-2; CATHL4, cathelicidin-4; FGA, fibrinogen alpha-chain precursor; FGB, fibrinogen beta-chain precursor; AHSG, alpha-2-HS-glycoprotein; HP, haptoglobin precursor; LTF, lactotransferrin; MIF, macrophage migration inhibitory factor; COTL1, coactosin-like protein; PRDX1, peroxiredoxin-1; ORM1, alpha-1-acid glycoprotein precursor; C3, complement C3 preproprotein; PGLYRP1, peptidoglycan recognition protein-1.
DISCUSSION
This study represents the first expression analysis of an Onchocerca spp. across the major lifecycle stages and at the immediate host-parasite interface of the nodule. A recurring theme of our analyses was the remarkable similarities between protein families identified by unbiased, proteomic methods in O. ochengi and those reported from O. volvulus as highly expressed according to classical approaches, such as representation in cDNA libraries from each stage. However, other discoveries emerging from this work, such as the differences in mitochondrion-related protein families dominating in each stage, were not anticipated. Moreover, quantification of nodule fluid proteins provided new insights into the immunomodulatory milieu surrounding the adult worms.
Bennuru et al. (26) achieved deeper coverage of the B. malayi proteome than we experienced in our study of O. ochengi. In all likelihood, this can be attributed to the fractionation protocol they employed. However, other factors may have contributed, such as the manual curation that accompanied the publication of the B. malayi genome (19), the utilization of a B. malayi EST dataset in addition to gene predictions from the genome (26), the application of a less stringent FDR (2% versus 1% in the current study), and the host contamination in O. ochengi samples that is more challenging to minimize than is the case for B. malayi (because of the nodular lifestyle of the former; see Experimental Procedures). In addition, there may be intrinsic differences in the dynamic range of protein expression between the two parasites; or a greater proportion of O. ochengi proteins may only be expressed in stages that were outside the scope of this study (mature Mf, L2 and L4). In general, the relative distribution of orthologous and non-orthologous proteins detected in each study across the lifecycle was similar, although vL3 was a significant outlier. This may reflect the challenges of obtaining O. ochengi vL3 in significant numbers for analysis, or fundamental differences in the complexity of the infective stage between the two species.
Proteins containing GST domains constituted one of the largest families that were enriched in AF and iuMf. The GSTs are a superfamily of multifunctional proteins that are ubiquitous across animals, plants and bacteria and are much studied in their cytosolic form because of their importance as phase II detoxification enzymes (130). In addition to detoxification, where typically they enzymatically conjugate glutathione with electrophilic, lipophilic, and nonpolar compounds including xenobiotics, GSTs have been reported to carry out a range of other functions. These include binding and transport of hydrophobic ligands (131); synthesis of eicosanoids with roles in immune modulation (132, 133); catabolism of amino acids (134); and inhibition of the Jun N-terminal kinase signaling pathway, thus protecting against hydrogen peroxide-induced apoptosis (135). Proposed functions of OvGST2 include the neutralization of lipid peroxidation products arising from immune-mediate damage to the parasite (77, 136). Conversely, OvGST1 is located in the outer hypodermal lamellae and cuticle, and is released in ESP from AF, suggesting it may have an immunomodulatory role at the host-parasite interface via the generation of prostanoids (137). Thus, our detection of an OvGST1 ortholog (nOo_09064) in NF is likely to reflect an equivalent function for this secreted GST in O. ochengi.
A striking feature of our study was the number of mitochondria-related proteins that were overrepresented in different stages, particularly in iuMf and vL3. Notably, it has been known for decades that most adult filariae depend primarily on fermentation of glucose for their energy requirements, whereas Mf also use oxidation of pyruvate obtained from catabolism of glucose or amino-acids (138, 139). The importance of aerobic respiration in iuMf is likely to place significant demands on mitochondrial function. Thus, as expected for an enzyme involved in protection of mitochondria from oxidative stress, deletion of the trxr-2 gene, which is mainly expressed in body wall and pharyngeal muscles in C. elegans, led to delayed development and reduced longevity under stress conditions (103). Moreover, inhibition of TrxR in a filarial parasite of cattle, Setaria cervi, impaired the motility and viability of adult worms and iuMf, and was associated with oxidative damage to lipids and proteins, leading to mitochondrial-mediated apoptosis (140). In B. malayi, many transcripts encoding mitochondrial metabolic enzymes were up-regulated in vL3 relative to L3 cultured in vitro for 2 days (20). During development in the insect vector, the transcriptional profile of B. malayi suggests that it primarily uses anaerobic dismutation of malate for its energy needs (22, 141), which occurs in the mitochondrial matrix (142). This process may be associated with an increase in the importation and processing of Krebs cycle enzymes in mitochondria, or vL3 may be “primed” to shift to oxidative phosphorylation for the energy-expensive processes of skin penetration and migration to the lymphatics in the definitive host (20). In this context, it may be informative to examine the interplay of mitochondria and Wolbachia during L3 to L4 development, as the latter replicate rapidly during this phase (68) and display high levels of expression for respiratory chain components (37).
Several of the mitochondrial proteins identified in our study have been proposed as drug targets, such as the nematode MPPs, which are poorly conserved relative to their mammalian counterparts (104). Moreover, the divergent TUFMs of nematodes, which exhibit unique C-terminal extensions, constitute attractive targets for highly specific anthelminthics (99). Recently, the gold-containing compound auranofin was identified as a lead adulticidal drug candidate that specifically inhibits filarial TrxR. Importantly, auranofin-treated AF of O. ochengi were found to be depleted of mitochondria (143).
A key discovery in NF was the presence of two TGF-β homologs. Bm-TGH-2 is a TGF-β family member that can be detected by Western blot in B. malayi ESP and can bind and signal through the mammalian TGF-β receptor (144). As TGF-β signaling can inhibit immune responses, suppressing T-cell proliferation and inducing regulatory T-cells, secretion of this filarial homolog has been proposed to have immunomodulatory effects. However, subsequent to the original identification of Bm-TGH-2, no proteomic analyses of B. malayi ESP have identified secreted TGF-B family members (23–25). In O. volvulus and O. ochengi, TGF-β family members were reported to be expressed in the basal layer of the cuticle, the intestine, and the reproductive tracts of male and female adult worms, including in the uterine and vaginal muscles, but no attempt to measure secretion was undertaken (145). In contrast, ESP from the gastrointestinal nematodes Heligmosomoides polygyrus and Teladorsagia circumcincta both contain TGF-β mimics that can signal through the mammalian TGF-β receptor, inducing regulatory T-cells in vitro (146). Although H. polygyrus also transcribes a TGF-β family member (147), this has been proposed to have a developmental role, and could not be detected in proteomic studies of H. polygyrus ESP (148, 149). Although it is unknown whether the O. ochengi TGF-β homologs identified here can signal through the mammalian TGF-β receptor in a similar manner to Bm-TGH-2, or simply have an internal developmental role associated with “spill-over” into the mammalian host, their presence in NF is suggestive of an active immunomodulatory function.
An initial analysis of the wOo proteome has been published, in which 122 proteins were identified from AF (37). The limited number of additional identifications achieved in the current study was anticipated, because the published analysis was based on a semipurified sample of bacteria, whereas no biological fractionation of O. ochengi was attempted here and Wolbachia proteomes have a substantial dynamic range (150). A comparison of the wOo proteome with that of its counterpart in B. malayi has been explored previously (37). It has been postulated from proteomic data obtained from Wolbachia within B. malayi, which was dominated by single-peptide identifications as in our study, that the endosymbionts exhibit extensive stage-specific expression during filarial development (26). Considering the stochastic nature of peptide identifications from very low abundance proteins, we find this scenario unlikely and in any case conflated by the substantial changes in Wolbachia density throughout the filarial lifecycle.
On the basis of the quantities of bovine antimicrobial proteins observed in NF, Wolbachia appears to be a major driver of innate immune responses in the nodule. Depletion of Wolbachia endosymbionts with antibiotics leads to a decline in neutrophilia in O. ochengi nodules and an influx of eosinophils, which degranulate on the parasite cuticle and ultimately kill the adult nematodes (36, 122). Wolbachia proteins that are known to activate neutrophils include WSP (151) and PAL (152), and GroEL may also have a role as there are precedents in other bacteria (153). Surprisingly, and in contrast with AF ESP from L. sigmodontis (43), we did not consistently detect Wolbachia proteins in NF. However, because WSP has been detected on the surface of B. malayi (154) and L. sigmodontis (43), symbiont proteins on the cuticle (rather than secreted products) may be a more important trigger for neutrophil activation in onchocerciasis. Both cathelicidins (124) and PGLYRP1 (155) have potent antibacterial activity, and might be liberated by neutrophils following activation of Toll-like receptors 2 and 4 by WSP (156). Importantly, although Wolbachia lacks a peptidoglycan-based cell wall (157), PGLYRP1 has been reported to kill other pathogens that lack peptidoglycan (158).
CONCLUSIONS
We have revealed a remarkable complexity and dynamism of the O. ochengi proteome throughout the major lifecycle stages, particularly with respect to mitochondrion-associated proteins, as well as the presence of TGF-β homologs in the O. ochengi secretome. In addition, the developmentally regulated protein families that have been identified in O. ochengi are represented by almost identical orthologs in O. volvulus, suggesting that their patterns of expression will be conserved in the human parasite. Although L3 have historically been the main focus of attention for vaccine design, our data on the overrepresentation of the peptidase M16 family suggests that it may be possible to design drugs to target this stage; whereas another potential source of vulnerability for both immunoprophylactic and chemotherapeutic approaches is embryogenesis. Indeed, mutated forms of cysteine proteinase inhibitor are highly effective at reducing female worm fertility in B. malayi (14) and microfilaraemia in L. sigmodontis (13), and immunologically targeting of triosephosphate isomerase can also impede embryogenesis in B. malayi (113). The evaluation of the ShK-domain protein as a vaccine in animal models is now a priority, and other components of the nodule secretome may constitute promising new diagnostic biomarkers if they can be shown to reach the peripheral circulation. In conclusion, the O. ochengi system in cattle will continue to play a critical role in our understanding not only of O. volvulus biology, but the means to its ultimate eradication.
Supplementary Material
Acknowledgments
We thank Youssouf Mouliom Mfopit (IRAD, Cameroon) for assistance in the harvesting of iuMf and NF; and Isaac Jato, Martin Manyo, Roland Enyong and Samuel Ngo (REFOTDE, Cameroon) for the collection of blood-fed blackflies. We also gratefully acknowledge the assistance of Catherine Hartley and Andrew Jones (University of Liverpool) with the collagenase digestion of nodules and the application of ProteoAnnotator to our dataset, respectively. We are indebted to Gaganjot Kaur and Georgios Koutsovoulos (University of Edinburgh) for assembly and annotation of the O. ochengi genome, and to Jim Geary (Michigan State University) and Timothy Geary (McGill University) for providing raw MS data for D. immitis from (48). We also thank Sara Lustigman (New York Blood Center), and James Cotton and Matthew Berriman (Wellcome Trust Sanger Institute), for early access to gene models from the O. volvulus genome.
Footnotes
Author contributions: S.D.A., D.X., S.W., P.A.E., D.W.T., J.M.W., V.N.T., and B.L.M. designed research; S.D.A., G.S.B., H.F.N., J.A.K., P.W.C., and B.L.M. performed research; S.D.A., D.X., R.K., E.J.L., H.J.M., M.L.B., and B.L.M. analyzed data; S.D.A., E.J.L., H.J.M., and B.L.M. wrote the paper.
* This study was supported by the 7th Framework Programme of the European Commission (project identifier HEALTH-F3-2010-242131). The funders played no part in the design, in the collection, analysis, and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Mf
- microfilariae
- AF
- adult female
- AM
- adult male
- vL3
- vector-derived third-stage infective larvae
- iuMf
- intrauterine microfilariae
- NF
- nodule fluid
- AEX
- anion exchange
- BR
- Britton-Robinson
- ESP
- excretory-secretory products
- WBE
- whole-body extracts
- Prx
- peroxiredoxin
- TGF
- transforming growth factor
- GST
- glutathione transferase
- MPP
- mitochondrial processing peptidase
- TrxR
- thioredoxin reductase
- PAG
- protein ambiguity group
- PAL
- peptidoglycan-associated lipoprotein
- PGLYRP1
- peptidoglycan recognition protein
- WSP
- Wolbachia surface protein
- wOo
- Wolbachia endosymbiont of Onchocerca ochengi.
REFERENCES
- 1. GBD 2013 Mortality and Causes of Death Collaborators (2015) Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385, 117–171 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4340604/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Plaisier A. P., Van Oortmarssen G. J., Remme J., and Habbema J. D. (1991) The reproductive lifespan of Onchocerca volvulus in West African savanna. Acta Trop. 48, 271–284 [DOI] [PubMed] [Google Scholar]
- 3. Schulz-Key H. (1990) Observations on the reproductive biology of Onchocerca volvulus. Acta Leiden 59, 27–44 [PubMed] [Google Scholar]
- 4. Anon. (2010) Working to overcome the global impact of neglected tropical diseases: First WHO report on neglected tropical diseases, In: Crompton D. W. T., and Peters P. (eds), World Health Organisation, Geneva [Google Scholar]
- 5. Crump A., Morel C. M., and Omura S. (2012) The onchocerciasis chronicle: from the beginning to the end? Trends Parasitol. 28, 280–288 [DOI] [PubMed] [Google Scholar]
- 6. Osei-Atweneboana M. Y., Eng J. K., Boakye D. A., Gyapong J. O., and Prichard R. K. (2007) Prevalence and intensity of Onchocerca volvulus infection and efficacy of ivermectin in endemic communities in Ghana: a two-phase epidemiological study. Lancet 369, 2021–2029 [DOI] [PubMed] [Google Scholar]
- 7. Nana-Djeunga H. C., Bourguinat C., Pion S. D., Bopda J., Kengne-Ouafo J. A., Njiokou F., Prichard R. K., Wanji S., Kamgno J., and Boussinesq M. (2014) Reproductive status of Onchocerca volvulus after ivermectin treatment in an ivermectin-naive and a frequently treated population from Cameroon. PLoS Negl. Trop. Dis. 8, e2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gardon J., Gardon-Wendel N., Demanga N., Kamgno J., Chippaux J. P., and Boussinesq M. (1997) Serious reactions after mass treatment of onchocerciasis with ivermectin in an area endemic for Loa loa infection. Lancet 350, 18–22 [DOI] [PubMed] [Google Scholar]
- 9. Mackenzie C. D., and Geary T. G. (2011) Flubendazole: a candidate macrofilaricide for lymphatic filariasis and onchocerciasis field programs. Expert Rev. Anti Infect. Ther. 9, 497–501 [DOI] [PubMed] [Google Scholar]
- 10. Taylor M. J., Hoerauf A., Townson S., Slatko B. E., and Ward S. A. (2014) Anti-Wolbachia drug discovery and development: safe macrofilaricides for onchocerciasis and lymphatic filariasis. Parasitology 141, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Longo M., Zanoncelli S., Colombo P. A., Harhay M. O., Scandale I., Mackenzie C., Geary T., Madrill N., and Mazue G. (2013) Effects of the benzimidazole anthelmintic drug flubendazole on rat embryos in vitro. Reprod. Toxicol. 36, 78–87 [DOI] [PubMed] [Google Scholar]
- 12. Hotez P. J., Bottazzi M. E., Zhan B., Makepeace B. L., Klei T. R., Abraham D., Taylor D. W., and Lustigman S. (2015) The Onchocerciasis Vaccine for Africa–TOVA–Initiative. PLoS Negl. Trop. Dis. 9, e0003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babayan S. A., Luo H., Gray N., Taylor D. W., and Allen J. E. (2012) Deletion of parasite immune modulatory sequences combined with immune activating signals enhances vaccine mediated protection against filarial nematodes. PLoS Negl. Trop. Dis. 6, e1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arumugam S., Wei J., Ward D., Abraham D., Lustigman S., Zhan B., and Klei T. R. (2014) Vaccination with a genetically modified Brugia malayi cysteine protease inhibitor-2 reduces adult parasite numbers and affects the fertility of female worms following a subcutaneous challenge of Mongolian gerbils (Meriones unguiculatus) with B. malayi infective larvae. Int. J. Parasitol. 44, 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boatin B. A., Toe L., Alley E. S., Nagelkerke N. J., Borsboom G., and Habbema J. D. (2002) Detection of Onchocerca volvulus infection in low prevalence areas: a comparison of three diagnostic methods. Parasitology 125, 545–552 [PubMed] [Google Scholar]
- 16. Burbelo P. D., Leahy H. P., Iadarola M. J., and Nutman T. B. (2009) A four-antigen mixture for rapid assessment of Onchocerca volvulus infection. PLoS Negl. Trop. Dis. 3, e438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gopal H., Hassan H. K., Rodriguez-Perez M. A., Toe L. D., Lustigman S., and Unnasch T. R. (2012) Oligonucleotide based magnetic bead capture of Onchocerca volvulus DNA for PCR pool screening of vector black flies. PLoS Negl. Trop. Dis. 6, e1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Globisch D., Moreno A. Y., Hixon M. S., Nunes A. A., Denery J. R., Specht S., Hoerauf A., and Janda K. D. (2013) Onchocerca volvulus-neurotransmitter tyramine is a biomarker for river blindness. Proc. Natl. Acad. Sci. U.S.A. 110, 4218–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J. E., Delcher A. L., Guiliano D. B., Miranda-Saavedra D., Angiuoli S. V., Creasy T., Amedeo P., Haas B., El-Sayed N. M., Wortman J. R., Feldblyum T., Tallon L., Schatz M., Shumway M., Koo H., Salzberg S. L., Schobel S., Pertea M., Pop M., White O., Barton G. J., Carlow C. K., Crawford M. J., Daub J., Dimmic M. W., Estes C. F., Foster J. M., Ganatra M., Gregory W. F., Johnson N. M., Jin J., Komuniecki R., Korf I., Kumar S., Laney S., Li B. W., Li W., Lindblom T. H., Lustigman S., Ma D., Maina C. V., Martin D. M., McCarter J. P., McReynolds L., Mitreva M., Nutman T. B., Parkinson J., Peregrin-Alvarez J. M., Poole C., Ren Q., Saunders L., Sluder A. E., Smith K., Stanke M., Unnasch T. R., Ware J., Wei A. D., Weil G., Williams D. J., Zhang Y., Williams S. A., Fraser-Liggett C., Slatko B., Blaxter M. L., and Scott A. L. (2007) Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li B. W., Rush A. C., Mitreva M., Yin Y., Spiro D., Ghedin E., and Weil G. J. (2009) Transcriptomes and pathways associated with infectivity, survival and immunogenicity in Brugia malayi L3. BMC. Genomics 10, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choi Y. J., Ghedin E., Berriman M., McQuillan J., Holroyd N., Mayhew G. F., Christensen B. M., and Michalski M. L. (2011) A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi. PLoS Negl. Trop. Dis. 5, e1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reference deleted in final version.
- 23. Hewitson J. P., Harcus Y. M., Curwen R. S., Dowle A. A., Atmadja A. K., Ashton P. D., Wilson A., and Maizels R. M. (2008) The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol. Biochem. Parasitol. 160, 8–21 [DOI] [PubMed] [Google Scholar]
- 24. Moreno Y., and Geary T. G. (2008) Stage- and gender-specific proteomic analysis of Brugia malayi excretory-secretory products. PLoS Negl. Trop. Dis. 2, e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennuru S., Semnani R., Meng Z., Ribeiro J. M., Veenstra T. D., and Nutman T. B. (2009) Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl. Trop. Dis. 3, e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bennuru S., Meng Z., Ribeiro J. M., Semnani R. T., Ghedin E., Chan K., Lucas D. A., Veenstra T. D., and Nutman T. B. (2011) Stage-specific proteomic expression patterns of the human filarial parasite Brugia malayi and its endosymbiont Wolbachia. Proc. Natl. Acad. Sci. U.S.A. 108, 9649–9654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris C. P., Evans H., Larsen S. E., and Mitre E. (2013) A comprehensive, model-based review of vaccine and repeat infection trials for filariasis. Clin. Microbiol. Rev. 26, 381–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michalski M. L., Griffiths K. G., Williams S. A., Kaplan R. M., and Moorhead A. R. (2011) The NIH-NIAID Filariasis Research Reagent Resource Center. PLoS Negl. Trop. Dis. 5, e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burchard G. D., Büttner D. W., and Bierther M. (1979) Electron microscopical studies on onchocerciasis. III. The Onchocerca nodule. Tropenmed. Parasitol. 30, 103–112 [PubMed] [Google Scholar]
- 30. Desjardins C. A., Cerqueira G. C., Goldberg J. M., Dunning Hotopp J. C., Haas B. J., Zucker J., Ribeiro J. M., Saif S., Levin J. Z., Fan L., Zeng Q., Russ C., Wortman J. R., Fink D. L., Birren B. W., and Nutman T. B. (2013) Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat. Genet. 45, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Godel C., Kumar S., Koutsovoulos G., Ludin P., Nilsson D., Comandatore F., Wrobel N., Thompson M., Schmid C. D., Goto S., Bringaud F., Wolstenholme A., Bandi C., Epe C., Kaminsky R., Blaxter M., and Maser P. (2012) The genome of the heartworm, Dirofilaria immitis, reveals drug and vaccine targets. FASEB J. 26, 4650–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie H., Bain O., and Williams S. A. (1994) Molecular phylogenetic studies on filarial parasites based on 5S ribosomal spacer sequences. Parasite 1, 141–151 [DOI] [PubMed] [Google Scholar]
- 33. Morales-Hojas R., Cheke R. A., and Post R. J. (2006) Molecular systematics of five Onchocerca species (Nematoda: Filarioidea) including the human parasite, O. volvulus, suggest sympatric speciation. J. Helminthol. 80, 281–290 [PubMed] [Google Scholar]
- 34. Trees A. J., Wood V. L., Bronsvoort M., Renz A., and Tanya V. N.. Animal models - Onchocerca ochengi and the development of chemotherapeutic and chemoprophylactic agents for onchocerciasis. Ann. Trop. Med. Parasitol. 92 Suppl 1:S175–S179, 1998 [PubMed] [Google Scholar]
- 35. Makepeace B. L., Jensen S. A., Laney S. J., Nfon C. K., Njongmeta L. M., Tanya V. N., Williams S. A., Bianco A. E., and Trees A. J. (2009) Immunisation with a multivalent, subunit vaccine reduces patent infection in a natural bovine model of onchocerciasis during intense field exposure. PLoS Negl. Trop. Dis. 3, e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hansen R. D., Trees A. J., Bah G. S., Hetzel U., Martin C., Bain O., Tanya V. N., and Makepeace B. L. (2011) A worm's best friend: recruitment of neutrophils by Wolbachia confounds eosinophil degranulation against the filarial nematode Onchocerca ochengi. Proc. R. Soc. B 278, 2293–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Darby A. C., Armstrong S. D., Bah G. S., Kaur G., Hughes M. A., Kay S. M., Koldkjaer P., Rainbow L., Radford A. D., Blaxter M. L., Tanya V. N., Trees A. J., Cordaux R., Wastling J. M., and Makepeace B. L. (2012) Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 22, 2467–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wahl G., Achukwi M. D., Mbah D., Dawa O., and Renz A. (1994) Bovine onchocercosis in north Cameroon. Vet. Parasitol. 52, 297–311 [DOI] [PubMed] [Google Scholar]
- 39. Tchakouté V. L., Graham S. P., Jensen S. A., Makepeace B. L., Nfon C. K., Njongmeta L. M., Lustigman S., Enyong P. A., Tanya V. N., Bianco A. E., and Trees A. J. (2006) In a bovine model of onchocerciasis, protective immunity exists naturally, is absent in drug-cured hosts, and is induced by vaccination. Proc. Natl. Acad. Sci. U.S.A. 103, 5971–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quintana J. F., Makepeace B. L., Babayan S. A., Ivens A., Pfarr K. M., Blaxter M., Debrah A., Wanji S., Ngangyung H. F., Bah G. S., Tanya V. N., Taylor D. W., Hoerauf A., and Buck A. H. (2015) Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasit. Vectors. 8, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schulz-Key H., and Karam M. (1984) Quantitative assessment of microfilariae and adults of Onchocerca volvulus in ethanol-fixed biopsies and nodules. Trans. R. Soc. Trop. Med. Hyg. 78, 157–159 [DOI] [PubMed] [Google Scholar]
- 42. Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L., Tate J., and Punta M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Armstrong S. D., Babayan S. A., Lhermitte-Vallarino N., Gray N., Xia D., Martin C., Kumar S., Taylor D. W., Blaxter M. L., Wastling J. M., and Makepeace B. L. (2014) Comparative analysis of the secretome from a model filarial nematode (Litomosoides sigmodontis) reveals maximal diversity in gravid female parasites. Mol. Cell. Proteomics 13, 2527–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate- a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. 57, 289–300 [Google Scholar]
- 45. Wisniewski J. R., Zougman A., and Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 46. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 47. Chawade A., Alexandersson E., and Levander F. (2014) Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 13, 3114–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Geary J., Satti M., Moreno Y., Madrill N., Whitten D., Headley S. A., Agnew D., Geary T., and Mackenzie C. (2012) First analysis of the secretome of the canine heartworm, Dirofilaria immitis. Parasit. Vectors. 5, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ternent T., Csordas A., Qi D., Gomez-Baena G., Beynon R. J., Jones A. R., Hermjakob H., and Vizcaino J. A. (2014) How to submit MS proteomics data to ProteomeXchange via the PRIDE database. Proteomics 14, 2233–2241 [DOI] [PubMed] [Google Scholar]
- 50. Parra G., Bradnam K., and Korf I. (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 51. Ghali F., Krishna R., Perkins S., Collins A., Xia D., Wastling J., and Jones A. R. (2014) ProteoAnnotator–open source proteogenomics annotation software supporting PSI standards. Proteomics 14, 2731–2741 [DOI] [PubMed] [Google Scholar]
- 52. Wu S., Zhu Z., Fu L., Niu B., and Li W. (2011) WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC. Genomics 12, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R. D., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Laugraud A., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., Mistry J., Mitchell A., Mulder N., Natale D., Orengo C., Quinn A. F., Selengut J. D., Sigrist C. J., Thimma M., Thomas P. D., Valentin F., Wilson D., Wu C. H., and Yeats C. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von M. C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petersen T. N., Brunak S., von H. G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 57. Bendtsen J. D., Jensen L. J., Blom N., von H. G., and Brunak S. (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 58. Krogh A., Larsson B., von H. G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 59. Duckert P., Brunak S., and Blom N. (2004) Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel 17, 107–112 [DOI] [PubMed] [Google Scholar]
- 60. Claros M. G., and Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 61. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 62. Altschul S. F., Wootton J. C., Gertz E. M., Agarwala R., Morgulis A., Schaffer A. A., and Yu Y. K. (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saitou N., and Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 65. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 66. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., and Higgins D. G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Page R. D. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 68. McGarry H. F., Egerton G. L., and Taylor M. J. (2004) Population dynamics of Wolbachia bacterial endosymbionts in Brugia malayi. Mol. Biochem. Parasitol. 135, 57–67 [DOI] [PubMed] [Google Scholar]
- 69. Gilbert J., Nfon C. K., Makepeace B. L., Njongmeta L. M., Hastings I. M., Pfarr K. M., Renz A., Tanya V. N., and Trees A. J. (2005) Antibiotic chemotherapy of onchocerciasis: in a bovine model, killing of adult parasites requires a sustained depletion of endosymbiotic bacteria (Wolbachia species). J. Infect. Dis. 192, 1483–1493 [DOI] [PubMed] [Google Scholar]
- 70. Joseph G. T., Huima T., Klion A., and Lustigman S. (2000) A novel developmentally regulated galectin of Onchocerca volvulus. Mol. Biochem. Parasitol. 106, 187–195 [DOI] [PubMed] [Google Scholar]
- 71. Klion A. D., and Donelson J. E. (1994) OvGalBP, a filarial antigen with homology to vertebrate galactoside-binding proteins. Mol. Biochem. Parasitol. 65, 305–315 [DOI] [PubMed] [Google Scholar]
- 72. Adam R., Kaltmann B., Rudin W., Friedrich T., Marti T., and Lucius R. (1996) Identification of chitinase as the immunodominant filarial antigen recognized by sera of vaccinated rodents. J. Biol. Chem. 271, 1441–1447 [DOI] [PubMed] [Google Scholar]
- 73. Wu Y., Egerton G., Underwood A. P., Sakuda S., and Bianco A. E. (2001) Expression and secretion of a larval-specific chitinase (family 18 glycosyl hydrolase) by the infective stages of the parasitic nematode, Onchocerca volvulus. J. Biol. Chem. 276, 42557–42564 [DOI] [PubMed] [Google Scholar]
- 74. Lustigman S., Zhang J., Liu J., Oksov Y., and Hashmi S. (2004) RNA interference targeting cathepsin L and Z-like cysteine proteases of Onchocerca volvulus confirmed their essential function during L3 molting. Mol. Biochem. Parasitol. 138, 165–170 [DOI] [PubMed] [Google Scholar]
- 75. Guiliano D. B., Hong X., McKerrow J. H., Blaxter M. L., Oksov Y., Liu J., Ghedin E., and Lustigman S. (2004) A gene family of cathepsin L-like proteases of filarial nematodes are associated with larval molting and cuticle and eggshell remodeling. Mol. Biochem. Parasitol. 136, 227–242 [DOI] [PubMed] [Google Scholar]
- 76. Lundquist E. A. (2006) Small GTPases, In: Greenwald I. (ed), WormBook, pp. 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liebau E., Wildenburg G., Brophy P. M., Walter R. D., and Henkle-Dührsen K. (1996) Biochemical analysis, gene structure and localization of the 24 kDa glutathione S-transferase from Onchocerca volvulus. Mol. Biochem. Parasitol. 80, 27–39 [DOI] [PubMed] [Google Scholar]
- 78. Liebau E., Wildenburg G., Walter R. D., and Henkle-Dührsen K. (1994) A novel type of glutathione S-transferase in Onchocerca volvulus. Infect. Immun. 62, 4762–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liebau E., Hoppner J., Muhlmeister M., Burmeister C., Luersen K., Perbandt M., Schmetz C., Buttner D., and Brattig N. (2008) The secretory omega-class glutathione transferase OvGST3 from the human pathogenic parasite Onchocerca volvulus. FEBS J. 275, 3438–3453 [DOI] [PubMed] [Google Scholar]
- 80. Liebau E., Eschbach M. L., Tawe W., Sommer A., Fischer P., Walter R. D., and Henkle-Dührsen K. (2000) Identification of a stress-responsive Onchocerca volvulus glutathione S-transferase (Ov-GST-3) by RT-PCR differential display. Mol. Biochem. Parasitol. 109, 101–110 [DOI] [PubMed] [Google Scholar]
- 81. Tanikawa N., Ohmiya Y., Ohkubo H., Hashimoto K., Kangawa K., Kojima M., Ito S., and Watanabe K. (2002) Identification and characterization of a novel type of membrane-associated prostaglandin E synthase. Biochem. Biophys. Res. Commun. 291, 884–889 [DOI] [PubMed] [Google Scholar]
- 82. Littler D. R., Brown L. J., Breit S. N., Perrakis A., and Curmi P. M. (2010) Structure of human CLIC3 at 2 Å resolution. Proteins 78, 1594–1600 [DOI] [PubMed] [Google Scholar]
- 83. Koonin E. V., Mushegian A. R., Tatusov R. L., Altschul S. F., Bryant S. H., Bork P., and Valencia A. (1994) Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain–study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 3, 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Armstrong L. C., Saenz A. J., and Bornstein P. (1999) Metaxin 1 interacts with metaxin 2, a novel related protein associated with the mammalian mitochondrial outer membrane. J. Cell. Biochem. 74, 11–22 [PubMed] [Google Scholar]
- 85. Xie J., Marusich M. F., Souda P., Whitelegge J., and Capaldi R. A. (2007) The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 581, 3545–3549 [DOI] [PubMed] [Google Scholar]
- 86. Berry K. L., Bulow H. E., Hall D. H., and Hobert O. (2003) A C. elegans CLIC-like protein required for intracellular tube formation and maintenance. Science 302, 2134–2137 [DOI] [PubMed] [Google Scholar]
- 87. Gretes M. C., Poole L. B., and Karplus P. A. (2012) Peroxiredoxins in parasites. Antioxid. Redox. Signal. 17, 608–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lu W., Egerton G. L., Bianco A. E., and Williams S. A. (1998) Thioredoxin peroxidase from Onchocerca volvulus: a major hydrogen peroxide detoxifying enzyme in filarial parasites. Mol. Biochem. Parasitol. 91, 221–235 [DOI] [PubMed] [Google Scholar]
- 89. Ghosh I., Eisinger S. W., Raghavan N., and Scott A. L. (1998) Thioredoxin peroxidases from Brugia malayi. Mol. Biochem. Parasitol. 91, 207–220 [DOI] [PubMed] [Google Scholar]
- 90. Ranjan M., Gruber J., Ng L. F., and Halliwell B. (2013) Repression of the mitochondrial peroxiredoxin antioxidant system does not shorten life span but causes reduced fitness in Caenorhabditis elegans. Free Radic. Biol. Med. 63, 381–389 [DOI] [PubMed] [Google Scholar]
- 91. Zipfel P. F., Schrum S., Bialonski A., and Buttner D. W. (1998) The peroxidoxin 2 protein of the human parasite Onchocerca volvulus: recombinant expression, immunolocalization, and demonstration of homologous molecules in other species. Parasitol. Res. 84, 623–631 [DOI] [PubMed] [Google Scholar]
- 92. Wilson W. R., Tuan R. S., Shepley K. J., Freedman D. O., Greene B. M., Awadzi K., and Unnasch T. R. (1994) The Onchocerca volvulus homologue of the multifunctional polypeptide protein disulfide isomerase. Mol. Biochem. Parasitol. 68, 103–117 [DOI] [PubMed] [Google Scholar]
- 93. Ko F. C., and Chow K. L. (2002) A novel thioredoxin-like protein encoded by the C. elegans dpy-11 gene is required for body and sensory organ morphogenesis. Development 129, 1185–1194 [DOI] [PubMed] [Google Scholar]
- 94. Chandrashekar R., Tsuji N., Morales T., Ozols V., and Mehta K. (1998) An ERp60-like protein from the filarial parasite Dirofilaria immitis has both transglutaminase and protein disulfide isomerase activity. Proc. Natl. Acad. Sci. U.S.A. 95, 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ichishita R., Tanaka K., Sugiura Y., Sayano T., Mihara K., and Oka T. (2008) An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J. Biochem. 143, 449–454 [DOI] [PubMed] [Google Scholar]
- 96. Kanazawa T., Zappaterra M. D., Hasegawa A., Wright A. P., Newman-Smith E. D., Buttle K. F., McDonald K., Mannella C. A., and van der Bliek A. M. (2008) The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 4, e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yasuda K., Hartman P. S., Ishii T., Suda H., Akatsuka A., Shoyama T., Miyazawa M., and Ishii N. (2011) Interrelationships between mitochondrial fusion, energy metabolism and oxidative stress during development in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 404, 751–755 [DOI] [PubMed] [Google Scholar]
- 98. Westermann B. (2010) Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin. Cell Dev. Biol. 21, 542–549 [DOI] [PubMed] [Google Scholar]
- 99. Ohtsuki T., and Watanabe Y. (2007) T-armless tRNAs and elongated elongation factor Tu. IUBMB Life 59, 68–75 [DOI] [PubMed] [Google Scholar]
- 100. Ohtsuki T., Sato A., Watanabe Y., and Watanabe K. (2002) A unique serine-specific elongation factor Tu found in nematode mitochondria. Nat. Struct. Biol. 9, 669–673 [DOI] [PubMed] [Google Scholar]
- 101. Joza N., Pospisilik J. A., Hangen E., Hanada T., Modjtahedi N., Penninger J. M., and Kroemer G. (2009) AIF: not just an apoptosis-inducing factor. Ann. N.Y. Acad. Sci. 1171, 2–11 [DOI] [PubMed] [Google Scholar]
- 102. Butler J. A., Mishur R. J., Bhaskaran S., and Rea S. L. (2013) A metabolic signature for long life in the Caenorhabditis elegans Mit mutants. Aging Cell 12, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li W., Bandyopadhyay J., Hwaang H. S., Park B. J., Cho J. H., Lee J. I., Ahnn J., and Lee S. K. (2012) Two thioredoxin reductases, trxr-1 and trxr-2, have differential physiological roles in Caenorhabditis elegans. Mol. Cells 34, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nomura H., Athauda S. B., Wada H., Maruyama Y., Takahashi K., and Inoue H. (2006) Identification and reverse genetic analysis of mitochondrial processing peptidase and the core protein of the cytochrome bc1 complex of Caenorhabditis elegans, a model parasitic nematode. J. Biochem. 139, 967–979 [DOI] [PubMed] [Google Scholar]
- 105. Berry E. A., De B. H., and Huang L. S. (2013) Unanswered questions about the structure of cytochrome bc1 complexes. Biochim. Biophys. Acta 1827, 1258–1277 [DOI] [PubMed] [Google Scholar]
- 106. Mossmann D., Meisinger C., and Vogtle F. N. (2012) Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819, 1098–1106 [DOI] [PubMed] [Google Scholar]
- 107. Lustigman S., Brotman B., Huima T., Prince A. M., and McKerrow J. H. (1992) Molecular cloning and characterization of onchocystatin, a cysteine proteinase inhibitor of Onchocerca volvulus. J. Biol. Chem. 267, 17339–17346 [PubMed] [Google Scholar]
- 108. Willenbucher J., Hofle W., and Lucius R. (1993) The filarial antigens Av33/Ov33–3 show striking similarities to the major pepsin inhibitor from Ascaris suum. Mol. Biochem. Parasitol. 57, 349–351 [DOI] [PubMed] [Google Scholar]
- 109. Erttmann K. D., and Gallin M. Y. (1996) Onchocerca volvulus: identification of cDNAs encoding a putative phosphatidyl-ethanolamine-binding protein and a putative partially processed mRNA precursor. Gene 174, 203–207 [DOI] [PubMed] [Google Scholar]
- 110. Gallin M. Y., Tan M., Kron M. A., Rechnitzer D., Greene B. M., Newland H. S., White A. T., Taylor H. R., and Unnasch T. R. (1989) Onchocerca volvulus recombinant antigen: physical characterization and clinical correlates with serum reactivity. J. Infect. Dis. 160, 521–529 [DOI] [PubMed] [Google Scholar]
- 111. Yamaguchi Y., Hasegawa Y., Honma T., Nagashima Y., and Shiomi K. (2010) Screening and cDNA cloning of Kv1 potassium channel toxins in sea anemones. Mar. Drugs 8, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Chhabra S., Chang S. C., Nguyen H. M., Huq R., Tanner M. R., Londono L. M., Estrada R., Dhawan V., Chauhan S., Upadhyay S. K., Gindin M., Hotez P. J., Valenzuela J. G., Mohanty B., Swarbrick J. D., Wulff H., Iadonato S. P., Gutman G. A., Beeton C., Pennington M. W., Norton R. S., and Chandy K. G. (2014) Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases. FASEB J. 28, 3952–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hewitson J. P., Ruckerl D., Harcus Y., Murray J., Webb L. M., Babayan S. A., Allen J. E., Kurniawan A., and Maizels R. M. (2014) The secreted triose phosphate isomerase of Brugia malayi is required to sustain microfilaria production in vivo. PLoS Pathog. 10, e1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Frank G. R., Wisnewski N., Brandt K. S., Carter C. R., Jennings N. S., and Selkirk M. E. (1999) Molecular cloning of the 22–24 kDa excretory-secretory 22U protein of Dirofilaria immitis and other filarial nematode parasites. Mol. Biochem. Parasitol. 98, 297–302 [DOI] [PubMed] [Google Scholar]
- 115. Constam D. B. (2014) Regulation of TGFbeta and related signals by precursor processing. Semin. Cell Dev. Biol. 32, 85–97 [DOI] [PubMed] [Google Scholar]
- 116. Rogalski T. M., Mullen G. P., Bush J. A., Gilchrist E. J., and Moerman D. G. (2001) UNC-52/perlecan isoform diversity and function in Caenorhabditis elegans. Biochem. Soc. Trans. 29, 171–176 [DOI] [PubMed] [Google Scholar]
- 117. Yang Y., Lee W. S., Tang X., and Wadsworth W. G. (2014) Extracellular matrix regulates UNC-6 (netrin) axon guidance by controlling the direction of intracellular UNC-40 (DCC) outgrowth activity. PLoS ONE 9, e97258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Vogel B. E., Muriel J. M., Dong C., and Xu X. (2006) Hemicentins: what have we learned from worms? Cell Res. 16, 872–878 [DOI] [PubMed] [Google Scholar]
- 119. Gotenstein J. R., Swale R. E., Fukuda T., Wu Z., Giurumescu C. A., Goncharov A., Jin Y., and Chisholm A. D. (2010) The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development 137, 3603–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Burket C. T., Higgins C. E., Hull L. C., Berninsone P. M., and Ryder E. F. (2006) The C. elegans gene dig-1 encodes a giant member of the immunoglobulin superfamily that promotes fasciculation of neuronal processes. Dev. Biol. 299, 193–205 [DOI] [PubMed] [Google Scholar]
- 121. Brattig N. W., Büttner D. W., and Hoerauf A. (2001) Neutrophil accumulation around Onchocerca worms and chemotaxis of neutrophils are dependent on Wolbachia endobacteria. Microbes Infect. 3, 439–446 [DOI] [PubMed] [Google Scholar]
- 122. Nfon C. K., Makepeace B. L., Njongmeta L. M., Tanya V. N., Bain O., and Trees A. J. (2006) Eosinophils contribute to killing of adult Onchocerca ochengi within onchocercomata following elimination of Wolbachia. Microbes Infect. 8, 2698–2705 [DOI] [PubMed] [Google Scholar]
- 123. Scocchi M., Wang S., and Zanetti M. (1997) Structural organization of the bovine cathelicidin gene family and identification of a novel member. FEBS Lett. 417, 311–315 [DOI] [PubMed] [Google Scholar]
- 124. Zanetti M. (2005) The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 7, 179–196 [PubMed] [Google Scholar]
- 125. Cooray R., Waller K. P., and Venge P. (2007) Haptoglobin comprises about 10% of granule protein extracted from bovine granulocytes isolated from healthy cattle. Vet. Immunol. Immunopathol. 119, 310–315 [DOI] [PubMed] [Google Scholar]
- 126. Pant S. D., Verschoor C. P., Schenkel F. S., You Q., Kelton D. F., and Karrow N. A. (2011) Bovine PGLYRP1 polymorphisms and their association with resistance to Mycobacterium avium ssp. paratuberculosis. Anim Genet. 42, 354–360 [DOI] [PubMed] [Google Scholar]
- 127. Roth J., Vogl T., Sorg C., and Sunderkotter C. (2003) Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 24, 155–158 [DOI] [PubMed] [Google Scholar]
- 128. Marti T., Erttmann K. D., and Gallin M. Y. (1996) Host-parasite interaction in human onchocerciasis: identification and sequence analysis of a novel human calgranulin. Biochem. Biophys. Res. Commun. 221, 454–458 [DOI] [PubMed] [Google Scholar]
- 129. Gottsch J. D., Eisinger S. W., Liu S. H., and Scott A. L. (1999) Calgranulin C has filariacidal and filariastatic activity. Infect. Immun. 67, 6631–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sheehan D., Meade G., Foley V. M., and Dowd C. A. (2001) Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Brophy P. M., and Pritchard D. I. (1994) Parasitic helminth glutathione S-transferases: an update on their potential as targets for immuno- and chemotherapy. Exp. Parasitol. 79, 89–96 [DOI] [PubMed] [Google Scholar]
- 132. Nicholson D. W., Ali A., Vaillancourt J. P., Calaycay J. R., Mumford R. A., Zamboni R. J., and Ford-Hutchinson A. W. (1993) Purification to homogeneity and the N-terminal sequence of human leukotriene C4 synthase: a homodimeric glutathione S-transferase composed of 18-kDa subunits. Proc. Natl. Acad. Sci. U.S.A. 90, 2015–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Meyer D. J., and Thomas M. (1995) Characterization of rat spleen prostaglandin H D-isomerase as a sigma-class GSH transferase. Biochem. J. 311, 739–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Board P. G., Baker R. T., Chelvanayagam G., and Jermiin L. S. (1997) Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem. J. 328, 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Tew K. D., and Ronai Z. (1999) GST function in drug and stress response. Drug Resist. Updat. 2, 143–147 [DOI] [PubMed] [Google Scholar]
- 136. Wildenburg G., Liebau E., and Henkle-Dührsen K. (1998) Onchocerca volvulus: ultrastructural localization of two glutathione S-transferases. Exp. Parasitol. 88, 34–42 [DOI] [PubMed] [Google Scholar]
- 137. Sommer A., Rickert R., Fischer P., Steinhart H., Walter R. D., and Liebau E. (2003) A dominant role for extracellular glutathione S-transferase from Onchocerca volvulus is the production of prostaglandin D2. Infect. Immun. 71, 3603–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Rew R. S., and Saz H. J. (1977) The carbohydrate metabolism of Brugia pahangi microfilariae. J. Parasitol. 63, 123–129 [PubMed] [Google Scholar]
- 139. Srivastava V. M., Saz H. J., and deBruyn B. (1988) Comparisons of glucose and amino acid use in adults and microfilariae of Brugia pahangi. Parasitol. Res. 75, 1–6 [DOI] [PubMed] [Google Scholar]
- 140. Tiwari S., Wadhawan M., Singh N., and Rathaur S. (2015) Effect of CDNB on filarial thioredoxin reductase : A proteomic and biochemical approach. J. Proteomics 113, 435–446 [DOI] [PubMed] [Google Scholar]
- 141. Reference deleted in final version.
- 142. Edwards C. B., Copes N., Brito A. G., Canfield J., and Bradshaw P. C. (2013) Malate and fumarate extend lifespan in Caenorhabditis elegans. PLoS ONE 8, e58345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bulman C. A., Bidlow C. M., Lustigman S., Cho-Ngwa F., Williams D., Rascon A. A. Jr., Tricoche N., Samje M., Bell A., Suzuki B., Lim K. C., Supakorndej N., Supakorndej P., Wolfe A. R., Knudsen G. M., Chen S., WILSON C., Ang K. H., Arkin M., Gut J., Franklin C., Marcellino C., McKerrow J. H., Debnath A., and Sakanari J. A. (2015) Repurposing auranofin as a lead candidate for treatment of lymphatic filariasis and onchocerciasis. PLoS Negl. Trop. Dis. 9, e0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Gomez-Escobar N., Gregory W. F., and Maizels R. M. (2000) Identification of tgh-2, a filarial nematode homolog of Caenorhabditis elegans daf-7 and human transforming growth factor beta, expressed in microfilarial and adult stages of Brugia malayi. Infect. Immun. 68, 6402–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Korten S., Buttner D. W., Schmetz C., Hoerauf A., Mand S., and Brattig N. (2009) The nematode parasite Onchocerca volvulus generates the transforming growth factor-beta (TGF-beta). Parasitol. Res. 105, 731–741 [DOI] [PubMed] [Google Scholar]
- 146. Grainger J. R., Smith K. A., Hewitson J. P., McSorley H. J., Harcus Y., Filbey K. J., Finney C. A., Greenwood E. J., Knox D. P., Wilson M. S., Belkaid Y., Rudensky A. Y., and Maizels R. M. (2010) Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-beta pathway. J. Exp. Med. 207, 2331–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. McSorley H. J., Grainger J. R., Harcus Y., Murray J., Nisbet A. J., Knox D. P., and Maizels R. M. (2010) daf-7-related TGF-beta homologues from trichostrongyloid nematodes show contrasting life-cycle expression patterns. Parasitology 137, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Hewitson J. P., Ivens A. C., Harcus Y., Filbey K. J., McSorley H. J., Murray J., Bridgett S., Ashford D., Dowle A. A., and Maizels R. M. (2013) Secretion of protective antigens by tissue-stage nematode larvae revealed by proteomic analysis and vaccination-induced sterile immunity. PLoS Pathog. 9, e1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Moreno Y., Gros P. P., Tam M., Segura M., Valanparambil R., Geary T. G., and Stevenson M. M. (2011) Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with next-generation sequencing transcriptomic information. PLoS Negl. Trop. Dis. 5, e1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Darby A. C., Christina G. A., Armstrong S. D., Hartley C. S., Xia D., Wastling J. M., and Makepeace B. L. (2014) Integrated transcriptomic and proteomic analysis of the global response of Wolbachia to doxycycline-induced stress. ISME. J. 8, 925–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bazzocchi C., Genchi C., Paltrinieri S., Lecchi C., Mortarino M., and Bandi C. (2003) Immunological role of the endosymbionts of Dirofilaria immitis: the Wolbachia surface protein activates canine neutrophils with production of IL-8. Vet. Parasitol. 117, 73–83 [DOI] [PubMed] [Google Scholar]
- 152. Tamarozzi F., Wright H. L., Johnston K. L., Edwards S. W., Turner J. D., and Taylor M. J. (2014) Human filarial Wolbachia lipopeptide directly activates human neutrophils in vitro. Parasite Immunol. 36, 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Argueta J. G., Shiota S., Yamaguchi N., Masuhiro Y., and Hanazawa S. (2006) Induction of Porphyromonas gingivalis GroEL signaling via binding to Toll-like receptors 2 and 4. Oral Microbiol. Immunol. 21, 245–251 [DOI] [PubMed] [Google Scholar]
- 154. Melnikow E., Xu S., Liu J., Li L., Oksov Y., Ghedin E., Unnasch T. R., and Lustigman S. (2011) Interaction of a Wolbachia WSP-like protein with a nuclear-encoded protein of Brugia malayi. Int. J. Parasitol. 41, 1053–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Liu C., Xu Z., Gupta D., and Dziarski R. (2001) Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 276, 34686–34694 [DOI] [PubMed] [Google Scholar]
- 156. Brattig N. W., Bazzocchi C., Kirschning C. J., Reiling N., Büttner D. W., Ceciliani F., Geisinger F., Hochrein H., Ernst M., Wagner H., Bandi C., and Hoerauf A. (2004) The major surface protein of Wolbachia endosymbionts in filarial nematodes elicits immune responses through TLR2 and TLR4. J. Immunol. 173, 437–445 [DOI] [PubMed] [Google Scholar]
- 157. Henrichfreise B., Schiefer A., Schneider T., Nzukou E., Poellinger C., Hoffmann T. J., Johnston K. L., Moelleken K., Wiedemann I., Pfarr K., Hoerauf A., and Sahl H. G. (2009) Functional conservation of the lipid II biosynthesis pathway in the cell wall-less bacteria Chlamydia and Wolbachia: why is lipid II needed? Mol. Microbiol. 73, 913–923 [DOI] [PubMed] [Google Scholar]
- 158. Guan R., Malchiodi E. L., Wang Q., Schuck P., and Mariuzza R. A. (2004) Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha. J. Biol. Chem. 279, 31873–31882 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.