Abstract
Prenatal hydronephrosis is a common condition that may spontaneously resolve after birth. However, this condition can result in renal damage and requires surgical correction in a number of cases. Preventing renal damage is paramount, but existing diagnostic technology is invasive, exposes infants to radiation, is costly, and is often indeterminate. A better understanding of the pathophysiology of renal obstruction as reflected in the urinary proteome may provide new insights into the disease that could potentially alter the clinical management of hydronephrosis. We performed a quantitative proteomics study of urine that was surgically obtained from eight clinically significant, unilaterally obstructed infants versus eight healthy controls, with the goal of identifying quantitatively varying proteins and the biological networks associated with them. Notably, urine was obtained from both the obstructed kidney and the bladder. Over 1100 proteins were identified, and a total of 76 quantitatively varying proteins were identified. Proteins involved in oxidative stress, inflammation, and renal disease pathways showed the most significant abundance differences. This study gives a deeper understanding of the critical proteomic changes associated with renal obstruction and represents the deepest proteomic profile of renal obstruction to date.
Currently 1–5% of all pregnancies are diagnosed prenatally with hydronephrosis or dilation of the kidney (1). Although many cases of hydronephrosis resolve, persistent hydronephrosis is often caused by a ureteropelvic junction obstruction (UPJO)1. UPJO or renal obstruction can result in abnormal renal development and loss of kidney function. Although renal obstruction always results in hydronephrosis, hydronephrosis does not always indicate clinically significant obstruction. Appropriately timed postnatal surgical intervention of clinically significant UPJO may prevent additional renal damage; however, identifying which children require surgical intervention versus observation is extremely clinically challenging.
The current clinical diagnostic workflow utilizes serial ultrasound and diuretic renography, which is invasive and exposes the infant or child to ionizing radiation. Furthermore, the current tests are subjective and inaccurate (2, 3) in determining clinically significant obstruction. These inaccuracies have resulted in a lack of clinical consensus on how to best identify which patients with hydronephrosis would benefit from surgical intervention.(1) Due to current challenges in UPJO patient stratification, there is a dire need for a sensitive, specific, and noninvasive standardized test that can identify the at-risk patients in need of surgical intervention.
Using urine as a reflection of the pathologic changes associated with UPJO and hydronephrosis has numerous advantages. Urine is readily available and may be obtained noninvasively and longitudinally. Urine is the proximal fluid of UPJO, as well as other urological diseases. Proximal fluids possess many potential advantages for revealing new pathology networks and insights into potential future biomarkers (4). In healthy individuals, urine obtained from the bladder is a combination of urine derived from both kidneys. However, in unilateral severe UPJO the “true” proximal fluid (urine directly from the obstructed kidney) will likely be significantly diluted in the bladder by the contribution from the nonobstructed kidney. We circumvent this issue by analyzing urine that was surgically obtained directly from the obstructed kidney, as well as urine from the bladder of UPJO patients representing the normal nonobstructed (contralateral) kidney. Although a test based on urine from the kidney would not be clinically feasible, studying the kidney-specific urinary proteome may provide new pathologic insights into UPJO that would not be obtainable on a study based on urine obtained from the bladder.
Previous studies have demonstrated that the urinary proteome may contain clinically significant biomarkers of UPJO (5–9). However, these studies either focused on the low molecular weight peptidome or achieved limited proteomic depth. In this work, we use large-scale proteomic methods to quantify the urinary proteomes of clinically unequivocal significant unilateral obstructed UPJO patients and compare them to healthy age- and sex-matched controls. Our goal was to perform an initial discovery study on this well-defined clinical cohort to identify potential pathologic mechanisms reflected in the urinary proteome. The unique advantage of our study was access to urine taken directly from a kidney undergoing surgical repair for UPJO. By surgically obtaining the most proximal fluid of UPJO (kidney urine), we were able to perform two comparisons to the control (1) from obstructed kidney urine and (2) urine from the bladder that represents the nonobstructed normal kidney and the systemic response to congenital unilateral UPJO. We hypothesized that the quantitation of proteomic changes in the urines of UPJO patients will enable patient stratification via the identification of biomarkers specific to patients in need of surgical intervention. Most importantly, identifying specific quantitative changes in the UPJO urinary proteome on a kidney-specific basis will lend further insight into the underlying pathological processes.
EXPERIMENTAL PROCEDURES
Patient Selection
All patients were identified using an IRB-approved protocol. We created a clinically defined, select infant cohort ≤ 2 yo with clinically significant severe unilateral UPJO and no other known medical history. Diseased patients had two inclusion criteria: (1) Society of Fetal Urology Grade 4 (scale 0–4) severe hydronephrosis on at least two serial kidney ultrasounds (US), and (2) a washout curve t1/2 ≥ 50 min (considered clinically definitive for obstruction) on a 99mTc mercaptoacetyltriglycine (MAG3) diuretic renography scan with furosemide delivered at F+20. Patients had negative voiding cystograms and no active or history of urinary tract infection. Cohort demographics are depicted in Table I. Normal controls were sex and age matched to each UPJO subject. Controls were identified from infants who were undergoing screening cystogram for a family history of vesicoureteral reflux. All normal controls had a normal kidney US, negative cystogram, no history of urinary tract infection, and no other comorbid condition. Each patient's clinical demographics are summarized in Table I.
Table I. A summary of the relevant clinical measurements for this cohort selection. Age in months, laterality of obstruction, grade of hydronephrosis by ultrasound (US), % function in each kidney, and t1/2 time on 99mTc mercaptoacetyltriglycine (MAG3) diuretic renogram are listed. A t1/2 is the measure of time it takes for of the radioisotope to be cleared from the kidney. A t1/2 > 20 minutes is considered to be pathologic for obstruction. UPJO patients selected for these studies all had significantly elevated t1/2 and persistent high grade hydronephrosis on serial US. Normal controls had no evidence of hydronephrosis. The patients studied were, based on current clinical parameters, unequivocally in need of surgical intervention.
| IEF STRIP | Sample | Gender | Age (mos) | Laterality | Hydronephrosis grade (0–4) | % Function R | % Function L | t1/2 MAG3 |
|---|---|---|---|---|---|---|---|---|
| B1,K1 | D1 | F | 6 | R | 4 | 46 | 54 | 54 |
| C1 | C1 | F | 1 | - | 0 | - | - | - |
| B2,K2 | D2 | M | 8 | L | 4 | 60 | 40 | 59 |
| C2 | C2 | M | 8 | - | 0 | - | - | - |
| B3,K3 | D3 | F | 7 | L | 4 | 50 | 50 | 100 |
| C3 | C3 | F | 11 | - | 0 | - | - | - |
| B4,K4 | D4 | M | 9 | R | 4 | 51 | 49 | 100 |
| C4 | C4 | M | 9 | - | 0 | - | - | - |
| B5,K5 | D5 | M | 3 | R | 4 | 53 | 47 | 100 |
| C5 | C5 | M | 5 | - | 0 | - | - | - |
| B6,K6 | D6 | M | 16 | L | 4 | 72 | 28 | 74 |
| C6 | C6 | M | 8 | - | 0 | - | - | - |
| B7,K7 | D7 | M | 10 | L | 4 | 51 | 49 | 68 |
| C7 | C7 | M | 10 | - | 0 | - | - | - |
| B8,K8 | D8 | M | 4 | R | 4 | 48 | 52 | 57 |
| C8 | C8 | M | 5 | - | 0 | - | - | - |
Sample Collection
UPJO Cohort
At the time of surgery using standard sterile technique, urine was collected by catheterization of the bladder (sample name = BU, n = 8). During surgical correction of the kidney, urine was atraumatically aspirated directly from the renal pelvis of the obstructed kidney prior to relief of the obstruction (sample name = KU, n = 8). Patients in this cohort were not on prophylactic antibiotics but received intravenous antibiotics at the time of surgery prior to incision. All patients had no solid food 6 h and no clear liquids 2 h prior to surgery.
Normal Cohort
During the screening cystogram, a urine sample was obtained at the time of catheterization of the bladder (sample name = CU, n = 8). No patients received anesthesia during the procedure. All patients were on low dose prophylactic antibiotics prior to the catheterization.
Sample Preparation
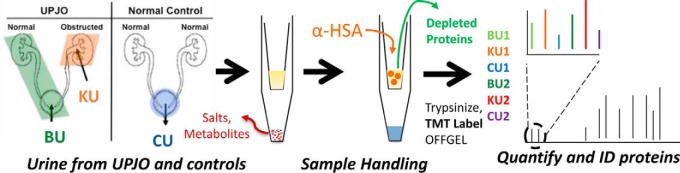
Samples were analyzed by urinalysis using a Siemens CLINITEK® status automated analyzer (Tarrytown, NY) to ensure there was no blood contamination or evidence of proteinuria. Furthermore, as part of standard clinical practice, an aliquot of all samples are cultured to ensure no evidence of infection. All samples were negative for infection, blood, and protein. All samples were centrifuged at 4500 g for 20 min to remove cellular debris, aliquoted, and frozen at -80 C in 1.5 ml centrifuge tubes (Eppendorf) until sample preparation by our previously published protocol (10). An overview of the sample preparation is shown in Fig. 1.
Fig. 1.
An overview of the sample preparation in which samples from BU, KU, or CU were prepared according to the spin-filter-based one-step protocol (10). Albumin was depleted, protein amounts were determined, and proteins were digested with trypsin. After TMT labeling, two groups of three samples each were mixed and analyzed by LC-MS/MS.
In brief, aliquots were thawed, desalted, and concentrated on 5000 mw cutoff filters. Samples were reduced and alkylated with iodoacetamide, and albumin was depleted with anti-HSA resin (Sartorius). Depleted samples were then quantified by the Bradford method, and an identical amount of protein from each sample was digested with sequencing-grade trypsin overnight (Promega).
Peptides were labeled with the Tandem Mass Tag (TMT-6) kits (Thermo) according to manufacturer's protocols. Peptides were then purified on Oasis HLB SPE cartridges. We mixed six labeled samples (two matched BU, KU, CU triplets) to create one study group. Each group was then separated into 24 fractions by IEF using an OFFGEL (Agilent) and dried using an SPD1010 speedvac (Thermo).
Mass Spectrometry and Database Searching
Fractionated, dried peptides were reconstituted in 5% FA/5% ACN, transferred to HPLC vials, and loaded onto an autosampler coupled to an LTQ-Orbitrap classic (Thermo) with a nanoflow UPLC system (Eksigent). The LC columns (15 cm × 100 μm inner diameter) were packed in house (Magic C18, 5 μm, 100 Å, Michrom BioResources, into PicoTips, New Objective, Woburn, MA). Samples were analyzed with a 60 min linear gradient (0–35% ACN with 0.2% formic acid) and a top six data-dependent acquisition method was utilized, with pulsed-Q dissociation. Normalized collision energy was 29%, and two microscans were averaged per MS/MS spectrum.
MGF files consisting of the 200 most intense fragment ions of each raw product ion spectrum were generated by an in-house script. MGFs were searched against the IPI Homo sapiens database (version 3.69, 87,130 target sequences) using Mascot version 2.2.04 (Matrix Science). The following search parameters were applied: Default charge states of 2+, 3+, and 4+ were used; trypsin (fully specific with one missed cleavage) was the protease utilized. A peptide mass tolerance of 10 ppm and a fragment ion search tolerance of 0.8 Dalton (Da) was permitted. Fixed modification on cysteine was carbamidomethyl and variable modifications were deamidation (N and Q), oxidation (M), TMT: 6-plex at N terminus and lysine residues and pyro-glutamic acid formation (Q). All data were searched against a combined target and decoy database. Peptide identifications were filtered to a 1% false discovery rate by the target-decoy approach, and all identified proteins were matched by two or more spectra. IPI listings were converted to UniGene IDs using an online tool (http://www.uniprot.org/), and protein isoforms were combined into a single UniGene ID for quantification. All raw data have been deposited in the MassIVE repository, with accession number MSV000079553.
Experimental Design and Statistical Rationale
The log2 values of the BU:CU and KU:CU reporter ions for each peptide spectral match (PSM) from a unique peptide were calculated. Individual PSM ratios were then normalized by the sample-wide mixing ratio. PSM ratios were grouped by gene ID, and the means of each gene were compared by one-way ANOVA to the remaining peptide mean. Globally, the quantitative PSM ratios were approximately normally distributed. The resulting p values for each gene ID were determined cohort-wide based on eight samples. p values were adjusted by the Benjamini–Hochberg correction, and the family-wise error rate (Q-value) was controlled at 5%. Furthermore, all statistically significant hits required a fold-change of 1.5 for BU:CU and 3.0 for KU:CU for further evaluation by pathway analysis.
Pathway Analysis
Statistically enriched pathways, toxicology results, and networks were identified by Ingenuity Pathways analysis software of identified proteins. Proteins of interest passing the above fold-changes from BU and KU quantification were analyzed separately to identify unique biological processes associated with BU and KU separately.
Western Blotting
Protein from urine samples was purified via Amicon 4–10K Centrifugal Filter Unit Clean-up (Millipore, Bedford, MA) and quantified using the Micro BCATM Assay Kit (Thermo Scientific, Rockford, IL). Equal amounts of protein from urine were used for each validated target (5 μg for HSP70 blots and 15 μg for GSTM1 blots) and were separated on NuPAGE 4–12% Bis-Tris gels (Life Technologies, Carlsbad, CA) at 125V along with HepG2 cell lysate in RIPA buffer as a positive control. Separated proteins were then transferred to a 0.45 μm InvitrolonTM PVDF membrane (Life Technologies, Carlsbad, CA). Membranes were blocked for 2 h using 5% dried nonfat milk in phosphate buffered serum/.05% Tween (PBS-T). Protein expression was determined by the following antibodies: HSP70 at a 1:10,000 dilution and GSTM1 at a 1:1,000 dilution (AbCam, Cambridge, MA) overnight at 4 °C in 5% bovine serum albumin (BSA) in PBS-T. Membranes were washed for 1 h with PBS-T and secondary goat anti-rabbit horseradish peroxidase (HRP) (Bio-Rad, Hercules, CA) was incubated with the membranes at room temperature for 1 h in 5% dried nonfat milk. The expression of each protein was determined by ECL Western Lightning (PerkinElmer Life and Analytical Sciences, Waltham, MA), and bands of interest corresponding to target proteins (HSP70 and GSTM1) were evaluated via quantitative densitometry (Image J, NIH). Considering the limited dynamic range of WB, we set a maximum fold-change of eightfold for calculations.
RESULTS
A total of 1113 unique proteins were identified (Table II), with similar numbers of proteins identified in each study group. Over 750 genes were quantifiable with three or more PSMs. Of those, 267 showed abundance differences that were statistically significant. As shown in supplementary materials, 74 were statistically significant in both KU and BU relative to CU. Furthermore, over 75% of all statistically significant proteins were identified in all samples (supplementary materials), meaning the varying proteins are commonly expressed.
Table II. A summary of the proteomic identifications per sample group. Sample groups averaged 660 proteins, 2165 unique peptides, and 6174 spectral matches. Two disease and control triplets were pooled per sample group.
| Sample group | Proteins | Genes | Peptides | Spectra |
|---|---|---|---|---|
| 1–2 | 650 | 539 | 2092 | 5727 |
| 3–4 | 666 | 546 | 2203 | 5762 |
| 5–6 | 628 | 491 | 1978 | 6442 |
| 7–8 | 695 | 552 | 2387 | 6768 |
| TOTALS (unique) | 1,113 | 810 | 4097 | 24699 |
The relationship between fold-change and q-value was visualized in volcano plots (Fig. 2) for each sample. BU samples are shifted toward higher q-values and lower fold-changes overall. Each of these shifts (larger fold-changes and smaller q-values) suggest that KU is more altered by UPJO than BU. It is clear that the KU proteome has been substantially altered by this disease, as expected for a proximal fluid. This finding highlights the value of analyzing urine directly from the obstructed kidney, as changes in KU are more apparent than those in BU.
Fig. 2.
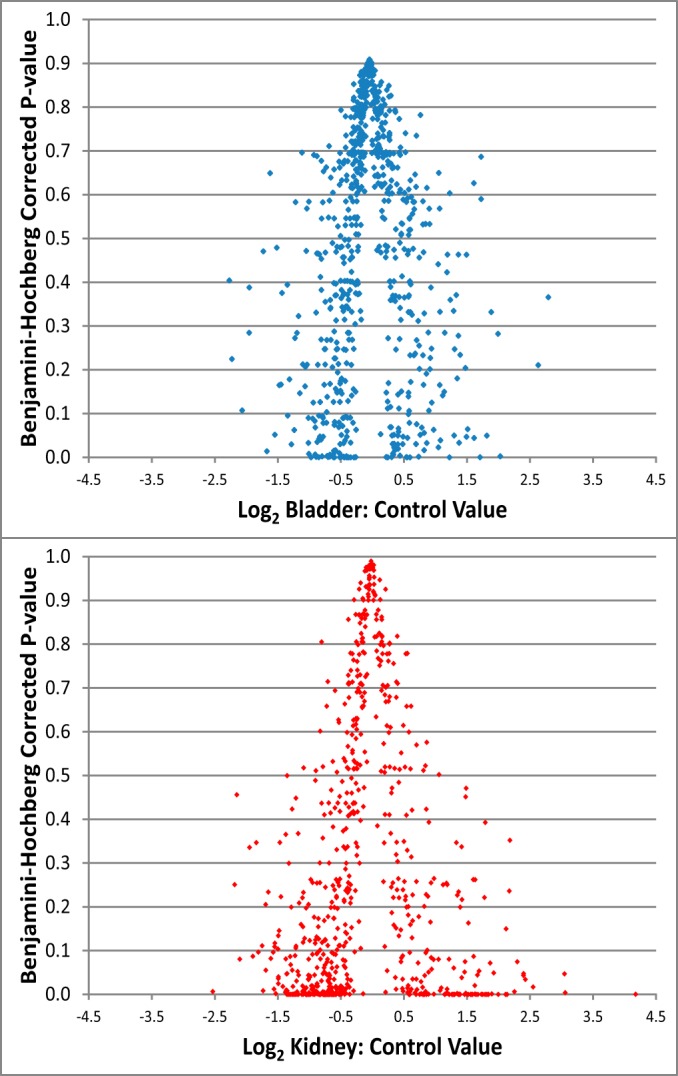
Volcano plots of the KC (A) and BC (B) ratios determined for all proteins quantified. The KC ratios were shifted toward smaller Q-values and larger fold changes overall, suggesting that KU samples were more altered than BU samples, as expected. This highlights the value of analyzing urine directly from the obstructed kidney.
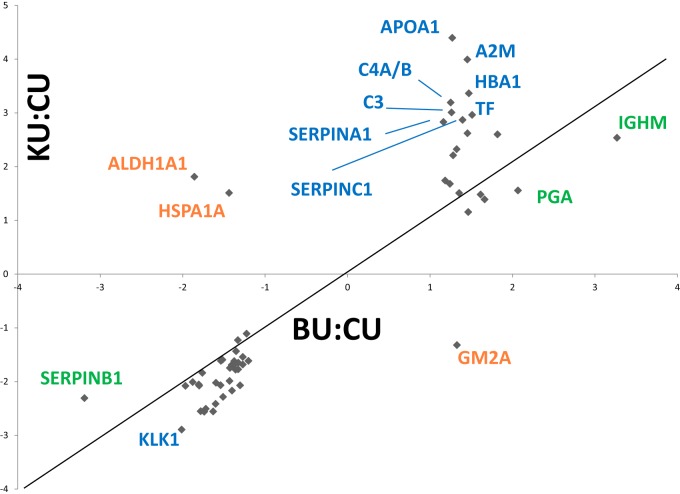
A total of 74 proteins were statistically significant in both KU:CU and BU:CU. The fold-changes of these proteins were plotted in Fig. 3 to visualize the quantitative relationship observed in these samples. While several of these proteins lie near the diagonal (e.g. similar fold-change in BU versus CU and KU versus CU), several others are off the diagonal, suggesting a preferential enrichment in one sample. For example, APOA1 is far more enriched in KU (4.4-fold enriched) than it is in BU (1.27-fold). These entries, although statistically significant in both samples, have pronounced differences in their regulation. In addition, a total of three proteins were up-/down-regulated in contrasting manner between the KU and BU samples. These included retinal dehydrogenase 1, heat shock 70 kDa protein 1A/1B, and ganglioside GM2 activator (GM2A), which are discussed in detail below. Heat shock 70 kDa protein 1A/1B and GM2A did not meet the initially applied fold cutoff for inclusion in the proteins of interest list. However, because of this unique quantitative relationship, these proteins were added to the proteins of interest list. In total, 76 proteins comprise the proteins of interest list.
Fig. 3.
Fold-changes of statistically significant proteins in the KU and BU samples are plotted. Although many proteins have similar fold-changes in both samples, there are several entries that are more enriched in one sample. In addition, off-diagonal proteins are differentially expressed in each sample.
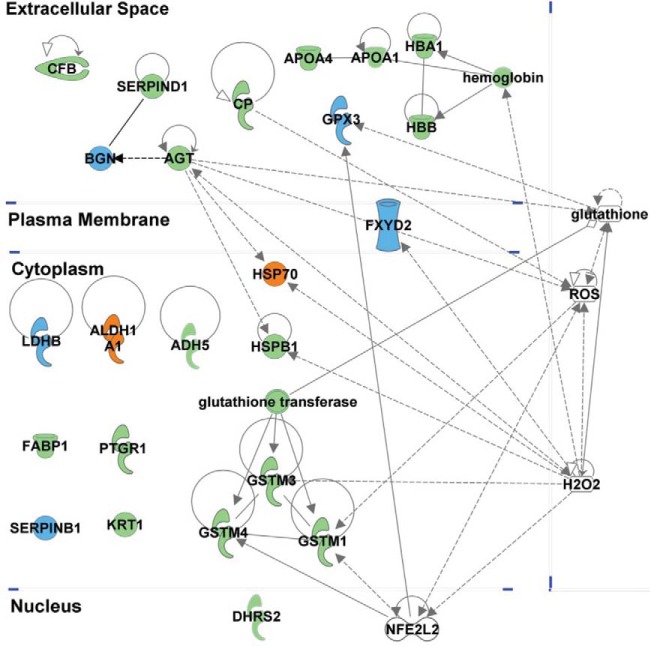
To determine the potential biological significance of the enriched and depleted proteins, we performed a bioinformatic analysis of the identified proteins with a specific focus on the statistically significant enriched or depleted markers using Ingenuity Pathways analysis software. Overall, several pathways were overrepresented among quantitatively varying proteins (supplementary materials). The overrepresented biological processes include oxidative stress/reactive oxygenated species (ROS) processing, fibrosis, and acute-phase inflammation reaction. Many of the proteins of interest are highly networked (Fig. 4).
Fig. 4.
The putative biomarker panel shows many highly networked proteins associated with oxidative stress. Entries shown in blue were down-regulated, while entries in green were up-regulated. The orange entries were differentially regulated in BU and KU samples. Highly networked small molecules and a regulatory protein, NFE2L2 are also shown to highlight underlying relationships.
The KU content of two proteins (GSTM1 and HSP70) were evaluated by Western blotting in order to validate the quantitative MS measurements and pathway analysis findings (Fig. 5). In correlation to the proteomic quantitative data and pathway analysis identifying ROS species activation, total (11) GSTM1 and HSP70 were both elevated in the KU as compared with CU by WB. This finding supports the idea of altered ROS processing in the obstructed kidney.
Fig. 5.
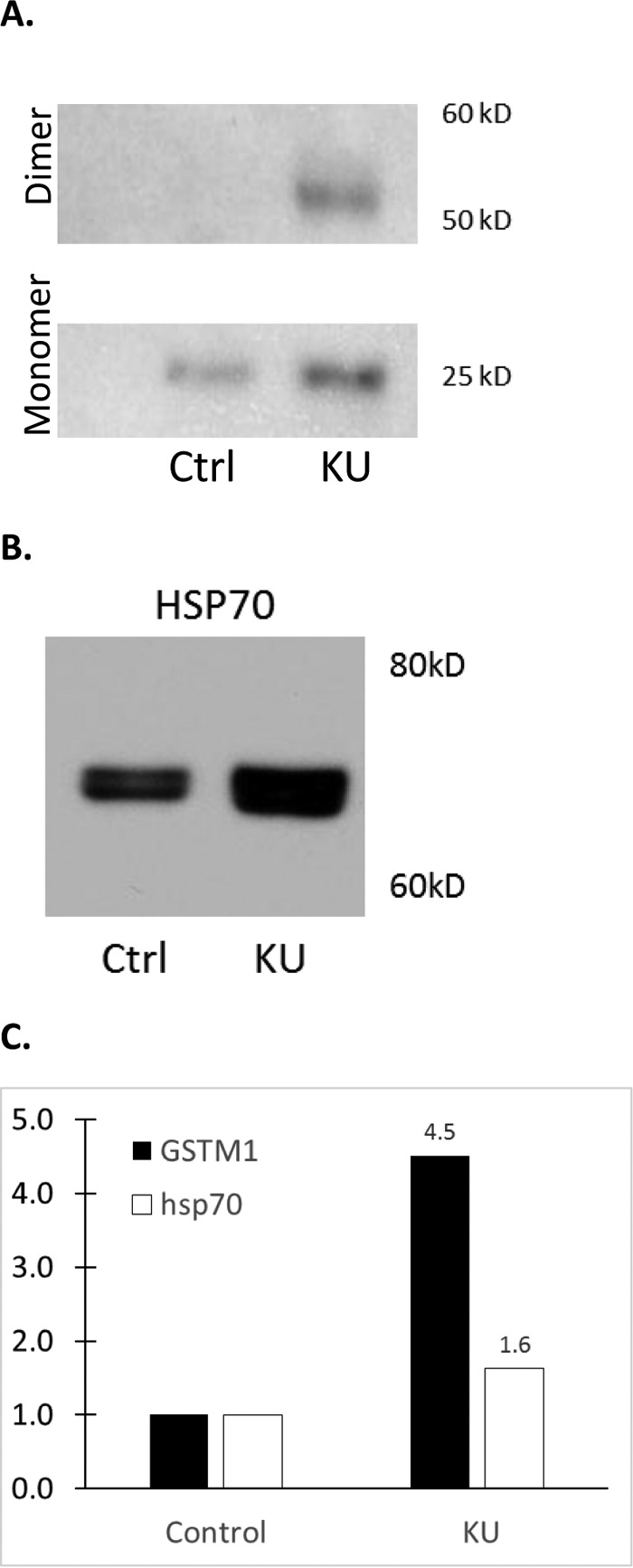
Western blotting validation of MS data. A representative blot is shown for (A) GSTM1, validating the increase in KU relative to CU observed by MS. In (B), validation of the observed increase of HSP70 in KU is shown. In (C), a bar graph showing the averaged changes of these two proteins in three samples by WB is shown.
DISCUSSION
In this study of the urinary proteome of infants with well-defined severe unilateral UPJO versus healthy age- and sex-matched controls, we identified 76 proteins of interest in UPJO from 1113 proteins identified in an unbiased quantitative discovery study of the obstructed urinary proteome. We demonstrated that (1) obstruction significantly changes the urinary proteome; (2) compensatory changes in the normal nonobstructed kidney may also be contributing to significant quantitative changes in the urinary proteome; and (3) proteins involved in oxidative stress play a critical role in the pathophysiology of UPJO and may be the basis for a clinical biomarker of UPJO.
Previous work also identified the potential value of analyzing urinary proteins in UPJO patient stratification.(5, 6, 8, 12) However, these studies were differently focused, exclusively analyzing the low molecular weight peptidome (< 10kDa) or had a low degree of proteomic coverage. Points of merit in the current study include: (1) very well defined, age- and sex-matched severe disease and control cohorts (Table I) to provide definitive insight into the pathophysiology of clinically significant renal obstruction; (2) unique access to urine directly from the diseased kidney (KU); and (3) the deepest profiling of the UPJO urinary proteome to date.
Currently, there is less clinical controversy about the need to intervene on patients with severe hydronephrosis, but there is no definitive consensus on the timing of surgical intervention, as many cases of severe hydronephrosis may still resolve without intervention. While there is a significant clinical need for markers to help with the management of lower grades of hydronephrosis, we specifically choose to study very severe obstruction to provide the most unbiased representation of the pathologic process of UPJO for this discovery study. Studying lesser degrees of hydronephrosis, although of high merit, would likely result in a cohort with varying clinical degrees of obstruction and physiology. Subsequently, a much larger number of patients would be required to identify any statistically relevant data, which potentially could preclude the logistics of an unbiased discovery-based proteomics study. Furthermore, the severe cohort has specific advantages over all animal models that are created postnatally and typically are complete obstruction models.
Although obtaining urine directly from the kidney is not clinically feasible, by studying kidney urine from patients with severe obstruction, we have the unique ability to study the most proximal body fluid for UPJO. This provides the purest insight into the pathology of UPJO and may have the highest likelihood of identifying potential clinically informative pathways or proteins when combined with deep proteomic profiling. Since diseases are often affected by an integrated pathway of proteins (13), achieving sufficient depth of proteomic data is critical to identifying the relevant pathways in a disease. With this in mind, we elected to perform an unbiased analysis of the entire urinary proteome.
This study has increased the number of proteins profiled from previous urinary UPJO proteomic studies by approximately an order of magnitude. In addition, many proteins that were implicated in obstructive nephropathy in a prior study (8) were also statistically significant in this study, with the same quantitative trends. Elevated proteins included angiotensin; albumin; keratin 8; keratin 9; lectin, galactoside-binding, soluble, 3 binding protein; macrophage inhibitory factor; myosin heavy chain 7; serpin peptidase inhibitor; clade A member 1; and transferrin. Epidermal growth factor was previously reported as down-regulated (14), and our study replicated that trend although statistical significance was not achieved in this study cohort. Similarly, prior studies evaluating BU also shared trends with KU quantitation. Glutathione S-transferase pi 1; GAPDH, and heat shock 27kDa protein 1 were all elevated in KU, as in prior work. Performing research on human subjects is challenging due to extensive heterogeneity between individuals. The fact that quantitative trends for proteins remained the same between different studies despite substantial differences in study design, patient selection, and analytical methodologies is quite striking and further supports the validity of the quantification.
The urine obtained directly from the obstructed kidney (KU) represents the most proximal fluid of UPJO and has the advantage of a higher concentration of the proteins of interest (15–17). UPJO inherently retards drainage from the obstructed kidney, so BU is expected to be overrepresented in urine filtered from the contralateral kidney, particularly in the case of severe obstruction. Subsequently, the protein profile identified in the bladder may more closely represent changes in the contralateral kidney or possibly systemic responses to unilateral UPJO, with minor diluted contributions from the obstructed KU. With this challenge in mind, a 1.5-fold-change cutoff among statistically significant BU hits was deemed appropriate for considering future putative BU biomarkers, whereas a more stringent threefold-change cutoff was required for putative KU biomarkers.
The potential dilution of proteins in BU also highlights the value of analyzing KU because, while the KU concentration of the undetermined biomarker may be high, the BU concentration could be lower, precluding quantitation in BU using discovery-phase proteomics. A good example of this is the quantitative profile of GSTM1. GSTM1 was only significantly elevated in KU. GSTM1 may only be elevated in the kidney due to the combination of a local kidney response and the mechanical nature of the obstruction precluding the detection in the bladder during severe obstruction. Alternatively, proteins with a similar KU and BU response may depict an overall systemic response to unilateral renal obstruction.
KU is obtained invasively and cannot be used as a screening biofluid. However, by identifying KU-specific proteins in cases of severe obstruction where the mechanical obstruction is the greatest, these potentially locally responsive proteins may become more evident in the bladder urine in cases with lesser degrees of obstruction or hydronephrosis. Additionally, since previous studies suggest that a large-scale quantitative proteomic approach may underestimate fold-changes (18–20), using independent and/or improved quantitative methods, such as directed MS, ELISA, or Western blotting, may find KU-exclusive targets are in fact altered in BU samples using other methodologies.
We specifically chose two proteins to validate in our study because of their specific measured quantitative patterns. GSTM1 was found elevated 18-fold in the KU relative to CU. HSP70 was 1.5-fold up in the kidney and 1.4-fold down in the bladder. Western blot evaluation of GSTM1 showed an increase of 4.5-fold in KU. MS results were similarly corroborated for the HSP70 WB, and a 1.6-fold increase in KU was observed.
Network analysis determined the biological processes of oxidative stress, fibrosis, and acute-phase reaction were overrepresented among the quantitatively varying proteins. Many of the proteins with the largest fold-changes are known oxidative stress response proteins (Fig. 4) and are regulated by NFE2L2, which is activated upon exposure to ROS to effect transcriptional changes in response to oxidative stress (21). Oxidative stress has been implicated in the pathogenesis of UPJO and animal models previously (22–24). A total of 24 statistically significant proteins were associated with oxidative stress, of which 13 are represented in the putative biomarker panel (Table III). For example, in accordance with the literature, HSP70 was significantly up-regulated in HUK, indicating a potential antioxidant protective effect during obstruction but conversely down-regulated in HUB (bladder), suggesting a different role in the compensatory mechanism of the contralateral kidney (25). Similar changes in HSP70 have been demonstrated with immunohistochemistry and Western blot analysis of the obstructed renal cortex from UUO rodent models (25), but this is the first human demonstration of quantitative changes of urinary HSP70 highlighting its position as a protein of interest in UPJO. Similarly, retinal dehydrogenase 1, which processes cytotoxic ROS, was also inversely expressed in BU and KU. Enrichment of these proteins in the KU samples may indicate an increased level of oxidative stress in the obstructed kidney. The other differentially expressed protein, GM2A, is known to act in concert with the established renal damage marker HEXA in the metabolic processing of glycolipids (26).
Table III. A list of selected biological functions associated with the 76 protein biomarker panel. The Benjamini–Hochberg corrected p values are listed (q-val), and the total number of proteins (from the 76 member putative biomarker panel) associated with each function is also listed.
| Function | q-val | Proteins |
|---|---|---|
| Inflammatory disease | 5.67E-09 | 40 |
| ROS metabolism | 5.27E-07 | 13 |
| Renal and urological disorder | 1.48E-05 | 13 |
| Adhesion of kidney cells | 1.53E-05 | 5 |
| Leukocyte migration | 6.35E-05 | 13 |
| Vasculogenesis | 6.02E-03 | 8 |
| Carbohydrate metabolism | 2.96E-03 | 9 |
The importance of the glutathione transferases in renal obstruction was recently reviewed (27). In our study, the antioxidant defense glutathione transferases (GSTM1, GSTM3, GSTM4) were significantly up-regulated in KU (up to 18-fold) suggesting an increased antioxidant protective role. However, in BU, other oxidative stress proteins such as glutathione peroxidase 3 (GPX3) were downregulated, implying a potential compensatory role of the contralateral normal kidney. DHRS2, which is specifically responsible for reducing several cytotoxic α-dicarbonyls generated by oxidative stress, was highly up-regulated in the kidney (28). FABP1 was also globally up-regulated, and reduces oxidative stress in vitro by processing reactive oxidized lipids (29). FXYD2, a sodium-potassium ion channel regulated by hydrogen peroxide and expressed in the proximal convoluted tubule, was globally downregulated (30). FXDY2 helps regulate sodium reabsorption in the kidney, and defects in FXDY2 have been shown to result in hypomagnesmia and hypocalciurea (31, 32). The observed decrease of FXDY2 in both BU and KU may suggest a bilateral functional dysregulation of the proximal tubule during UPJO. While small molecules such as glutathione, hydrogen peroxide, and nitric oxide are not observable in proteomic studies, the gene expression pattern suggests an increase in oxidative stress in both kidneys. Notably, Biglycan (BGN) is down-regulated over fivefold in the BU samples, and an increase in nitric oxide concentration is known to down-regulate its expression in mesenchymal cells in vitro (33). BGN also protects mesenchymal cells from TNFα–induced cell death in vitro, so the loss of BGN in the BU of UPJ patients may indicate loss of a protective effect, which may signal the beginning of renal damage in these patients (33).
The role of oxidated lipids in UPJO has not been well-established, but proteins involved in the metabolic processing of lipids were identified in this study. FABP1 was shown to protect against ischemia-induced damage in an animal model and is a promising marker of human ischemia (34). FABP1 was over threefold enriched in the BU samples. Furthermore, FABP1 has been shown to irreversibly bind cytotoxic lipid metabolites (35). It is hypothesized that after binding, the protein is transferred to urinary space, protecting the epithelial cell from further damage. This may be the effect underlying the threefold enrichment in BU samples observed in this study.
Excessive leukocyte elastase has been shown to cause cellular death in lung epithelia by inducing toxic oxidative stress (36). SERPINB1, a leukocyte elastase inhibitor, irreversibly destroys the proteolytic capability of leukocyte elastase. The observed loss of SERPINB1 in the kidneys may enable further damage by leukocyte-elastase mediated oxidative stress.
Taken in concert, these data indicate that renal obstruction disrupts the normal balance of oxidative stress elements in both the obstructed kidney and contralateral kidney as compared with control urine, and these changes are reflected quantitatively in the urinary proteome. While oxidative stress underlies many renal conditions, a coordinated quantitative pattern of a small group of proteins is more likely to be specific to a particular underlying pathology/condition. Furthermore, when combined with relevant clinical data, we anticipate that validated proteomic changes may have the potential to be leveraged as a clinically significant biomarker in the future.
This discovery study analyzes the urinary proteins associated with UPJO with an unbiased approach to identify useful potential biomarkers and to understand more about the pathophysiology of this disease. The data presented are the deepest urinary quantitative proteomic profile of this disease to date, quantifying proteins from over 750 genes. Statistically significant proteins were associated with several biological processes that are previously known or hypothesized to affect UPJO pathophysiology or the pathophysiology of animal models of renal obstruction. Further filtering the statistically significant proteins yielded a list of 76 putative biomarkers to be assessed for predictive power in future cohorts. These were highly networked, and were associated with oxidative stress, glutathione metabolism, and immune response. The predictive power of these markers will be evaluated in larger independent cohorts via directed MS studies.
Supplementary Material
Acknowledgments
We would like to thank all members of the Proteomics Center at Boston Children's Hospital for their assistance. We also thank the Department of Urology at Boston Children's Hospital for their continued support.
Footnotes
Author contributions: R.S.L. designed the research; J.W.F., S.A.K., A.C.B., and A.R.V. performed the research; H.S. contributed new reagents or analytic tools; J.W.F., P.S.C., A.R.V., and R.S.L. analyzed data; J.W.F. and R.S.L. wrote the paper; and H.S. contributed MS and proteomics expertise.
* National Institutes of Health grants DK096238 supported this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
1 The abbreviations used are:
- UPJO
- ureteropelvic junction obstruction
- BU
- bladder urine
- KU
- kidney urine
- CU
- control urine
- PSM
- peptide spectral match.
REFERENCES
- 1. Lee R. S., Cendron M., Kinnamon D. D., and Nguyen H. T. (2006) Antenatal hydronephrosis as a predictor of postnatal outcome: A meta-analysis. Pediatrics 118, 586–593 [DOI] [PubMed] [Google Scholar]
- 2. Taylor A., Garcia E. V., Binongo J. N., Manatunga A., Halkar R., Folks R. D., and Dubovsky E. (2008) Diagnostic performance of an expert system for interpretation of 99mTc MAG3 scans in suspected renal obstruction. J. Nucl. Med. 49, 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bao J., Manatunga A., Binongo J. N., and Taylor A. T. (2011) Key variables for interpreting 99mTc-mercaptoacetyltriglycine diuretic scans: Development and validation of a predictive model. AJR Am. J. Roentgenol. 197, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Teng P. N., Bateman N. W., Hood B. L., and Conrads T. P. (2010) Advances in proximal fluid proteomics for disease biomarker discovery. J. Proteome Res. 9, 6091–6100 [DOI] [PubMed] [Google Scholar]
- 5. Decramer S., Wittke S., Mischak H., Zürbig P., Walden M., Bouissou F., Bascands J. L., and Schanstra J. P. (2006) Predicting the clinical outcome of congenital unilateral ureteropelvic junction obstruction in newborn by urinary proteome analysis. Nat. Med. 12, 398–400 [DOI] [PubMed] [Google Scholar]
- 6. Decramer S., Bascands J. L., and Schanstra J. P. (2007) Non-invasive markers of ureteropelvic junction obstruction. World J. Urol. 25, 457–465 [DOI] [PubMed] [Google Scholar]
- 7. Caubet C., Lacroix C., Decramer S., Drube J., Ehrich J. H., Mischak H., Bascands J. L., and Schanstra J. P. (2010) Advances in urinary proteome analysis and biomarker discovery in pediatric renal disease. Pediatr. Nephrol. 25, 27–35 [DOI] [PubMed] [Google Scholar]
- 8. Mesrobian H. G., Mitchell M. E., See W. A., Halligan B. D., Carlson B. E., Greene A. S., and Wakim B. T. (2010) Candidate urinary biomarker discovery in ureteropelvic junction obstruction: A proteomic approach. J. Urol. 184, 709–714 [DOI] [PubMed] [Google Scholar]
- 9. Madsen M. G., Norregaard R., Frøkiaer J., and Jørgensen T. M. (2011) Urinary biomarkers in prenatally diagnosed unilateral hydronephrosis. J. Pediatr. Urol. 7, 105–112 [DOI] [PubMed] [Google Scholar]
- 10. Vaezzadeh A. R., Briscoe A. C., Steen H., and Lee R. S. (2010) One-step sample concentration, purification, and albumin depletion method for urinary proteomics. J. Proteome Res. 9, 6082–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hornby J. A., Luo J. K., Stevens J. M., Wallace L. A., Kaplan W., Armstrong R. N., and Dirr H. W. (2000) Equilibrium folding of dimeric class mu glutathione transferases involves a stable monomeric intermediate. Biochemistry 39, 12336–12344 [DOI] [PubMed] [Google Scholar]
- 12. Decramer S., Zürbig P., Wittke S., Mischak H., Bascands J. L., and Schanstra J. P. (2008) Identification of urinary biomarkers by proteomics in newborns: Use in obstructive nephropathy. Contrib. Nephrol. 160, 127–141 [DOI] [PubMed] [Google Scholar]
- 13. Schadt E. E. (2009) Molecular networks as sensors and drivers of common human diseases. Nature 461, 218–223 [DOI] [PubMed] [Google Scholar]
- 14. Taha M. A., Shokeir A. A., Osman H. G., Abd El-Aziz Ael A., and Farahat S. E. (2007) Pelvi-ureteric junction obstruction in children: The role of urinary transforming growth factor-beta and epidermal growth factor. BJU Int. 99, 899–903 [DOI] [PubMed] [Google Scholar]
- 15. Alatas F., Alatas O., Metintas M., Colak O., Harmanci E., and Demir S. (2001) Diagnostic value of CEA, CA 15–3, CA 19–9, CYFRA 21–1, NSE and TSA assay in pleural effusions. Lung Cancer 31, 9–16 [DOI] [PubMed] [Google Scholar]
- 16. Kishi T., Grass L., Soosaipillai A., Scorilas A., Harbeck N., Schmalfeldt B., Dorn J., Mysliwiec M., Schmitt M., and Diamandis E. P. (2003) Human kallikrein 8, a novel biomarker for ovarian carcinoma. Cancer Res. 63, 2771–2774 [PubMed] [Google Scholar]
- 17. Rosty C., Christa L., Kuzdzal S., Baldwin W. M., Zahurak M. L., Carnot F., Chan D. W., Canto M., Lillemoe K. D., Cameron J. L., Yeo C. J., Hruban R. H., and Goggins M. (2002) Identification of hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein I as a biomarker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res. 62, 1868–1875 [PubMed] [Google Scholar]
- 18. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., and Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ow S. Y., Noirel J., Salim M., Evans C., Watson R., and Wright P. C. (2010) Balancing robust quantification and identification for iTRAQ: Application of UHR-ToF MS. Proteomics 10, 2205–2213 [DOI] [PubMed] [Google Scholar]
- 20. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., and Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]
- 21. Huang H. C., Nguyen T., and Pickett C. B. (2000) Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. U.S.A. 97, 12475–12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeh C. H., Chiang H. S., Lai T. Y., and Chien C. T. (2011) Unilateral ureteral obstruction evokes renal tubular apoptosis via the enhanced oxidative stress and endoplasmic reticulum stress in the rat. Neurourol. Urodyn. 30, 472–479 [DOI] [PubMed] [Google Scholar]
- 23. Klein J., Kavvadas P., Prakoura N., Karagianni F., Schanstra J. P., Bascands J. L., and Charonis A. (2011) Renal fibrosis: insight from proteomics in animal models and human disease. Proteomics 11, 805–815 [DOI] [PubMed] [Google Scholar]
- 24. Klein J., Gonzalez J., Miravete M., Caubet C., Chaaya R., Decramer S., Bandin F., Bascands J. L., Buffin-Meyer B., and Schanstra J. P. (2011) Congenital ureteropelvic junction obstruction: Human disease and animal models. Int. J. Exp. Pathol. 92, 168–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rinaldi Tosi M. E., Bocanegra V., Manucha W., Gil Lorenzo A., and Vallés P. G. (2011) The Nrf2-Keap1 cellular defense pathway and heat shock protein 70 (Hsp70) response. Role in protection against oxidative stress in early neonatal unilateral ureteral obstruction (UUO). Cell Stress Chaperones 16, 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sandhoff K. (2001) The GM2-gangliosidoses and the elucidation of the beta-hexosaminidase system. Adv. Genet. 44, 67–91 [DOI] [PubMed] [Google Scholar]
- 27. Manucha W., and Vallés P. (2008) Hsp70/nitric oxide relationship in apoptotic modulation during obstructive nephropathy. Cell Stress Chaperones 13, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shafqat N., Shafqat J., Eissner G., Marschall H. U., Tryggvason K., Eriksson U., Gabrielli F., Lardy H., Jörnvall H., and Oppermann U. (2006) Hep27, a member of the short-chain dehydrogenase/reductase family, is an NADPH-dependent dicarbonyl reductase expressed in vascular endothelial tissue. Cell Mol. Life Sci. 63, 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamamoto T., Noiri E., Ono Y., Doi K., Negishi K., Kamijo A., Kimura K., Fujita T., Kinukawa T., Taniguchi H., Nakamura K., Goto M., Shinozaki N., Ohshima S., and Sugaya T. (2007) Renal L-type fatty acid–binding protein in acute ischemic injury. J. Am. Soc. Nephrol. 18, 2894–2902 [DOI] [PubMed] [Google Scholar]
- 30. Wetzel R. K., Pascoa J. L., and Arystarkhova E. (2004) Stress-induced expression of the gamma subunit (FXYD2) modulates Na,K-ATPase activity and cell growth. J. Biol. Chem. 279, 41750–41757 [DOI] [PubMed] [Google Scholar]
- 31. Farman N., Fay M., and Cluzeaud F. (2003) Cell-specific expression of three members of the FXYD family along the renal tubule. Ann. N.Y. Acad. Sci. 986, 428–436 [DOI] [PubMed] [Google Scholar]
- 32. Meij I. C., Koenderink J. B., van Bokhoven H., Assink K. F., Groenestege W. T., de Pont J. J., Bindels R. J., Monnens L. A., van den Heuvel L. P., and Knoers N. V. (2000) Dominant isolated renal magnesium loss is caused by misrouting of the Na(+),K(+)-ATPase gamma-subunit. Nat. Genet. 26, 265–266 [DOI] [PubMed] [Google Scholar]
- 33. Schaefer L., Beck K. F., Raslik I., Walpen S., Mihalik D., Micegova M., Macakova K., Schonherr E., Seidler D. G., Varga G., Schaefer R. M., Kresse H., and Pfeilschifter J. (2003) Biglycan, a nitric oxide-regulated gene, affects adhesion, growth, and survival of mesangial cells. J. Biol. Chem. 278, 26227–26237 [DOI] [PubMed] [Google Scholar]
- 34. Park J. M., and Bloom D. A. (1998) The pathophysiology of UPJ obstruction. Current concepts. Urol. Clin. North Am. 25, 161–169 [DOI] [PubMed] [Google Scholar]
- 35. Bennaars-Eiden A., Higgins L., Hertzel A. V., Kapphahn R. J., Ferrington D. A., and Bernlohr D. A. (2002) Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J. Biol. Chem. 277, 50693–50702 [DOI] [PubMed] [Google Scholar]
- 36. Aoshiba K., Yasuda K., Yasui S., Tamaoki J., and Nagai A. (2001) Serine proteases increase oxidative stress in lung cells. Am. J. Physiol. Lung Cell Mol. Physiol. 281, L556–L564 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.