Abstract
Comprehensive characterization of signaling in pancreatic ductal adenocarcinoma (PDAC) promises to enhance our understanding of the molecular aberrations driving this devastating disease, and may identify novel therapeutic targets as well as biomarkers that enable stratification of patients for optimal therapy. Here, we use immunoaffinity-coupled high-resolution mass spectrometry to characterize global tyrosine phosphorylation patterns across two large panels of human PDAC cell lines: the ATCC series (19 cell lines) and TKCC series (17 cell lines). This resulted in the identification and quantification of over 1800 class 1 tyrosine phosphorylation sites and the consistent segregation of both PDAC cell line series into three subtypes with distinct tyrosine phosphorylation profiles. Subtype-selective signaling networks were characterized by identification of subtype-enriched phosphosites together with pathway and network analyses. This revealed that the three subtypes characteristic of the ATCC series were associated with perturbations in signaling networks associated with cell-cell adhesion and epithelial-mesenchyme transition, mRNA metabolism, and receptor tyrosine kinase (RTK) signaling, respectively. Specifically, the third subtype exhibited enhanced tyrosine phosphorylation of multiple RTKs including the EGFR, ERBB3 and MET. Interestingly, a similar RTK-enriched subtype was identified in the TKCC series, and 'classifier' sites for each series identified using Random Forest models were able to predict the subtypes of the alternate series with high accuracy, highlighting the conservation of the three subtypes across the two series. Finally, RTK-enriched cell lines from both series exhibited enhanced sensitivity to the small molecule EGFR inhibitor erlotinib, indicating that their phosphosignature may provide a predictive biomarker for response to this targeted therapy. These studies highlight how resolution of subtype-selective signaling networks can provide a novel taxonomy for particular cancers, and provide insights into PDAC biology that can be exploited for improved patient management.
Pancreatic ductal adenocarcinoma (PDAC)1 remains one of the most deadly solid cancers, characterized by extremely poor survival rates and limited therapeutic options (1). Postoperative treatment of patients is largely limited to chemotherapeutics, such as gemcitabine, nab-paclitaxel or Folfirinox, although the addition of the EGFR-directed kinase inhibitor erlotinib to gemcitabine results in a modest improvement in patient survival (2). In addition, patient stratification for therapy remains in its infancy. These factors highlight an urgent need to better understand the molecular mechanisms that contribute to PDAC development, progression and heterogeneity.
Over the last two decades, a deeper understanding of the genetic and molecular basis of cancer has led to the development of targeted therapeutics and personalized treatment strategies that combine such approaches with companion biomarkers. This paradigm has yet to be successfully applied to PDAC, which likely explains its poor overall response to adjuvant therapy. Although almost all PDACs harbor activating mutations in KRAS, and inactivating mutations in TP53, SMAD4 and CDKN2A occur at a frequency of > 30% (3, 4), mutations in these genes are not associated with clinically 'actionable' phenotypes. Evidence for the existence of different molecular phenotypes of PDAC has been found through exploration of the genomic landscape of this disease. Exome sequencing identified an unsuspected role for axon guidance pathway genes in ∼20% of PDAC patients (3), whereas whole genome sequencing of PDAC specimens provided the basis for classification into four subtypes based upon patterns of genomic structural variation (4). In the latter study, a subtype of patients characterized by unstable genomes and/or a BRCA mutational signature was demonstrated to have increased sensitivity to platinum-based therapy. In addition, transcript profiling subclassifies PDAC into subtypes exhibiting contrasting histopathogical characteristics, mutation patterns and patient outcome (5) as well as differential sensitivity to erlotinib and gemcitabine (6), and a gene expression signature corresponding to the EGFR pathway correlates with erlotinib responsiveness in primary PDAC xenografts (7). These genomic and transcriptomic studies strongly suggest that a greater understanding of the phenotypic heterogeneity of PDAC may lead to improved patient stratification for therapy.
A variety of proteomics approaches have been applied in the search for diagnostic or prognostic biomarkers for PDAC, although this work has yet to achieve clinical impact (8). In a recent study, proteomic and phosphoproteomic interrogation of PDAC versus normal tissue via a LC-MS/MS workflow identified several protein kinases exhibiting increased expression in tumor tissue, including HIPK1 and MLCK (9). This suggests that a comprehensive survey of deregulated signaling in PDAC might represent a fruitful strategy for identification of therapeutic targets and disease subclassification. Furthermore, it is well-established that tyrosine kinase signaling pathways can be deregulated at several different levels in PDAC. In addition to the near-universal presence of activating KRAS mutations (3), these include: overexpression of particular RTKs, including EGFR, ERBB3 and AXL (10–13); increased activation of nonreceptor tyrosine kinases, such as Src (14); and enhanced activation of the phosphatidylinositol 3-kinase (PI3K) pathway because of AKT2 amplification (15) or PTEN loss (16). However, to date, interrogation of tyrosine kinase signaling in PDAC has been limited to candidate-based studies, and an unbiased, global survey of tyrosine phosphorylation patterns in this malignancy has yet to be reported. In the current study, we have addressed this knowledge gap by undertaking global MS-based phosphotyrosine profiling (17, 18) across two large, independent PDAC cell line cohorts, revealing novel PDAC subgroups characterized by contrasting tyrosine phosphorylation patterns, signaling networks, and sensitivity to erlotinib. This new molecular taxonomy for PDAC provides important insights into disease mechanisms and highlights potential biomarkers to help guide patient stratification for therapy.
EXPERIMENTAL PROCEDURES
Cell Culture
Two cohorts of pancreatic cancer cell lines were used. Cohort 1 cell lines (termed the ATCC Cohort) were purchased from and authenticated by the American Type Culture Collection (ATCC) (AsPC-1, BxPC-3, CFPAC-1, Capan-1, Capan-2, HPAC, HPAF-II, Hs700T, Hs766T, Panc 02.03, Panc 03.27, Panc 04.03, Panc05.04, Panc 08.13, Panc 10.05, Panc-1, PL45, MiaPaca-2, SU.86.86, SW1990) and were used and cultured according to ATCC protocols. Cohort 2 cell lines (termed the TKCC Cohort) were isolated from primary patient-derived pancreatic ductal adenocarcinoma xenografts in house (TKCC 2.1, TKCC 04, TKCC 05, TKCC 06, TKCC 07, TKCC 09, TKCC 10, TKCC 12, TKCC 14, TKCC 15, TKCC 16, TKCC 17, TKCC 18, TKCC 19, TKCC 22, TKCC 26, TKCC 27) (4). Animal experimentation for cell line generation was approved by the Garvan Institute/St Vincent's Hospital Animal Ethics Committee (approval number: ARA 12/21). Culture conditions used are summarized in supplemental Table S1.
Phosphoproteomic Profiling
Generation of Protein Lysates
Cells were plated, allowed to reach ∼70% confluence, and placed into base culture medium, without the addition of serum or additional growth factors, for 6 h prior to lysate collection. This was undertaken in order to minimize differences in signaling because of culture conditions and enable comparison of the inherent signaling network properties of the different cell lines. For each cell line, two independent biological replicates of ∼2 × 108 cells were generated. ATCC cells were homogenized in 8 m Urea lysis buffer containing 20 mm HEPES (pH 8), 2.5 mm sodium pyrophosphate, 1 mm β-glycerol phosphate, 1 mm sodium orthovanadate and the reducing agent 1 mm tris (2-carboxyethyl) phosphine (TCEP). The procedure for TKCC cells was similar except the lysis buffer was based on 6 m Guanidine hydrochloride containing 50 mm Tris-HCL (pH 8). Samples were sonicated on ice, cleared by centrifugation, then alkylated for 45 min with 4 mm iodoacetamide (in the dark) at room temperature (RT). These lysates were subsequently used to conduct pTyr profiling, as well as Western blotting.
Preparation of Lysates for Mass Spectrometry Analysis
20 mg (TKCC panel) or 30 mg (ATCC panel) of cell line lysate was diluted to 1 mm guanidine hydrochloride or urea respectively with 50 mm ammonium bicarbonate, and digested at 1:200 (w/w) with Lysyl Endopeptidase (WAKO Chemicals, Cape Charles, VA, USA) for 4 h (RT) and then at 1:100 (w/w) overnight (RT) with TPCK treated trypsin (Worthington, Lakewood, NJ, USA, ratio of 1:100). Two “spike-in” heavy peptides with alanine (al) C13N15 modifications were added prior to tC-18 clean-up (Waters, Milford, MA, USA). These peptides were EF1A1 (EHA(al)LLApYTLGVK) (500 fM) and MAPK14 (HTDDEMTGpYVA(al)TR) (50 pm) (Mimotopes, Nottinghill, Vic, Aus).
Phosphopeptide Immunoprecipitation
Purified peptides were lyophilized overnight and dissolved in immunoaffinity purification (IAP) buffer (50 mm MOPS (pH 7.4) with 10 mm sodium phosphate dibasic, 130 mm sodium chloride and either 0.5% (v/v) Nonidet P-40 (ATCC panel) or 1% (v/v) n-octyl-b-d-glucopyranoside (TKCC panel)). For the ATCC panel, Sepharose 4B beads pre-conjugated to 100 μg of PY100 antibody (Cell Signaling Technology, Danvers, MA, USA) were then added. Peptides were incubated with antibody overnight at 4 °C on a rotating apparatus. For the TKCC panel, the flowthrough from the PY100 IP was collected and immunoprecipitated again with 150 μg of Purified Mouse Anti-Phosphotyrosine (pY20) antibody (BD Transduction Labs, San Jose, CA, USA). Peptides exhibiting nonspecific binding were removed by three washes with IAP buffer and three washes with dH2O. Bound peptides were eluted with 0.15% (v/v) trifluoroacetic acid (TFA) in 40% (v/v) acetonitrile.
LC-MS/MS, Identification and Quantification
For MS analyses, dissolved peptides were separated by nano-LC using an Ultimate 3000 HPLC and autosampler system (Thermo Scientific, Waltham, MA, USA), and mass spectra were acquired on Q Exactive Plus for the TKCC Cohort samples or Orbitrap Velos for the ATCC Cohort samples. Samples were desalted by loading onto a C18 pre-column (100 μm, 2 cm column; particle size 5 μm; pore size 100 Å; Acclaim PepMap RSLC, Thermo Scientific), and separated on a 12 cm 75 μm ID analytical column pulled to an internal diameter of 5 μm by a P-2000 laser puller (Sutter Instruments Co, Novato, CA, USA) packed with C18 Magic reverse phase material, using a Dionex Ultimate 3000 LC system. For the Q Exactive, peptides were eluted at 250 nL/min using a gradient of acetonitrile in 0.1% (v/v) aqueous formic acid as follows: 2–10% in 1 min, 10–26% in 27 min, 26–34% in 2 min, and 34–80% in 5 min. The eluent was directed into a nano-electrospray ion source (Nanospray Flex, Thermo Scientific) with a spray voltage of 1.7 kV. Survey scans in the mass range of 375–1600 m/z were acquired with a resolution of 70,000 at m/z 200 and an AGC target of 3e6 ions (max IIT 120 ms). The top 12 most intense ions (ion selection threshold > 1000 ions, charge state 2–5, preferred peptide match, exclude isotopes) were sequentially isolated (isolation window 1.8 m/z) at a target of 1e5 ions (max IIT 100 ms), and fragmented in the HCD cell (normalized collision energy 27). MS/MS spectra were acquired in the Orbitrap mass analyzer at a resolution of 17,500 at m/z 200 and ions were excluded from selection for a further 15 s. For the Orbitrap Velos, peptides were electrosprayed directly into the MS using a spray voltage of 1.8 kV, survey scans in the mass range of 350–1750 m/z were acquired with a resolution of 60,000 at m/z 400 and an AGC target of 1e6 ions. The top 10 most intense ions (ion selection threshold > 500 ions, charge state ≥2) were sequentially isolated (isolation window 2.5 m/z) at a target of 1e5 ions and fragmented in the linear ion trap (normalized collision energy 30), and ions were excluded from selection for a further 30 s.
MS Data Analysis
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (19) via the PRIDE partner repository with the data set identifier PXD003198 (http://www.ebi.ac.uk/pride/archive/login Username: reviewer72200@ebi.ac.uk, Password: Mta7c1N4). Raw MS data were analyzed using MaxQuant software version 1.1.1.25 (20). Database searching was performed using the Andromeda search engine integrated into the MaxQuant environment against the human UniProt database release 2010_10 (35,073 entries), concatenated with known contaminants as well as the reversed sequences of all entries (21). Protein, peptide, and site FDRs were controlled at a maximum of 1%, individualized precursor mass tolerances were applied as described (20), fragment mass tolerance was 20 ppm (default). The MaxQuant search included the fixed modification of cysteine carbamidomethyl and methionine oxidation, acetyl (Protein N-term) and phosphorylation (STY), and was limited to a maximum of 2 missed cleavages. For completeness, label-free data underwent a post MaxQuant processing step to extract all MS quantitated peaks from MaxQuant results files independent of MS/MS identification. All data were analyzed as log2 values.
Normalization and Imputation of Label-free Data with “Spike-in” Standard Heavy Peptides
The label-free intensity values for the EEF1A1 and MK14 heavy peptides in each cell line were averaged and a subsequent normalization factor generated. Label-free MS intensity values for each pTyr site quantified in the data set were divided by the appropriate normalization factor. Imputation of missing values, which is necessary prior to more advanced bioinformatic analyses, was performed using K-Nearest Neighbor (22) on normally distributed data.
Bioinformatics and Statistical Analysis
Data analyses were performed using Microsoft Office Excel, R statistical programming language (Bioconductor packages) and the bioinformatics platform Perseus (Max Planck Institute of Biochemistry) version 1.2.0.19.
Stepwise Hierarchical Clustering and Classifier List Prediction
A stepwise approach was developed to identify stable clustering topology. Firstly, hierarchical clustering was performed multiple times with an incremental inclusion of phosphorylation sites (ordered by variance across cell lines) each time. It started from using the top 10 most variable and continued until the top 412 variable phosphorylation sites were included for ATCC data set and the top 433 sites for TKCC. The sites were included on the basis of each phosphorylation site having greater variance than the average variance of all sites. By comparison of individual stepwise clustering results, the most frequent clustering topology was then identified. The random forest classifier (19) for subtypes of each cell line cohort was built respectively with a two-step approach by the R package randomForest 4.6–10. In the first step, a classifier was built based on the pTyr sites that were used in the stepwise clustering analysis. In the second step, only the most predictive pTyr sites with a mean decrease accuracy greater than 1.5 in the first classifier were selected, and used to build a refined classifier.
Subtype-Specific Site Identification, Pathway Enrichment and Protein-Protein Interaction Network Analyses
To identify subtype-specific sites, a multiple-sample ANOVA test was performed with multiple test correction. This identifies pTyr sites that have statistically significant (p < 0.05) specificity to one of the subtypes. This was followed by pairwise t-tests (p < 0.05) to identify subtype-specific high and subtype-specific low pY sites, representing those sites showing high and low relative tyrosine phosphorylation in a subtype-specific manner. KOBAS was used to perform pathway enrichment analysis (23). The hypergeometric test was selected to test statistical enrichment of KEGG and Reactome pathways, and the p values were corrected for multiple comparisons. The protein-protein interactions among proteins of interest were retrieved from the Protein Interaction Network Analysis (PINA) platform (24), and kinase-substrate relationships were downloaded from the PhosphoSitePlus database. The networks were generated and visualized using PINA4MS, a Cytoscape plugin for PINA.
Cell Viability Assay
Cell viability was assayed in biological triplicate (n = 3) directly using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA). Cells were plated in full medium into a 96-well plate at 1.5 × 103 cells/well. Erlotinib (Symansis, Timaru, NZ), or vehicle (DMSO) were added the next day (Day 0) and the assay was undertaken at Day 5. All drugs were dissolved in DMSO and control cells received vehicle only. Significance was assessed by one-way ANOVA across treatments (p < 0.05 considered as significant). Dose-response relationships were analyzed with GraphPad Prism.
Antibodies and Western Blot Analysis
Immunoblotting and densitometry analyses were performed in biological duplicate (n = 2) as previously described (17). Protein extracts were separated using 4–12% Bis-Tris precast gels (Invitrogen, Carlsbad, CA, USA) and were processed according to the manufacturer's protocol. Primary antibodies used in this study were pY1189 ErbB3, ErbB3, pY1148-EGFR, pY1173-EGFR, pY1192-EGFR, Met (Cell Signaling Technology); EGFR, pY1234-MET, hnRNPs C1/C2, U and K and RMBX (Santa Cruz Biotechnology, Dallas, TX, USA); FAK (BD Transduction Laboratories); beta-Actin (Sigma-Aldrich, St Louis, MO, USA) and hnRNP A2B1, H, and R (Abcam, Cambridge, UK).
DNA Extraction and Somatic Mutation Mapping
DNA was extracted using the Qiagen Allprep Kit in accordance with the manufacturer's instructions. Mutation screening of OncoCarta Panels v1.0, v2.0 and v3.0 (Sequenom, San Diego, CA, USA) was conducted by the manufacturer to detect and quantify mutation frequencies in the cell line panel.
RESULTS
Determination of the PDAC Tyrosine Phosphoproteome
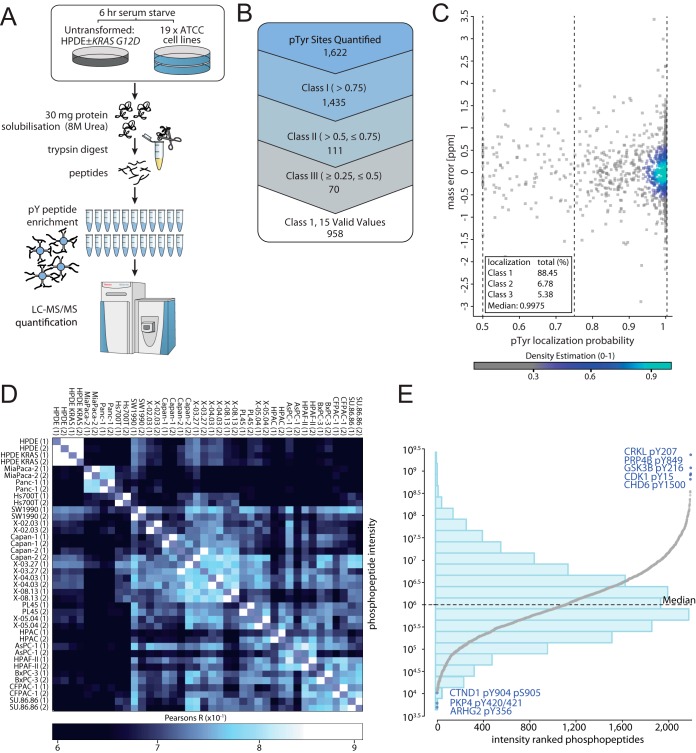
Tyrosine phosphorylation profiling was undertaken across 19 PDAC cell lines sourced from the American Type Culture Collection (ATCC) (supplemental Table S1), as well as normal pancreatic duct epithelial (HPDE) cells and HPDE cells expressing oncogenic KRAS (25). Tryptic peptides underwent immunoaffinity enrichment coupled with a high-resolution mass spectrometry (HR MS using LC-MS/MS) (Fig. 1A). A comprehensive list of 1622 pTyr sites in peptide sequences derived from 797 nonredundant proteins were quantified (Fig. 1B) (supplemental Table S2). Based on the MaxQuant derived phosphorylation site localization probability score, 89% of sites (1,435) were identified as having a high confidence localization probability of > 0.75 (class 1) (Fig. 1C). MS experiments were performed in duplicate with high biological replicate technical reproducibility (Fig. 1D). The phosphopeptide intensities showed a normal distribution over ∼6 orders of magnitude (Fig. 1E). Approximately 50% of the phosphoproteins identified exhibited a gene ontology cellular compartment classification localizing them at the plasma membrane (supplemental Fig. S1A). In terms of molecular function, prominent categories were protein kinase, phosphatase and receptor activity, consistent with signaling functions (Supplemental Fig. S1B). However, a novel observation was that ∼15% of the proteins identified exhibited RNA binding activity (supplemental Fig. S1B).
Fig. 1.
Phosphoproteomics workflow and overall results. A, The shot-gun phosphoproteomics workflow. B, Tyrosine phosphorylation sites quantified in the ATCC cell line cohort phosphotyrosine (pTyr) proteomics screen according to localization probability (LP) and valid values, including class 1 (> 0.75 LP), class 2 (> 0.5, ≤ 0.75 LP) and class 3 (≥ 0.25, ≤ 0.5 LP). The term “valid values” refers to the presence of a MS quantitated peak in the MaxQuant results files for a given phosphorylation site. Six sites have a localization probability of less than 0.25 and are not listed within the figure. C, Visualization of the localization probability distribution and mass error for pTyr sites quantified in the ATCC data set. D, ATCC cell line biological replicate technical reproducibility according to Pearsons correlation (R). E, Phosphopeptides ranked by respective intensities (log10), with overlaid histogram visualization. The bar heights of the histogram estimate the density of phosphorylation site intensities at the indicated intensity intervals (23 bins in total) that are shown on the y axis.
Subclassification of PDAC Cell Lines Based on Tyrosine Phosphorylation Patterns
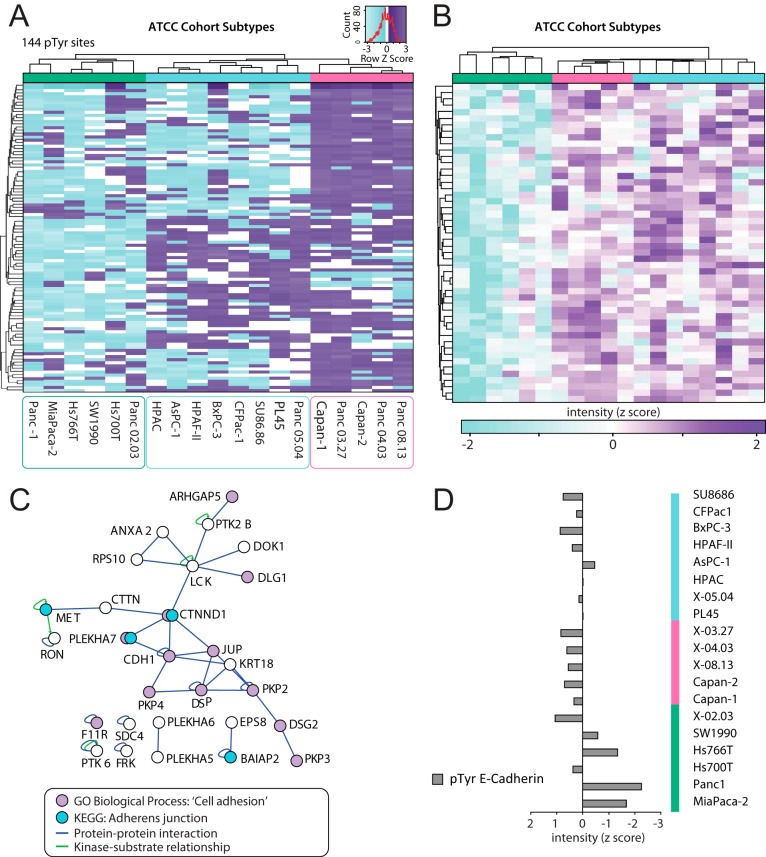
The tyrosine phosphorylation profiles of the ATCC PDAC cell lines were compared using a normalization (using standard heavy peptides) and bioinformatics pipeline, the latter consisting of four major focal points (supplemental Fig. S1C and S1D). We chose to utilize only the most accurate 958 class I pTyr sites quantified by total ion current (TIC) in at least 75% of cell lines (Fig. 1B), with imputation of missing values based on k-nearest neighbor performed on the normally distributed and normalized data prior to further bioinformatics analysis. First, to identify whether the tyrosine phosphoproteome can group the cell lines into reproducible subtypes, we conducted a stepwise resampling of the phosphorylation sites to isolate the 10 most frequent hierarchical clustering topologies. The most frequently occurring topology (Fig. 2A) was identified to occur 58% of the time. This topology includes two broad clusters, with the second broad cluster splitting into two subclusters containing greater than two lines. For the purpose of downstream bioinformatic analysis, the three main cell line clusters were defined as three subtypes. The ATCC subtype 1 (green bar) contains lines MiaPaca-2, Panc-1, Hs700T, Hs766T, Panc 02.03 and SW1990. ATCC subtype 2 (pink bar) contains lines Capan-1, Capan-2, Panc 03.27, Panc 04.03 and Panc 08.13 and ATTC subtype 3 (blue bar) contains lines PL45, Panc 05.04, HPAC, AsPC-1, HPAF-II, BxPC-3, CFPac-1 and SU86.86.
Fig. 2.
Identification of ATCC cell line cohort subtypes and function-based analysis. A, The 19 ATCC cell lines reproducibly form 3 subtypes (See colored bars, subtype 1 is green, 2 is pink and 3 is blue) using stepwise unsupervised hierarchical clustering. The heatmap shows the clustering topology of the subtype specific sites identified in a supervised manner by ANOVA. B, Unsupervised hierarchical clustering of ATCC subtype 1 (green bar) specific phosphosites with low relative phosphorylation compared with subtype 2 (pink bar) and 3 (blue bar). C, Isolated protein-protein interaction network of ATCC subgroup 1 specific phosphosites with low relative phosphorylation. Protein-protein (blue lines) and kinase-substrate (green lines) interactions are displayed. Blue circles indicate phosphoproteins enriched in this subtype that are involved in the adherens junction pathway, and purple circles represent proteins characterized by the GOBP term 'cell adhesion' (GO:0007155). D, E-cadherin (CADH1) tyrosine phosphorylation at pTyr sites 753/754/755 from a single peptide for each cell line, and then normalized by median centering (z score).
To determine whether somatic mutations in PDAC were relevant to the identified subtypes, all cell lines were assessed using the high-throughput Oncocarta mutation profiling panels (Sequenom). Thirty-nine cancer genes with known mutation profiles in cancer, totalling 495 individual somatic mutations, were screened (supplemental Table S3). The predominant mutations identified were located in KRAS (Gly12Cys, Gly12Asp, Gly12Val, Gly61His) and TP53 (Arg248Trp, Arg248Gln, Gly245Ser, Arg273His) (supplemental Fig. S2A, supplemental Table S3). Additional mutations in CTNNB1 (Ser37Phe) (in Hs700T cells) and FBXW7 (Arg465Cys) (in AsPc-1 cells) were also detected. These analyses also confirmed that BxPC-3 and Hs700T do not contain a KRAS mutation. Because the Oncocarta panel did not screen for all known TP53 mutations, we complemented this analysis by a survey of the literature and appropriate databases (http://p53.free.fr/Database/Cancer_cell_lines/Pancreatic_cancer.html, http://p53.iarc.fr/CellLines.aspx) (supplemental Table S3). This revealed that the majority of the ATCC cell lines exhibit mutant TP53. Overall, there does not appear to be a correlation between the somatic mutation profiles of the cell lines and their allocated subtypes.
Identification of Segregation-driving Phosphosites
To identify subtype-specific pTyr sites, an ANOVA-based approach was implemented and applied. It identified 144 pTyr sites that had statistically significant specificity (p < 0.05) to one of the three subtypes (Fig. 2A, supplemental Fig. S1C). All subtype-specific phosphoproteins then underwent combined bioinformatic analyses to identify whether particular signaling pathways or protein-protein interaction networks were characteristic of the different ATCC subtypes.
ATCC subtype 1 was characterized by hypophosphorylation of 65 pTyr sites exhibiting higher phosphorylation across ATCC subtype 2 and 3 cell lines (Fig. 2B). When the function of proteins harboring these sites was considered, there was a significant enrichment for proteins involved in formation and regulation of cell-cell adheren junctions (AJs) and tight junctions (TJs) (p = 0.002 and 0.06 respectively) (Fig. 2C). Isolation of proteins characterized by the Gene Ontology Biological Processes (GOBP) term 'cell adhesion' (GO:0007155) identified 12 subtype 2 and 3 specific proteins involved in this biological process. Further interrogation and analysis at the phosphoproteome and protein expression level indicated that subtype 1 cells exhibited decreased E-cadherin tyrosine phosphorylation (Fig. 2D) and expression (supplemental Fig. S2B), as well as reduced KRAS expression and increased levels of vimentin (supplemental Fig. S2B). Because AJs and TJs are structures important for maintaining epithelial cell architecture and polarity, KRAS expression is characteristic of PDAC cells with an epithelial phenotype (26), and E-cadherin and vimentin expression are associated with epithelial and mesenchymal cells respectively, these data indicate that ATCC subtype 1 PDAC cells exhibit characteristics of epithelial to mesenchyme transition (EMT) .
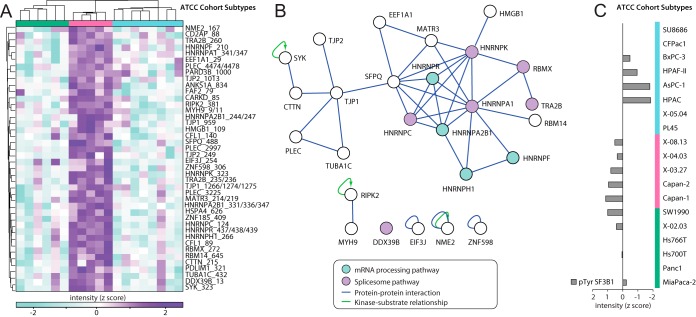
ATCC subtype 2 was found to contain 54 up-regulated subtype-specific pTyr sites, (Fig. 3A), a far greater number than seen in subtype 1 and 3 (1 and 7 up-regulated pTyr sites respectively). The sites enriched in ATCC subtype 2 exhibited a striking enrichment for proteins involved in the mRNA processing and spliceosome pathways, with corrected p values of 0.000003 and 0.000030 respectively. Mapping protein-protein interactions among these phosphoproteins identified the presence of interaction 'hubs' that centered on specific members of these pathways (Fig. 3B). Differences in the abundance of a particular phosphopeptide might reflect altered relative phosphorylation of the corresponding protein, and/or changes in total protein levels. Western blotting revealed that expression of hnRNPs C, R, H, and K, as well as RBMX, was similar across the entire panel (supplemental Fig. S2C), indicating that the increased tyrosine phosphorylation of these proteins in Subtype 2 represents enhanced relative phosphorylation rather than an alteration in protein abundance. However, in the case of hnRNPA2B1, increased expression in subtype 2 may contribute to increased yield of hnRNPA2B1-derived phosphopeptides.
Fig. 3.
Identification of a subtype of ATCC cell line cohort enriched for RNA processing function. A, Unsupervised hierarchical clustering of ATCC subtype 2 (pink bar) specific sites with high relative phosphorylation. B, The ATCC subtypes 2 specific sites with high phosphorylation were analyzed for protein-protein (blue lines) and kinase-substrate (green lines) interactions. Blue circles indicate phosphoproteins enriched in this subtype that are involved in mRNA processing, and purple circles those in the splicosome pathway. C, SF3B1 tyrosine phosphorylation at pTyr sites 70 and 84 were summated for each cell line, and then normalized by median centering (z score).
Splicing factor 3B subunit 1 (SF3B1), a splicing component of the U2 snRNP complex, is mutated in 4–5% of PDAC patients (3). By quantification of 2 pTyr sites on SF3B1, we identified an ATCC subtype 2-specific pattern of elevated SF3B1 tyrosine phosphorylation (Fig. 3C). Although the role of these phosphorylation events are unclear at present, their increased abundance in ATCC subtype 2 provides further evidence that dysregulated mRNA stability or processing characterizes this PDAC phenotypic subgroup.
RTK Expression and Activation in the ATCC Subgroups
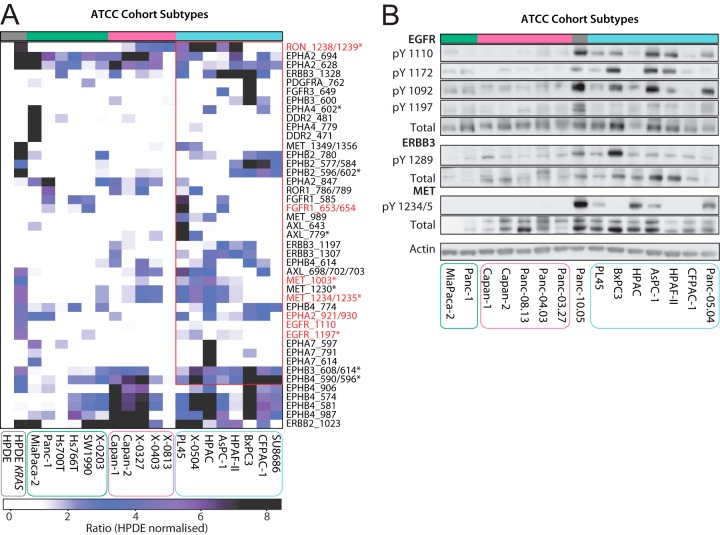
Given the importance of oncogenic RTK signaling in carcinogenesis, we determined the association between all RTKs quantified in the ATCC data set and the identified subtypes. We first normalized the HPDE KRAS and ATCC cell line panel data to matched site intensities from HPDEs, which we utilized as a 'normal' cell line control. We then isolated the relative TIC for all RTK pTyr sites. This revealed that expression of active KRAS in HPDEs results in markedly enhanced site-selective tyrosine phosphorylation of several RTKs, including RON, EPHA2, MET and ERBB2 (Fig. 4A). In addition, this identified that ATCC subtype 3 shows increased pTyr abundance of key oncogenic RTKs (Fig. 4A). Using a two-sample t test, 15 RTK pTyr sites exhibited significantly enhanced phosphorylation in subtype 3 versus the other subgroups. These included sites from the RTKs EGFR, MET, RON, EPHA4, EPHB2/3/4, and DDR2. Western blotting using phosphospecific and total antibodies against EGFR, MET and ErbB3 confirmed these data and demonstrated that both increased RTK expression and relative phosphorylation contribute to the observed pattern of RTK tyrosine phosphorylation across the panel (Fig. 4B).
Fig. 4.
Kinase activation as a basis for ATCC subtype 3 characterization. A, LC-MS/MS quantified RTK tyrosine phosphorylation across the ATCC cell lines. Phosphosite intensities are normalized using HPDE cell line site-specific phosphorylation. t test significant sites (p < 0.05) are indicated by an asterisk (*). Activation sites identified are highlighted by red text. B, ATCC cell lines RTK activity was characterized by Western blotting using EGFR, ErbB3 and MET phosphotyrosine specific antibodies. ATCC cell lines are divided into subtype 1 (green), 2 (pink) and 3 (blue). Gray bar: unclassified cell line.
Subtype Validation Using a Second Cell Line Cohort
PDAC is a highly heterogeneous cancer (3). Therefore, it was important to determine whether the three subtypes identified in the ATCC cell line panel could be detected in an independent PDAC cell line panel generated by our group from patient-derived xenograft models. Application of the phosphotyrosine profiling methodology to this panel of 17 PDAC cell lines, termed “The Kinghorn Cancer Centre” (TKCC) series, in biological duplicate achieved a depth of 1220 quantified pTyr sites, 90% of which were Class 1 sites with strong biological replicate technical reproducibility (Fig. 5A, supplemental Fig. S3A and supplemental Table S2). In total, 1833 class 1 phosphosites were identified in the ATCC and TKCC data sets. A nonredundant list of the 25 most abundant tyrosine phosphopeptides in each PDAC cell line cohort was generated, identifying phosphosites of possible significance to PDAC pathogenicity (supplemental Fig. S3B). Our study also identified over 400 novel phosphorylation sites (supplemental Fig. S3C, supplemental Table S3).
Fig. 5.
Subclassification of an independent PDAC cell line cohort based on tyrosine phosphorylation patterns. A, Tyrosine phosphorylation sites identified and quantified in the TKCC cell line cohort phosphotyrosine (pTyr) proteomics screen according to localization probability (LP) and valid values, including class 1 (> 0.75 LP), class 2 (>0.5, < 0.75 LP) and class 3 (≥ 0.25, ≤ 0.5 LP). B, The 17 TKCC cell lines reproducibility form 3 subtypes (See colored bars, subtype 1 is purple, 2 is orange and 3 is blue) using stepwise unsupervised hierarchical clustering. The heatmap shows the identified topology using the subtype specific sites identified in a supervised manner using ANOVA. C, A protein-protein interaction network formed by TKCC subtype 3 cell lines (blue bar), with phosphoproteins (purple nodes) connected via protein-protein (blue lines) or by kinase-substrate (green lines) relationships. Phosphoproteins enriched in this subtype for axon guidance pathways and ErbB signaling are highlighted with blue and green shading respectively. D, LC-MS/MS quantified RTK tyrosine phosphorylation across the TKCC cell lines according to the identified subtypes (See colored bars, subtype 1 is purple, 2 is orange and 3 is blue). Activation sites identified are highlighted by red text.
Using our bioinformatics pipeline (supplemental Fig. S1C), stepwise unsupervised hierarchical clustering of class 1 TKCC pTyr sites was conducted. A more relaxed data filtering approach was applied to this data set, to allow for the hypothesis of 3 subtypes being present. Thus we required greater than 1/3rd (6 of the 17) of the cell lines to be quantified for a given site. Interestingly, as with the ATCC cohort, unsupervised hierarchical clustering of the TKCC cohort also revealed 3 subgroups including 2 broad clusters, one of which contained 2 subclusters (Fig. 5B). We therefore defined the TKCC cell line cohort subtypes as follows; TKCC subtype 1 (purple bar) with cell lines TKCC 14, 26, 19, 16, 09, 10, and 15. TKCC subtype 2 (orange bar) contains TKCC lines 05, 27, and 22 and TKCC subtype 3 (blue bar) contains lines 06, 12, 07, 04, 18, 2.1, and 17. As for the ATCC cohort, there was no obvious relationship between these subtypes and the mutation status of KRAS or TP53 (supplemental Table S3).
Next we identified subtype-specific pTyr sites present in the TKCC cohort, identifying 383 pTyr sites using the ANOVA-based approaches. TKCC subtypes 1 and 2 were identified to only have down-regulated subtype-specific pTyr sites (101 and 73 sites respectively), whereas TKCC subtype 3 was identified to only have up-regulated subtype-specific pTyr sites (209 sites). We then utilized pathway analysis to characterize these subtype-specific sites, and identified enrichment for Ephrin (p = 0.04) and EGFR (p = 0.05) signaling in TKCC subtype 3. However, unlike the ATCC cohort, no enrichment for mRNA regulatory function was found within the 3 TKCC subtypes. Within the up-regulated subtype 3 sites we identified a dense network of protein-protein interactions, with strong representation from the axon guidance and EGFR signaling pathways (Fig. 5C). Isolation and mapping of the relative TIC of all RTK pTyr sites within the TKCC data set identified enhanced phosphorylation of many RTKs in TKCC subtype-3 (Fig. 5D). Tyrosine residues exhibiting enhanced phosphorylation included activation sites on EGFR, EPHA2, DDR1, FGFR1, INSR, MERTK, MET, and RON, indicating that increased activity of these RTKs characterizes this subtype.
Prediction of Subtypes Using a Random Forest-derived Signature
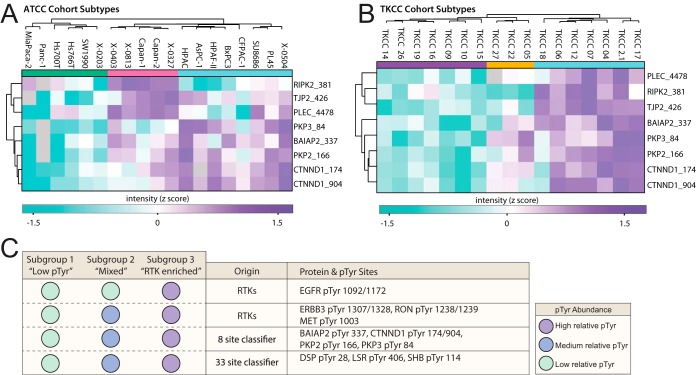
Next, we set out to identify, in a supervised manner, phosphosites that distinguish the three subtypes present in both the ATCC and TKCC series. We first isolated “classifier sites” for each cohort. These are sites identified using Random Forest (RF) models, based on having a high power to predict the defined topological subgroups (Mean Decrease Accuracy score of ≥ 1.5). Testing of these sites (ATCC, 69 sites and TKCC, 78 sites) using the left-out samples during the construction process of each RF model (out-of-bag approach) estimated that they predict the subtype topology of the corresponding cohort without error. We then tested the predictive power of the “classifier sites” in the alternate data set to where they were identified. The 33 ATCC classifier sites found in the TKCC cell lines data set could predict the TKCC topology with an accuracy of 94%. The 59 TKCC classifier sites quantified in the ATCC data set could predict the ATCC topology with an accuracy of 84%. Therefore, the 33 ATCC “classifier sites” represent the most robust signature to predict PDAC subgroups. The “classifier sites” from each cohort were then compared to identify the “common classifier sites” (Fig. 6A and 6B). These 8 sites predict the TKCC topology with an accuracy of 94%, and the ATCC topology with an accuracy of 89%. Therefore, we have identified a signature of 8 sites with a high predictive power across two PDAC cell line cohorts. Interestingly, this classifier does not include RTK phosphorylation sites characteristic of the “RTK-enriched” subgroup in the two cell line cohorts, indicating that the 3 subgroups identified in the independent cell line panels share additional conserved features. Specifically, subtypes 1, 2 and 3 in each panel clearly exhibit low, medium and high phosphorylation, respectively, of BAIAP2 Y337, PKP3 Y84, PKP2 Y166, CTNND1 Y174, and CTNND1 Y904. We note that although specific sites in RIPK2, TJP2 and PLEC are present in the classifier, their subtype-selective relative tyrosine phosphorylation differs between the two panels. A summary of phosphorylation sites that exhibit subtype-selective patterns of phosphorylation that are conserved across the two different cohorts is provided in Fig. 6C.
Fig. 6.
A random forest-derived phosphosite signature distinguishes PDAC subtypes. A, Clustering of the eight “common classifier sites” segregates the ATCC cell line cohort into subtypes (See colored bars, subtype 1 is green, 2 is pink and 3 is blue). B, Clustering of the eight “common classifier sites” segregates the TKCC cell line cohort into subtypes (See colored bars, subtype 1 is purple, 2 is orange and 3 is blue). C, A summary table of pTyr sites with consistent abundance profiles for a given subtype across the two PDAC cell line cohorts. These include subtypes; “Low pTyr” (subtype 1 shown by green in ATCC, purple in TKCC), “Mixed pTyr” (subtype 2 shown by pink in ATCC, orange in TKCC) and “RTK enriched” (subtype 3 shown by blue for both ATCC and TKCC).
Sensitivity of the identified PDAC subgroups to the targeted therapeutic, erlotinib
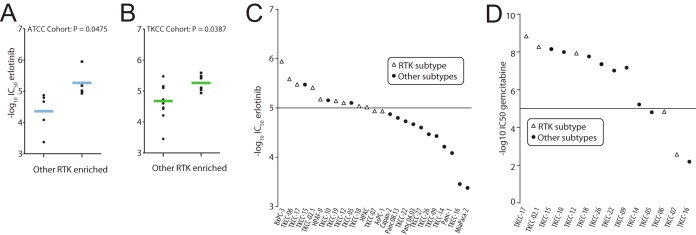
Given the increased expression and/or activation of EGFR and ErbB3 in ATCC and TKCC Subtype 3 (referred to as the RTK-enriched subtypes) and the clinical relevance of the small molecule EGFR inhibitor erlotinib in PDAC (27), we tested selected cell lines from each cohort for sensitivity to this EGFR kinase inhibitor. Indeed, both the ATCC and TKCC RTK-enriched subgroups exhibited enhanced sensitivity to erlotinib (Fig. 7A–7C) and therefore an increased dependence on EGFR kinase activity for cell proliferation. Importantly, this was not mirrored in the sensitivity profile of the TKCC cohort for the PDAC therapeutic gemcitabine (Fig. 7D), indicating a drug-selective effect. These data indicate that a phosphosignature characteristic of the RTK-enriched subtype could be used as a predictive biomarker for erlotinib sensitivity, and highlight a potential clinical application for this novel PDAC subclassification.
Fig. 7.
EGFR signaling dependence in the RTK-enriched PDAC subtypes. A, Mean scatter plot of IC50 values for erlotinib-treated ATCC cell lines that are either from “RTK enriched” or “other” subtypes. p values represent statistics using two-tailed unpaired t test. B, Mean scatter plot of IC50 values for erlotinib treated TKCC cell lines that are either from “RTK enriched” or “other” subtypes. p values represent statistics using two-tailed unpaired t test. C, IC50 values in negative log10 of drug concentration are plotted for each TKCC or ATCC cell line tested with erlotinib. Open triangles are “RTK enriched” cell lines and circles are lines from all alternate subtypes. D, IC50 values in negative log10 of drug concentration are plotted for each TKCC cell line tested with gemcitabine. Open triangles are “RTK enriched” cell lines and circles are TKCC subtype 1 and 2 cell lines.
DISCUSSION
In this paper we interrogate, for the first time, the heterogeneity of PDAC at the level of protein tyrosine phosphorylation. Our study identifies ∼1800 tyrosine phosphorylation sites across two large, independent PDAC cell line cohorts, provides new insights into dysregulated pathways and processes in PDAC, greatly extends our knowledge regarding the suite of RTKs activated in this disease, and also enables a novel subclassification based on tyrosine phosphorylation profiles. In addition, it identifies a PDAC subgroup characterized by high-level phosphorylation of multiple RTKs that exhibits increased sensitivity to erlotinib, providing 'proof-of-principle' that this approach could be exploited for development of predictive biomarkers.
A previous study based on gene expression profiling subclassified PDAC into three subtypes: classical, quasimesenchymal (QM), and exocrine-like, exhibiting differences in patient outcomes and therapeutic responses (6). Only the former two subtypes could be detected among human PDAC cell lines. In contrast, in silico microdissection of gene expression profiles resolved only two tumor subtypes, “classical” (equivalent to the namesake subtype identified by Collisson et al.) and “basal” (28). These workers suggested that the exocrine-like phenotype identified by Collisson et al. may reflect contamination from normal tissue, and that the “mesenchymal” component of the QM subtype might be derived from tumor stroma (28). However, a recent subclassification based on transcriptomics has identified four subtypes: squamous (corresponding to QM), aberrantly differentiated endocrine exocrine (ADEX) (corresponding to exocrine), pancreatic progenitor (classical) and immunogenic (5). Comparing our study to that of Collisson et al., there is not a simple relationship between any of the three subtypes we identified and the gene expression-defined classical or QM subtypes. For example, although some of our ATCC subtype 1 lines group in the QM subtype, this association is not perfect, because Panc 03–27, a Subtype 2 line, is also categorized as QM. In addition, the classical subtype contains examples of both subtype 2 and 3 cell lines from our study. Moreover, because all the PDAC cell lines characterized in the Moffitt et al. study (the majority of which are included in our ATCC panel) were assigned to a single subtype, basal, our approach is clearly resolving subtypes not detected by these workers. Also of note, for the ATCC panel, the tyrosine phosphorylation-defined subtypes do not exhibit an obvious relationship to patterns of mutation of known cancer-associated genes, including KRAS and TP53. Similarly, the different TKCC subtypes were not related to KRAS and TP53 status. Consequently, tyrosine phosphorylation profiling is providing a novel subclassification that is not mirrored in those obtained via transcriptomics or consideration of mutation patterns.
The identification of phosphosite classifiers enabled us to determine that the three subgroups resolved in each cell line cohort share distinctive characteristics in terms of tyrosine phosphorylation patterns. For example, subtypes 1, 2, and 3 in each panel clearly exhibit low, medium and high phosphorylation, respectively, of BAIAP2 Y337, PKP3 Y84, PKP2 Y166, CTNND1 Y174, and CTNND1 Y904. Interestingly, BAIAP2 regulates cytoskeletal organization, whereas PKP3, PKP2, and CTNND1 localize to or regulate cell-cell adhesions. The functional role of the latter three proteins is of particular interest given that a Sleeping Beauty mutagenesis screen for genes associated with PDAC development demonstrated an important role for the adherens and tight junction signaling pathways, and implicated CTNND1 as a tumor suppressor gene in this malignancy (29). Indeed, low expression of CTNND1 in a PDAC patient cohort was associated with poor survival (29). An interesting possibility is that the differential phosphorylation of proteins associated with cell-cell adhesions reflects, at least in part, modulation of the axon guidance pathway, a key regulator of cell-cell junction integrity and signaling that is strongly implicated in PDAC (3). Supporting this hypothesis, the phosphorylation of MET and several Eph receptors, which represent upstream regulators of this pathway, is markedly elevated in subtype 3.
One pathway commonly altered in PDAC is mRNA splicing, with mutations in splicing factors SF3B1 or RBM10 occurring in 4–5% of patients (3, 30). Interestingly, our analysis identified an alternative mechanism for dysregulation of RNA metabolism in PDAC, with markedly increased tyrosine phosphorylation of SF3B1, as well as many other proteins involved in regulation of RNA metabolism, including hnRNPC, H and K, and RBMX, being detected in ATCC subtype 2. Because aberrant expression of particular hnRNPs is associated with a switch in mRNA splicing of particular oncogenes or tumor suppressors toward pro-tumorigenic isoforms (31, 32), an attractive hypothesis is that the enhanced tyrosine phosphorylation of these regulators of RNA metabolism also positively impacts on oncogenic signaling pathways. In support of this, tyrosine phosphorylation of hnRNPs K and A2B1 is known to affect either their binding to, or the translation of, target mRNAs (33–35). The cell lines that characterize ATCC subtype 2 therefore present powerful models to test this hypothesis, as well as potential therapeutic opportunities. With regard to the latter, the splicing factors themselves could be targeted with small molecule inhibitors such as spliceostatin A (36). Alternative strategies include inhibition of upstream tyrosine kinases or downstream oncogenic pathways. However, subtype-selective phosphorylation of proteins associated with RNA metabolism was not detected in the TKCC cohort, and the reason for this is not clear at present. A potential explanation is the heterogeneity of PDAC, such that cell lines with this signature are under-represented in the TKCC cohort. Another possibility, which is not mutually exclusive, is that in the corresponding TKCC subgroup, alternative mechanisms act to dysregulate mRNA processing and/or the pathways that lie downstream.
An additional conserved feature of the molecular taxonomy provided by our phosphotyrosine profiling was the increased tyrosine phosphorylation of numerous RTKs, including EGFR, ERBB2, ERBB3, MET, FGFR1, and AXL, as well as several EphA and EphB receptors, in subtype 3 of both cell line panels. Additional lines of evidence support important roles for many of these receptors in PDAC pathogenesis. For example, signaling by the EGFR is required early in PDAC development (37), focal amplifications of MET, FGFR1, and ERBB2 occur in PDAC genomes (4) and contrasting levels of AXL activation can occur at different metastatic sites from the same PDAC patient (38). In addition, high expression of EphA7 or EphB2 is associated with poor prognosis in PDAC (39, 40). However, our study provides the first demonstration that a PDAC subtype exists that exhibits co-activation of many of these RTKs. This finding has important implications for the rational design of targeted therapies. In other cancers, co-activation of multiple RTKs is associated with resistance to treatment with individual TKIs, necessitating use of combination approaches using several TKIs or multikinase inhibitors (17, 41). Consequently, although subtype 3 cell lines exhibit a greater inhibition of proliferation upon treatment with the EGFR TKI erlotinib than those from other subtypes, and the subtype 3 cell lines HPAC and PL45 are sensitive to knockdown of MET and AXL, respectively (42), it remains possible that combined targeting of subtype 3-enriched RTKs will achieve a greater efficacy. This possibility is supported by a recent study demonstrating that the efficacy of MEK1 inhibition in PDAC is limited by compensatory signaling by multiple RTKs to the PI3K pathway, and that drug resistance can be overcome by combined targeting of EGFR, ERBB2, PDGFRA, and AXL (12). Importantly, the detailed information provided by our study will assist the design of therapeutic approaches directed at effective RTK blockade, and also fuel further studies that characterize crosstalk between particular RTKs in PDAC.
In the clinical trial that demonstrated the efficacy of erlotinib and gemcitabine co-treatment in PDAC, improved response to erlotinib was not associated with EGFR expression levels (27). This is consistent with our data indicating that EGFR levels are not consistently higher in subtype 3 cell lines, versus those from the other subgroups. Instead, it appears to be the tyrosine phosphorylation pattern associated with subtype 3, rather than EGFR expression, that is associated with increased sensitivity to this drug. Of note, although loss of p53 function is associated with a decreased dependence on the EGFR in PDAC (43), the increased sensitivity of ATCC and TKCC subtype 3 (the RTK subtypes) to erlotinib cannot be explained by TP53 mutational status. In addition, it is unrelated to K-Ras 'addiction' (26), because two out of three K-Ras-addicted cell lines in the ATCC cohort (Panc-08–13 and Capan-1) fall outside of the erlotinib-sensitive subtype. One component of the erlotinib sensitivity “signature” could be ERBB3 phosphorylation, because expression of this receptor is positively correlated with the erlotinib response of PDAC cell lines (44, 45). Although our study was directed toward identification of novel PDAC phenotypic subgroups and phosphorylation-based classifiers for these, rather than drug response signatures, our findings suggest that phosphosignatures of response to targeted therapies in PDAC could be formally derived and applied to patient stratification for therapy. This could be implemented via use of RPPAs or targeted MS approaches. Ultimately this could lead to improved management of patients suffering from this devastating disease.
Supplementary Material
Acknowledgments
We thank the Australian Pancreatic Cancer Genome Initiative (APGI) and all their members for their continuing support. Membership of the APGI is summarized below.
Garvan Institute of Medical Research: Andrew V. Biankin1, Amber L. Johns1, Amanda Mawson1, David K. Chang1, Christopher J. Scarlett1, Mary-Anne L. Brancato1, Sarah J. Rowe1, Skye L. Simpson1, Mona Martyn-Smith1, Michelle T. Thomas1, Lorraine A. Chantrill1, Venessa T. Chin1, Angela Chou1, Mark J. Cowley1, Jeremy L. Humphris1, Marc D. Jones1, R. Scott Mead1, Adnan M. Nagrial1, Marina Pajic1, Jessica Pettit1, Mark Pinese1, Ilse Rooman1, Jianmin Wu1, Jian Tao1, Renee DiPietro1, Clare Watson1, Rachel Wong1, Andreia V. Pinho1, Marc Giry-Laterriere1, Roger J. Daly1, Elizabeth A. Musgrove1, Robert L. Sutherland1; Queensland Center for Medical Genomics, Institute for Molecular Bioscience: Sean M. Grimmond2, Nicola Waddell2, Karin S. Kassahn2, David K. Miller2, Peter J. Wilson2, Ann-Marie Patch2, Sarah Song2, Ivon Harliwong2, Senel Idrisoglu2, Craig Nourse2, Ehsan Nourbakhsh2, Suzanne Manning2, Shivangi Wani2, Milena Gongora2, Matthew Anderson2, Oliver Holmes2, Conrad Leonard2, Darrin Taylor2, Scott Wood2, Christina Xu2, Katia Nones2, J. Lynn Fink2, Angelika Christ2, Tim Bruxner2, Nicole Cloonan2, Felicity Newell2, John V. Pearson2; Royal North Shore Hospital: Jaswinder S. Samra3, Anthony J. Gill3, Nick Pavlakis3, Alex Guminski3, Christopher Toon3; Bankstown Hospital: Andrew V. Biankin4, Ray Asghari4, Neil D. Merrett4, David K. Chang4, Darren A. Pavey4, Amitabha Das4; Liverpool Hospital: Peter H. Cosman5, Kasim Ismail5, Chelsie O'Connor5; Westmead Hospital: Vincent W. Lam6, Duncan McLeod6, Henry C. Pleass6, Arthur Richardson6, Virginia James6; Royal Prince Alfred Hospital: James G. Kench7, Caroline L. Cooper7, David Joseph7, Charbel Sandroussi7, Michael Crawford7, James Gallagher7; Fremantle Hospital: Michael Texler8, Cindy Forrest8, Andrew Laycock8, Krishna P. Epari8, Mo Ballal8, David R. Fletcher8, Sanjay Mukhedkar8; Sir Charles Gairdner Hospital: Nigel A. Spry9, Bastiaan DeBoer9, Ming Chai9; St John of God Healthcare: Nikolajs Zeps10, Maria Beilin10, Kynan Feeney10; Royal Adelaide Hospital: Nam Q. Nguyen11, Andrew R. Ruszkiewicz11, Chris Worthley11, Chuan P. Tan11, Tamara Debrencini11; Flinders Medical Centre: John Chen12, Mark E. Brooke-Smith12, Virginia Papangelis12; Greenslopes Private Hospital: Henry Tang13, Andrew P. Barbour13; Envoi Pathology: Andrew D. Clouston14, Patrick Martin14; Princess Alexandria Hospital: Thomas J. O'Rourke15, Amy Chiang15, Jonathan W. Fawcett15, Kellee Slater15, Shinn Yeung15, Michael Hatzifotis15, Peter Hodgkinson15; Austin Hospital: Christopher Christophi16, Mehrdad Nikfarjam16, Angela Mountain16; Victorian Cancer Biobank17.
1The Kinghorn Cancer Centre, Garvan Institute of Medical Research, 370 Victoria Street, Darlinghurst, Sydney, New South Wales 2010, Australia; 2Queensland Center for Medical Genomics, Institute for Molecular Bioscience, University of Queensland, St Lucia, Queensland 4072, Australia; 3Royal North Shore Hospital, Westbourne Street, St Leonards, New South Wales 2065, Australia; 4Bankstown Hospital, Eldridge Road, Bankstown, New South Wales 2200, Australia; 5Liverpool Hospital, Elizabeth Street, Liverpool, New South Wales 2170, Australia; 6Westmead Hospital, Hawkesbury and Darcy Roads, Westmead, New South Wales 2145, Australia; 7Royal Prince Alfred Hospital, Missenden Road, Camperdown, New South Wales 2050, Australia; 8Fremantle Hospital, Alma Street, Fremantle, Western Australia 6959, Australia; 9Sir Charles Gairdner Hospital, Hospital Avenue, Nedlands, Western Australia 6009, Australia; 10St John of God Healthcare, 12 Salvado Road, Subiaco, Western Australia 6008, Australia; 11Royal Adelaide Hospital, North Terrace, Adelaide, South Australia 5000, Australia; 12Flinders Medical Centre, Flinders Drive, Bedford Park, South Australia 5042, Australia; 13Greenslopes Private Hospital, Newdegate Street, Greenslopes, Queensland 4120, Australia; 14Envoi Pathology, 1/49 Butterfield Street, Herston, Queensland 4006, Australia; 15Princess Alexandria Hospital, Cornwall Street & Ipswich Road, Woolloongabba, Queensland 4102, Australia; 16Austin Hospital, 145 Studley Road, Heidelberg, Victoria 3084, Australia; 17Victorian Cancer Biobank, 1 Rathdowne Street, Carlton, Victoria 3053, Australia.
Footnotes
Author contributions: ESH undertook MS-based phosphotyrosine profiling across the ATCC cohort as well as Western blot analyses and erlotinib sensitivity assays for this panel. AMN and MP generated cell lines and subsequent cell lysates for the TKCC cohort, and AMN characterized erlotinib sensitivity across this panel. S-PS undertook phosphotyrosine profiling for the TKCC cohort. GML facilitated tissue culture processes. ESH, FH, LGH, DKC, AVB and RJD contributed to experimental design and project planning. Bioinformatic analyses were primarily undertaken by JW, with bioinformatics contributions and figure generation performed by ESH. ESH, JW and RJD prepared the manuscript, with input from RGP and AY. Overall project coordination was undertaken by RJD.
* This work was supported by a Program Grant (535903) and Project Grant (1083896) from the National Health and Medical Research Council (NHMRC) of Australia. RJD and AY are supported by NHMRC Fellowships (1058540 and 1044041, respectively). RGP and AY are recipients of a NHMRC Program Grant (1037320). ESH is the recipient of a Research Scholar Award (10/RSA/1-28) from the Cancer Institute of New South Wales and an Australian Postgraduate Award from the University of New South Wales. JW is supported by SRF for ROCS, State Education Ministry (SEM), and open research grant (2016), State Key Laboratory of Protein and Plant Gene Research.
 This article contains supplemental material.
This article contains supplemental material.
Authors declare no conflict of interest.
1 The abbreviations used are:
- PDAC
- pancreatic ductal adenocarcinoma
- AJs
- adheren junctions
- ATCC
- American Type Culture Collection
- EMT
- epithelial to mesenchyme transition
- HPDE
- human pancreatic duct epithelial
- IAP
- immunoaffinity purification
- LC-MS/MS
- Liquid chromatography-tandem mass spectrometry
- MDA
- mean decrease accuracy
- MS
- mass spectrometry
- PCA
- principal component analysis
- PINA
- Protein Interaction Network Analysis
- pTyr
- phosphotyrosine
- QM
- quasimesenchymal
- RF
- Random Forest
- RT
- Room temperature
- RTK
- receptor tyrosine kinase
- TBST
- Tris Buffered Saline with Tween
- TCEP
- tris (2-carboxyethyl) phosphine
- TFA
- trifluoroacetic acid
- TJs
- tight junctions
- TKCC
- The Kinghorn Cancer Centre.
REFERENCES
- 1. Vogelzang N. J., Benowitz S. I., Adams S., Aghajanian C., Chang S. M., Dreyer Z. E., Janne P. A., Ko A. H., Masters G. A., Odenike O., Patel J. D., Roth B. J., Samlowski W. E., Seidman A. D., Tap W. D., Temel J. S., Von Roenn J. H., and Kris M. G. (2012) Clinical cancer advances 2011: Annual Report on Progress Against Cancer from the American Society of Clinical Oncology. J. Clin. Oncol. 30, 88–109 [DOI] [PubMed] [Google Scholar]
- 2. Paulson A. S., Tran Cao H. S., Tempero M. A., and Lowy A. M. (2013) Therapeutic advances in pancreatic cancer. Gastroenterology 144, 1316–1326 [DOI] [PubMed] [Google Scholar]
- 3. Biankin A. V., Waddell N., Kassahn K. S., Gingras M. C., Muthuswamy L. B., Johns A. L., Miller D. K., Wilson P. J., Patch A. M., Wu J., Chang D. K., Cowley M. J., Gardiner B. B., Song S., Harliwong I., Idrisoglu S., Nourse C., Nourbakhsh E., Manning S., Wani S., Gongora M., Pajic M., Scarlett C. J., Gill A. J., Pinho A. V., Rooman I., Anderson M., Holmes O., Leonard C., Taylor D., Wood S., Xu Q., Nones K., Fink J. L., Christ A., Bruxner T., Cloonan N., Kolle G., Newell F., Pinese M., Mead R. S., Humphris J. L., Kaplan W., Jones M. D., Colvin E. K., Nagrial A. M., Humphrey E. S., Chou A., Chin V. T., Chantrill L. A., Mawson A., Samra J. S., Kench J. G., Lovell J. A., Daly R. J., Merrett N. D., Toon C., Epari K., Nguyen N. Q., Barbour A., Zeps N., Kakkar N., Zhao F., Wu Y. Q., Wang M., Muzny D. M., Fisher W. E., Brunicardi F. C., Hodges S. E., Reid J. G., Drummond J., Chang K., Han Y., Lewis L. R., Dinh H., Buhay C. J., Beck T., Timms L., Sam M., Begley K., Brown A., Pai D., Panchal A., Buchner N., De Borja R., Denroche R. E., Yung C. K., Serra S., Onetto N., Mukhopadhyay D., Tsao M. S., Shaw P. A., Petersen G. M., Gallinger S., Hruban R. H., Maitra A., Iacobuzio-Donahue C. A., Schulick R. D., Wolfgang C. L., Morgan R. A., Lawlor R. T., Capelli P., Corbo V., Scardoni M., Tortora G., Tempero M. A., Mann K. M., Jenkins N. A., Perez-Mancera P. A., Adams D. J., Largaespada D. A., Wessels L. F., Rust A. G., Stein L. D., Tuveson D. A., Copeland N. G., Musgrove E. A., Scarpa A., Eshleman J. R., Hudson T. J., Sutherland R. L., Wheeler D. A., Pearson J. V., McPherson J. D., Gibbs R. A., and Grimmond S. M. (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Waddell N., Pajic M., Patch A. M., Chang D. K., Kassahn K. S., Bailey P., Johns A. L., Miller D., Nones K., Quek K., Quinn M. C., Robertson A. J., Fadlullah M. Z., Bruxner T. J., Christ A. N., Harliwong I., Idrisoglu S., Manning S., Nourse C., Nourbakhsh E., Wani S., Wilson P. J., Markham E., Cloonan N., Anderson M. J., Fink J. L., Holmes O., Kazakoff S. H., Leonard C., Newell F., Poudel B., Song S., Taylor D., Waddell N., Wood S., Xu Q., Wu J., Pinese M., Cowley M. J., Lee H. C., Jones M. D., Nagrial A. M., Humphris J., Chantrill L. A., Chin V., Steinmann A. M., Mawson A., Humphrey E. S., Colvin E. K., Chou A., Scarlett C. J., Pinho A. V., Giry-Laterriere M., Rooman I., Samra J. S., Kench J. G., Pettitt J. A., Merrett N. D., Toon C., Epari K., Nguyen N. Q., Barbour A., Zeps N., Jamieson N. B., Graham J. S., Niclou S. P., Bjerkvig R., Grutzmann R., Aust D., Hruban R. H., Maitra A., Iacobuzio-Donahue C. A., Wolfgang C. L., Morgan R. A., Lawlor R. T., Corbo V., Bassi C., Falconi M., Zamboni G., Tortora G., Tempero M. A., Gill A. J., Eshleman J. R., Pilarsky C., Scarpa A., Musgrove E. A., Pearson J. V., Biankin A. V., and Grimmond S. M. (2015) Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bailey P., Chang D. K., Nones K., Johns A. L., Patch A. M., Gingras M. C., Miller D. K., Christ A. N., Bruxner T. J., Quinn M. C., Nourse C., Murtaugh L. C., Harliwong I., Idrisoglu S., Manning S., Nourbakhsh E., Wani S., Fink L., Holmes O., Chin V., Anderson M. J., Kazakoff S., Leonard C., Newell F., Waddell N., Wood S., Xu Q., Wilson P. J., Cloonan N., Kassahn K. S., Taylor D., Quek K., Robertson A., Pantano L., Mincarelli L., Sanchez L. N., Evers L., Wu J., Pinese M., Cowley M. J., Jones M. D., Colvin E. K., Nagrial A. M., Humphrey E. S., Chantrill L. A., Mawson A., Humphris J., Chou A., Pajic M., Scarlett C. J., Pinho A. V., Giry-Laterriere M., Rooman I., Samra J. S., Kench J. G., Lovell J. A., Merrett N. D., Toon C. W., Epari K., Nguyen N. Q., Barbour A., Zeps N., Moran-Jones K., Jamieson N. B., Graham J. S., Duthie F., Oien K., Hair J., Grutzmann R., Maitra A., Iacobuzio-Donahue C. A., Wolfgang C. L., Morgan R. A., Lawlor R. T., Corbo V., Bassi C., Rusev B., Capelli P., Salvia R., Tortora G., Mukhopadhyay D., Petersen G. M., Australian Pancreatic Cancer Genome, I., Munzy D. M., Fisher W. E., Karim S. A., Eshleman J. R., Hruban R. H., Pilarsky C., Morton J. P., Sansom O. J., Scarpa A., Musgrove E. A., Bailey U. M., Hofmann O., Sutherland R. L., Wheeler D. A., Gill A. J., Gibbs R. A., Pearson J. V., Waddell N., Biankin A. V., and Grimmond S. M. (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 [DOI] [PubMed] [Google Scholar]
- 6. Collisson E. A., Sadanandam A., Olson P., Gibb W. J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G. E., Jakkula L., Feiler H. S., Ko A. H., Olshen A. B., Danenberg K. L., Tempero M. A., Spellman P. T., Hanahan D., and Gray J. W. (2011) Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jimeno A., Tan A. C., Coffa J., Rajeshkumar N. V., Kulesza P., Rubio-Viqueira B., Wheelhouse J., Diosdado B., Messersmith W. A., Iacobuzio-Donahue C., Maitra A., Varella-Garcia M., Hirsch F. R., Meijer G. A., and Hidalgo M. (2008) Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res. 68, 2841–2849 [DOI] [PubMed] [Google Scholar]
- 8. Roesli C. (2015) Recent advances in proteomically subtyping pancreatic ductal adenocarcinomas and their potential clinical impact. Expert Rev. Proteomics 12, 5–8 [DOI] [PubMed] [Google Scholar]
- 9. Britton D., Zen Y., Quaglia A., Selzer S., Mitra V., Lobetaner C., Jung S., Bohm G., Schmid P., Prefot P., Hoehle C., Koncarevic S., Gee J., Nicholson R., Ward M., Castellano L., Stebbing J., Zucht H. D., Sarker D., Heaton N., and Pike I. (2014) Quantification of pancreatic cancer proteome and phosphorylome: indicates molecular events likely contributing to cancer and activity of drug targets. PloS One 9, e90948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friess H., Yamanaka Y., Kobrin M. S., Do D. A., Buchler M. W., and Korc M. (1995) Enhanced erbB-3 expression in human pancreatic cancer correlates with tumor progression. Clin. Cancer Res. 1, 1413–1420 [PubMed] [Google Scholar]
- 11. Koorstra J. B., Karikari C. A., Feldmann G., Bisht S., Rojas P. L., Offerhaus G. J., Alvarez H., and Maitra A. (2009) The Axl receptor tyrosine kinase confers an adverse prognostic influence in pancreatic cancer and represents a new therapeutic target. Cancer Biol. Therapy 8, 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pettazzoni P., Viale A., Shah P., Carugo A., Ying H., Wang H., Genovese G., Seth S., Minelli R., Green T., Huang-Hobbs E., Corti D., Sanchez N., Nezi L., Marchesini M., Kapoor A., Yao W., Francesco M. E., Petrocchi A., Deem A. K., Scott K., Colla S., Mills G. B., Fleming J. B., Heffernan T. P., Jones P., Toniatti C., DePinho R. A., and Draetta G. F. (2015) Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer Res. 75, 1091–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ueda S., Ogata S., Tsuda H., Kawarabayashi N., Kimura M., Sugiura Y., Tamai S., Matsubara O., Hatsuse K., and Mochizuki H. (2004) The correlation between cytoplasmic overexpression of epidermal growth factor receptor and tumor aggressiveness: poor prognosis in patients with pancreatic ductal adenocarcinoma. Pancreas 29, e1–8 [DOI] [PubMed] [Google Scholar]
- 14. Nagaraj N. S., Smith J. J., Revetta F., Washington M. K., and Merchant N. B. (2010) Targeted inhibition of SRC kinase signaling attenuates pancreatic tumorigenesis. Mol. Cancer Therapeutics 9, 2322–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruggeri B. A., Huang L., Wood M., Cheng J. Q., and Testa J. R. (1998) Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol. Carcinogenesis 21, 81–86 [PubMed] [Google Scholar]
- 16. Foo W. C., Rashid A., Wang H., Katz M. H., Lee J. E., Pisters P. W., Wolff R. A., Abbruzzese J. L., Fleming J. B., and Wang H. (2013) Loss of phosphatase and tensin homolog expression is associated with recurrence and poor prognosis in patients with pancreatic ductal adenocarcinoma. Human Pathol. 44, 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hochgrafe F., Zhang L., O'Toole S. A., Browne B. C., Pinese M., Porta Cubas A., Lehrbach G. M., Croucher D. R., Rickwood D., Boulghourjian A., Shearer R., Nair R., Swarbrick A., Faratian D., Mullen P., Harrison D. J., Biankin A. V., Sutherland R. L., Raftery M. J., and Daly R. J. (2010) Tyrosine phosphorylation profiling reveals the signaling network characteristics of Basal breast cancer cells. Cancer Res. 70, 9391–9401 [DOI] [PubMed] [Google Scholar]
- 18. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., and Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 19. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 21. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 22. Hrydziuszko O., and Viant M. R. (2012) Missing values in mass spectrometry based metabolomics: an undervalued step in the data processing pipeline. Metabolomics 8, 161–174 [Google Scholar]
- 23. Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C. Y., and Wei L. (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cowley M. J., Pinese M., Kassahn K. S., Waddell N., Pearson J. V., Grimmond S. M., Biankin A. V., Hautaniemi S., and Wu J. (2012) PINA v2.0: mining interactome modules. Nucleic Acids Res. 40, D862–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qian J., Niu J., Li M., Chiao P. J., and Tsao M. S. (2005) In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K-ras oncogenic activation in pancreatic carcinogenesis. Cancer Res. 65, 5045–5053 [DOI] [PubMed] [Google Scholar]
- 26. Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., and Settleman J. (2009) A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moore M. J., Goldstein D., Hamm J., Figer A., Hecht J. R., Gallinger S., Au H. J., Murawa P., Walde D., Wolff R. A., Campos D., Lim R., Ding K., Clark G., Voskoglou-Nomikos T., Ptasynski M., Parulekar W., and National Cancer Institute of Canada Clinical Trials, G. (2007) Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 25, 1960–1966 [DOI] [PubMed] [Google Scholar]
- 28. Moffitt R. A., Marayati R., Flate E. L., Volmar K. E., Loeza S. G., Hoadley K. A., Rashid N. U., Williams L. A., Eaton S. C., Chung A. H., Smyla J. K., Anderson J. M., Kim H. J., Bentrem D. J., Talamonti M. S., Iacobuzio-Donahue C. A., Hollingsworth M. A., and Yeh J. J. (2015) Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genetics 47, 1168–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mann K. M., Ward J. M., Yew C. C., Kovochich A., Dawson D. W., Black M. A., Brett B. T., Sheetz T. E., Dupuy A. J., Australian Pancreatic Cancer Genome, I., Chang D. K., Biankin A. V., Waddell N., Kassahn K. S., Grimmond S. M., Rust A. G., Adams D. J., Jenkins N. A., and Copeland N. G. (2012) Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 109, 5934–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Witkiewicz A. K., McMillan E. A., Balaji U., Baek G., Lin W. C., Mansour J., Mollaee M., Wagner K. U., Koduru P., Yopp A., Choti M. A., Yeo C. J., McCue P., White M. A., and Knudsen E. S. (2015) Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Golan-Gerstl R., Cohen M., Shilo A., Suh S. S., Bakacs A., Coppola L., and Karni R. (2011) Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 71, 4464–4472 [DOI] [PubMed] [Google Scholar]
- 32. Lefave C. V., Squatrito M., Vorlova S., Rocco G. L., Brennan C. W., Holland E. C., Pan Y. X., and Cartegni L. (2011) Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 30, 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ostareck-Lederer A., Ostareck D. H., Cans C., Neubauer G., Bomsztyk K., Superti-Furga G., and Hentze M. W. (2002) c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 22, 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ostrowski J., Schullery D. S., Denisenko O. N., Higaki Y., Watts J., Aebersold R., Stempka L., Gschwendt M., and Bomsztyk K. (2000) Role of tyrosine phosphorylation in the regulation of the interaction of heterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners. J. Biol. Chem. 275, 3619–3628 [DOI] [PubMed] [Google Scholar]
- 35. White R., Gonsior C., Kramer-Albers E. M., Stohr N., Huttelmaier S., and Trotter J. (2008) Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J. Cell Biol. 181, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maguire S. L., Leonidou A., Wai P., Marchio C., Ng C. K., Sapino A., Salomon A. V., Reis-Filho J. S., Weigelt B., and Natrajan R. C. (2015) SF3B1 mutations constitute a novel therapeutic target in breast cancer. J. Pathol. 235, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perera R. M., and Bardeesy N. (2012) Ready, set, go: the EGF receptor at the pancreatic cancer starting line. Cancer cell 22, 281–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim M. S., Zhong Y., Yachida S., Rajeshkumar N. V., Abel M. L., Marimuthu A., Mudgal K., Hruban R. H., Poling J. S., Tyner J. W., Maitra A., Iacobuzio-Donahue C. A., and Pandey A. (2014) Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol. Cell. Proteomics 13, 2803–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giaginis C., Tsourouflis G., Zizi-Serbetzoglou A., Kouraklis G., Chatzopoulou E., Dimakopoulou K., and Theocharis S. E. (2010) Clinical significance of ephrin (eph)-A1, -A2, -a4, -a5 and -a7 receptors in pancreatic ductal adenocarcinoma. Pathol. Oncol. Res. 16, 267–276 [DOI] [PubMed] [Google Scholar]
- 40. Lu Z., Zhang Y., Li Z., Yu S., Zhao G., Li M., Wang Z., Wang Q., and Yang Y. (2012) Overexpression of the B-type Eph and ephrin genes correlates with progression and pain in human pancreatic cancer. Oncol. Lett. 3, 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stommel J. M., Kimmelman A. C., Ying H., Nabioullin R., Ponugoti A. H., Wiedemeyer R., Stegh A. H., Bradner J. E., Ligon K. L., Brennan C., Chin L., and DePinho R. A. (2007) Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318, 287–290 [DOI] [PubMed] [Google Scholar]
- 42. Kothari V., Wei I., Shankar S., Kalyana-Sundaram S., Wang L., Ma L. W., Vats P., Grasso C. S., Robinson D. R., Wu Y. M., Cao X., Simeone D. M., Chinnaiyan A. M., and Kumar-Sinha C. (2013) Outlier kinase expression by RNA sequencing as targets for precision therapy. Cancer Discovery 3, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Navas C., Hernandez-Porras I., Schuhmacher A. J., Sibilia M., Guerra C., and Barbacid M. (2012) EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell 22, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frolov A., Schuller K., Tzeng C. W., Cannon E. E., Ku B. C., Howard J. H., Vickers S. M., Heslin M. J., Buchsbaum D. J., and Arnoletti J. P. (2007) ErbB3 expression and dimerization with EGFR influence pancreatic cancer cell sensitivity to erlotinib. Cancer Biol. Therapy 6, 548–554 [DOI] [PubMed] [Google Scholar]
- 45. Liles J. S., Arnoletti J. P., Tzeng C. W., Howard J. H., Kossenkov A. V., Kulesza P., Heslin M. J., and Frolov A. (2010) ErbB3 expression promotes tumorigenesis in pancreatic adenocarcinoma. Cancer Biol. Therapy 10, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.