Abstract
The principle of shotgun proteomics is to use peptide mass spectra in order to identify corresponding sequences in a protein database. The quality of peptide and protein identification and quantification critically depends on the sensitivity and specificity of this assignment process. Many peptides in proteomic samples carry biochemical modifications, and a large fraction of unassigned spectra arise from modified peptides. Spectra derived from modified peptides can erroneously be assigned to wrong amino acid sequences. However, the impact of this problem on proteomic data has not yet been investigated systematically. Here we use combinations of different database searches to show that modified peptides can be responsible for 20–50% of false positive identifications in deep proteomic data sets. These false positive hits are particularly problematic as they have significantly higher scores and higher intensities than other false positive matches. Furthermore, these wrong peptide assignments lead to hundreds of false protein identifications and systematic biases in protein quantification. We devise a “cleaned search” strategy to address this problem and show that this considerably improves the sensitivity and specificity of proteomic data. In summary, we show that modified peptides cause systematic errors in peptide and protein identification and quantification and should therefore be considered to further improve the quality of proteomic data annotation.
Mass spectrometry has matured to a level where it is able to assess the complexity of the human proteome (1). The typical workflow of a shotgun proteomic experiment involves digestion of proteins into peptides. The resulting peptide mixtures are then analyzed by tandem mass spectrometry in order to obtain the mass of the peptide and the fragmentation pattern. Algorithms such as Mascot (2), Andromeda (3) or Sequest (4) then identify peptides by matching these data to protein databases. Although these algorithms are routinely used in hundreds of proteomic studies, minimizing false-positive and false-negative identifications during the database search remains an important challenge. Recently, deep proteomic studies identified >10,000 proteins in mammalian cell lines (5, 6), and large scale studies across several tissues identified more than 80% of the expected human proteome (7, 8). This is a major achievement and provides a valuable resource for the community. However, the extent of false protein identifications in these data sets is under debate (9, 10) and subject to ongoing research and refinement (11).
Peptide sequence assignments can lead to false-positive identifications from at least three different sources: (1) low-quality spectra (12), (2) imperfect data processing algorithms (e.g. errors in charge state determination (13), monoisotopic peak identification etc.), or (3) the use of incomplete database search space (13, 14). In the latter case, the correct match is not contained in the search space, for example because of incomplete protein annotation or the occurrence of unexpected biochemical modifications. Because spectra cannot be matched to the correct sequence, they can be erroneously assigned to a different peptide in the database.
The identification of peptides with modifications is particularly challenging: On the one hand, allowing for multiple possible modifications in a standard database search leads to a combinatorial expansion that dramatically increases the search space (15). On the other hand, when a specific modification is not considered, peptides carrying this modification cannot be correctly identified. Modifications can be introduced in vivo (e.g. phosphorylation, ubiquitination), in vitro during sample preparation (e.g. carbamidomethylation, carbamylation) or both (e.g. deamidation, acetylation, methylation). It is estimated that every unmodified peptide is accompanied by ∼10 modified versions that are typically less abundant (16). Therefore, deeper and deeper coverage of the proteome is expected to lead to more and more spectra derived from modified peptides. This makes modified peptides a particularly vexing problem in deep proteomic studies. For example, a recent article reported that at least one third of all unassigned spectra represent modified peptides (17). Hence, modified peptides are a systematic source of false-negative identifications (i.e. type II errors). The global impact of modified peptides on false-positive identifications (i.e. type I errors) in deep proteomic data sets has not yet been assessed.
Here, we used a combination of different database search strategies to systematically investigate this problem. We find that about half of false positive hits can be because of modified peptides. These misidentifications give rise to erroneous protein identification and quantification. Eliminating these false positive hits substantially improves the quality of data annotation. In summary, we identify modified peptides as a systematic source of biases in protein identification and quantification in deep proteomic data sets and outline a strategy to minimize type I errors caused by modified peptides.
EXPERIMENTAL PROCEDURES
MaxQuant Output
The Following Output Files Were Used from MaxQuant (18) Software Version 1.5.2.8
evidence.txt - This file contains all information on the identified peptides, including peptide sequence, protein ID, modification status, search score, m/z, mass-error, charge, etc. A unique identifier is given that links each peptide spectrum match (PSM)1 to other files.
apl-files - These files correspond to the mgf peaklist format (mascot generic format) and list features for each MS/MS scan, including precursor m/z, precursor charge state, fragment m/z and corresponding intensities. Apl-files contain all information necessary for the Andromeda search engine to process the scan. Apl-files are written by MaxQuant after precursor mass calibration.
msms.txt - This file contains additional information on identified fragment matches from MS/MS spectra, including e.g. fragment intensities, mass deviations, etc.
allPeptides.txt - This file contains information on features, including identified and non-identified peptides. Additional peptide identifications from the dependent peptides search (implementation of ModifiComb (19) for MaxQuant software) are reported in this file.
Sample Collection and Preparation
The proteomic data for HeLa was published previously (5) and downloaded from proteomicsDB. The proteomic data for HEK293 was published previously (20) and generated as described. Briefly, cells were grown in Dulbecco's Modified Eagle Medium (Life Technologies, California, USA). Lysis was performed in 50 mm ammonium bicarbonate buffer (pH 8.0) containing 2% SDS and 0.1 m DTT. Sulfhydryl groups were alkylated by adding iodoacetamide to a final concentration of 0.25 m and incubation for 20 min. Proteins were precipitated according to Wessel and Fluegge (21), resuspended in 6 m urea/2 M thiourea/10 mm HEPES and digested into peptides using Lys-C (3 h) and Trypsin (overnight, diluted 4× with 50 mm ABC). Peptides were then acidified, desalted and subjected to isoelectric focusing (IEF) for fractionation.
LC-MS/MS and Data Analysis
Peptides from proteomic samples were desalted using stage tip purification and subsequently analyzed by online liquid-chromatography tandem mass-spectrometry on a Q-Exactive (ThermoFisher, Massachusetts, USA) instrument using nano-electrospray ionization. Resolution was set to 70,000 and 17,500 for full and fragments scans respectively. Data was acquired with “fast” settings as described (22). The proteomic raw data for HEK293 has been uploaded to the Pride archive and is accessible under the project identifier “PXD002389.”
The synthetic peptides IESSIQSLQDLSK (Grid2), IESLSSQLSNLQK (Lmnb1), and IESLSSQLSNLEK were ordered from Biosyntan GmbH (Berlin, Germany), suspended in Buffer containing 5% acetonitrile and 0.1% formic acid and then analyzed by LC-MS/MS as described above.
Peptides from proteomic samples were identified from MS/MS spectra by searching against the recent Uniprot human database (2014–10, 88,840 protein sequences) using MaxQuant version 1.5.2.8. For the standard search carbamidomethyl (Cys) was set as fixed, oxidation (Met) and acetylation (protein N-term) as variable modifications for both data sets investigated.
Unknown modifications were identified by the “dependent peptides” setting implemented in MaxQuant version 1.5.2.8 in a standard search. The implemented algorithm performs spectrum matching to identify modified peptides in an unbiased manner (19). If an unidentified spectrum matches an identified spectrum the mass shift (corresponding to the modification of the peptide) of the theoretical and observed precursor mass and the matched sequence will be reported. Modified peptides will be only identified if they are derived from an already identified unmodified peptide (see also Results section).
Modified peptides were extracted from allPeptides.txt along with the ΔM mass shift between base and dependent peptides. Abundant modifications based on these results from the ModifiComb algorithm were selected for further consideration (= deamidation, carbamylation and methylation for the HEK293 data set; = deamidation, loss of ammonia, dehydration and oxidation for the HeLa data set). To identify amino acid preferences for these modifications, modified peptide sequences along with amino acid preferences for the above mentioned modifications were extracted from allPeptides.txt. Site specificities were estimated by counting the modified residues for each predicted modified amino acid. In cases, when the ModifiComb algorithm identified multiple amino acids as potential modification sites, the count was divided by the number of different amino acids reported. Finally, modifications on amino acids that are chemically impossible (as i.e. modifications on inert side-chains) or on chemically disfavored amino acids were excluded.
According to this, raw-files from the HEK293 data set were analyzed with one of the following selected variable modifications (site specificities and approximated global search space increase relative to the standard search): deamidation (Asn, Gln anywhere - 7.2), methylation (Lys, Glu, Asp anywhere - 44.7), and carbamylation (any N terminus - 2.0). The combined search for the HEK293 data set included all variable modifications mentioned above (236.2). For the control search an arbitrary, but according to the ModifiComb estimation non-existent, mass-shift of −11.01 was allowed to occur on Asn and Gln residues. Raw files from Nagaraj et al. were searched separately with one of the following variable modifications: deamidation (Gln, Asn - 7.2), dehydration (Glu, Asp anywhere; any n-terminus - 36.2), ammonia loss (Gln anywhere; any N terminus - 6.5) and oxidation (Trp, Tyr anywhere - 2.4). The search space increase was estimated empirically on an in silico tryptic digest by considering a maximum of two missed cleavages and up to five modified sites per peptide (the constraint used by MaxQuant). Computational time was read out from MQ-file runningTimes.txt. All MQ runs were performed on an Intel Xeon CPU X5560 with 2.8 Ghz and 64 GB memory using 24 threads. The following settings were used for all searches: A maximum of two missed cleavages was allowed. Enzyme specificity was set to Trypsin/P, meaning that cleavage is allowed to occur between lysine or arginine and proline. PSM and protein FDR was set to 1. Minimal peptide length was set to 7 amino acids and the main search peptide tolerance was set to 4.5 ppm. Second peptide option was set to off. Modified peptide identifications with an Andromeda search score greater than 40 and a delta score (that gives the score difference between the best and second best matching candidate for a MS/MS scan, as is MQ standard settings) greater than 6 were allowed. All searches for a given data set were based on one set of Andromeda peak list files (apl-files).
Data Processing and Statistical Rational
Identification of Conflicting PSMs
In general, the significance of PSM and protein identifications was estimated by a target-decoy-search strategy (23). The target decoy search strategy is designed to control false positives (type I errors). Spectra were searched against a concatenated database, which contains all candidate proteins plus control proteins with pseudo-reverted sequences as previously described (18). Spectra are matched to candidate peptide sequences and are assigned a score by the Andromeda search engine (18).
Text file outputs from MaxQuant were processed using Python and R scripts. PSMs were extracted from the evidence.txt.
As has been shown by Savitski et al. (2015), Andromeda scores have higher significance levels with increasing peptide length. To normalize for this we used the previously published protocol for length-normalization of Andromeda scores (11) (LScore). Briefly, and as described by Savitski et al. (11), all PSMs of the same length were binned in score intervals of one and smoothed by a moving average with a window size of five. The local FDR in each score bin was calculated by dividing the number of decoy PSMs by the number of target PSMs and the resulting distribution was smoothed using a moving average with a window size of five. The minimum score over all bins with a local FDR less than 0.05 was used as local peptide length-dependent cut-off. The Andromeda score was then divided by the local peptide length-dependent cut-off to yield the LScore. PSMs were then sorted according to their LScore in decreasing order. If indicated, PSMs were additionally filtered to a given FDR cut-off of 0.01 (1%) based on the target decoy approach. This procedure truncates the sorted list of PSMs at the point where the fraction of decoy hits to total hits exceeds the cut-off (global PSM FDR cut-off).
Each PSM reporting a deamidated, methylated or carbamylated (for HeLa data set: deamidated, dehydrated, oxidized (Trp, Tyr) or reported with loss of ammonia), peptide was compared with the corresponding PSM of the same MS/MS scan in the standard search. PSMs with different reported sequences in the standard search and one of the searches with the selected modification (e.g. deamidation, methylation, carbamylation, etc.) were labeled as “conflicting.”
The cumulative empirical false positive rates were simulated based on the data transformations proposed by Elias and Gygi, 2007 for a concatenated search (23). Briefly, and as described by Elias and Gygi, estimated correct identifications were calculated by subtracting twice the number of decoy PSMs from total PSMs at a given Andromeda Score. Incorrect identifications were estimated by doubling the number of decoy PSMs returned at given Andromeda Score. The cumulative false positive rate was estimated by dividing estimated incorrect by estimated incorrect identifications larger than a given Andromeda Score threshold.
MSMS Extraction
The assigned “conflicting” spectrum reported as IESSIQSLQDLSK from a standard search was plotted and the following annotations as given in msms.txt were added: fragment-matches, score, mass, modifications and sequence for the two different search variants. Unassigned fragment peaks were extracted from apl-files. The spectrum identified as IESLSSQLSNLQK from the search with deamidation was plotted opposing the identification from the standard search. The MSMS spectra from synthesized spectra were extracted from Thermo XCalibur Qual Browser.
Comparison of Decoy Database Hits
Decoy database hits were extracted from the evidence.txt of the standard search. Decoy hits in the standard search identified as conflicting PSMs were compared with the remaining decoy database hits (other decoy hits). The LScore and the Intensities of the decoy hits were compared at no PSM FDR threshold and 0.01, respectively. The significance of the difference between both populations was evaluated by applying a two-sample Kolmogorov-Smirnov test as implemented in R (ks.test).
Protein Filtering
For the analysis at the protein identification level we first filtered the list of identified spectra as described above to a PSM FDR = 0.01 for the standard search and to no PSM FDR threshold for the searches with modification using LScores. Proteins identified by conflicting PSMs (PSMs with different reported sequences in standard search and in searches with additional variable modifications) and normal PSMs (with one reported sequence only) were assigned as “conflicting and other PSMs.” Proteins identified only by conflicting PSMs were assigned as “conflicting PSM(s) only.”
Where indicated, we additionally applied FDR filtering at the protein level. To this end, we extracted the best scoring protein for each PSM from the evidence Table in the evidence.txt file (column: “Leading razor protein”). We assigned to each of these best scoring proteins a protein PEP by multiplying the PEPs of the contained peptide sequences. Only the best (lowest) PEP of all spectra that were assigned to an individual peptide was considered (18). The list of identified proteins was then sorted according to the protein PEP (in increasing order) and a cut-off at the desired FDR was applied using the target decoy approach explained above. Note that the protein PEP was only used to rank the list of identified proteins and had no other statistical use beyond that.
Identifications assigned as “conflicting and other PSM(s)” and “conflicting PSMs only” were compared with the identifications in the protein FDR filtered list. To estimate protein abundance, we calculated protein iBAQ values from all evidences (standard search result) in the evidence.txt table and a cleaned version (conflicting PSMs removed). iBAQ values are calculated by dividing summed peak intensities by the total number of theoretically observable peptides, as previously described (24). Proteins assigned to a conflicting spectrum in the search with the selected variable modification were denoted “source” and proteins in the standard search “target” protein.
Protein and Peptide Abundances for GRID2 and LMNB1
Protein abundances of GRID2, LMNB1 and of GRID2-derived peptides were downloaded from the website www.humanproteomemap.org as RGB color codes (additive color model, using red, green and blue channels) on 01/22/2015 and reassembled into a heatmap using R.
Peptide Identifications as a Function of FDR Threshold
PSMs were extracted from evidence.txt and LScores were calculated as described above. Identifications were sorted according to their LScores for the standard search, the cleaned standard search (i.e. conflicting PSMs that were identified as described above removed) and the combined search. The number of identified spectra or non-redundant peptide sequences as a function of the PSM FDR (decoy hits/total hits) threshold was calculated from the resulting lists.
Where indicated, the confidence of the identified modified peptides was additionally assessed as a function of the PTM FDR. Therefore, modified PSMs were sorted according to their LScore and FDR filters were applied on this subgroup. The resulting PTM FDR filtered lists were compared with the identifications from the standard search (filtered at a PSM FDR = 0.01). The number of conflicting PSMs that was explained by PTM FDR filtered modified PSMs was plotted as a function of the applied PTM FDR.
RESULTS
Estimation of Peptide Modification Frequency
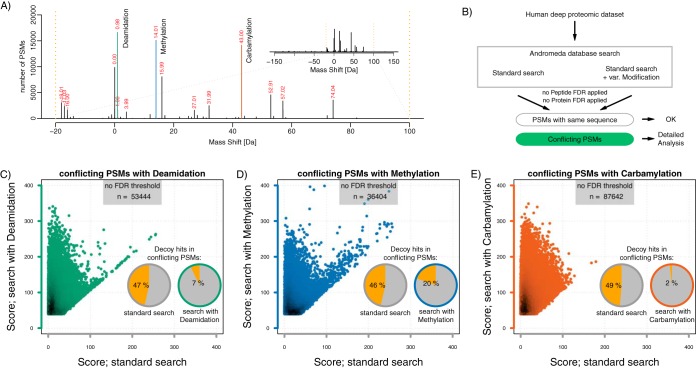
First, to obtain an overview of peptide modifications, we analyzed a deep proteomic data set from HEK293 cells (20). To systematically identify peptide modifications in this sample we used the ModifiComb algorithm (19) implemented in MaxQuant (18). Similar to other algorithms ModifiComb can detect modifications in an unbiased manner (15). First, a standard database search is performed to identify unmodified “base” peptides. Second, ModifiComb matches spectra that could not be matched in the first round to these base peptides. If they show consistent mass shifts at the MS and MS/MS level, they are identified as modified variants (“dependent peptides”). Applying this algorithm to our data set we found that deamidation (D) was most frequent, followed by methylation (M) and carbamylation (C) (see Fig. 1A). In total, this analysis identified ∼12% of unassigned MS2 spectra as derived from modified peptides. Thus, our analysis corroborates the previous finding that modified peptides are a significant source of false-negative identifications (17).
Fig. 1.
Modified peptides as a source of false spectra identification. A, Distribution of most frequent mass shifts of dependent peptides in the region from −20 to 100 Da. A view on the wider mass region is shown in the insert B, Raw data files were analyzed without (standard search) or with additional selected variable modifications: deamidation (Gln, Asn), methylation (Lys, Glu, Asp) or carbamylation (peptide N-term). All searches included methionine oxidation and protein N-terminal acetylation as variable modifications. C–E, Pairwise comparison of Andromeda scores from conflicting PSMs in a search with and without deamidation, methylation or carbamylation, as indicated. The fraction of decoy database matches among conflicting PSMs is given for either search variant in the pie charts.
Modifications Cause False-positive Peptide Identification
It has been shown for individual spectra that modified peptides can be misassigned to wrong amino acid sequences when the modification is not considered during the search (13, 14). However, the global impact of modified peptides on false positive identifications has not yet been investigated. We hypothesized that a standard search that does not take frequent modifications into account will misidentify some modified peptides as unmodified versions of other peptides. To assess this possibility we focused on the three most frequent modifications (D, M, C). We carried out four separate database searches: one standard search and three additional searches, each including D, M or C as an individual variable modification (see Fig. 1B), with the site specificities suggested by ModifiComb. We first wanted to select all spectra that could potentially be misidentified. To this end, we compared the entire set of peptide identifications between searches. Because score thresholds (at a given FDR threshold) can vary depending on whether or not modifications are considered, we first performed all searches without FDR filtering. All spectra that were (1) assigned to an unmodified peptide in the standard search and (2) reported to be modified in one of the other searches were then selected as conflicting peptide spectrum matches (PSMs). We next assessed the characteristics of these matches. We first had a look at the distribution of search scores. Conflicting PSMs had consistently higher scores in the search with the modification than in the standard search (see Fig. 1C–1E). This is expected because the search space for the search with the modification contains both unmodified and modified peptides. The only way for a spectrum to be assigned to a modified sequence is if that peptide scores better than any of the unmodified peptides.
We then asked which of the searches explained conflicting identifications better. Therefore, we had a look at the fraction of decoy database hits among conflicting identifications in either search variant. We used this as a proxy to estimate the total number false positives in these subsets. In the case of 50% decoy hits in a specific subset, all target and decoy matches are considered to be random (that is, false positive). Among conflicting identifications, we found almost 50% of decoy matches in the standard search, which indicates that conflicting identifications are almost entirely false positive hits in the standard search. In the searches with the respective modification the fraction of decoy database hits was on average six times lower, suggesting that most of these hits are correct. These data indicate that most of the conflicting identifications represent modified peptides that were misidentified in the standard search. As an additional control we also used an arbitrary non-existent mass-shift for the same kind of analysis (see supplemental Fig. S1). As expected, the fraction of decoy hits for conflicting PSMs was roughly 50% for both, the modified search with an arbitrary mass-shift and the unmodified search. We conclude that modified peptides are a specific source of false discoveries in deep proteomic data sets. Analysis of an independent deep proteomic data set looked overall similar (see supplemental Fig. S2A–S2E).
In a typical data analysis procedure for shotgun proteomics one only cares about spectra that survive a defined FDR cut-off. This is typically done with the target-decoy database search strategy (23, 25): The general idea is that searches are performed against a concatenated database which contains all candidate proteins (“target” part) plus control proteins with reversed, shuffled or randomized sequences (“decoy” part). Score cut-offs are then selected in order to adjust the fraction of false positive peptide spectrum matches (PSMs) to a user-defined value. We therefore investigated how many misidentifications by modified peptides survive a 0.01 FDR cut-off. Applying this cut-off to the results of the standard search markedly reduced the number of conflicting matches (from 166,073 to 8,552; see also supplemental Fig. S3). This result shows that FDR filtering via the target-decoy approach is an efficient means to reduce misidentifications by modified peptides.
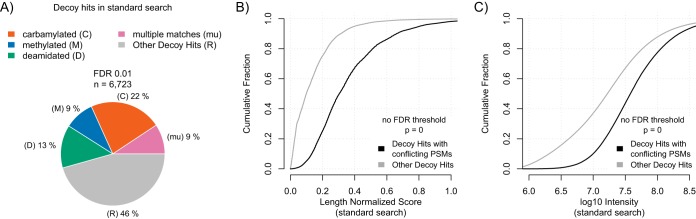
We next asked how many of the remaining false positive hits after FDR filtering could be attributed to modified peptides. To answer this question, we had a closer look at all decoy hits that survived the 0.01 FDR filtering at the PSM level. These decoy hits are a proxy for false positive hits in the target database (23). We observed that 54% of decoy matches that survived the 0.01 FDR cut-off in a standard search were conflicting identifications (see Fig. 2A). Thus, about 50% of remaining false positives at FDR = 0.01 appear to be because of modified peptides. In an independent data set this fraction was smaller but still substantial (∼20%, see supplemental Fig. S2F).
Fig. 2.
Characteristics of conflicting matches. A, Fraction of conflicting (deamidation: D, methylation: M, carbamylation: C) PSMs among all decoy database hits from a standard search at PSM FDR 0.01. Decoy database hits that were explained by conflicting peptides in more than one of the searches are denoted as “mu” (multiple matches). Cumulative LScore (B) and intensity (C) distribution of decoy database hits from a standard search at no PSM FDR threshold. The population of decoy database hits was split into hits that were caused by conflicting peptides and other hits. p values were derived from the Kolmogorov-Smirnov test.
It is recommended to estimate the confidence of modified peptide identifications with a so-called subgroup FDR (that is, FDR filtering applied only on the subgroup of modified PSMs) rather than a global FDR (that is, FDR filtering applied on modified and unmodified PSMs combined) (26, 27). In order to further assess the confidence of conflicting PSMs as modified peptides, we additionally filtered the list of conflicting PSMs by such subgroup FDRs (see supplemental Fig. S4A). The stringent subgroup FDR filtering (to 0.01) reduced the percentage of conflicting PSMs among false positives in the standard search from 54% to 44% (see supplemental Fig. S4B). Thus, the vast majority (81%) of the conflicting PSMs are confidently identified modified peptides. Because our analysis only considers the three most frequent modifications, the true number of false positive hits because of modified peptides is expected to be even higher.
False positive identifications can result from different sources. Therefore, we next asked if false positives caused by conflicting PSMs are systematically different from other false positive hits. We found that decoy hits caused by conflicting PSMs had significantly higher search scores than other decoy hits (see Fig. 2B). Moreover, peptides with conflicting PSMs had significantly higher intensities (see Fig. 2C). Therefore, these misassignments are expected to have a stronger impact on protein quantification than random hits (see also below). We conclude that conflicting PSMs are a particularly problematic source of false positive identifications.
Erroneous Protein Identification and Quantification by Modified Peptides
In shotgun proteomics, peptide level data is used to identify and quantify proteins. Therefore, we next asked how much misidentified peptides2 affect the protein level. Protein inference is not a trivial task and there is some controversy about how peptide level data should be translated to the protein level (25). With respect to identification, it is not clear if and how the target-decoy strategy should also be employed at the protein level. Most recent studies apply FDR filters at both the peptide and the protein level. However, this general practice has been challenged for deep proteomic studies (11, 28). Several recent papers therefore did not use protein level filtering (7, 8).
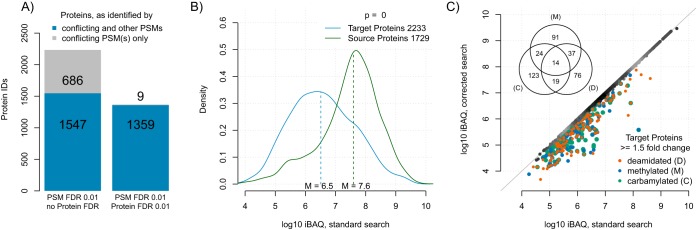
Without protein level filtering, 686 proteins were exclusively identified by misassigned modified peptides (see Fig. 3A). This corresponds to about 6% of all proteins. Thus, modified peptides are a considerable source of false protein identifications. Modified peptides were also misassigned to 1547 additional proteins, which were also represented by other peptides. Although in these cases the wrong peptides will not lead to false protein identifications, they can still affect protein quantification. As expected, applying a 0.01 protein FDR cut-off reduced the number of false positive protein identifications below 1% (see Fig. 3A). Hence, FDR filtering at the protein level is an efficient means to control false positive protein identifications caused by modified peptides.
Fig. 3.
Modified peptides as a source of false protein identification and quantification. A, Protein identifications because of misidentified modified peptides in a standard search. The number of false identifications is given at a PSM FDR cut-off at 0.01 with and without additional protein FDR cut-off at 0.01. B, Density plot of protein log10 iBAQ values as estimates of protein abundances. Source proteins of modified peptides and proteins that were misidentified by modified peptides (target proteins) are depicted. Dashed lines indicate the median of the respective distribution. C, Pairwise comparison of abundance estimates of target proteins in a standard search and a search where conflicting PSMs were removed (corrected). Target proteins that were overestimated by modified peptides ≥1.5-fold-change are highlighted as indicated. Decoy database hits were removed for all subfigures. A cut-off at PSM FDR 0.01 and no protein FDR-threshold was applied for subfigures b and c.
Next, we wanted to assess the impact of misassigned modified peptides on protein quantification. To this end, we investigated the abundance of proteins with conflicting PSMs (see Fig. 3B). We found that source proteins (that is, proteins which give rise to modified peptides) were on average more than 10 times more abundant than target proteins (that is, proteins with misassigned modified peptides). Thus, most modified peptides are derived from abundant proteins and are misassigned to proteins of lower abundance. Consequently, without filtering at protein level, the abundance of ∼380 target proteins (5%) was systematically overestimated in the standard search (see Fig. 3C). Again, FDR filtering at the protein level reduced the number of proteins overestimated in abundance to ∼220 (3%). We conclude that modified peptides give rise to systematic false protein identification and overestimation of protein abundance.
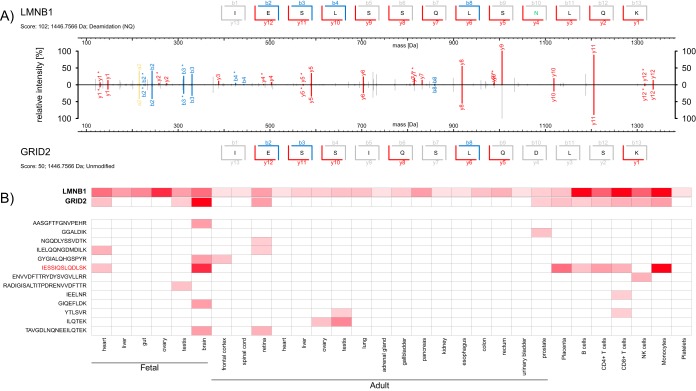
To illustrate the problem, we present the example of a deamidated peptide derived from the nuclear laminar protein LMNB1 (see Fig. 4A). The standard search misidentifies this spectrum as an isobaric peptide derived from the glutamate-receptor GRID2. GRID2 is selectively expressed in Purkinje cells (29). In contrast, the Human Proteome Map (8) (www.humanproteomemap.org) reports a more widespread expression pattern because of the same misidentified peptide (see Fig. 4B). Because of this misidentification the expression profile of GRID2 resembles that of LMNB1 - the source protein of the deamidated peptide. We validated these findings using synthetic peptides (see supplemental Fig. S5). This example shows that misidentified spectra derived from modified peptides already entered public data repositories and caused misleading information about protein expression profiles.
Fig. 4.
False identification of a LMNB1 peptide as GRID2-derived peptide results in a wrong protein expression profile. A, MS/MS spectrum derived from a deamidated LMNB1 peptide (upper panel, deamidated amino acid highlighted in green) misidentified as an unmodified isobaric GRID2 peptide (lower panel). Fragment ions with water or ammonia loss are marked with '*'. B, Expression pattern of LMNB1 and GRID2 proteins (top) and GRID2-derived peptides (bottom) according to www.humanproteomemap.org. The deamidated LMNB1 peptide that was erroneously identified as unmodified GRID2 peptide is highlighted in red.
Targeted Removal of Conflicting Peptide Spectral Matches
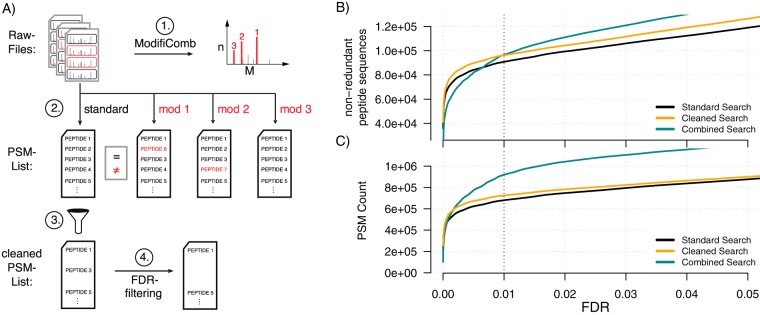
The data presented so far identifies modified peptides as a significant source of false-positive hits with considerable impact on protein identification and quantification. Therefore, we thought about possible strategies to solve this problem. The simplest idea would be to allow for different variable modifications in the standard database search. However, because of the combinatorial expansion of the search space, this procedure increases the score thresholds for significant peptide identifications and may thus decrease the total number of identifications at a given FDR (15). Based on our results we propose an alternative strategy, which involves four steps (see Fig. 5A). First, an unbiased algorithm such as ModifiComb is employed to identify the most prevalent modifications in a specific sample. Second, a standard database search (with relaxed FDR cut-offs) and several parallel searches with individual variable modifications are performed. Each of these additional searches only considers an individual variable modification to limit search space expansion. Third, spectra with conflicting identifications and higher scores in the modified form are not replaced by the results from the secondary searches but instead removed from the standard search. Finally, this cleaned data set is used to adjust the protein and/or peptide FDR to the desired level based on the number of decoy hits.
Fig. 5.
Targeted removal of conflicting spectra. A, Database search strategy to identify and discard false identifications derived from modified peptides. Standard search and searches with modification were performed without protein and PSM FDR threshold. Spectra that gave rise to conflicting sequence information were removed (cleaned search). The results were compared with the uncorrected standard search and a search where the variable modifications D+M+C were considered in a single pass (combined). The number of identified spectra (B) and non-redundant peptide sequences (C) is plotted as a function of the PSM FDR.
To assess the performance of this “cleaned search” approach, we counted the number of identified spectra and unique peptide sequences that could be assigned at a given FDR and compared it to the results of the standard search (that is, without cleaning). We then compared the number of non-redundant peptide sequences (Fig. 5B) and PSM counts (Fig. 5C) that could be assigned as a function of the FDR threshold. Removing conflicting matches improved the performance relative to the standard search at all depicted FDR thresholds (compare yellow and black curves in Fig. 5B and 5C). At an FDR of 0.01, 44,358 additional spectra and 5447 additional unique peptide sequences could be identified in the cleaned data set. This corresponds to a ∼6% increase in the total number of assigned peptide sequences. The increase in coverage is remarkable, especially because we only removed conflicting PSMs from the standard search and did not add new identifications. The improved performance is thus solely because of the targeted removal of dubious hits. Conversely, if we keep the number of assigned spectra constant (673,237 at 0.01 FDR in the standard search), we reduce the effective FDR to 0.54%. In either case, the targeted removal of mismatches derived from modified peptides substantially improves the quality of data annotation. Again, analysis of an independent data set yielded overall similar findings (see supplemental Fig. S2G). To further assess the validity of our “cleaned search” approach, we estimated false positive rates (FPR) based on the data transformations proposed by Elias and Gygi, 2007 (23) (see supplemental Fig. S6A, S6B). We calculated the FPR for the cleaned standard search and the standard search. We found that both FPRs were overall in accordance with each other (see supplemental Fig. S6C). Thus, the targeted removal of conflicting spectra does not influence our FPR estimates.
For comparison, we also used a “combined search” that allows for all additional variable modifications (D + M + C) in a single search. At the level of identified spectra, this strategy outperformed the other approaches at most FDR cut-offs (green curve, Fig. 5C). This is expected because a lot of spectra derived from modified peptides that are false negatives in the other searches can now be assigned. However, the increase in identified PSMs does not directly translate to an increase in coverage at the level of non-redundant peptide sequences (green curve, Fig. 5B): At high stringency cut-offs the combined search identified less peptides than the other methods. At more and more relaxed FDR cut-offs the combined search first outperformed the standard search and later also the cleaned search approach. Thus, the relative performance of the combined search depends on the FDR threshold. There is also a practical limitation of the combined search method: Searching for many variable modifications in parallel greatly increases database search time. In our case, the theoretical search space increase was 236-fold, and we empirically observed a 30-fold increase in search time using MaxQuant.
DISCUSSION
Shotgun proteomics is stepping up to explain the diversity and complexity of the entire human proteome (1); a development that comes with more and more extensive data sets. As for all high-throughput technologies, the shotgun approach is associated with specific sources of errors and systematic biases. Here, we identified modified peptides as a systematic source of false positive peptide and protein identification. First, we show that thousands of peptides and hundreds of proteins are misidentified because of modifications that are not considered in a standard database search. We show that false positives caused by modified peptides are more abundant and have higher scores than other false-positives, which make them particularly problematic. Moreover, we demonstrate that these false positives affect protein quantification and lead to wrong protein expression profiles. Finally, we outline a database search strategy that alleviates this problem and considerably improves the quality of data annotation.
Identifying and minimizing systematic errors is essential for comprehensive and high quality proteomics. Known sources of false positive identifications include poor quality spectra (12), errors in primary data processing (13) and incomplete database search space (13, 14). How much these factors contribute to false positive identifications is unclear. In our data set, about half of false positives were because of deamidated, carbamylated, and methylated peptides. It is known that the prevalence of modifications in a proteomic sample is affected by the sample preparation protocol (16). Consistently, in another data set, different modifications predominated and the fraction of false-positives because of the top four modifications was lower (∼18%, see supplemental Fig. S2F). Because we only considered the most abundant modifications, the impact of all modifications on false positive identifications is likely higher in both cases. Thus, modifications can account for a large fraction of false positive identifications. This alone strongly emphasizes the need for a solution to this problem. Moreover, we observed that misidentified modified peptides are significantly more abundant and have higher search scores than other false positives. Thus, they have a bigger impact on the data than random matches. For example, they cause systematic overestimation of protein abundance and lead to wrong protein expression profiles.
False positive protein identification by modified peptides was almost completely eliminated by stringent protein FDR filtering (see Fig. 3A). As there is no current consensus on if and how protein FDR filtering should be applied to deep proteomic data sets (11, 28, 30), some researchers chose not to filter at the protein level (7, 8) whereas others filtered their data sets (5, 6, 17). Our results suggest that protein level filtering is important to limit false-positive identifications.
Although stringent FDR filtering at the peptide and protein level can limit the number of false positive identifications, it is not the best strategy to deal with the problem of modified peptides. Ultimately, data quality depends on the quality of the PSMs generated by the search engine: A high fraction of misassigned spectra leaves less space for correct identifications after the filtering step. It is therefore desirable to remove false identifications by modified peptides in the first place, before any FDR filtering. This strategy is in line with MS/MS data reduction methods as i.e. removal of unidentifiable spectra (31), removal of background spectra (32), removal of PSMs with high mass error (33), removal of spectra with ambiguous charge state (34) or removal of spectra in the context of iterative searches (35). We demonstrated that the statistical power of the analysis is substantially improved when systematic sources of false positives are considered (see Fig. 5B, 5C, supplemental Fig. S2G).
It is generally recommended to limit the number of variable modifications in a database search as much as possible (15). The reason for this is that allowing for multiple modifications dramatically increases the search space and therefore the chance for random hits. In fact, using a small custom database specific for the sample of interest increases the statistical power and gives superior results (36, 37). It has even been argued recently that mass spectrometrists should only search for peptides they care about and neglect other peptides, even though they are present in the sample (38). In contrast to this suggestion, we show that it is important to make sure that the search space is comprehensive. Specifically, it is essential that most experimentally observed peptide fragmentation spectra have corresponding matches in the search space and can thus be correctly assigned. Therefore, even though a researcher may not be interested in a specific modification, it is important to consider it during the search if it is abundant in the sample. Otherwise, some of the corresponding spectra will be misassigned and introduce significant biases.
We outline a simple strategy how modifications can be taken into account without increasing the search space (Fig. 5A). The general idea is to perform parallel searches on the same raw data: one standard search and several individual searches with specific variable modifications. The spectra of identified modified peptides are then removed from the standard search. This “cleaned search” approach does not increase the search space in the standard search. Importantly, we find that this workflow considerably improves the quality of data annotation. We would also like to point out that this approach cannot only be used to reduce problems caused by modified peptides. For example, the same strategy could be used to eliminate false positive hits caused by abnormally cleaved peptides (13), peptides which undergo in-source fragmentation (17) or peptides derived from “contaminating” proteins that are not of interest in a specific experimental setting.
An obvious disadvantage of our approach is that it discards modified peptides and therefore neglects potentially useful information. The information loss can be considerable because at least one third of unassigned spectra are estimated to be because of modified peptides (17). It could be an option to replace the dubious identifications in the standard search with modified peptides identified in the modified search. However, with respect to FDR control this would yield a complicated situation: The search space of the modified search is larger. Hits in the modified search therefore have different significance levels than hits in the standard search at identical scores. Furthermore, identifying modified peptides and basing quantification on them is not always useful for two reasons. First, most modified peptides are present in sub-stoichiometric amounts relative to their unmodified variants (16). This means that identifying them does not lead to an increase in independent sequence information, because they often represent variants of already identified unmodified peptides. Second, modified peptides are not well suited for protein quantification because their abundance can be affected by biological and/or chemical factors. Especially in methods like iTRAQ/TMT, where peptides are labeled after digestion (39), it can readily happen that modifications are introduced differently during sample handling. The advantage of our cleaned search approach is that it increases the coverage of unmodified peptides which are more reliable for protein quantification.
The fact that some spectra derived from modified peptides can be mistakenly assigned to unmodified sequences could also be integrated into some recently published search strategies (such as the cascaded (40) or ISPTM (35) search approach): Instead of accepting significant unmodified spectra after the first search round one could keep a selected set or all of them in the pool for the following searches. Ambiguous identifications could then be compared between the different searches to decide whether the identification(s) should be discarded or accepted. Another alternative strategy would be to modify the ModifiComb (19) approach: Instead of comparing significantly identified peptides to those below the threshold, one could compare the significant peptides to all peptides and devise a scoring scheme to decide when a significantly identified peptide is more likely to be a modified version of another peptide. Yet another option could be not to allow different modifications on the same peptide during the database search. This would prevent the combinatorial expansion of the search space and could be implemented in search engines. In any case, our results indicate that it is important to systematically consider modified peptides in future proteomic studies.
Supplementary Material
Acknowledgments
We thank Erik McShane, Daniel Perez-Hernandez, Russ Hodge, and three unknown reviewers for valuable comments on this manuscript.
Footnotes
Author contributions: M.S. designed research; B.B. and H.Z. performed research; B.B. and H.Z. analyzed data; B.B., H.Z., and M.S. wrote the paper.
* B.B. is funded by the ViroSign project of the Federal Ministry of Education and Research (BMBF).
 This article contains supplemental material.
This article contains supplemental material.
2 We use the term “misidentified” because most conflicting PSMs appear to be false positive hits in the standard search. Note that this is a simplifying assumption because not all peptides with conflicting PSMs are necessarily misidentified in the standard search.
1 The abbreviations used are:
- PSM(s)
- peptide spectrum match(es)
- A
- ammonia loss
- C
- carbamylation
- D
- deamidation
- FDR
- false discovery rate
- H
- dehydration
- HEK293
- human embryonic kidney 293 cells
- iBAQ
- intensity based absolute quantification
- M
- methylation
- O
- oxidation
- PEP
- posterior error probability
- RGB
- red, green and blue color model.
REFERENCES
- 1. Nilsson T., Mann M., Aebersold R., Yates J. R. 3rd, Bairoch A., and Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 2. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 3. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 4. Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 5. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., and Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geiger T., Wehner A., Schaab C., Cox J., and Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111 014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., Mollenhauer M., Slotta-Huspenina J., Boese J. H., Bantscheff M., Gerstmair A., Faerber F., and Kuster B. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
- 8. Kim M. S., Pinto S. M., Getnet D., Nirujogi R. S., Manda S. S., Chaerkady R., Madugundu A. K., Kelkar D. S., Isserlin R., Jain S., Thomas J. K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N. A., Balakrishnan L., Advani J., George B., Renuse S., Selvan L. D., Patil A. H., Nanjappa V., Radhakrishnan A., Prasad S., Subbannayya T., Raju R., Kumar M., Sreenivasamurthy S. K., Marimuthu A., Sathe G. J., Chavan S., Datta K. K., Subbannayya Y., Sahu A., Yelamanchi S. D., Jayaram S., Rajagopalan P., Sharma J., Murthy K. R., Syed N., Goel R., Khan A. A., Ahmad S., Dey G., Mudgal K., Chatterjee A., Huang T. C., Zhong J., Wu X., Shaw P. G., Freed D., Zahari M. S., Mukherjee K. K., Shankar S., Mahadevan A., Lam H., Mitchell C. J., Shankar S. K., Satishchandra P., Schroeder J. T., Sirdeshmukh R., Maitra A., Leach S. D., Drake C. G., Halushka M. K., Prasad T. S., Hruban R. H., Kerr C. L., Bader G. D., Iacobuzio-Donahue C. A., Gowda H., and Pandey A. (2014) A draft map of the human proteome. Nature 509, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ezkurdia I., Vazquez J., Valencia A., and Tress M. (2014) Analyzing the first drafts of the human proteome. J Proteome Res 13, 3854–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serang O., and Kall L. (2015) Solution to Statistical Challenges in Proteomics Is More Statistics, Not Less. J. Proteome Res. 14, 4099–4103 [DOI] [PubMed] [Google Scholar]
- 11. Savitski M. M., Wilhelm M., Hahne H., Kuster B., and Bantscheff M. (2015) A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Mol. Cell. Proteomics 14, 2394–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Resing K. A., Meyer-Arendt K., Mendoza A. M., Aveline-Wolf L. D., Jonscher K. R., Pierce K. G., Old W. M., Cheung H. T., Russell S., Wattawa J. L., Goehle G. R., Knight R. D., and Ahn N. G. (2004) Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal. Chem. 76, 3556–3568 [DOI] [PubMed] [Google Scholar]
- 13. Chen Y., Zhang J., Xing G., and Zhao Y. (2009) Mascot-derived false positive peptide identifications revealed by manual analysis of tandem mass spectra. J. Proteome Res. 8, 3141–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stevens S. M. Jr., Prokai-Tatrai K., and Prokai L. (2008) Factors that contribute to the misidentification of tyrosine nitration by shotgun proteomics. Mol. Cell. Proteomics 7, 2442–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahrne E., Muller M., and Lisacek F. (2010) Unrestricted identification of modified proteins using MS/MS. Proteomics 10, 671–686 [DOI] [PubMed] [Google Scholar]
- 16. Nielsen M. L., Savitski M. M., and Zubarev R. A. (2006) Extent of modifications in human proteome samples and their effect on dynamic range of analysis in shotgun proteomics. Mol. Cell. Proteomics 5, 2384–2391 [DOI] [PubMed] [Google Scholar]
- 17. Chick J. M., Kolippakkam D., Nusinow D. P., Zhai B., Rad R., Huttlin E. L., and Gygi S. P. (2015) A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat. Biotechnol. 33, 743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 19. Savitski M. M., Nielsen M. L., and Zubarev R. A. (2006) ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics 5, 935–948 [DOI] [PubMed] [Google Scholar]
- 20. Eravci M., Sommer C., and Selbach M. (2014) IPG strip-based peptide fractionation for shotgun proteomics. Methods Mol. Biol. 1156, 67–77 [DOI] [PubMed] [Google Scholar]
- 21. Wessel D., and Flugge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 22. Kelstrup C. D., Young C., Lavallee R., Nielsen M. L., and Olsen J. V. (2012) Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J. Proteome Res. 11, 3487–3497 [DOI] [PubMed] [Google Scholar]
- 23. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 24. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 25. Nesvizhskii A. I. (2010) A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics 73, 2092–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chalkley R. J. (2013) When target-decoy false discovery rate estimations are inaccurate and how to spot instances. J. Proteome Res. 12, 1062–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fu Y. (2012) Bayesian false discovery rates for post-translational modification proteomics. Stat. Interface 5, 47–59 [Google Scholar]
- 28. Reiter L., Claassen M., Schrimpf S. P., Jovanovic M., Schmidt A., Buhmann J. M., Hengartner M. O., and Aebersold R. (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics 8, 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Araki K., Meguro H., Kushiya E., Takayama C., Inoue Y., and Mishina M. (1993) Selective expression of the glutamate receptor channel delta 2 subunit in cerebellar Purkinje cells. Biochem. Biophys. Res. Commun. 197, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 30. Gupta N., Bandeira N., Keich U., and Pevzner P. A. (2011) Target-decoy approach and false discovery rate: when things may go wrong. J. Am. Soc. Mass. Spectrom. 22, 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flikka K., Martens L., Vandekerckhove J., Gevaert K., and Eidhammer I. (2006) Improving the reliability and throughput of mass spectrometry-based proteomics by spectrum quality filtering. Proteomics 6, 2086–2094 [DOI] [PubMed] [Google Scholar]
- 32. Junqueira M., Spirin V., Santana Balbuena T., Waridel P., Surendranath V., Kryukov G., Adzhubei I., Thomas H., Sunyaev S., and Shevchenko A. (2008) Separating the wheat from the chaff: unbiased filtering of background tandem mass spectra improves protein identification. J. Proteome Res. 7, 3382–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hsieh E. J., Hoopmann M. R., MacLean B., and MacCoss M. J. (2010) Comparison of database search strategies for high precursor mass accuracy MS/MS data. J. Proteome Res. 9, 1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadygov R. G., Eng J., Durr E., Saraf A., McDonald H., MacCoss M. J., and Yates J. R. 3rd (2002) Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1, 211–215 [DOI] [PubMed] [Google Scholar]
- 35. Huang X., Huang L., Peng H., Guru A., Xue W., Hong S. Y., Liu M., Sharma S., Fu K., Caprez A. P., Swanson D. R., Zhang Z., and Ding S. J. (2013) ISPTM: an iterative search algorithm for systematic identification of post-translational modifications from complex proteome mixtures. J. Proteome Res. 12, 3831–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X., Slebos R. J., Wang D., Halvey P. J., Tabb D. L., Liebler D. C., and Zhang B. (2012) Protein identification using customized protein sequence databases derived from RNA-Seq data. J. Proteome Res. 11, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappe J., Gevaert K., and Van Damme P. (2013) Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12, 1780–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noble W. S. (2015) Mass spectrometrists should search only for peptides they care about. Nat. Methods 12, 605–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson A., Schafer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., and Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 40. Kertesz-Farkas A., Keich U., and Noble W. S. (2015) Tandem Mass Spectrum Identification via Cascaded Search. J. Proteome Res. 14, 3027–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.