Abstract
Quantitative mass spectrometry data necessitates an analytical pipeline that captures the accuracy and comprehensiveness of the experiments. Currently, data analysis is often coupled to specific software packages, which restricts the analysis to a given workflow and precludes a more thorough characterization of the data by other complementary tools. To address this, we have developed PyQuant, a cross-platform mass spectrometry data quantification application that is compatible with existing frameworks and can be used as a stand-alone quantification tool. PyQuant supports most types of quantitative mass spectrometry data including SILAC, NeuCode, 15N, 13C, or 18O and chemical methods such as iTRAQ or TMT and provides the option of adding custom labeling strategies. In addition, PyQuant can perform specialized analyses such as quantifying isotopically labeled samples where the label has been metabolized into other amino acids and targeted quantification of selected ions independent of spectral assignment. PyQuant is capable of quantifying search results from popular proteomic frameworks such as MaxQuant, Proteome Discoverer, and the Trans-Proteomic Pipeline in addition to several standalone search engines. We have found that PyQuant routinely quantifies a greater proportion of spectral assignments, with increases ranging from 25–45% in this study. Finally, PyQuant is capable of complementing spectral assignments between replicates to quantify ions missed because of lack of MS/MS fragmentation or that were omitted because of issues such as spectra quality or false discovery rates. This results in an increase of biologically useful data available for interpretation. In summary, PyQuant is a flexible mass spectrometry data quantification platform that is capable of interfacing with a variety of existing formats and is highly customizable, which permits easy configuration for custom analysis.
Technological advances in quantitative MS-based proteomics now permit measurement of the abundance of tens of thousands of molecules in complex biological systems in a global fashion. MS-based quantitative analysis is traditionally achieved through two methodologies: label-free or those involving labeling of samples for multiplexed analysis. Label-free quantification uses either the number of spectra or the precursor ion intensity of the detected analytes to provide abundance estimates (1, 2). Label-based quantification is based on introducing known mass differences between samples and then comparing the relative abundance of the ions of interest that differ by the known masses. The mass difference has traditionally been introduced into peptides through chemical labeling of samples with various stable isotope-containing “tags” or by metabolically incorporating isotopically labeled amino acids. Two popular chemical modification strategies are isobaric tag for relative and absolute quantification (iTRAQ)1 and tandem mass tags (TMT), which rely on fragmentation of the reporter tag for quantification (3, 4). For in vivo incorporation of isotopic labels, stable isotope labeling by amino acids in cell culture (SILAC) is commonly used in which isotopically labeled amino acids, usually arginine and lysine, are incorporated into proteins as they are synthesized (5).
The utility of quantitative MS data is highly dependent on the accuracy and comprehensiveness of the tools used for analysis. The first step in quantitative proteomic data analysis is to associate spectral information of MS and MS/MS scans with peptide sequences, followed by quantification of the identified peptides. This provides two avenues to increase the rate of quantification: an increase in spectral assignments and/or an increase in the fraction of scans that are quantified. To increase the number of spectral assignments, common approaches include the integration of multiple database search engines, iterative searches, wide-tolerance searches, and custom database searches (6–13). The major benefit of these approaches is an increase in the number of spectra assigned to peptides. Although increasing the number of spectral assignments allows for a greater number of quantitative measurements, it does not address a more fundamental issue that programs used for quantitation only quantify a subset of the available data. Analogous to database search engines, each program used for quantitation has its own method for quantifying data. Thus, data quantified by one program may be missed by another and there may be inherent limitations that are common to several programs.
To address these issues, we have developed PyQuant as a new framework for quantitative analysis of MS data. PyQuant is a versatile, cross-platform quantitation tool that can be used in conjunction with existing data analysis frameworks or as a quantification node for a minimal, light-weight mass spectrometry data analysis pipeline. PyQuant accepts a variety of input formats, including the mzML and pepXML formats, several commonly used proteomic search engines and allows for a generic tab delimited input for search engines that are not natively supported. Additionally, it is compatible with widely used quantitative MS methods such as metabolic labeling (e.g. SILAC, NeuCode, 15N, and 18O) and chemical tagging (e.g. iTRAQ and TMT) and allows users to define custom labeling and data quantitation strategies. PyQuant can serve as a simple, stand-alone quantitation program following peptide assignment of many search algorithms such as Comet or X!Tandem (14, 15). This allows the user to have a minimal, easily deployed pipeline for mass spectrometry data analysis. Also, PyQuant can serve as a post-processor of data analyzed with existing frameworks such as MaxQuant (16), Proteome Discoverer, or the Trans-Proteomic Pipeline (TPP) (17), which allows for an additional, independent algorithm to verify existing quantification values as well as quantify data excluded by other algorithms.
MATERIALS AND METHODS
Software Development and the Availability of PyQuant
PyQuant is a command line driven program developed using the Python programming language, and is compatible with both Python 2.7+ and Python 3.5+. For GUI-based access, PyQuant can be deployed using the Wooey framework [https://github.com/wooey/Wooey/]. The web based interface uses jQuery, DataTables, pako, and c3 for visualization. Installation instructions, source code, and guides for PyQuant are available at https://pandeylab.github.io/pyquant/ and it is compatible with most major operating systems.
Although PyQuant is capable of quantifying at any MSn levels and is highly customizable, for simplicity we describe PyQuant's algorithm by stepping through two common quantification scenarios: a traditional proteomics data analysis pipeline, in which the XIC of a precursor ion is quantified, and an experiment utilizing chemical tags such as iTRAQ, where the MS2 scan is used to identify and provide quantification of a peptide. However, PyQuant can be configured to quantify at any MS level, which allows it to be used with emerging mass spectrometry strategies such as MS3-based quantification. Finally, we adopt the term analyte to comprise the physical entity of interest, such as a peptide and we use the term ion species to refer to a distinct m/z value. The analyte may be comprised of one or more ion species, such as the monoisotopic peak as well as peaks resulting from the inclusion of naturally occurring isotopes.
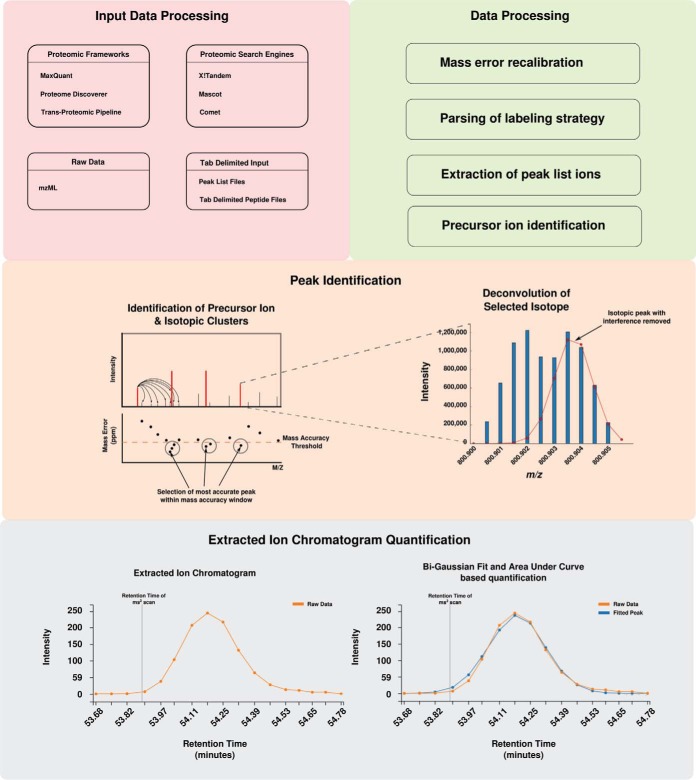
The steps of PyQuant can be broken down into several distinct steps: input data processing, peak picking, and quantification of the extracted ion chromatogram (XIC) (Fig. 1). Because of PyQuant's flexibility, there are many ways peaks can be assigned and selected. Thus, we cover first how PyQuant processes different types of data, and end with a step common to all algorithms, quantification of the XIC.
Fig. 1.
Overview of PyQuant's data processing algorithm.
Input Data Processing
PyQuant accepts a variety of data as input. For data that has been processed with other frameworks, PyQuant currently supports the pepXML files created by the TPP, msf files produced by Proteome Discoverer, and evidence and ms_ms text files produced by MaxQuant. For search engines, PyQuant can parse the output of X!Tandem (15), search engines that produce pepXML files such as Comet (14) and Mascot (18) if the ms_parser library is installed. To provide compatibility with an assortment of programs, PyQuant can also be supplied with a generic tab delimited file providing information on spectral assignments. Lastly, PyQuant is able to operate solely on raw mass spectrometry data in the mzML format.
Processing of Raw Data
In the absence of any provided annotation, PyQuant will quantify all MS1 scans with a corresponding MS2 scan. This can be used to perform simple analysis of raw data, such as identifying intense peaks that may represent sample contaminants, unspecified modifications, or interesting biological findings such as novel or nontryptic peptides that were missing from a database search. PyQuant can also be provided with target ions to search for in raw data, which can be used to identify missing values between replicates or perform targeted searches for post-translational modifications through the use of signature ions.
Processing of Mixed Resolution Data
It is sometimes useful to acquire scans at different resolution levels, such as in NeuCode labeling. To assist with this analysis, PyQuant allows a user to define a minimal resolution power of a scan to be considered for quantification. This allows the user to quantify scans using this time saving data acquisition strategy, which is becoming more popular as the resolving power of mass spectrometers increases.
Processing of Peptide Spectral Assignments
After being provided with the output of a given search engine or platform, PyQuant begins by parsing result files for peptide spectral assignments (PSMs). Following this, PyQuant checks whether a labeling strategy has been defined. For known data formats that define the labeling strategies, such Proteome Discoverer's msf file or X!Tandem's XML output, any user defined labeling strategy is automatically parsed and applied. However, for cases where this information is not embedded in the file, or if a vendor format changes, the labeling strategy can be supplied as a simple tab delimited file. Next, because the mass error changes as a function of the m/z of an ion, the error between the theoretical peptide's mass and the observed mass is plotted and fit to a spline function (supplemental Fig. S1). This spline is used to offset the machine drift with higher m/z values, which would otherwise make it difficult to identify values at a higher m/z within a given error tolerance. PyQuant then identifies the precursor ions of PSMs, and if a labeling strategy has been defined, PyQuant searches for corresponding labeled peaks in the quantification scans irrespective of whether these peaks have been fragmented. This helps to identify and quantify peaks that were not selected for fragmentation.
At this point, PyQuant has identified precursor ions and any applicable labeled peaks from the PSMs. If the user chooses to quantify scans at the MS1 level, PyQuant identifies the isotopic cluster of each precursor ion. However, because each isotopic cluster may be comprised of a mixture of overlapping ion species, the profile for each ion is fitted to a Gaussian mixture model to remove any interfering ions (supplemental Fig. S2). Following this, the peaks for each isotopic cluster are integrated to provide an abundance measure for each ion species detected. Finally, this process is repeated for neighboring scans until all ion species comprising an analyte can no longer be identified in two subsequent scans.
Quantifying the extracted ion chromatogram
To measure the abundance of a given analyte, the extracted ion chromatogram (XIC) is generated by extracting intensities from each isotopic peak of an ion species and plotting them against their respective retention times. For each XIC, multiple Bi-Gaussian peaks are fit to the data to accurately model the ion species of interest as well as any contaminating ions. A Bi-Gaussian peak was chosen because it better models the skewness of elution profiles as compared with a Gaussian distribution (supplemental Fig. S3). To avoid over-fitting the data with multiple Bi-Gaussian distributions, the number of Bi-Gaussian peaks modeling the data is determined using the Bayesian information criterion (BIC) (19), which guards against over-fitting by penalizing additional parameters. Following this, the mean and standard deviations of each peak is compared with the retention time that the initial scan was identified at, and peaks not containing the retention time are excluded. This routine is performed for all isotopic clusters and labeled pairs found for a given ion species. Next, because the elution profile for isotopic clusters and their isotopically labeled pairs should be similar, the peak shape of all isotopic clusters are compared and outlier peaks are identified and excluded by the minimum covariance determinant method (supplemental Fig. S4A–S4C) (20). Lastly to measure the abundance for each ion, the area under the XIC is integrated.
Confidence Estimates of Quantification
PyQuant uses several approaches to quantify data. However, this can result in the inclusion of noisy, unreliable data. To guard against this, PyQuant employs several methods to provide an estimate of the accuracy of quantification. PyQuant uses a machine learning algorithm trained with a manually curated data set of “good” and “poor” fits to provide a confidence measurement of the quality of fits. This takes into account variables such as the signal to noise ratio, the intensity of an analyte, the width of an analyte's retention time, and the density of the fitted peak to assess how accurate a given measurement is. Within the data set of known ratios, selecting only high confidence fits results in a tighter distribution of quantified peptides (supplemental Fig. S5). Lastly, because there is no substitute for manual validation, PyQuant provides an interactive HTML based output that allows a user to filter values, assign cutoffs, and manually inspect every isotope selected and the XIC.
Data Output and Visualization
PyQuant provides two files for the user. One is a tab delimited file that can be easily viewed in excel or processed further in a given data analysis pipeline. The other is a HTML file that offers an interactive browser-based exploration of the data. PyQuant uses several open source JavaScript libraries to provide an intuitive user interface with advanced data table manipulation. Additionally, PyQuant provides detailed graphics depicting which isotopes were selected in each scan, the XIC with corresponding peak fits, and integration values for every ion species. This allows the user to make an informed choice on whether an ion deemed significant by their chosen criteria, such as a fold change between samples, is truly significant or merely an algorithmic error.
Preparation of Peptide Samples for Standard MS Data Set
A lysine and arginine auxotrophic E. coli strain, SLE1, was purchased from the Caenorhabditis Genetics Center (CGC) and grown in M9 Minimal Media (Cold Spring Harbor Protocols). Each culture was grown with a mixture of SILAC isotopes at the desired mixing ratio, lysed, and fractionated via SDS-PAGE. Gel pieces were excised and destained with 40 mm ammonium bicarbonate and 40% acetonitrile (destain buffer) at room temperature. Following destaining, samples were reduced by incubation with 5 mm dithiothreitol (DTT) for 10 min at 60 °C. After reduction, samples were alkylated by 10 mm iodoacetoamide for 20 min. Following reduction and alkylation, samples were washed with destain buffer and incubated with 100% acetonitrile on ice until dehydrated. After dehydration, samples were resuspended in 10 μg/ml trypsin until the samples were rehydrated. After this, excess trypsin was removed and the sample was digested overnight. Elution of digested peptides was performed by adding 80% acetonitrile with 0.1% trifluoroacetic acid (TFA) and incubated at 25 °C on a shaker for 20 min. After 20 min, the supernatant was removed and the eluted peptides were lyophilized and stored at −20 °C until LC-MS/MS analysis.
For labeling of C. elegans, E. coli strain SLE1 was purchased from the CGC and labeled with light and heavy amino acids. Light cultures were grown in LB and heavy cultures were grown in M9 Minimal Media (Cold Spring Harbor Protocols) with the isotopically labeled amino acids Arginine-10 and Lysine-8. Each culture was grown overnight, and centrifuged at 8000 × g for 10 min at 4 °C. For each 250 ml of culture pelleted, 50 ml of S. Media (21) was added and the pellet was resuspended. For culturing of C. elegans, 50 ml of resuspended E. coli was added to a 1 L flask with C. elegans and 10 μg/ml of Nystatin to prevent fungal growth. The culture was monitored and when the bacteria was almost clear (∼3 days) the C. Elegans were isolated in accordance with previously described methods (21). C. elegans were resuspended in 9 m urea containing a protease inhibitor mixture (Roche #10711400 Branford, CT), 50 mm beta-galactosidase, 25 mm sodium fluoride, and 1 mm sodium orthovandate. Next, the resuspension was tip sonicated and viewed under a microscope following sonication until the C. elegans were sufficiently broken apart. Following this, the lysate was cleared by centrifugation at 14,000 × g for 20 min at 4 °C and protein concentration was measured via BCA. 250 μg from light and heavy C. elegans were combined for a total of 500 μg. The lysate was then alkylated with 10 mm of iodoacetamide and reduced with 5 mm of dithiothreitol. Following this, the lysate was diluted to a final urea concentration of 3 m and digested with Lys-c for 4 h at room temperature. After digestion with Lys-c, the lysate was diluted again to a final urea concentration of 1.5 m and digested overnight with trypsin. After confirming digestion efficiency, the sample was acidified with 1% TFA and centrifuged at 2000 × g for 5 min at room temperature. The supernatant was then loaded onto a Sep-Pak cleanup C18 column (Waters, Cat#WAT051910 Milford, MA) equilibrated with 0.1% TFA. Columns were washed with 12 ml of 0.1% TFA and peptides were eluted with 6 ml of 40% acetonitrile with 0.1% TFA. Eluted peptides were then lyophilized and subjected to basic reverse phase liquid chromatography and then mass spectrometry analysis.
LC-MS/MS
Peptide samples were analyzed on an LTQ-Orbitrap Elite mass spectrometer (Thermo Electron, Bremen, Germany) interfaced with Easy-nLC II nanoflow liquid chromatography systems (Thermo Scientific, Odense, Southern Denmark). The peptide digests from each fraction were reconstituted in Solvent A (0.1% formic acid) and loaded onto a trap column (75 μm × 2 cm) packed in-house with Magic C18 AQ (Michrom Bioresources, Inc., Auburn, CA) (5 μm particle size, pore size 100 Å) at a flow rate of 5 μl/min with solvent A (0.1% formic acid in water). Peptides were resolved on an analytical column (75 μm × 20 cm) at a flow rate of 350 nl min-1 using a linear gradient of 7–30% solvent B (0.1% formic acid in 95% acetonitrile) over 60 min. Mass spectrometry analysis was carried out in a data dependent manner with full scans (350–1,800 m/z) acquired using an Orbitrap mass analyzer at a mass resolution of 120,000 in Elite at 400 m/z. The twenty most intense precursor ions from a survey scan were selected for MS/MS from each duty cycle and detected at a mass resolution of 15,000 at a m/z of 400 in the Orbitrap analyzer. All the tandem mass spectra were produced by higher-energy collision dissociation (HCD) method. Dynamic exclusion was set for 30 s with a 10 p.p.m. mass window. The automatic gain control for full FT MS was set to 1 million ions and for FT MS/MS was set to 0.05 million ions with a maximum ion injection times of 100 ms and 200 ms, respectively. Lock-mass from ambient air (m/z 445.120025) was used for the internal calibration.
Data Analysis
Data analysis on the Proteome Discoverer platform (version 2.0) was performed using MASCOT (version 2.2.0) (22) and SEQUEST (23) as the search algorithms. For both MaxQuant and Proteome Discoverer, the search parameters allowed for two missed cleavages; carbamidomethylation at cysteine as a fixed modification; N-terminal acetylation, deamidation at asparagine and glutamine, oxidation at methionine, and the appropriate SILAC labeling provided as variable modifications. For C. elegans data, the variable modifications of phosphorylation at serine, threonine and tyrosine were specified. MS data was acquired on the LTQ-Orbitrap Elite mass spectrometer, the monoisotopic peptide tolerance was set to 10 ppm and MS/MS tolerance to 0.1 Da. The false discovery rate was set to 1% at the peptide level.
For analysis of MaxQuant and Proteome Discoverer data (supplemental Tables S1 and S2), PyQuant was provided with the output of each respective program to perform an additional round of quantification. PyQuant was provided with the msf output of Proteome Discoverer and mzML files of the raw data. For C. elegans, the additional parameter of “–spread” was supplied; otherwise the default parameters were retained (precursor mass error of 5 ppm and isotope-selection error of 2.5 ppm). For MaxQuant, the ms_ms.txt table was provided as input.
Data Availability
The raw mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD003327 (24).
RESULTS AND DISCUSSION
The goal of PyQuant is to provide a scalable, versatile framework for MS-based quantitative analyses. To demonstrate the various analyses PyQuant enables and its novel capabilities, we used publicly available data sets that utilize SILAC (25), NeuCode (26), 15N, and MS3 based TMT technologies (27).
PyQuant Accurately Quantifies a Variety of Quantitative MS Data
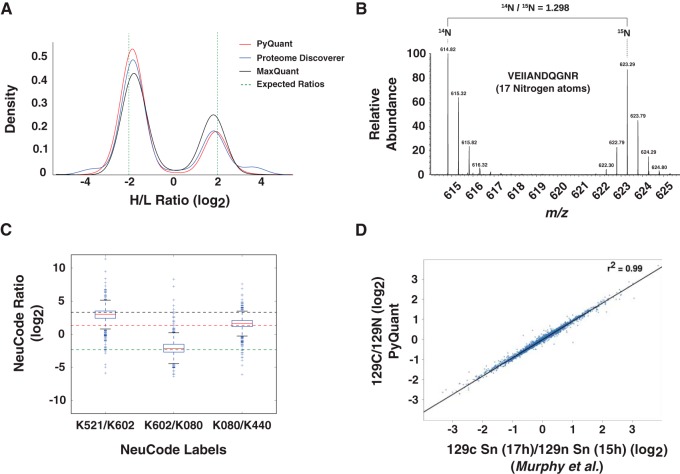
To assess PyQuant against a SILAC sample with a known isotopic mixture, three separate E. coli cultures were grown with a mixture of light, medium and heavy arginine and lysine amino acids in different ratios (Table I). Following overnight growth, the cultures were lysed and processed for LC-MS/MS as described under materials and methods. The raw data from sample 3 which contained all labels in known amounts was analyzed with two existing platforms, MaxQuant and Proteome Discoverer in addition to PyQuant (supplemental Table S3 and S4 with additional protein level measurements for Proteome Discoverer in supplemental Table S5). As shown in Fig. 2A, PyQuant was able to recapitulate the known sample ratios, and consistently matched SILAC ratios given by Proteome Discoverer and MaxQuant. Thus, PyQuant is able to match these existing algorithms in quantifying data of known ratios.
Table I. Samples grown with known ratios of isotopically labeled amino acids.
| Mixing ratios |
||||||
|---|---|---|---|---|---|---|
| Lys-0 | Lys-4 | Lys-8 | Arg-0 | Arg-6 | Arg-10 | |
| Sample 1 | 4 | 0 | 1 | 1 | 0 | 4 |
| Sample 2 | 1 | 0 | 1 | 1 | 0 | 1 |
| Sample 3 | 1 | 2 | 4 | 4 | 2 | 1 |
Fig. 2.
PyQuant correctly quantifies various quantitative mass spectrometry data methods. A, Two known mixtures of SILAC isotopes in E. coli is quantified with three different platforms: PyQuant (red line), MaxQuant (black line), and Proteome Discover (blue line). The expected log2 ratios are −2, and 2, which are indicated by a dashed green line. All three programs are centered on approximately the same ratios, and have similar peak widths. B, PyQuant is able to correctly quantify 15N labeled data. The raw MS1 spectrum of a peptide from a sample comprised of both 15N and 14N labeled peptides is plotted. The ratio of the two species as calculated by PyQuant is shown above, which agrees with the manual interpretation of the relative abundance levels. C, A multiplexed experiment that is labeled with NeuCode is shown. The box-plots indicate the distribution of peptides quantified by PyQuant, and the dashed lines indicate known mixture ratios. As shown, the median value, represented by a red line within the box-plot, for each ratio measured is approximately equal to the known mixing ratio. Thus, PyQuant is capable of reproducing the known mixtures of isotopes in the experiment. D, PyQuant is used to quantify a TMT 10-plex experiment using MS3 fragmentation for quantification. On the x axis are values obtained by Murphy et al. in their study, and on the y axis are the same values obtained by PyQuant. As shown, the values are nearly indistinguishable, providing evidence that PyQuant is capable of MS3-based quantification.
Although SILAC uses labeling of select amino acids, a similar technique, 15N, labels organisms by the metabolic incorporation of 15N isotopes into any nitrogen containing amino acids (28). To evaluate PyQuant's ability to handle 15N-based quantitative MS data, a 15N labeled mouse liver was compared with an unlabeled mouse liver. Equal amounts of proteins extracted from each mouse liver were mixed and processed for LC-MS/MS as described under materials and methods. Following peptide assignment with Proteome Discoverer, the raw data and search results were processed with PyQuant to quantify the relative abundance of 15N to 14N containing peptides. Because of MaxQuant and Proteome Discoverer's inability to quantify 15N data, to evaluate the performance of PyQuant, spectra were manually interrogated and compared with the ratio provided by PyQuant, which confirmed that PyQuant provides values that are consistent with manual interpretation (Fig. 2B).
A recently developed in vivo labeling technique is NeuCode, where various combinations of 13C, 15N, and 2H are incorporated into an amino acid in order to label a cell or organism (29). Because of the number of carbon, nitrogen, and hydrogen atoms in an amino acid, the number of combinations of these isotopes results in marked increase in multiplexing capabilities, with up to 36 different mass combinations for lysine alone. Currently, no available software supports quantification of NeuCode isotopes. Thus, we tested PyQuant's ability to quantify this type of data. An experiment with known mixtures of NeuCode labeled lysates was analyzed with PyQuant (26). As shown in Fig. 2C, PyQuant was capable of quantifying NeuCode data and the median quantification values matched the known isotopic mixtures of each NeuCode reagent.
For chemical tagging methods, two common methods are iTRAQ and TMT. Each method uses isobaric tags that do not cause a mass shift between samples that can be detected in MS1 scans, but upon fragmentation the tags provide the relative abundance of each labeled sample. Because both methods enable multiplexed comparisons of samples that may not be metabolically labeled, such as clinical samples, it has been widely employed in the quantitative proteomics field. However, one complication which arises in MS2 based quantification is interference from co-fragmentation of background ions, which can systematically skew relative abundance measurements (30). A solution to this complication is the use of MS3 for quantification as opposed to MS2, which reduces the chance of co-fragmentation of unwanted ions from MS/MS (31). We assessed PyQuant's ability to quantify at the MS3 level by comparing values derived from PyQuant to previously published MS3 based quantifications (Fig. 2D) (27). This revealed that PyQuant was nearly identical to the previously published values and is capable of MS3 based quantification.
Increased Quantification Rates by PyQuant
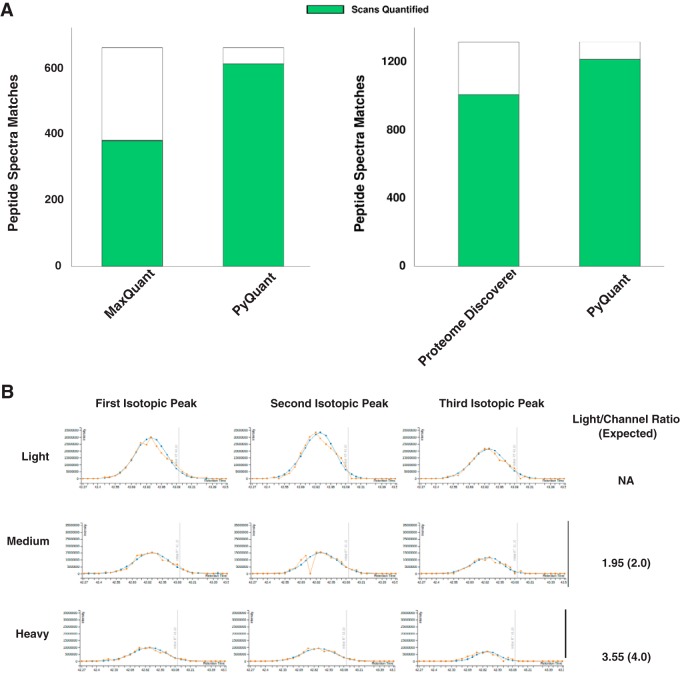
Because different peaks could be picked by different quantitation programs, employing multiple programs should increase the proportion of peptide ions that are quantified. In the E. coli data used above, we wished to determine the extent to which PyQuant could increase quantification. As shown in Fig. 3A, when analyzing a simple E. coli mixture, PyQuant increased the rate of quantification by 25 and 45% when compared with Proteome Discoverer and MaxQuant, respectively. To determine how confident the quantification of these peptides missed by other programs was, we evaluated many of peptides which MaxQuant and Proteome Discoverer failed to quantify in PyQuant's output, and determined most of them were accurate (Fig. 3B). In addition to this manual analysis, we evaluated the general distribution of peptides quantified only by PyQuant to see how well they agreed with the expected Heavy:Light peptide ratios (supplemental Fig. S6). This revealed that for the majority of peptides that were not quantified by MaxQuant but were quantified by PyQuant, most values reported by PyQuant were of the expected Heavy:Light ratios (supplemental Fig. 6A). For peptides not quantified by Proteome Discoverer but quantified by PyQuant, there was a broader distribution of peptide ratios with ∼58% of the data being within 1 log2 of its expected Heavy:Light ratio (supplemental Fig. 6B). This highlights that for some data, PyQuant may be overly optimistic in its ability to quantify data. For users to fully evaluate the quality of the data, PyQuant can generate an interactive HTML output that allows the user to view the isotopic clusters selected, the raw XIC, and the fitted XIC for each isotope.
Fig. 3.
PyQuant is capable of increasing the quantification rates of MaxQuant and Proteome Discover. A, PSMs from MaxQuant were extracted and analyzed with PyQuant, which resulted in a 45% increase in the number of spectra quantified as compared with MaxQuant alone. Similarly, PSMs from Proteome Discoverer were analyzed with PyQuant, which resulted in a 25% increase in the number of spectra quantified as compared with Proteome Discover. B, The quantification by PyQuant of a peptide which was not quantified by MaxQuant is shown. The extracted ion chromatogram for each label is robust with multiple isotopomers identified for every label. The quantified ratio as compared with the expected ratio for each channel is shown on the right.
PyQuant Can Quantify Isotopically Labeled Peptides with Unexpected Isotopic Patterns
SILAC was initially developed for cell culture systems but is increasingly used for labeling model organisms for global, quantitative analysis of protein abundance. A potential complication of SILAC in organisms is the metabolic conversion of experimentally introduced labeled amino acids into other amino acids (32, 33). Normally, the incorporation of naturally occurring carbon and nitrogen isotopes results in the spread of a peptide from its monoisotopic mass, with each combination of isotopes appearing as a distinct species in a mass spectrum. Without amino acid conversion, this pattern can be theoretically calculated and used to identify which peaks correspond to a peptide of interest. However, when there is an additional source of labeled isotopes, such as metabolic conversion of labeled amino acids, the isotopic cluster widens and deviates significantly from the theoretical distribution, thus complicating the interpretation of protein abundance.
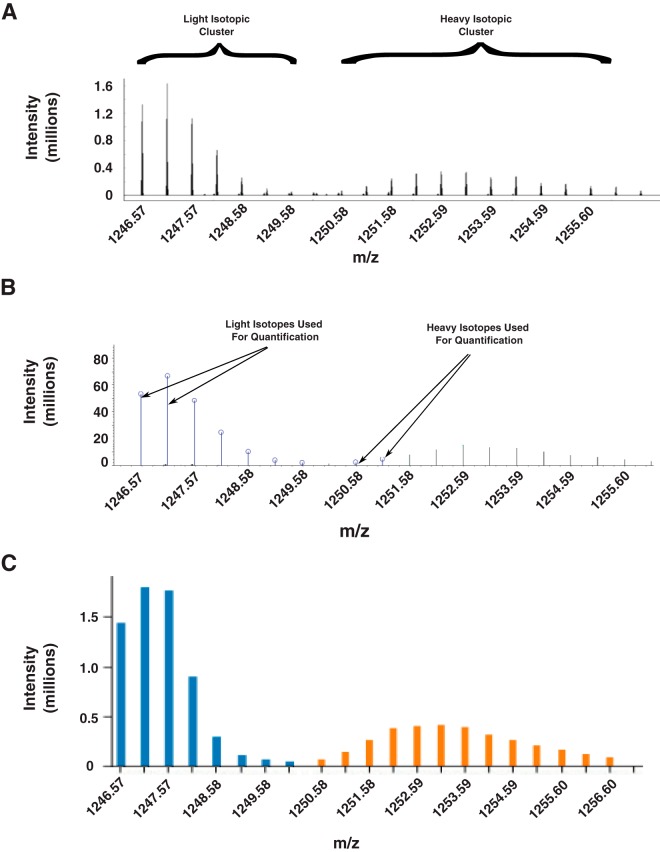
To evaluate how isotopic conversions impact the quantification of existing programs, we labeled the nematode C. elegans with heavy and light amino acids by growing them in the presence of labeled bacteria. As shown in Fig. 4A, the use of arginine as an isotopic label resulted in the spread of the heavy isotopic cluster, which deviated significantly from the isotopic distribution of the unlabeled peak. Notably, this peak pattern deviates from the known arginine to proline conversion that creates distinct satellite distributions of peptides containing proline derived from isotopically labeled arginine (33, 34). Instead, this peak distribution is consistent with a previously described pattern resulting from the arginase pathway, in which Arg-10 is converted to numerous amino acids including aspartate, asparagine, methionine, lysine, and threonine (32). This data was quantified with PyQuant, Proteome Discoverer, and MaxQuant. We found that Proteome Discover and MaxQuant routinely underestimated the heavy label's abundance, resulting in a systematic bias in the SILAC ratios reported. We hypothesize this may be because of the reliance of these algorithms on matching corresponding peaks from each label's isotopic distribution thereby ignoring peaks that deviate significantly from the theoretical distribution of the isotopic cluster (Fig 4B). Although restricting the data to known theoretical distributions is useful for removing contaminating peaks, in cases such as this, it is an incorrect assumption. Thus, to correctly quantify labels that have undergone catabolism, parameters in PyQuant can be set to not enforce the theoretical distribution on labeled data. With this option, PyQuant correctly identifies and quantifies the entire isotopic envelope (Fig. 4C). Thus, PyQuant can be particularly useful in correcting complications arising from metabolic pathways and permits labeling of organisms with minimal loss of quantitative information.
Fig. 4.
PyQuant quantifies isotopic profiles resulting from amino acid conversions. A, C. elegans sample was labeled with Arginine-10, which was metabolically converted to other amino acids. As seen, this results in a spread of the heavy isotopic cluster as compared with the light isotopic cluster. B, Selection of isotopes from light and heavy isotopic envelope by Proteome Discoverer. Proteome discoverer excludes several isotopes from the light and heaavy isotopic envelope. This results in Proteome Discover quantifying only a subset of the heavy data, and systematically under-reporting Heavy/Light SILAC ratios. C, Selection of isotopes by PyQuant. Most of the ions of both the light (shown in blue) and heavy (shown in orange) isotopic envelopes are selected for quantification by PyQuant.
Targeted Ion Quantification with PyQuant
In conventional data-dependent acquisition experiments, the most abundant ions are chosen for fragmentation. Although this approach is useful in most cases, it can lead to non-intuitive results in complex samples. For instance, if many ions species exist in a given scan window that are of similar intensities, the mass spectrometer will randomly select a set number of ions to fragment. In replicate studies, this has the effect that ions selected in one sample may not be chosen for fragmentation in the other sample. This is commonly observed in the sometimes poor overlap in PSMs between replicates. To overcome this issue, PyQuant can perform targeted searches for peptides identified in one replicate that are not fragmented, but present in the MS1 spectra of another replicate.
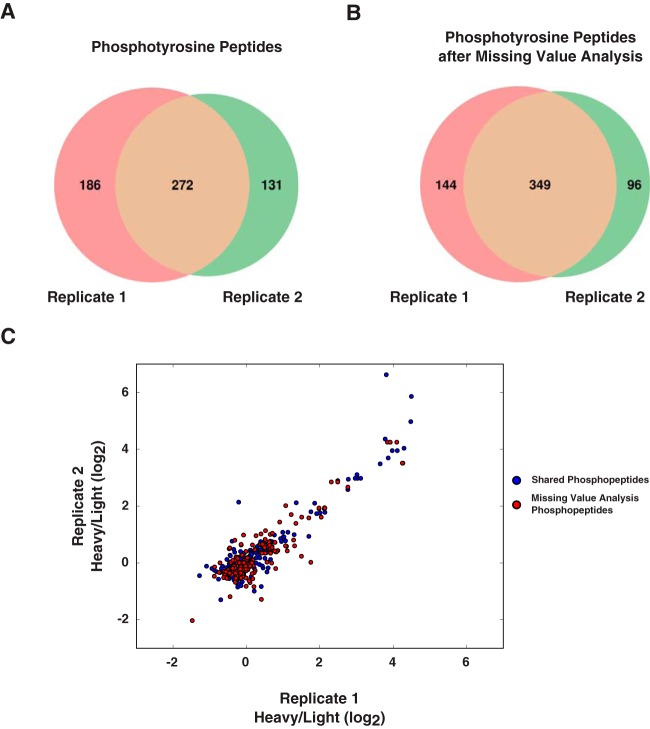
To determine how well a targeted search can increase the concordance between replicates, we applied PyQuant's missing value analysis to a phosphotyrosine enrichment experiment. A recent study evaluated the changes in phosphorylation levels as a function of Thymic stromal lymphopoietin (TSLP) signaling (25). Upon re-analysis of this data, we found between two replicates, one replicate contained 186 phosphotyrosine peptides unique to it and the second replicate contained 131 phosphotyrosine peptides unique to it (Fig. 5A). We performed a targeted search for these missing peptides on the raw data of each replicate with PyQuant. This targeted search provided replicate information for 77 of these phosphopeptides, which increased the concordance of quantified phosphopeptides from 46% to 59% between the replicates (Fig. 5B). Lastly, we plotted the phosphopeptide fold changes for each replicate with and without the missing value analysis. This revealed that many peptides followed the same general trend of phosphopeptide abundance between the two samples (Fig. 5C).
Fig. 5.
Filling in missing values between replicate studies. A, The Venn diagram shows the overlap of hyperphosphorylated peptides from two replicate phosphotyrosine enrichments. Over half of the data is not replicated between the two samples. B, After performing a missing value search with PyQuant on peptides unique to each replicate, peptides which were not fragmented in a given replicate are quantified. This increase is shown by the increased overlap between the two replicates. C, The SILAC ratios of phosphopeptides between each replicate. Each axis represents the SILAC ratios of a particular replicate. Blue circles correspond to phosphopeptides fragmented and quantified in both replicates (the overlap in A). Red circles represent the values recovered from PyQuant's missing value analysis. Each red circle was fragmented in only one replicate, but by using the elution time and precursor ion, the ion is able to be quantified irrespective of its fragmentation. As shown, the red and blue circles have a similar trend between the two replicates, indicating the missing values follow a similar trend as the ions fragmented in both replicates.
It is important to note that this approach does have limitations, as many ions can appear at identical m/z values and retention times. However, in well controlled experiments, we believe this option in conjunction with PyQuant's detailed visual outputs is a powerful tool for adding confidence to measured peptides which are considered significant in one replicate, but missing in another replicate.
We envision PyQuant as a useful complement to existing tools that can be used to provide an additional measure of confidence for quantified values as well as to quantify ions that may be omitted by some programs. Similarly, other tools may capture data that PyQuant misses. Thus, the ability of tools to operate together more easily facilitates the integration of multiple tools for a comprehensive analysis of a sample. For simple analysis, PyQuant can be used in a plug-and-play fashion with a variety of data sources and provides a simple, easily parsed output. This allows the creation of and easy deployment of minimal, light weight data analysis pipelines. Lastly, because PyQuant decouples the quantification of mass spectrometry data from the traditional peptide spectrum identification and quantification pipeline, PyQuant opens new avenues for novel and highly customized quantification strategies on a variety of data types.
Supplementary Material
Footnotes
Author contributions: C.J.M., M.K., and A.P. designed research; C.J.M. performed research; C.J.M. contributed new reagents or analytic tools; C.J.M., M.K., C.N., and A.P. analyzed data; C.J.M., M.K., and A.P. wrote the paper.
* This study was supported by NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036, http://proteomics.cancer.gov).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- iTRAQ
- isobaric tag for relative and absolute quantification
- TMT
- tandem mass tags
- PSM
- peptide spectral assignments
- XIC
- extracted ion chromatogram.
REFERENCES
- 1. Liu H., Sadygov R. G., and Yates J. R. 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 2. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 3. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., and Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 4. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., and Hamon C. (2003) Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 5. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 6. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., and Nesvizhskii A. I. (2011) iProphet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwon T., Choi H., Vogel C., Nesvizhskii A. I., and Marcotte E. M. (2011) MSblender: A probabilistic approach for integrating peptide identifications from multiple database search engines. J. Proteome Res. 10, 2949–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alves G., Wu W. W., Wang G., Shen R. F., and Yu Y. K. (2008) Enhancing peptide identification confidence by combining search methods. J. Proteome Res. 7, 3102–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tharakan R., Edwards N., and Graham D. R. M. (2010) Data maximization by multipass analysis of protein mass spectra. Proteomics 10, 1160–1171 [DOI] [PubMed] [Google Scholar]
- 10. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 11. Chick J. M., Kolippakkam D., Nusinow D. P., Zhai B., Rad R., Huttlin E. L., and Gygi S. P. (2015) A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat. Biotechnol. 33, 743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Renuse S., Chaerkady R., and Pandey A. (2011) Proteogenomics. Proteomics 11, 620–630 [DOI] [PubMed] [Google Scholar]
- 13. Wang X., Slebos R. J., Wang D., Halvey P. J., Tabb D. L., Liebler D. C., and Zhang B. (2012) Protein identification using customized protein sequence databases derived from RNA-Seq data. J. Proteome Res. 11, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eng J. K., Jahan T. A., and Hoopmann M. R. (2013) Comet: An open-source MS/MS sequence database search tool. Proteomics 13, 22–24 [DOI] [PubMed] [Google Scholar]
- 15. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 16. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 17. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., and Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 19. Schwarz G. (1978) Estimating the dimension of a model. Ann. Stat. 6, 461–464 [Google Scholar]
- 20. Rousseeuw P. J., and Driessen K. V. (1999) A fast algorithm for the minimum covariance determinant estimator. Technometrics 41, 212–223 [Google Scholar]
- 21. Stiernagle T. (2006) Maintenance of C. elegans. Wormbook, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 23. Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 24. no J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhong J., Kim M.-S., Chaerkady R., Wu X., Huang T.-C., Getnet D., Mitchell C. J., Palapetta S. M., Sharma J., O'Meally R. N., Cole R. N., Yoda A., Moritz A., Loriaux M. M., Rush J., Weinstock D. M., Tyner J. W., and Pandey A. (2012) TSLP Signaling network revealed by SILAC-based phosphoproteomics. Mol. Cell. Proteomics 11, M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merrill A. E., Hebert A. S., MacGilvray M. E., Rose C. M., Bailey D. J., Bradley J. C., Wood W. W., el, Masri M., Westphall M. S., Gasch A. P., and Coon J. J. (2014) NeuCode Labels for relative protein quantification. Mol. Cell. Proteomics 13, 2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murphy J. P., Stepanova E., Everley R. A., Paulo J. A., and Gygi S. P. (2015) Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae. Mol. Cell. Proteomics 14, 2454–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Washburn M. P., Ulaszek R., Deciu C., Schieltz D. M., and Yates J. R. (2002) Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 74, 1650–1657 [DOI] [PubMed] [Google Scholar]
- 29. Hebert A. S., Merrill A. E., Bailey D. J., Still A. J., Westphall M. S., Strieter E. R., Pagliarini D. J., and Coon J. J. (2013) Neutron-encoded mass signatures for multiplexed proteome quantification. Nat. Methods 10, 332–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., and Wright P. C. (2009) iTRAQ Underestimation in simple and complex mixtures: “The Good, the Bad and the Ugly.” J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]
- 31. Ting L., Rad R., Gygi S. P., and Haas W. (2011) MS3 eliminates ratio distortion in isobaric labeling-based multiplexed quantitative proteomics. Nat. Methods 8, 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borek W. E., Zou J., Rappsilber J., and Sawin K. E. (2015) Deletion of genes encoding arginase improves use of “heavy” isotope-labeled arginine for mass spectrometry in fission yeast. PLoS ONE 10, e0129548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Hoof D., Pinkse M. W., Oostwaard D. W., Mummery C. L., Heck A. J., and Krijgsveld J. (2007) An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat. Methods 4, 677–678 [DOI] [PubMed] [Google Scholar]
- 34. Ong S. E., Kratchmarova I., and Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD003327 (24).