Abstract
Thyroid hormones modulate not only multiple functions in vertebrates (energy metabolism, central nervous system function, seasonal changes in physiology, and behavior) but also in some non-vertebrates where they control critical post-embryonic developmental transitions such as metamorphosis. Despite their obvious biological importance, the thyroid hormone precursor protein, thyroglobulin (Tg), has been experimentally investigated only in mammals. This may bias our view of how thyroid hormones are produced in other organisms. In this study we searched genomic databases and found Tg orthologs in all vertebrates including the sea lamprey (Petromyzon marinus). We cloned a full-size Tg coding sequence from western clawed frog (Xenopus tropicalis) and zebrafish (Danio rerio). Comparisons between the representative mammal, amphibian, teleost fish, and basal vertebrate indicate that all of the different domains of Tg, as well as Tg regional structure, are conserved throughout the vertebrates. Indeed, in Xenopus, zebrafish, and lamprey Tgs, key residues, including the hormonogenic tyrosines and the disulfide bond-forming cysteines critical for Tg function, are well conserved despite overall divergence of amino acid sequences. We uncovered upstream sequences that include start codons of zebrafish and Xenopus Tgs and experimentally proved that these are full-length secreted proteins, which are specifically recognized by antibodies against rat Tg. By contrast, we have not been able to find any orthologs of Tg among non-vertebrate species. Thus, Tg appears to be a novel protein elaborated as a single event at the base of vertebrates and virtually unchanged thereafter.
Keywords: molecular evolution, thyroid, thyroid hormone, Xenopus, zebrafish, metamorphosis, thyroglobulin
Introduction
Of the many signaling molecules that participate in the regulation of human physiology and are evolutionarily conserved, thyroid hormones (THs)4 are among the most puzzling (1). Curiously, these hormones control different processes in different species (2). In many amphibians as well as in teleost fishes and some invertebrate chordates, THs trigger metamorphosis, a spectacular life-history transition (3). THs have also been implicated in the control of seasonality, a process during which animals adapt their reproductive function and behavior to the annual change of photoperiod (4). In mammals, THs regulate crucial physiological functions including oxidative metabolism, heart rate, and thermogenesis (5–8), which is of considerable relevance because nearly 10% of the human population may be at risk of developing a thyroid-related disease during their lifespan (9).
THs are produced in two forms: T4, which harbors four iodine atoms and behaves as a hormonal precursor, and T3, with three iodine atoms, representing the active hormone that binds the receptor. In addition to their unusual diversity of functions, THs are also unusual in their mode of production. First, the reason for a requirement of iodine in a compound with hormonal activity (the only one to our knowledge) remains a mystery (10). Second, with a molecular mass of <0.8 kDa and based upon a structure that engages only two tyrosyl residues, THs are synthesized by vertebrate thyrocytes from a large dimeric protein named thyroglobulin (Tg) with a monomer molecular mass of 330 kDa and containing nearly 2750 residues (11, 12).
Given the importance of THs in controlling major life history transitions in many species and the complexity of the Tg protein, it is striking to realize that only mammalian Tgs have been subject to detailed investigation (11, 13–15). Besides mammals, all other available Tg sequences are only predictions from genome assembly or EST data, and even these are known only from Gnathostomes. As a consequence, it remains unknown how the TH precursor protein emerged during evolution. Based on biochemical characterization, there have been a few reports of possible Tg orthologs in invertebrate chordates (16), but these assignments have remained controversial (17). Surprisingly, no complete Tg has yet been characterized in Xenopus or zebrafish, two major model systems that have been used to decipher the role of THs in physiology and development (8, 18).
Tg is a multidomain protein that harbors different modules, from N to C termini: a signal peptide, four Tg type 1 repeats (Tg1), a linker region, six additional Tg1 repeats, a hinge region, three Tg type 2 repeats (Tg2), one Tg1 repeat, five Tg type 3 repeats (Tg3) and a CholinEsterase-Like (ChEL) domain (11, 12). The signal peptide addresses the newly synthesized protein into the endoplasmic reticulum where it is immediately removed, whereas the repeat domains dictate the protein structure (19). The ChEL domain is implicated in conformational maturation and export of newly synthesized Tg through the secretory pathway to the thyroid follicle lumen (19, 20) as well as in the de novo formation of T3 (21, 22). During its complex trafficking, Tg undergoes many post-translational modifications critical for its biological function (23).
In mammals, newly synthesized Tg is first glycosylated at many sites (24) and folded via the formation of numerous intradomain disulfide bonds. Spacing between the cysteine residues has allowed for the identification of distinct Tg repeat domains controlling proper folding of the protein (11). Tg then forms a 660-kDa homodimer before its secretion, which it requires for thyroid hormonogenesis (12, 25–27). Once in the follicle lumen, many Tg tyrosines are iodinated as a consequence of thyroid peroxidase activity, and specific iodotyrosine pairs are “coupled” to form THs within the Tg protein (22, 28). In particular, Tyr-5 and Tyr-130 (for clarity, all amino acid positions cited in this paper use the numbering system based upon the human mature protein with the signal peptide cleaved) are, respectively, known as an acceptor-donor pair for coupling and are the most important hormonogenic site, yielding nearly half of the T4 synthesized within Tg (29, 30). Several other secondary hormonogenic tyrosines have also been identified (e.g. Tyr-847, -973, -1291, -1448, -2554, -2568, and -2747), with the last one being the most important site for de novo T3 formation (31).
Many iodinated Tg molecules (bearing embedded THs) remain for an extended period within the thyroid follicle lumen, allowing for the long term storage of iodine within the colloid of the thyroid follicles. Thyroid-stimulating hormone stimulates endocytic uptake of Tg from the follicular lumen by thyrocytes followed by Tg delivery to lysosomes for extensive proteolytic cleavage, resulting in the liberation of THs into the bloodstream (32, 33). Overall, TH synthesis is energetically very costly as it requires the synthesis and endocytic destruction of two very large Tg monomers to produce only one or two TH molecules.
Herein, we have examined the extent of conservation of Tg organization and structure across the whole vertebrate clade. We have cloned and expressed Tg both from western clawed frog (Xenopus tropicalis) and zebrafish (Danio rerio) and demonstrate that these are secreted proteins. Moreover, to complete the analysis, we searched genomic data accumulated in lamprey (Petromyzon marinus) and in genome sequences outside of vertebrates, with the thought that identifying functionally relevant regions of Tg may help in the search for distant orthologs. Remarkably, our findings strongly suggest that the mechanism of Tg synthesis is conserved throughout the vertebrates, but there is no clear sign of Tg presence outside vertebrates. Thus, all evidence suggests that the Tg modular structure appeared at the base of vertebrates and has remained conserved since then.
Results
Cloning and Expression of Xenopus and Zebrafish Full-length Tg
Using RT-PCR with primers designed from the partial predicted sequences available in complete genome data, we were able to isolate a full-length zebrafish (D. rerio) Tg cDNA clone that encodes a 2733-amino acid protein harboring a very similar organization to that of human Tg (Fig. 1, A and B).
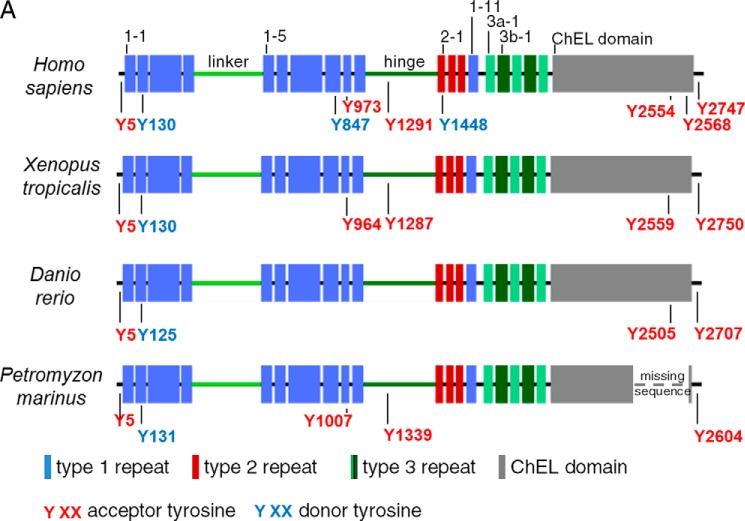
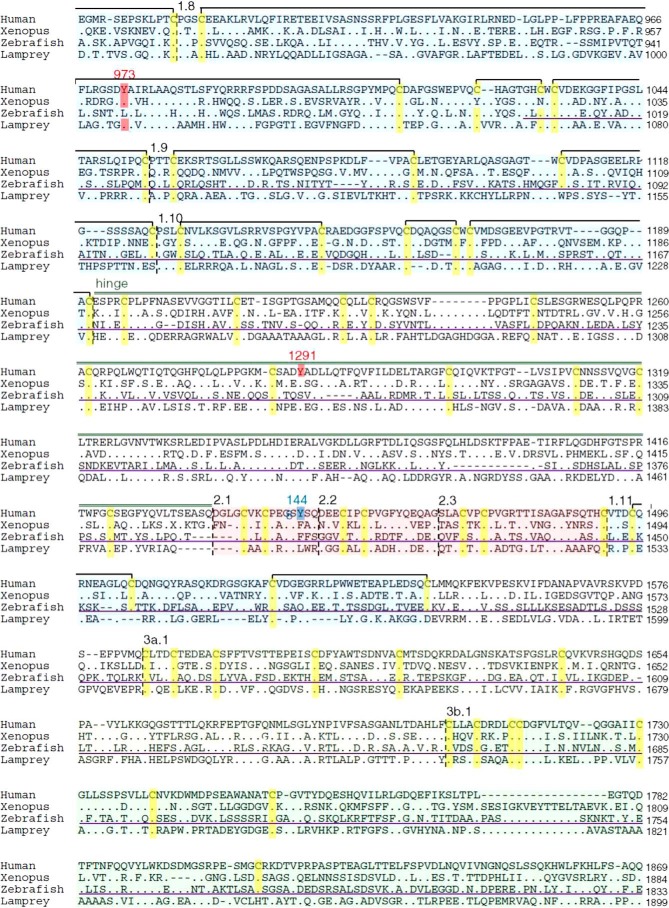
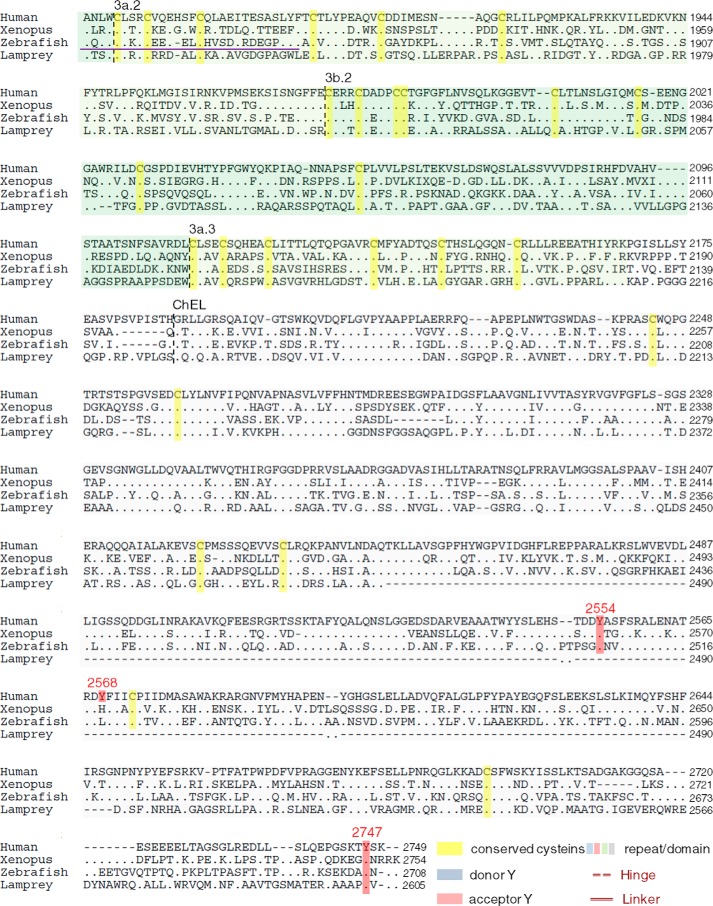
FIGURE 1.
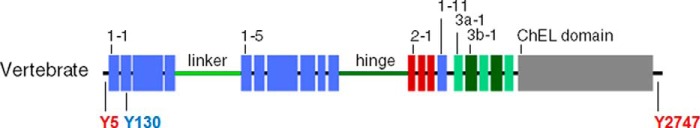
Schematic organization of human, Xenopus, zebrafish, and lamprey Tgs. A, Tg1 repeats are shown by blue boxes, Tg2 repeats by red boxes, Tg3 repeats by green boxes, and ChEL domain by gray boxes. The linker and the hinge are represented by a light green and dark green line, respectively. The main hormonogenic tyrosines are represented with their positions in each sequence. B, alignment of human, Xenopus, zebrafish, and lamprey Tg. Tg1 repeats are highlighted in light blue, Tg2 repeats are in red, Tg3 repeats are in green, and ChEL domains are in gray. The linker region is indicated by a light green line, and the hinge region is indicated by a dark green line. The donor tyrosines are highlighted in blue, and the acceptor tyrosines are in red. Conserved cysteines are highlighted in yellow with presumptive disulfide bonds of Tg1 repeats shown as black brackets at the top of each line. The 5′-end of Xenopus retrieved after mass sequencing is underlined in orange. The RNA probes for in situ hybridization assays are underlined in purple.
Using the same strategy, we isolated a western clawed frog (X. tropicalis) Tg cDNA clone that was incomplete at its 5′-end. This cDNA starts at an exon whose position is conserved and encodes a peptide sequence (EYQL … ) that includes the putative conserved hormonogenic Tyr-5 but did not encode any immediate upstream sequence corresponding to a signal peptide. We called this Xenopus Tg cDNA “incomplete” and extended the 5′-end of the clone using a combined approach of PCR amplification and massive sequencing (Fig. 1B). We obtained a sequence that did seem to correspond to an upstream initiating open reading frame, encoding two potential initiator methionine residues: MAPVLLSTMTHRFIYTIFCVFRIIAAIETEYQL. When analyzed by the SignalP 4.1 program, the second of these two methionines predicts the generation of a functional signal peptide with a satisfactory cleavage site, preserving the position of hormonogenic Tyr-5, ultimately encoding a 2772 residue protein.
Given the recent suggestion of extrathyroidal expression of Tg variants (34), we used zebrafish and Xenopus-specific probes to examine Tg mRNA expression during embryogenesis in both species. Expression of tg in zebrafish was evaluated by whole mount in situ hybridization (ISH) at four different stages of development (Fig. 2A, supplemental Fig. S1A). Because the Tg gene encodes a very large mRNA as well as the possibility of extrathyroidal expression (34), we employed two different probes corresponding to different Tg regions, one at the 5′ end of the transcript and the other in the middle of the transcript. At 24 h post-fertilization (hpf), no expression was observed. At 48 hpf, Tg was expressed in one localized spot in the pharyngeal region of the fish. At 120 hpf, the expression pattern increased in size and adopted an elongated shape but remained exclusive to the pharyngeal region. This corresponds to the organization of the teleost thyroid gland that, in contrast to the well defined gland of amniotes, is scattered in several nodules to form a chain-like pattern along the pharyngeal region. The expression pattern was identical at 144 hpf, and the same pattern was observed with both probes, rendering unlikely any major extrathyroidal expression of an alternative transcript.
FIGURE 2.
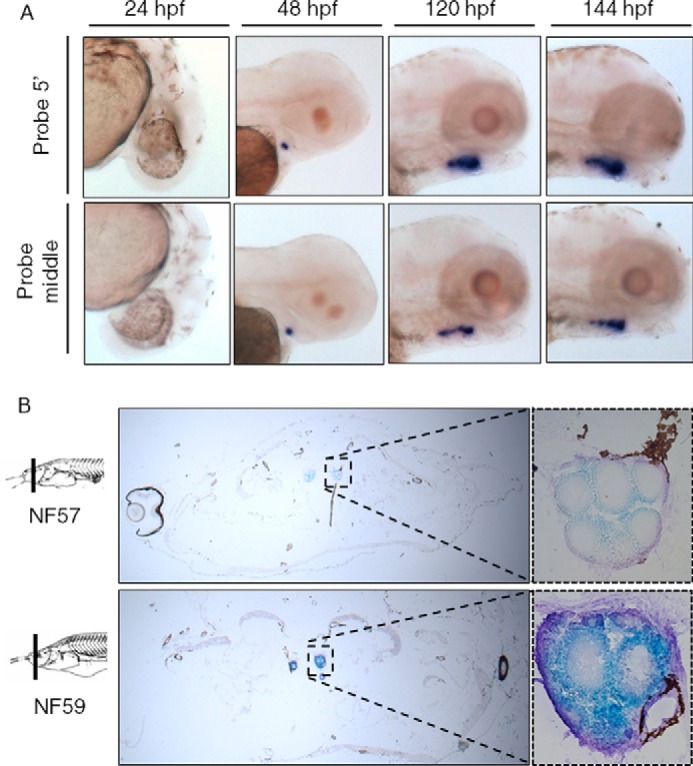
Expression of Tg in zebrafish and Xenopus revealed by ISH. A, for zebrafish embryo experiments, two probes were used, in the 5′ part of the Tg (upper panel) and in the middle (lower panel). ISH were performed at 24, 48, 120, and 144 hpf as indicated. In situ labeling appears in purple. B, for Xenopus tadpole sections, probes were used at stage NF57 and NF59 according to the Nieuwkoop and Faber's classification (73). In situ labeling appears in blue. Negative controls with sense probe show no staining (supplemental Fig. S1).
In the Xenopus tadpole, as in zebrafish, Tg was found to be expressed exclusively in thyrocytes of the thyroid gland (Fig. 2B, supplemental Fig. S1B). ISH on cross-sections revealed expression at Nieuwkoop Faber stages NF57 and NF59. Although ISH is not a quantitative method, the difference in labeling tends to indicate that, as expected, the thyroid is more active at stage NF59 (climax of metamorphosis) than NF57 (onset of metamorphosis), consistent with increased TH levels described at NF59 (35).
Characterization of Recombinant Zebrafish and Xenopus Tg Proteins
As Tg secretion is known to be mandatory for its biological function in vertebrates (23), we wished to investigate the secretability of recombinant Tg from Xenopus and zebrafish. The cDNAs encoding mouse and zebrafish Tg were expressed in 293T cells. Secretion into the cell culture medium was collected for 24 h, the cells were lysed, and both were analyzed by SDS-PAGE and Western blotting with a polyclonal antibody against rodent Tg (Fig. 3A). As expected, there was very strong immunoreactivity of mouse Tg, which was secreted efficiently from cells (C) to medium (M), and the secreted Tg exhibited a slower (higher) mobility consistent with modification of N-linked glycans by Golgi processing enzymes (see below). Zebrafish Tg was also immunoreactive, indicating preserved structural elements of rodent Tg contained within Zebrafish Tg, although the signal strength was weaker than for mouse Tg (Fig. 3A). Nevertheless, zebrafish Tg was clearly also secreted, and the protein exhibited a slower (higher) mobility in the medium than in the cell lysate (Fig. 3A, lanes 5 and 6).
FIGURE 3.
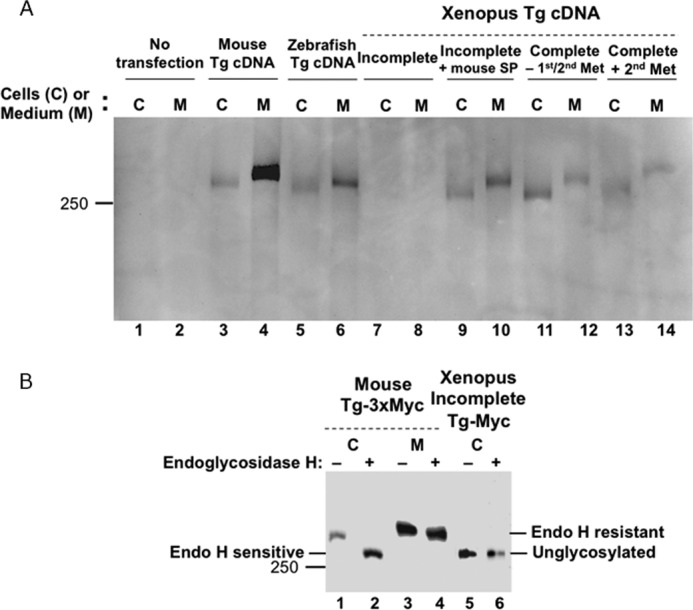
Expression and secretion of zebrafish and Xenopus Tg. A, recombinant Tgs of the indicated species were expressed in 293T cells (C) and secreted into the medium (M). Mouse Tg was used as a positive control (lanes 3 and 4). After a 1-day collection of media, both cell lysates and media were analyzed by SDS-PAGE and Western blotting with a polyclonal anti-rodent Tg antibody. Note that secreted Tg has a slower (higher) mobility because of carbohydrate processing that occurs as Tg migrates through the Golgi complex. Incomplete Xenopus Tg lacks a signal peptide, and the protein is poorly expressed (lanes 7 and 8). Adding a mouse signal peptide (SP) rescues its expression and secretion of the incomplete Xenopus Tg (lanes 9 and 10). Full-length Xenopus Tg is expressed and secreted (lanes 11 and 12). Initiating Tg translation with the second methionine of the Xenopus signal peptide is sufficient for expression and secretion (lanes 13 and 14). B, glycosylation analysis of mouse Tg-myc and the incomplete Xenopus Tg-myc by endoglycosidase H digestion proves that intracellularly, mouse Tg begins within the endoplasmic reticulum (endoglycosidase H-sensitive, lanes 1 and 2) and traverses the Golgi complex where the protein becomes endoglycosidase H-resistant lanes 3 and 4). In contrast, the incomplete Xenopus Tg is unglycosylated (lanes 5 and 6), indicating failure to enter the endoplasmic reticulum. Both cell lysates and media were analyzed by SDS-PAGE and Western blotting with a polyclonal anti-Myc antibody; 10 times more protein from the cells expressing the incomplete Xenopus Tg was loaded to detect these bands.
We next examined expression of the incomplete Xenopus Tg cDNA that included the putative conserved hormonogenic Tyr-5 but did not encode any apparent signal peptide. When transfected in 293T cells, this incomplete Xenopus Tg protein was poorly expressed in either the cells or medium (Fig. 3A, lanes 7 and 8). To better understand the role of the signal peptide in the stability and trafficking of Tg, we performed endoglycosidase H digestion. For mouse Tg, the protein starts off intracellularly as an endoglycosidase H-sensitive species (Fig. 3B, lanes 1 and 2) and is converted to a secreted protein that has a slower (higher) mobility that is endoglycosidase H-resistant (lanes 3 and 4). By contrast, the incomplete Xenopus Tg cDNA encodes a protein that is unglycosylated even before endoglycosidase H digestion (Fig. 3, B lane 5 and 6), indicating that this protein never enters the secretory pathway (as a consequence of the absence of a signal peptide). Thus, as for other secretory proteins (36) the N-terminal signal peptide fulfills a critical conserved function in mediating intracellular trafficking and secretion.
Because we suspected the absence of the signal peptide as a primary reason for the lack of Tg secretion, the mouse Tg signal peptide was engineered in-frame at the 5′-end of the Xenopus Tg open reading frame. With this, Tg protein expression was increased, and secretion was obtained (Fig. 3A, lane 10). After we succeeded in extending the 5′-end of the incomplete Xenopus Tg cDNA clone, we expressed in 293T cells the complete Tg cDNA bearing both potential initiator methionyl residues and found that the encoded Tg protein was well expressed and secreted (Fig. 3A, lanes 11 and 12). Because we expected that the second methionyl residue would lead to a functional signal peptide, we mutated (truncated) our cDNA to delete the first potential initiator methionine and found that this Tg protein was also well expressed and well secreted (Fig. 3A, lanes 13 and 14). Moreover, sequence analysis predicted that the signal peptides of human, Xenopus and zebrafish Tg are all enriched in α helix (supplemental Fig. S2), indicating a conservation of their structure despite divergence in amino acid sequences. These data support the concept that Zebrafish Tg and Xenopus Tg, like mammalian Tgs, are secretable proteins.
This result seemed surprising as Xenopus Tg cDNA encodes a protein that lacks the second disulfide bond of the Tg repeat 1–8 because of substitution of two Cys residues (Fig. 1B). This disulfide bond had been presumed to be essential for the structural integrity of the Tg type 1 repeat domain (23). To test its importance, we mutagenized these Cys residues within the context of full-length mouse Tg (Fig. 4). Surprisingly, the mTg-C1043Y-C1050N double mutant Tg was well expressed and secreted (Fig. 4, lane 10); indeed, even the single cysteine mutants mTg-C1043Y or mTg-C1050N (which prevent formation of the same disulfide bond but leave an unpaired Cys residue) were secreted (Fig. 4, lane 6 and 8). Thus, we conclude that the second Tg1–8 disulfide bond is not required for Tg secretion.
FIGURE 4.
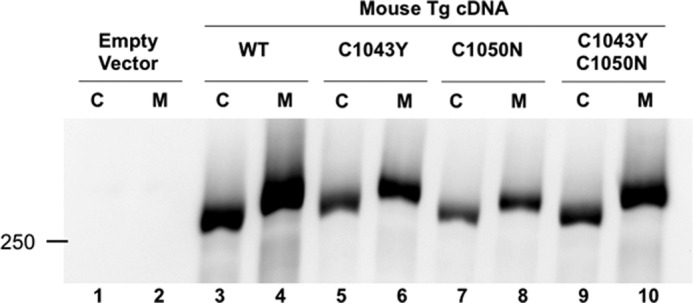
Role of the Tg1–8 repeat in the secretion of Tg. Cysteines forming the second disulfide bond of Tg repeat 1–8 were mutated individually or in a pair within mouse Tg. Both cell lysates (C) and media (M) were analyzed by SDS-PAGE and Western blotting with a polyclonal anti rodent Tg antibody. Neither the single mutations (lanes 5 and 6 and lanes 7 and 8) nor the double mutation (lanes 9 and 10) block expression or the secretion of the Tg protein.
A Tg Sequence in Lamprey
Early biochemical evidence has suggested the existence in lamprey (P. marinus) of a large iodinated protein present in the endostyle and/or thyroid gland (37–39). Nevertheless, the corresponding clones have never been isolated. We, therefore, searched for Tg orthologs in basal vertebrates (chondrichthyes and agnathas, for which several genomes are available). We found a clear candidate for lamprey Tg, including an excellent signal peptide (MRTSPLLPATTTLYLVLWIGTISALY) predicted by the SignalP.4.1 program. The retrieved sequence encodes a partial 2605-amino acid protein that contains the various N-terminal Tg repeats as well as the C terminus of the ChEL domain. However, this sequence misses part of the ChEL domain that is not present in the genome data (Fig. 1A), creating a sequence gap present between residues 2453 and 2632 of the lamprey protein (Fig. 1B).
Structural Conservation Coupled with Sequence Divergence of Vertebrate Tgs
Comparing the organization of Tgs in human, Xenopus, zebrafish, and lamprey (Fig. 1A) clearly shows that Tg in each of these species contains all of the domains found in mammalian Tg (11). Indeed, the Tg in each of these species harbors 11 Tg type-1, 3 Tg type-2, and 5 Tg type-3 domains. In contrast to previous reports (17) we observed that the seventh and ninth Tg1 repeats are also present in zebrafish (although the Tg1–7 repeat does seem to be absent in Percomorph fishes; supplemental Fig. S3, A and B). In addition, Tg3 domains, subcharacterized as type-3a and type-3b (called Tg3a and Tg3b, respectively), appear to be present in each of these species. Moreover, Xenopus, zebrafish, and lamprey Tgs exhibit a linker and a hinge region in the middle of the protein and a ChEL domain at the C-terminal end. These regions are of similar size to the mammalian one, with preservation of conserved cysteines. Taken together, these data indicate a strong conservation of the molecular architecture of Tg throughout the vertebrates.
The conserved regional/domain organization of Tg is not reflected at the level of amino acid sequence conservation, as the encoded Tg proteins are quite divergent in their primary structure. Indeed the overall sequence conservation of Tg between human and other species is: Xenopus 48%, zebrafish 43%, and lamprey 31% (note that the lamprey value is marginally affected by missing sequence from part of the ChEL domain). Thus, at this level, the Tg proteins are actually quite divergent.
Prediction of the secondary structures suggests that Tg from each of the four species shares multiple β-sheets and α-helices (supplemental Fig. S4), with a pattern of β-sheet located at the beginning of the Tg1 repeats and α-helix located at the end. The linker region is also predicted to be well structured with 30% α-helix and 10% β-sheet. An amphipathic helix is predicted at the end of the linker with five negatively charged amino acids on one side and five hydrophobic residues on the other (supplemental Fig. S4, red square). This amphipathic helix could have a role in Tg interaction with other partners. Comparison of the Tg sequence with crystallized proteins on PDB identified the Tg motif CWCV in the proteins P41ICF (accession code 1L3H) and ILGF4 (accession code 2DSR), which utilize this motif to form two distinct disulfide bonds (involving an additional upstream and downstream cysteine that are not contained within the motif). This motif is particularly well conserved in the sequences studied and only absent in repeats Tg1–2 of lamprey and Tg1–8 and Tg1–10 of Xenopus. Consequently, this repeated motif is likely to be similarly engaged in forming disulfide bonds within Tg and, thus, serving as the backbone for proper Tg folding.
Striking Conservation of Key Residues Highlights the Functional Conservation of Tgs
The general sequence divergence of the various Tg proteins renders even more striking the strong conservation of key residues known to be important for Tg function. These key amino acids include, in particular, the hormonogenic tyrosines (22, 29). The two tyrosines mainly involved in TH synthesis in mammals, Tyr-5 and Tyr-130 (human numbering), are strictly conserved in Xenopus (Tyr-5 and Tyr-130), zebrafish (Tyr-5 and Tyr-125), and lamprey (Tyr-5 and Tyr-131). Of the other hormonogenic residues, Tyr-2747 (the primary site for T3 formation in mammals; Ref. 21) is also strictly conserved in the three species (Tyr-2750 in Xenopus, Tyr-2707 in zebrafish, and Tyr-2604 in lamprey). Tyr-2554 is conserved in Xenopus and zebrafish (respectively, Tyr-2559 and Tyr-2505) but falls into a gap in the predicted lamprey sequence. Tyr-1291, which is considered to have a lesser role in TH synthesis, is conserved in zebrafish and lamprey (respectively Tyr-1287 and Tyr-1339) but not in Xenopus. Other hormonogenic tyrosines Tyr-847, -973, -1291, -1448, and -2568 that have been described with minor activities in human Tg were not conserved (Fig. 1B). Therefore, in summary, the most potent hormonogenic tyrosines found in human (Tyr-5, -130, and -2747) are well conserved in Xenopus, zebrafish, and lamprey, four divergent species of vertebrates, suggesting that formation of THs is mechanistically conserved throughout the vertebrates.
A complete sequence alignment of the four Tgs highlights that 90% of the cysteines are found at orthologous positions in human, Xenopus, zebrafish, and lamprey Tg, whereas only 14% of the amino acids overall are conserved between the four primary structures (Fig. 1B). The observation of two missing cysteine partners in Tg repeat 1–8 (Fig. 1B) suggests co-evolution at these two positions. In zebrafish, there are also two other missing cysteines (at position 343 and 1262), but despite these minor differences, the organization of the disulfide bond network within Tg proteins appears tightly conserved among the Gnathostomes.
Interestingly in lamprey, there are additional missing cysteines at positions 114 and 120 (repeat Tg1–2), 1146 and 1166 (repeat Tg1–9), and 1558 and 1569 (Tg1–11 repeat) that are thought to be disulfide partners in human Tg (Fig. 1B) suggesting, again, co-evolution of these residues. Additionally, lamprey Tg appears to lacks Cys-388 (linker) and Cys-589 (repeat Tg1–5), but this does not seem to affect the predicted secondary structure. In contrast, of the 16 identifiable N-linked glycosylation sites in human Tg (24), only 2 are conserved through all of the four species studied, whereas 8 others are strictly human-specific. Finally, it is worth noting that the catalytic site of the ChEL domain is not conserved as an active esterase in any of the Tgs, suggesting that loss of catalytic activity in this domain is ancient.
Phylogenetic Analysis of Tg Sequences
Amino acid sequences of Tg were used to build an ML (Maximum Likelihood) tree using both experimental plus predicted sequences (Fig. 5). The tree exhibits the overall expected topology, with preserved phylogenetic relationships between the major vertebrate clades (40). Most of the nodes are based on strong statistical support (>0.900, Fig. 5).
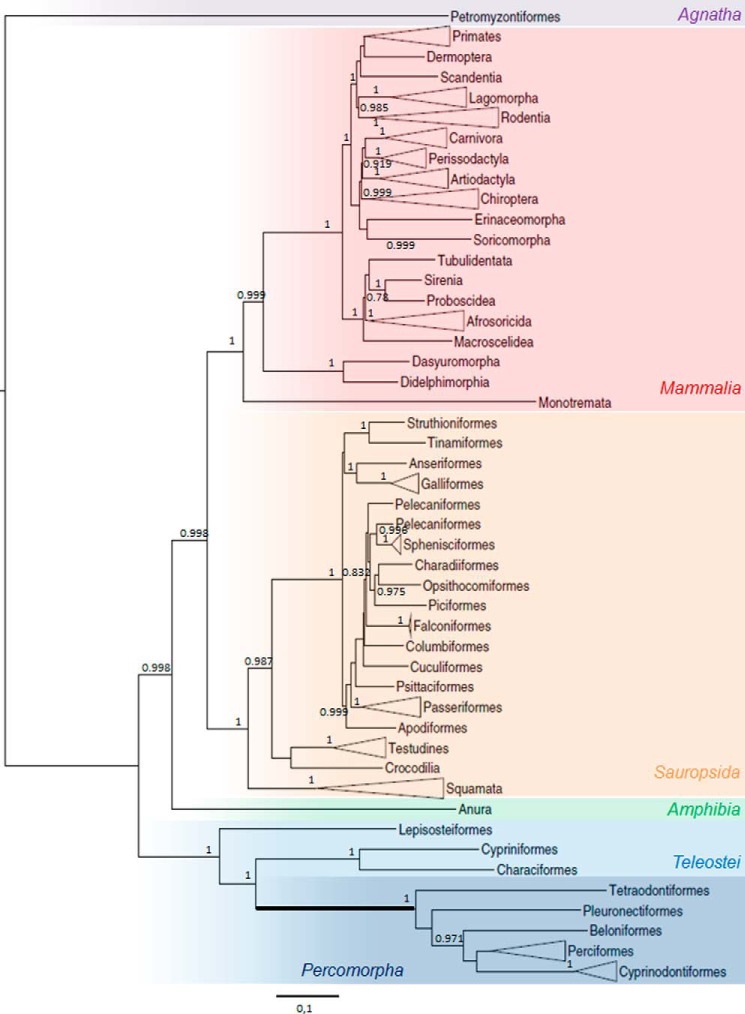
FIGURE 5.
Tg phylogeny. Phylogenetic tree built by maximum likelihood using experimentally determined Tg amino acid sequences as well as predicted sequences. Statistical approximate likelihood ratio test (aLRT) branch supports are indicated when >0.7. Agnathes are highlighted in purple, teleosts are in light blue, percomorphs are in dark blue, amphibians are in green, sauropsidas are in orange, and mammals are in red. The bold line supports the percomorphs.
In this phylogeny, we noted a very long branch (highlighted in bold, Fig. 5) supporting the Percomorpha, indicating accelerated Tg evolution within this group. This acceleration happened in a structurally conserved context as, once again, the cysteine residues and the modular structure of Tg remained (with the exception of the Tg1–7 repeat; supplemental Fig. S3, A and B). This acceleration might be linked to the fast radiation of Percomorpha, consistent with the rapid gene evolution that has already been described is this group (41).
Given the modular structure of Tg, we also computed individual trees using the isolated domains (namely, Tg1 only, Tg2, Tg3, Linker, Hinge, and ChEL) based on the cloned sequences (supplemental Table S4A). Overall, all the trees calculated from individual domains exhibit the same topology as that seen in the tree based on the full-length protein, indicating strong structural conservation of the entire protein (supplemental Fig. S5).
Tg Is Not Found Outside of Vertebrates
Some data have been interpreted as suggesting the existence of an amphioxus (Branchiostoma lanceolatum) Tg (16). Nevertheless, upon extensive search of the amphioxus genome, we found no evidence of a Tg ortholog (42). Similarly, we were not able to retrieve any Tg-like sequence in the most recent versions of urochordate, echinoderm, and protostomes genomes. To explore this further, we built a phylogeny of sequences encoding Tg type-1 repeats (supplemental Fig. S6A). None of the type-1 sequences we tested (including some not previously analyzed in the literature: Capitella teleta and Crassostrea gigas) clusters with any of the type-1 repeats of Tg. The data indicate that the proteins harboring Tg1 repeats in these species are not orthologs of vertebrate Tg.
In Strongylocentrotus purpuratus and tunicate species, we retrieved several Tg2-like sequences as EGF GCC2-GCC3 domains (according to NCBI identification). Again, these type-2 repeats retrieved in non-vertebrate species do not cluster with Tg in a phylogenetic tree (supplemental Fig. S6B). Therefore, we conclude that these proteins are also not orthologs of vertebrate Tg. We did not retrieve any Tg3-like sequences outside of vertebrates. Altogether, these results indicate that there are no clear orthologs of Tg outside of vertebrates.
Discussion
In this study we have examined evolution of the Tg gene and observed that all of the multiple domains of Tg are conserved throughout all the vertebrates studied. Although the overall primary structures are quite divergent between the Tg of human, Xenopus, zebrafish, and lamprey, we find that the hormonogenic tyrosines and most cysteine residues are extremely well conserved (Fig. 6). The conservation of both domain structure and cysteine residues is consistent with secondary structural predictions that are similar between human, Xenopus, zebrafish, and lamprey. Taken together, these observations show Tg evolution to be unusual as it exhibits a wide span of sequence divergence and yet retains a selected subset of specific amino acids that are highly conserved, which is enough to ensure conservation of structure and function.
FIGURE 6.
Prototypic vertebrate thyroglobulin. Vertebrate common features are shown: Tg1 repeats are shown in blue boxes, Tg2 repeats are in red boxes, Tg3 repeat is green boxes, and ChEL domain is by gray boxes. The linker and the hinge are represented by a light green and dark green line, respectively. The main hormonogenic tyrosines found in the investigated vertebrate species are represented with their positions in human numbering.
Conservation of Selected Tg Residues
The tyrosines at the orthologous positions of human Tyr-5 and Tyr-130 (but not the tyrosyl residues in between) are strikingly conserved in all identified Tgs, enforcing the idea that they are likely to work together in all vertebrates. It is known that these two residues form the major hormonogenic site in mammals, with a preference for T4 (30, 43). Given the three-dimensional structure imposed by the cysteines and the resulting disulfide bonds, it is probable that Tg folds in a way that brings Tyr-5 and Tyr-130 into close proximity. Thus, there is likely to be co-evolution of Tyr-5 and Tyr-130 within Tg, as mutation of one of the two would likely be counter-selected because this would adversely affect hormone production.
The tyrosines Tyr-2554 and Tyr-2747 are also conserved in all the species we studied (except in lamprey Tg in which Tyr-2554 ortholog falls into the unsequenced part of the gene). This suggests that these two tyrosines are evolutionarily significant hormonogenic sites (Fig. 6). This contrasts with the internal hormonogenic tyrosines Tyr-847, -973, -1291, -1448, and -2568 that are poorly conserved in the species we studied. This absence of conservation is consistent with biochemical evidence suggesting that in mammals those tyrosines are not the most widely used for TH synthesis (31). Tg is known to undergo conformational changes in response to different physiological conditions (44–46). It may be possible that in special situations different hormonogenic sites are used, thereby allowing Tg to optimize hormone production, as in iodide deficiency. In this case, it is conceivable that evolution of cysteines or other structural features, along with neighboring hormonogenic tyrosines, might have occurred in parallel to produce these secondary sites of TH formation. Given that one often overlooked function of Tg is iodine storage (23), it may also be possible that some of these sites are more important for this function than for hormone production. If so, then one could anticipate that these “storage” sites are less positionally conserved than the hormonogenic sites as they do not require the precise spatial organization necessary for the coupling reaction to occur.
Conservation of Tg Cysteines
The cysteine residues are highly conserved, with 90% found at the same positions between human, Xenopus, zebrafish, and lamprey Tg. In particular, the motif CWCV found in Tg1 repeats is well conserved. These amino acids are known to form intradomain disulfide bonds that are important for Tg conformation. Interestingly, most cysteine loss between human, Xenopus, zebrafish, and lamprey corresponds to loss of specific disulfide pairs (i.e. two Cys residues e.g. Cys-114 and Cys-120). Note that the absence of one cysteine means loss of a potential disulfide bond, which is expected to relieve selection pressure on the other cysteine, allowing its mutation. Why some specific disulfide bonds appear less important than others will require a three-dimensional structural analysis of the Tg protein. In contrast, the amino acids supporting Tg glycosylation are less conserved, allowing more variability across species.
Conservation of Domains in Vertebrate Tg
It is quite striking that the domain composition of Tg is very much conserved, suggesting a strong selective pressure on the overall organization of the protein inherited from the vertebrate common ancestor (Fig. 6). For this reason it was surprising that an N-terminal signal peptide domain had not previously been described in the Tg sequences from Xenopus or zebrafish. In this study we clearly demonstrated that both Xenopus and zebrafish Tg have conserved the signal peptide domain, and indeed, the secretion properties of Tg from these species are similar to those from mammals (Figs. 3 and 4). Moreover, iodinated proteins have been identified in the thyroid gland lumen of Xenopus laevis (47) and zebrafish (48). Together with our present findings, these data strongly suggest that Tg iodination and TH synthesis occurs in a manner quite similar to that in mammals. Additionally, both the hinge and linker segments that separate the Tg repeats are present in all the species examined, suggesting that they have an important role in protein structure. We predict that both of these segments are structured in α helixes and β sheets and that this molecular architecture is conserved between human, Xenopus, zebrafish, and lamprey (Fig. 1B and supplemental Fig. S4). In particular, the hinge segment between Tg1–10 and Tg2–1 repeats is likely to be structured rather than representing an uncoiled domain. This also applies within the Tg repeats; not only are the three types of repeats (11) present in the Tg of all vertebrates, but also their number and order from N-terminal to C-terminal is consistent. Moreover, within repeat Tg1–3, Tg1–7, and Tg1–8, there are also additional sequences of consistent size (Fig. 1B) (11) that are present in human, Xenopus, zebrafish, and lamprey, suggesting that these regions are required for the biological function of Tg.
It is worth noting that although Tg1 repeats and the ChEL domain can be found in other proteins (e.g. nidogen, acetylcholinesterase), the hinge and linker segments do not resemble a functional domain of any other known proteins. This calls for a deeper investigation of the biological role of those two segments, which could be crucial to understanding the evolution of Tg, especially given that they articulate precisely between distinct repeats in the overall Tg structure.
How Has Tg Evolved?
Lamprey can shed light on the evolution of the Tg protein according to its basal phylogenetic position among vertebrates. The strong conservation of Tg in lampreys when compared with the gnathostomes Tgs is noticeable given the fact that the two clades diverged >500 million years ago, including the divergence in the thyroid gland organization of the lampreys (49). Indeed, in the pharyngeal area of the lamprey larva (the ammocoete), there is an exocrine secretion of iodoprotein containing THs (50) through the endostyle, the longitudinal ciliated groove on the ventral wall of the pharynx. The endostyle may be considered as the equivalent of the thyroid gland as it expresses TTF1/Nkx-2.1, a transcription factor that, along with Pax-8, is required for thyroid specification (51). During metamorphosis, the endostyle becomes reorganized into a true endocrine thyroid gland (37, 50, 52). As the immunolabeling of lamprey Tg remains controversial (52), it remains unclear whether Tg expression starts in the ammocoete endostyle or only in the adult when the gland is fully formed. This point will be critical for further understanding the evolution of TH synthesis. Indeed, a very similarly organized endostyle, with TTF1/Nkx-2.1 expression (53) and iodine accumulation (54), is also found in invertebrate chordates such as amphioxus (55). Thus, it is striking that we are not able to identify any orthologs of Tg outside of vertebrates where THs are known to be important, such as amphioxus (56) or Ciona (57), leaving open the question of the TH precursor protein(s) in these species.
The various domains of Tg are more ancient than Tg itself (17, 58, 59). By studying the evolution of the Tg1 repeats, previous investigations (59) found that these repeats are indeed present in all bilaterian taxa (61). Even if many proteins were found to contain several Tg1 repeats (17, 59) and, to a lesser extent, Tg2 repeats in amphioxus and sea urchin, the identification of those candidates as Tg orthologs remains doubtful. In addition, the ChEL domain is present in many proteins with very different activities, such as acetylcholinesterase itself and also other esterases and (noncatalytic) neuroligins (58).
Our results strongly suggest that the Tg design is a vertebrate-specific novelty originating at the base of this phylum and has undergone only marginal subsequent modification. A peculiarity of vertebrates that is likely to explain the presence of Tg is the thyroid gland itself. Indeed, Tg undergoes a complex trafficking from the thyrocyte to the follicular lumen. Both the signal peptide and the correct folding of Tg is essential for that trafficking (60). The follicular structure is also required as the central repository of iodotyrosines and THs within the body, and as these stores are maintained as organified iodide within Tg, it would be interesting to assess the question of co-evolution of thyroid follicular Tg storage with the iodine storage function.
Alternatively, given the modular structure of Tg and because cysteines form disulfide bonds within the modules, one might speculate the existence of step-by-step modular additions that led to vertebrate Tg. One hypothesis could be that an ancestral gene encoding an iodine storage protein harboring a Tg repeat-like structure allowed intronic sequences to become expressed as linker and hinge segments as part of the Tg coding sequence (41). In the vertebrate lineage, involvement of these sequences in TH synthesis and iodide storage would then prevent them from being lost. Thus starting from a protein with low efficiency, selective forces could have favored the emergence of Tg as observed today. Efforts are still needed to establish or refute the existence of such an early divergent Tg precursor in species such as Amphioxus.
Conclusion and Perspectives
Tg is a peculiar protein with a highly conserved structure suggesting strong selective forces for its preservation among vertebrates. Based on our analysis of a representative mammal, amphibian, teleost fish, and a basal vertebrate, it appears that all domains are conserved throughout the vertebrates. Remarkably, no orthologs of Tg outside of vertebrates have been found. Hence, we support the idea that Tg is a vertebrate novelty. Thus, how THs are synthesized in invertebrate chordates remains a mystery. Understanding the requirement for Tg in TH synthesis may also require investigating the lamprey Tg expression pattern before the endostyle matures into the adult thyroid gland.
Experimental Procedures
Cloning and Expression of Xenopus and Zebrafish Tg cDNAs
RNA from the head of Xenopus (X. tropicalis) and total RNA from zebrafish (D. rerio) were used for retro-transcription using the Super-Script III (Invitrogen) following the manufacturer's protocol.
The cloning of both genes was performed in two steps, the first one for sequencing and sequence checking and the second one for cloning (supplemental Table S1). (i) Primers were designed following the gene predictions available in GenBankTM (XM_689200.4 and XM_002939515.1 for zebrafish and Xenopus, respectively) to obtain 1000-bp-long overlapping amplicons. Amplicons were sequenced by the Sanger method (72) in both directions. For both genes, the full-length sequences were reconstructed from these overlapping partial sequences. (ii) Primers were designed following the sequencing result to obtain an about 2000-bp-long amplicon with 15 overlapping nucleotides at each end of the primers. PCRs were performed with the Phusion High-Fidelity polymerase (Thermo Scientific) following the manufacturer's recommendations for the temperature and time of the various PCR steps (Supplemental Table S1). For each gene, four amplicons were obtained, assembled, and cloned in pSG5 plasmid between EcoRI (5′) and BglII (3′) cloning site by In-Fusion cloning (Thermo Scientific) following the manufacturer's recommendations.
Determination of the 5′-end of Xenopus Tg
We failed to clone the 5′-end of the Xenopus Tg with the method described above. Thus, we used the Teloprime cDNA Amplification kit from Lexogen to find the missing 5′-end encoding the signal peptide. To initiate the reverse transcription step, we used either the reverse transcription primer (RTP) primer from the Lexogen kit or a reverse primer (external primer, supplemental Table S1) designed in the 5′ part of the known Xenopus sequence. The kit was then used following the manufacturer's recommendations. The full-length and partial double-stranded cDNAs obtained were used as the template in all the combinations of PCR and nested PCR amplification events using the forward primer and one of the reverse primers (supplemental Table S1). PCR were performed in 25 μl using 5 units of AmpliTaq Gold DNA Polymerase (Thermo), 2 mm MgCl2, 200 μm concentrations of each dNTP, 0.5 μm primer, and 1 μl of cDNA. The PCR mixture was denatured at 98 °C for 5 min followed by 20 or 40 cycles of 30s at 98 °C, 30s at 55 °C, and 60s at 72 °C. PCR amplification was achieved with a final elongation step of 7 min at 72 °C. In addition to these first amplification round, PCR re-amplifications were conducted using all the combinations of PCR and nested PCR primers by using diluted (1/20) first PCR reactions as template. PCR conditions were the same as described above with 30 cycles of amplification. 24 PCR products were obtained, purified using the Nucleospin. An Ion Torrent library was built using 100 ng of the with the amplicon mix following the manufacturer's protocol (Prepare Amplicon Libraries without Fragmentation Using the Ion Plus Fragment Library kit, Thermofisher). Sequencing of the library was performed resulting from those PCRs sequenced on of a 318v2 chip using an Ion Torrent PG. We compared the sequencing resulting in 618,754 reads that were assembled to determine a consensus sequence corresponding to the 5′ missing extremity. We compared the consensus sequence consensus with the Xenopus genome available on Ensembl to identify the 5′-end of the Xenopus Tg. This region was then chemically synthetized and incorporated in the previous incomplete clone to generate a complete clone encoding the full-length Xenopus Tg.
Search for Tg Sequences in Basal Vertebrate Genomes
A prediction for Tg (ENSPMAG 00000001187) was recovered from the lamprey genome assembly available on ENSEMBL (49). The sequences similarity with Tg was investigating by BLAST. The AUGUSTUS method (61) was run on the containing scaffold GL476337, containing the predicted gene, to improve the protein prediction and increase the sequence length on both N-terminal and C-terminal ends.
Search for Tg Sequences in Non-vertebrate Genomes
To investigate Tg sequences in non-vertebrate genomes, we searched for Tg1, Tg2, or Tg3 repeats using TBLASTN and TBLASTX (62) against some available genomes of tunicates, cephalochordates, echinoderms, hemichordates, mollusks, annelids, cnidarians, and xenocoelomorphs (supplemental Table S2). To assess if the retrieved sequences were orthologs of Tg-domains and not from other proteins, we compare the retrieved sequences with Tg domain of other protein such as Nidogen, SMOC, or TACSTD (supplemental Table S3).
Phylogenetic Analysis of Vertebrate Tg
Different datasets were used for phylogenetic analyses. The first one includes the available amino acid sequences of human, dog, cow, rat, mouse, (supplemental Table S4A), Xenopus, and zebrafish (from the present study), which have been experimentally described and cloned. The second includes also 133 predicted sequences of other vertebrate species available in GenBankTM (supplemental Table S4B) or in Ensembl. All the Tg amino acid sequences were aligned using MUSCLE 3.8 (63). Trees were generated using the maximum likelihood method under a JTT substitution matrix plus an eight-category gamma rate correction (α estimated) with the proportion of invariant sites estimated. Full-length trees were calculated using both cloned and predicted sequences. Trees of specific domains were calculated using cloned sequences. Branch support was estimated with 1000 bootstrap replicates only for the tree with cloned sequences. Approximate likelihood ratio test (aLRT) was used for larger trees with both cloned and predicted proteins (64). Both alignments and tree calculations were performed using the Seaview v4.5 software (65).
Prediction of Secondary Structures and Protein Features
The presence of a signal peptide was investigated using the SignalP 4.1 method (66). Prediction of the secondary structure and identification of α-helices, β-sheet, or coiled region were performed with the SOPMA (67), PHD (68), and Predator (69) methods.
ISH in Zebrafish and Xenopus
Two different probes were tested in zebrafish corresponding to respectively the 5′ (probe 1: 1709 bp long, positions 42–602 in the amino acid sequence; Fig. 1B) and the middle (probe 2: 1994 bp long, positions 1202–1862 in the amino acid sequence) of the Tg coding sequence. One probe was tested in Xenopus in the 5′ of the sequence (1031 bp long, position 148–485). ISHs were performed as previously described (70, 71).
Plasmids and Mutagenesis
Mouse Tg with C-terminal 3×-myc epitope tag was cloned into the pcDNA3.1 expression vector. A cDNA fragment encoding a single-myc epitope tag was introduced in-frame at the C terminus of the incomplete Xenopus Tg cDNA using two primers named Myc1 and Myc2. To introduce into the incomplete Xenopus Tg cDNA the sequence encoding the signal peptide of mouse Tg or that from the complete Xenopus Tg, two KpnI restriction sites were created using four primers: KpnI 1F, KpnI 1R, KpnI 2F, and KpnI 2R. A mouse Tg signal peptide fragment was generated by PCR using primers TgM 1 and TgM 2 and full-length mouse Tg cDNA as a template. The sequence encoding the complete Xenopus Tg signal peptide fragment named Tg spf with the first and seceond methionine residues was introduced via the In-Fusion HD Cloning kit (Clontech). The first methionine was then mutagenized using primers Mut 1F and Mut1R. The mouse Tg-C1043Y and Tg-C1050N mutants were created using two sets of primers, respectively: C1043YF/C1043R and C1050YR/C1050YF. All mutagenesis was performed with the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent). Sequences of primers used for these various steps described above are listed in supplemental Table S5.
Cell Culture and Transfection
293T cells were cultured (37 °C, 5% CO2) in 12-well plates in DMEM with 10% fetal bovine serum and penicillin (100 units/ml) and streptomycin (100 μg/ml) (Thermo Fisher Scientific). Unless otherwise specified, 1 μg of plasmid DNA was transfected per well using Lipofectamine 2000 (Thermo Fisher Scientific) as per the manufacturer's instructions.
Analysis of Tg Protein Synthesis and Secretion
For detection of recombinant Tg expression, at 16 h after transfection, fresh medium was placed in each well and collected for 24 h. The cells were lysed in radioimmune precipitation assay buffer with complete protease inhibitor mixture (Roche Applied Science). For immunoblotting analysis, 2.5 μg of cell lysate proteins per lane and a proportional fraction of conditioned media were resolved by SDS 6% PAGE and electrotransferred to nitrocellulose. Bands were visualized by Western blotting with rabbit anti-myc (Immunology Consultants Laboratory) or rabbit anti Tg with horseradish peroxidase-conjugated secondary antibody (Jackson Immunochemicals) using SuperSignal West Pico Chemiluminescent Substrate (Thermo-Fisher). For endoglycosidase H digestion (New England BioLabs), 10 μg of cell lysate protein and a proportional fraction of media were boiled for 10 min in 0.5% SDS plus 40 mm DTT. After cooling, the samples were digested with 50 units/μl endoglycosidase H in 50 mm sodium acetate, pH 6, or mock-digested for 1 h at 37 °C before SDS-PAGE and immunoblotting.
Author Contributions
G. H., Y. M., P. A., and V. L. wrote the paper. V. L. and P. A. designed the study. G. H. cloned the genes and performed the phylogeny analysis. Y. M. performed the Western blot experiments. M. T., J.-B. F., and B. D. performed the in situ hybridization. T. L. provided assistance in phylogeny analysis. B. G. and S. H. performed the mass sequencing and its analysis. G. D. performed the structural analysis. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Max Telford who kindly investigated xenocoelomorph genomes for Tg1 repeats as well as Sylvie Mazan and Hector Escriva, who granted us access to their chondrichthyes and amphioxus databases. We thank François Penin for fruitful discussion about protein structure. We are grateful to Marie Sémon, Gabriel Markov, and Masatoshi Matsunami for suggestions on phylogeny building as well as the Pôle Scientifique de Modélisation numérique (PSMN) of the ENS de Lyon for computing resources. We thank Pauline Salis for critical review. The sequencing of the 5′ Tg of Xenopus was performed in the Sequencing Platform of the IGFL.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK40344 (to P. A.). We also thank the Ecole Normale Supérieure de Lyon (ENS) and CNRS for financial support. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S5 and Figs. S1–S6.
- TH
- thyroid hormone
- Tg
- thyroglobulin
- Tg1
- Tg2, Tg3, Tg types 1, 2, and 3, respectively
- ChEL
- cholinesterase-like
- ISH
- in situ hybridization
- hpf
- hours post-fertilization.
References
- 1. Holzer G., and Laudet V. (2015) Thyroid hormones: a triple-edged sword for life history transitions. Curr. Biol. 25, R344–R347 [DOI] [PubMed] [Google Scholar]
- 2. Tata J. (2013) The road to nuclear receptors of thyroid hormone. Biochim. Biophys. Acta 1830, 3860–3866 [DOI] [PubMed] [Google Scholar]
- 3. Laudet V. (2011) The origins and evolution of vertebrate metamorphosis. Curr. Biol. 21, R726–R737 [DOI] [PubMed] [Google Scholar]
- 4. Dardente H., Hazlerigg D. G., and Ebling F. J. (2014) Thyroid hormone and seasonal rhythmicity. Front. Endocrinol. (Lausanne) 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mullur R., Liu Y.-Y., and Brent G. A. (2014) Thyroid hormone regulation of metabolism. Physiol. Rev. 94, 355–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williams G. R. (2014) Role of thyroid hormone receptor-α1 in endochondral ossification. Endocrinology 155, 2747–2750 [DOI] [PubMed] [Google Scholar]
- 7. Gong D. W., He Y., Karas M., and Reitman M. (1997) Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, β3-adrenergic agonists, and leptin. J. Biol. Chem. 272, 24129–24132 [DOI] [PubMed] [Google Scholar]
- 8. Little A. G., Kunisue T., Kannan K., and Seebacher F. (2013) Thyroid hormone actions are temperature-specific and regulate thermal acclimation in zebrafish (Danio rerio). BMC Biol. 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hollowell J. G., Staehling N. W., Flanders W. D., Hannon W. H., Gunter E. W., Spencer C. A., and Braverman L. E. (2002) Serum TSH, T4, and thyroid antibodies in the United States population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J. Clin. Endocrinol. Metab. 87, 489–499 [DOI] [PubMed] [Google Scholar]
- 10. Miller A. E., and Heyland A. (2013) Iodine accumulation in sea urchin larvae is dependent on peroxide. J. Exp. Biol. 216, 915–926 [DOI] [PubMed] [Google Scholar]
- 11. Malthiéry Y., and Lissitzky S. (1987) Primary structure of human thyroglobulin deduced from the sequence of its 8448 base complementary DNA. Eur. J. Biochem. 165, 491–498 [DOI] [PubMed] [Google Scholar]
- 12. van de Graaf S. A., Ris-Stalpers C., Pauws E., Mendive F. M., Targovnik H. M., and de Vijlder J. J. (2001) Up to date with human thyroglobulin. J. Endocrinol. 170, 307–321 [DOI] [PubMed] [Google Scholar]
- 13. Kim P. S., Ding M., Menon S., Jung C. G., Cheng J. M., Miyamoto T., Li B., Furudate S., and Agui T. (2000) A missense mutation G2320R in the thyroglobulin gene causes non-goitrous congenital primary hypothyroidism in the WIC-rdw rat. Mol. Endocrinol. 14, 1944–1953 [DOI] [PubMed] [Google Scholar]
- 14. Lee J.-Y., Uzuka Y., Tanabe S., Sarashina T., Suzuki H., and Sato M. (2007) Cloning and characterization of canine thyroglobulin complementary DNA. Domest. Anim. Endocrinol. 32, 178–189 [DOI] [PubMed] [Google Scholar]
- 15. Mercken L., Simons M. J., Swillens S., Massaer M., and Vassart G. (1985) Primary structure of bovine thyroglobulin deduced from the sequence of its 8,431-base complementary DNA. Nature 316, 647–651 [DOI] [PubMed] [Google Scholar]
- 16. Monaco F., Dominici R., Andreoli M., de Pirro R., and Roche J. (1981) Thyroid hormone formation in thyroglobulin synthesized in the amphioxus (Branchiostoma lanceolatum pallas). Comp. Biochem. Physiol. B 70, 341–343 [Google Scholar]
- 17. Belkadi A., Jacques C., Savagner F., and Malthièry Y. (2012) Phylogenetic analysis of the human thyroglobulin regions. Thyroid Res. 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown D. D., and Cai L. (2007) Amphibian metamorphosis. Dev. Biol. 306, 20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J., and Arvan P. (2011) Repeat motif-containing regions within thyroglobulin. J. Biol. Chem. 286, 26327–26333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park Y. N., and Arvan P. (2004) The acetylcholinesterase homology region is essential for normal conformational maturation and secretion of thyroglobulin. J. Biol. Chem. 279, 17085–17089 [DOI] [PubMed] [Google Scholar]
- 21. Dunn J. T., Anderson P. C., Fox J. W., Fassler C. A., Dunn A. D., Hite L. A., and Moore R. C. (1987) The sites of thyroid hormone formation in rabbit thyroglobulin. J. Biol. Chem. 262, 16948–16952 [PubMed] [Google Scholar]
- 22. Lamas L., Anderson P. C., Fox J. W., and Dunn J. T. (1989) Consensus sequences for early iodination and hormonogenesis in human thyroglobulin. J. Biol. Chem. 264, 13541–13545 [PubMed] [Google Scholar]
- 23. Di Jeso B., and Arvan P. (2016) Thyroglobulin from molecular and cellular biology to clinical endocrinology. Endocr. Rev. 37, 2–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang S. X., Pollock H. G., and Rawitch A. B. (1996) Glycosylation in human thyroglobulin: location of the N-linked oligosaccharide units and comparison with bovine thyroglobulin. Arch. Biochem. Biophys. 327, 61–70 [DOI] [PubMed] [Google Scholar]
- 25. Van Herle A. J., Vassart G., and Dumont J. E. (1979) Control of thyroglobulin synthesis and secretion (first of two parts). N. Engl. J. Med. 301, 239–249 [DOI] [PubMed] [Google Scholar]
- 26. Van Herle A. J., Vassart G., and Dumont J. E. (1979) Control of thyroglobulin synthesis and secretion (second of two parts). N. Engl. J. Med. 301, 307–314 [DOI] [PubMed] [Google Scholar]
- 27. Kim P. S., and Arvan P. (1991) Folding and assembly of newly synthesized thyroglobulin occurs in a pre-Golgi compartment. J. Biol. Chem. 266, 12412–12418 [PubMed] [Google Scholar]
- 28. Ris-Stalpers C., and Bikker H. (2010) Genetics and phenomics of hypothyroidism and goiter due to TPO mutations. Mol. Cell. Endocrinol. 322, 38–43 [DOI] [PubMed] [Google Scholar]
- 29. Dunn A. D., Corsi C. M., Myers H. E., and Dunn J. T. (1998) Tyrosine 130 is an important outer ring donor for thyroxine formation in thyroglobulin. J. Biol. Chem. 273, 25223–25229 [DOI] [PubMed] [Google Scholar]
- 30. Palumbo G., Gentile F., Condorelli G. L., and Salvatore G. (1990) The earliest site of iodination in thyroglobulin is residue number 5. J. Biol. Chem. 265, 8887–8892 [PubMed] [Google Scholar]
- 31. Ohmiya Y., Hayashi H., Kondo T., and Kondo Y. (1990) Location of dehydroalanine residues in the amino acid sequence of bovine thyroglobulin. Identification of “donor” tyrosine sites for hormonogenesis in thyroglobulin. J. Biol. Chem. 265, 9066–9071 [PubMed] [Google Scholar]
- 32. Brix K., Lemansky P., and Herzog V. (1996) Evidence for extracellularly acting cathepsins mediating thyroid hormone liberation in thyroid epithelial cells. Endocrinology 137, 1963–1974 [DOI] [PubMed] [Google Scholar]
- 33. Rubio I. G., and Medeiros-Neto G. (2009) Mutations of the thyroglobulin gene and its relevance to thyroid disorders. Curr. Opin. Endocrinol. Diabetes Obes. 16, 373–378 [DOI] [PubMed] [Google Scholar]
- 34. Wu H., Suzuki S., Sellitti D. F., Doi S. Q., Tanigawa K., Aizawa S., Akama T., Kawashima A., Mishima M., Ishii N., Yoshida A., Hisatome I., Koles N. L., Katoh R., and Suzuki K. (2009) Expression of a thyroglobulin (Tg) variant in mouse kidney glomerulus. Biochem. Biophys. Res. Commun. 389, 269–273 [DOI] [PubMed] [Google Scholar]
- 35. Leloup J., and Buscaglia M. (1977) La triiodothyronine, hormone de la métamorphose des Amphibiens. C. R. Acad. Sci. (Paris) 284, 2261–2263 [Google Scholar]
- 36. Baïsse B., Galisson F., Giraud S., Schapira M., and Spertini O. (2007) Evolutionary conservation of P-selectin glycoprotein ligand-1 primary structure and function. BMC Evol. Biol. 7, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suzuki S., and Kondo Y. (1973) Thyroidal morphogenesis and biosynthesis of thyroglobulin before and after metamorphasis in the lamprey, Lampetra reissneri. Gen. Comp. Endocrinol. 21, 451–460 [DOI] [PubMed] [Google Scholar]
- 38. Wright G. M., Filosa M. F., and Youson J. H. (1980) Immunocytochemical localization of thyroglobulin in the transforming endostyle of anadromous sea lampreys, Petromyzon marinus L., during metamorphosis. Gen. Comp. Endocrinol. 42, 187–194 [DOI] [PubMed] [Google Scholar]
- 39. Monaco F., Andreoli M., La Posta A., and Roche J. (1978) Thyroglobulin biosynthesis in a larval (ammocoete) and adult freshwater lamprey (Lampetra planeri Bl). Comp. Biochem. Physiol. B 60, 87–91 [DOI] [PubMed] [Google Scholar]
- 40. Shen X.-X., Liang D., Wen J.-Z., and Zhang P. (2011) Multiple genome alignments facilitate development of npcl markers: a case study of tetrapod phylogeny focusing on the position of turtles. Mol. Biol. Evol. 28, 3237–3252 [DOI] [PubMed] [Google Scholar]
- 41. Cortesi F., Musilová Z., Stieb S. M., Hart N. S., Siebeck U. E., Malmstrøm M., Tørresen O. K., Jentoft S., Cheney K. L., Marshall N. J., Carleton K. L., and Salzburger W. (2015) Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc. Natl. Acad. Sci. U.S.A. 112, 1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paris M., Brunet F., Markov G. V., Schubert M., and Laudet V. (2008) The amphioxus genome enlightens the evolution of the thyroid hormone signaling pathway. Dev. Genes Evol. 218, 667–680 [DOI] [PubMed] [Google Scholar]
- 43. Dunn J. T., and Dunn A. D. (1999) The importance of thyroglobulin structure for thyroid hormone biosynthesis. Biochimie 81, 505–509 [DOI] [PubMed] [Google Scholar]
- 44. Dunn J. T., and Ray S. C. (1975) Changes in the structure of thyroglobulin following the administration of thyroid-stimulating hormone. J. Biol. Chem. 250, 5801–5807 [PubMed] [Google Scholar]
- 45. Berg G., and Ekholm R. (1975) Electron microscopy of low iodinated thyroglobulin molecules. Biochim. Biophys. Acta 386, 422–431 [DOI] [PubMed] [Google Scholar]
- 46. Formisano S., Di Jeso B., Acquaviva R., Consiglio E., and Palumbo G. (1983) Calcium-induced changes in thyroglobulin conformation. Arch. Biochem. Biophys. 227, 351–357 [DOI] [PubMed] [Google Scholar]
- 47. Neuenschwander P. (1972) Uhrastruktur und Jodaufnahme der Schilddriise bei Larven des Krallenfrosches (Xenopus laevis Daud.). Z. Zellforsch. 130, 553–574 [PubMed] [Google Scholar]
- 48. Brown D. (1997) The role of thyroid hormone in zebrafish and axolotl development. Proc. Natl. Acad. Sci. U.S.A. 94, 13011–13016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith J. J., Kuraku S., Holt C., Sauka-Spengler T., Jiang N., Campbell M. S., Yandell M. D., Manousaki T., Meyer A., Bloom O. E., Morgan J. R., Buxbaum J. D., Sachidanandam R., Sims C., Garruss A. S., et al. (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 45, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Youson J. H. (1997) Is lamprey metamorphosis regulated by thyroid hormones? Am. Zool. 37, 441–460 [Google Scholar]
- 51. Kimura S. (1996) Thyroid-specific enhancer-binding protein role in thyroid function and organogenesis. Trends Endocrinol. Metab. 7, 247–252 [DOI] [PubMed] [Google Scholar]
- 52. Kluge B., Renault N., and Rohr K. (2005) Anatomical and molecular reinvestigation of lamprey endostyle development provides new insight into thyroid gland evolution. Dev. Genes Evol. 215, 32–40 [DOI] [PubMed] [Google Scholar]
- 53. Ogasawara M., Shigetani Y., Suzuki S., Kuratani S., and Satoh N. (2001) Expression of thyroid transcription factor-1 (TTF-1) gene in the ventral forebrain and endostyle of the agnathan vertebrate Lampetra japonica. Genesis 30, 51–58 [DOI] [PubMed] [Google Scholar]
- 54. Barrington E. J. W. (1958) The localization of organically bound iodine in the endostyle of amphioxus. J. Mar. Biol. Assoc. U.K. 37, 117–126 [Google Scholar]
- 55. Hiruta J., Mazet F., Yasui K., Zhang P., and Ogasawara M. (2005) Comparative expression analysis of transcription factor genes in the endostyle of invertebrate chordates. Dev. Dyn. 233, 1031–1037 [DOI] [PubMed] [Google Scholar]
- 56. Paris M., Escriva H., Schubert M., Brunet F., Brtko J., Ciesielski F., Roecklin D., Vivat-Hannah V., Jamin E. L., Cravedi J.-P., Scanlan T. S., Renaud J.-P., Holland N. D., and Laudet V. (2008) Amphioxus postembryonic development reveals the homology of chordate metamorphosis. Curr. Biol. 18, 825–830 [DOI] [PubMed] [Google Scholar]
- 57. Patricolo E., Cammarata M., and D'Agati P. (2001) Presence of thyroid hormones in ascidian larvae and their involvement in metamorphosis. J. Exp. Zool. 290, 426–430 [DOI] [PubMed] [Google Scholar]
- 58. Krejci E., Duval N., Chatonnet A., Vincens P., and Massoulié J. (1991) Cholinesterase-like domains in enzymes and structural proteins: functional and evolutionary relationships and identification of a catalytically essential aspartic acid. Proc. Natl. Acad. Sci. U.S.A. 88, 6647–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Novinec M., Kordis D., Turk V., and Lenarcic B. (2006) Diversity and evolution of the thyroglobulin type-1 domain superfamily. Mol. Biol. Evol. 23, 744–755 [DOI] [PubMed] [Google Scholar]
- 60. Arvan P., Kim P. S., Kuliawat R., Prabakaran D., Muresan Z., Yoo S. E., and Abu Hossain S. (1997) Intracellular protein transport to the thyrocyte plasma membrane: potential implications for thyroid physiology. Thyroid 7, 89–105 [DOI] [PubMed] [Google Scholar]
- 61. Sommerfeld D., Lingner T., Stanke M., Morgenstern B., and Richter H. (2009) AUGUSTUS at MediGRID: adaption of a bioinformatics application to grid computing for efficient genome analysis. Future Gener. Comput. Syst. 25, 337–345 [Google Scholar]
- 62. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Edgar R. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Anisimova M., Gil M., Dufayard J.-F. F., Dessimoz C., and Gascuel O. (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gouy M., Guindon S., and Gascuel O. (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 [DOI] [PubMed] [Google Scholar]
- 66. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 67. Geourjon C., and Deléage G. (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11, 681–684 [DOI] [PubMed] [Google Scholar]
- 68. Rost B., Sander C., and Schneider R. (1994) PHD, an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 10, 53–60 [DOI] [PubMed] [Google Scholar]
- 69. Frishman D., and Argos P. (1996) Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 9, 133–142 [DOI] [PubMed] [Google Scholar]
- 70. Fini J. B., Le Mével S., Palmier K., Darras V. M., Punzon I., Richardson S. J., Clerget-Froidevaux M. S., and Demeneix B. A. (2012) Thyroid hormone signaling in the Xenopus laevis embryo is functional and susceptible to endocrine disruption. Endocrinology 153, 5068–5081 [DOI] [PubMed] [Google Scholar]
- 71. Gibert Y., Samarut E., Pasco-Viel E., Bernard L., Borday-Birraux V., Sadier A., Labbé C., Viriot L., and Laudet V. (2015) Altered retinoic acid signalling underpins dentition evolution. Proc. Biol. Sci. 282, 20142764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smith L. M., Sanders J. Z., Kaiser R. J., Hughes P., Dodd C., Connell C. R., Heiner C., Kent S. B. H., and Hood L. E. (1986) Fluorescence detection in automated DNA sequence analysis. Nature 321, 674–679 [DOI] [PubMed] [Google Scholar]
- 73. Faber J., and Nieuwkoop P. D. (1994) Normal Table of Xenopus Laevis (Daudin), Garland Publishing, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.