Abstract
β-Thalassemia (β-Thal) is one of the most common genetic diseases in the world. The generation of patient-specific β-Thal-induced pluripotent stem cells (iPSCs), correction of the disease-causing mutations in those cells, and then differentiation into hematopoietic stem cells offers a new therapeutic strategy for this disease. Here, we designed a CRISPR/Cas9 to specifically target the Homo sapiens hemoglobin β (HBB) gene CD41/42(−CTTT) mutation. We demonstrated that the combination of single strand oligodeoxynucleotides with CRISPR/Cas9 was capable of correcting the HBB gene CD41/42 mutation in β-Thal iPSCs. After applying a correction-specific PCR assay to purify the corrected clones followed by sequencing to confirm mutation correction, we verified that the purified clones retained full pluripotency and exhibited normal karyotyping. Additionally, whole-exome sequencing showed that the mutation load to the exomes was minimal after CRISPR/Cas9 targeting. Furthermore, the corrected iPSCs were selected for erythroblast differentiation and restored the expression of HBB protein compared with the parental iPSCs. This method provides an efficient and safe strategy to correct the HBB gene mutation in β-Thal iPSCs.
Keywords: cell therapy, CRISPR/Cas, genetic disease, hemoglobin, induced pluripotent stem cell (iPS cell) (iPSC)
Introduction
β-Thalassemia (β-Thal)2 is one of the most common genetic diseases worldwide (1–3). The point mutations or small deletions in the HBB gene that affect mRNA transcription, splicing, or translation eventually lead to deficiency in β-hemoglobin and cause β-Thal. In China, CD41/42(−CTTT), CD17(A→T), and IVS2–654(C→T) constitute frameshift or point mutations in β-globin and are three of the most common β-Thal mutations (3). The 4-bp deletion (−CTTT) at CD41/42 represents the most common β-Thal mutation in Southeast Asia and is shared with the Southern Chinese (4). Currently, hematopoietic stem cell (HSC) transplantation is the only way to cure β-Thal; however, the source of human leukocyte antigen-matched healthy donors is limited. Induced pluripotent stem cells (iPSCs) derived from patient somatic cells, which can self-renew indefinitely without losing the ability to differentiate into all cell types (5–9) and hold great promise for regenerative medicine, represent an ideal cell population for in situ correction of disease-causing mutations (10, 11). Generation of β-Thal patient iPSCs, correction of the mutations housed in those iPSCs, and subsequent differentiation into HSCs offer an opportunity for autologous transplantation for disease treatment (12–18). Recently, the development of gene editing to correct the HBB mutation in β-Thal iPSCs followed by differentiation of the corrected iPSCs into HSCs offered a new therapeutic option for those who do not have a bone marrow match to that of potential donors (12, 16, 17).
Recently, the RNA-guided Cas9 nuclease from microbial clustered regularly interspaced short palindromic repeats (CRISPR) was used to facilitate efficient genome engineering in eukaryotic cells by specifying a 20-nucleotide-targeting sequence within its template RNA (18, 19). However, Cas9-mediated genome editing via non-homologous end joining or homology-directed repair in mammalian cells in the presence of donor vector DNA can repair damaged or mutated DNA and facilitate sequence editing (19).
Traditional methods of gene correction rely on antibiotic selection and then removal of the marker, which is a lengthy process that increases the risk of generating additional genome instability in human iPSCs (20). CRISPR/Cas9-induced genome editing was recently performed in β-Thal iPSCs using a piggyBac or donor vector as the repair template (15, 16). Previous studies demonstrated that single strand oligodeoxynucleotides (ssODNs) could be used as templates to generate point mutations and short sequence insertions in human cells and animal models (21–23); however, no studies have been reported showing the efficacy of using ssODNs as a template to correct the HBB mutation in β-Thal patient-specific iPSCs.
Here, we describe correction of the HBB CD41/42(−CTTT) mutation in β-Thal iPSCs by mutation-specific CRISPR/Cas9 gRNA and ssODNs. After correction, the iPSCs still retained pluripotency and multidifferentiation ability. Whole-exome sequencing showed that the mutation load associated with gene editing in the iPSCs was minimal. When the corrected iPSCs differentiated into erythrocytes, we verified the restoration of HBB protein expression. Our results indicated that the combination of ssODNs with CRISPR/Cas9-mediated genome editing was capable of correcting a human genetic mutation and may be useful for future therapeutic applications.
Results
The gRNA Design and Activity Assay
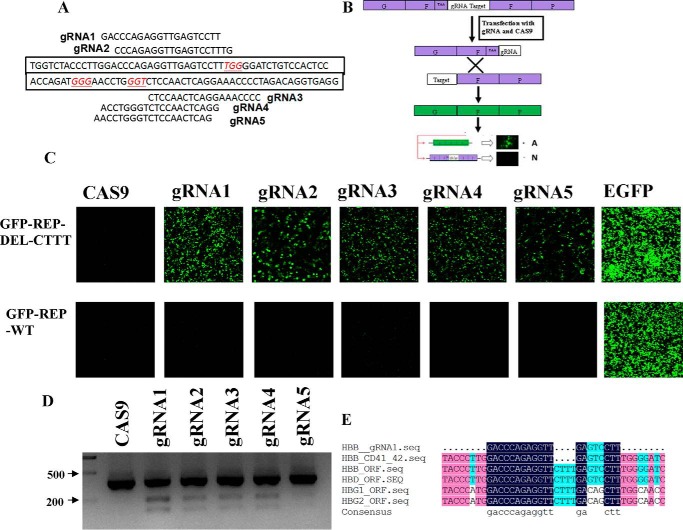
To correct the HBB gene CD41/42(−CTTT) mutation in β-Thal iPSCs in situ, we designed five gRNAs that recognized the specific CD41/42 mutation sequence of HBB gene (Fig. 1A). To test the specific cleavage activity of the designed gRNAs, we constructed two GFP reporter vectors that included the target sequence: one contained the mutation sequence, named GFP-REP-del-CTTT, and the other contained the wild-type sequence (named GFP-REP-WT) associated with the HBB gene at the CD41/42 site. Two 250-bp segments of the GFP-coding sequence flanking each side of the CRISPR gRNA-targeting sequence and the 5′ end containing the stop codon were amplified (Fig. 1B). Once the target sequence was cleaved by Cas9, the duplicated homologous sequence recombined into a full-length GFP, which could be detected to evaluate the efficiency of the designed gRNAs. Thus, we transfected the GFP-REP-del-CTTT reporter together with the Cas9 gRNAs into 293T cells and observed that the GFP signals were significantly increased compared with that of the GFP reporter transfected with only Cas9. These results demonstrated that the gRNAs efficiently targeted the reporter sequence, whereas the GFP-REP-WT transfected along with the Cas9 gRNAs showed no activity. This indicated that the mutation-specific gRNAs were unable to bind to the wild-type sequence, suggesting that the gRNA binding to the mutation sequence was specific (Fig. 1C).
FIGURE 1.
Site-specific gRNA design and activity assay. A, the target sequences of five gRNAs of the mutated HBB gene CD41/42 site. B, schematic of the GFP reporter assay. C, top, fluorescence images of 293T cells co-transfected with Cas9 gRNA and GFP-REP-del-CTTT. Bottom, fluorescence images of the co-transfected cells 48 h later. D, T7 endonuclease 1 assay to assess Cas9 gRNA activity in β-iPS-41/42 cells. E, the alignment of the gRNA1 target sequence (seq), HBB CD41/42 mutation, HBB WT, HBD, HBG1, and HBG2 sequences.
To further confirm the Cas9 gRNA cleavage of the genome from β-iPS-41/42 cells, the Cas9 gRNA vector was transfected for 1 day and underwent puromycin selection for another 2 days. Then the genomic DNA was extracted and PCR-amplified, and the T7 endonuclease 1 assay was performed, revealing that gRNA1 showed the highest activity (Fig. 1D). Because HBB, HBD, HBG1, and HBG2 are highly homologous, we aligned the mRNA sequences with the CD41/42 mutation sequence and the gRNA1 sequence; the alignment showed that there is a 4-base gap between the gRNA1 sequence and the wild-type sequence at the CD41/42 site (Fig. 1E).
Picking and Sequencing Corrected Clones
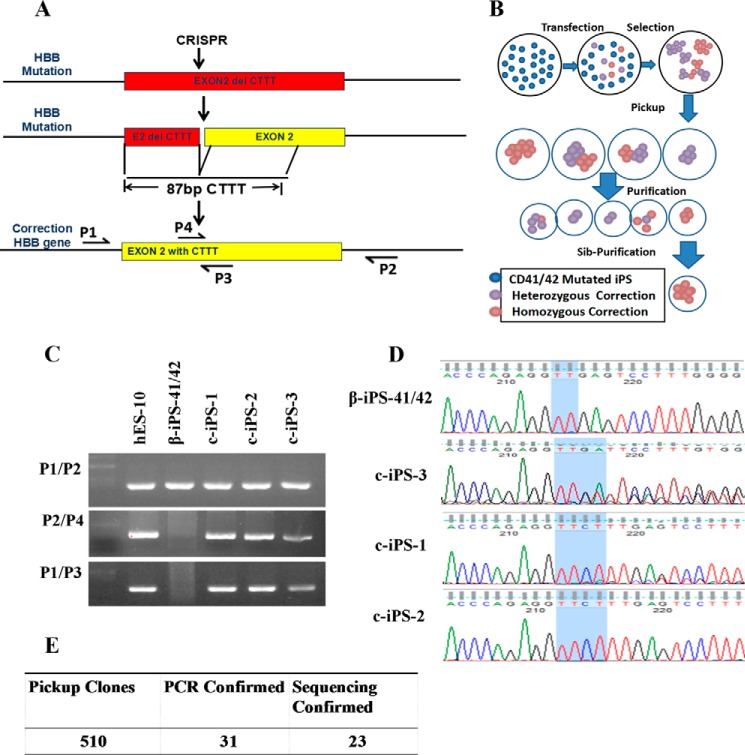
To correct the CTTT deletion in HBB gene in β-Thal iPSCs, the β-iPS-41/42 cells were transfected with ssODNs and Cas9 gRNA1 vectors for 1 day followed by puromycin (0.5 μg/ml) selection for 2 days (Fig. 2, A and B). To obtain the corrected clone, we selected it from hundreds of parental cells and used PCR analysis to detect the corrected clones. We designed a common primer pair (P1 and P2) and an allele-specific primer pair (P3 and P4), which distinguish the mutation and corrected DNA sequences, and they were used to screen for the gene-corrected clones. Only on-target homologous recombination events are detected using this strategy when primers P1 and P3 or P2 and P4 were combined because the PCR products included the donor sequence and genomic DNA sequences beyond the homologous recombination junction (Fig. 2C). The PCR products of the P1 and P2 primers were used for sequencing to confirm the gene correction (Fig. 2D). The results showed that some clones had heterozygous corrections, such as c-iPS-3, and some had homozygous corrections. We chose two homologous clones, c-iPS-1 and c-iPS-2, for use in subsequent experiments. Of the 510 clones tested, the PCR assay demonstrated that 31 clones showed the specific bands; among them, sequencing confirmed that 23 were corrected clones (Fig. 2E).
FIGURE 2.
Gene correction of the β-iPS-41/42 cell mutation. A, schematic overview of the gene-targeting strategy for the human HBB CD41/42 mutation. B, schematic overview of the gene-corrected clone purification method. Sib-Purification means sibling purification. C, PCR results showing the corrected clones. Primers P1 and P2 flank ssODNs and amplified a 414-bp band; primers P3 and P4 specifically anneal to the corrected gene sequence, which contained the CTTT sequence. D, sequencing results of the CD41/42 mutation site before and after gene correction. β-iPS-41/42 cells showed just TT. The c-iPS-3 clone results indicate heterozygous correction and the CTTT mutation. Both c-iPS-1 and c-iPS-2 clones showed homozygous correction. E, of 510 clones, 31 demonstrated successful gene correction. After sequencing, 23 remained corrected.
Pluripotency of Gene-corrected β-Thal iPSCs
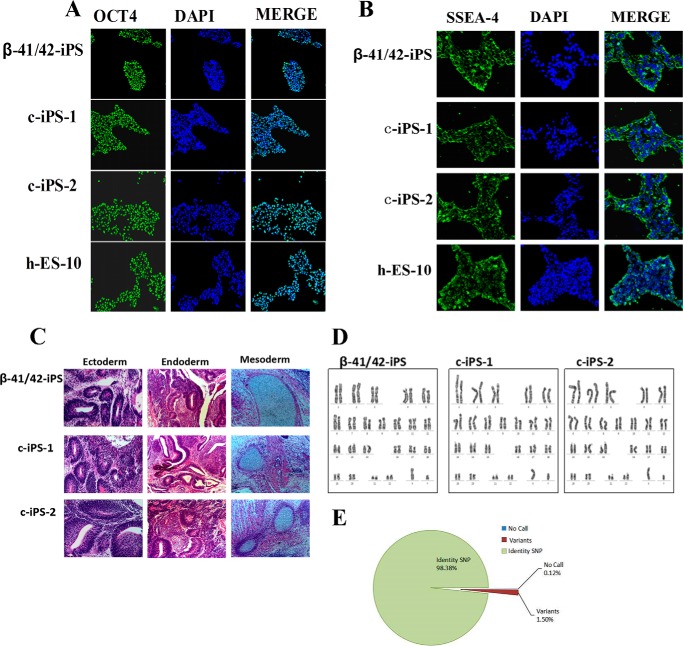
To characterize the pluripotency of the gene-corrected β-iPS-41/42 clones, immunostaining was performed to detect pluripotent gene expression. The results showed that typical pluripotent markers, such as OCT4 and SSEA-4, were expressed in both the c-iPS-1 and c-iPS-2 clones, indicating that they maintained pluripotency gene correction (Fig. 3, A and B). Injection of 107 gene-corrected β-Thal iPSCs into immunodeficient mice resulted in initiation of teratoma formation displaying all three germ layers (Fig. 3C), demonstrating that these cells maintained their pluripotency well after gene targeting. To confirm that all three cell lines were similarly derived from the same origin, we compared 749,157 SNP sites among the β-iPS-41/42, c-iPS-1, and c-iPS-2 cells to determine their identity. As shown in Fig. 3E, 98.38% of the interrogated SNP probes on the array were consistent, and 0.12% failed to be detected. Only 1.5% of the SNP sites appeared to have a genotype different from others. These results suggested that the c-iPS-1, c-iPS-2, and β-iPS-41/42 cells shared the same origin (Fig. 3E).
FIGURE 3.
Pluripotency of gene-corrected β-Thal iPSCs. A and B, immunostaining for OCT4 and SSEA-4 showed that the β-iPS-41/42 cells and the two corrected iPSC clones (c-iPS-1 and c-iPS-2) retained pluripotency. hES-10 was a positive control. C, hematoxylin and eosin staining of teratomas derived from the β-iPS-41/42 cells and the gene-corrected c-iPS-1 and c-iPS-2 cells. D, normal karyotypes of β-iPS-41/42 cells and the gene-corrected c-iPS-1 and c-iPS-2 clones. E, pie chart showing the ratio of the identities among the three iPS cell lines according to 749,157 genome-wide SNP probes.
The Whole-exome Sequence and Copy Number Variation (CNV) Detection
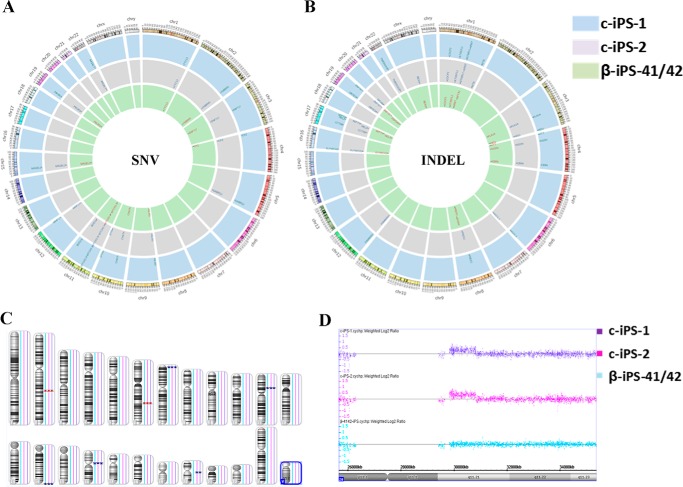
One important concern of CRISPR in gene editing is off-target cleavage because it might introduce extra mutations into the genome, especially into the exome regions. To investigate the effects of gene correction on exome integrity in iPSCs, exome sequencing and high resolution CNV and SNP genotyping (Affymetrix Cytoscan HD array) were performed on the β-iPS-41/42, c-iPS-1, and c-iPS-2 cells. First, we detected the predicted off-target sites in the exome; the data showed no obvious mutations in these sites compared with parental cells (supplemental Fig. 1 and supplemental Table 1). Using stringent criteria to eliminate bias from the sequencing process, we detected 15, 15, and 10 single nucleotide variants (SNVs) when comparing c-iPS-1, c-iPS-2, and β-41/42-iPS cell lines with the hg19 reference human genome (Fig. 4A and supplemental Table 2). Five SNVs in the exome regions of GABRG2, SPRN, BUD13, MMP17, and A4GALT were introduced into c-iPS-1 and c-iPS-2 cells, whereas the remaining 10 SNVs were present in all three iPSCs lines, suggesting that they were carried over from the originating patient genome or as a consequence of iPSC induction. However, our analysis also detected small insertions and deletions (indels) (Fig. 4B). As compared with β-41/42-iPS, 11 and 12 indels were generated during gene editing in c-iPS-1 and c-iPS-2, respectively. These indel-related genes are presented in supplemental Table 2.
FIGURE 4.
The whole-exome sequencing of the parental and gene-corrected iPSCs. A, single nucleotide variations in iPSCs by exome sequencing. B, insertions or deletions in the gene-corrected iPSCs by exome sequencing. C, genomic representation demonstrates the difference in the number of parental and gene-corrected iPSCs. Blue boxes represent amplifications, and red boxes represent deletions with sizes varying by CNV coverage. D, Chromosome Analysis Suite (ChAS) 2.0 data browser representation of the only de novo microduplication of chromosome 20 in gene-corrected iPSCs compared with its parental cells.
Fig. 4C shows a map of the CNVs identified in the samples. As compared with β-41/42-iPS, there was only one de novo CNV detected in both gene-corrected cell lines, and it involved a 990-kb segment located in the area of Chr20q21,11. The other seven CNVs were inherited from the parental cells. The induced variant region contained 30 genes, including two OMIM genes, COX4I2 (OMIM607976) and MYLK2 (OMIM606566), and others such as HM13, ID1, BCL2L1, TPX2, FOXS1, PDRG1, HCK, and PLAGL2. Mutations in the OMIM genes are associated with disease; however, the effect of the gene dosage increase is currently unknown.
We also observed that not only all SNV and CNV candidates but also 75% of indels appeared in both corrected lines, suggesting that the mutations were not induced in a random manner. Gene Ontology (GO) shows the functions of these genes (supplemental Table 2); however, indel calling exhibits a high false-positive rate, which is a common technical difficulty associated with current forms of whole-genome sequencing (WGS). Subsequent analysis was required to filter out most of the false-positive candidates to estimate the number of real indels.
The Differentiation of iPSCs to HSCs and Erythrocytes
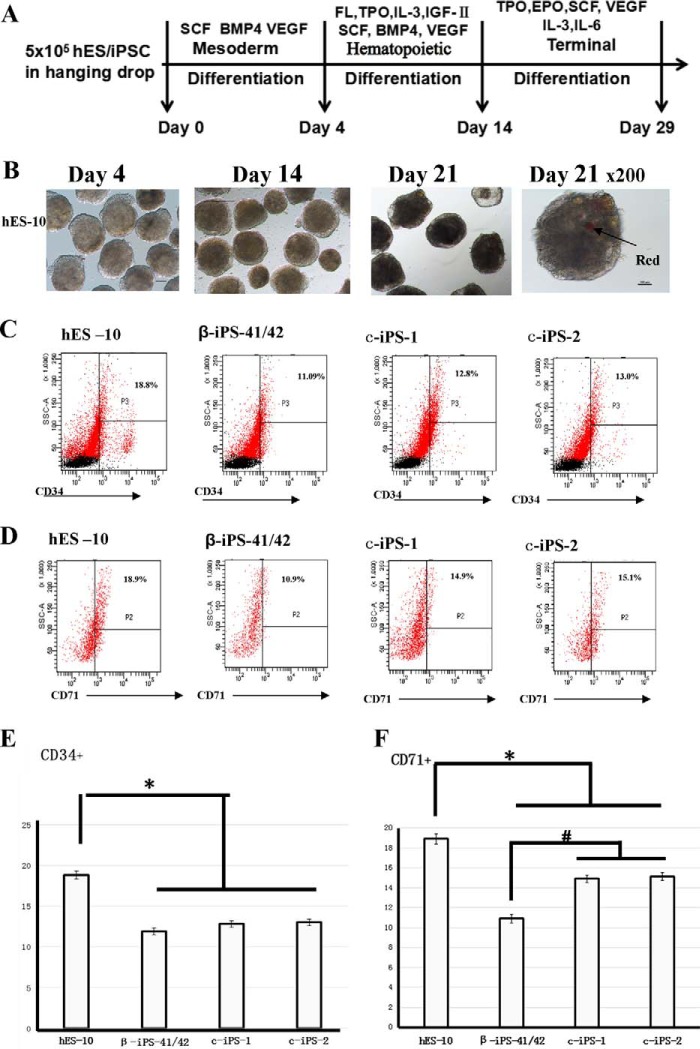
To determine whether the correction of disease-causing mutations in β-Thal iPSCs restored the function of the HBB gene in the form of expression of full-length β-globin protein, we used an embryoid body (EB) strategy in combination with a chemically defined system to examine the hematopoietic differentiation efficiency of human embryonic stem cells (hESCs) (hESC line 10 (hES-10)), β-iPS-41/42, and the two corrected iPS cell lines c-iPSC-1 and c-iPSC-2 into HSCs and erythrocytes. The schematic diagram of the protocol for hematopoietic differentiation is shown in Fig. 5A. The kinetic change of EB morphology was imaged during the time course of EB formation; as time passed, EBs took shape, became larger, and matured (Fig. 5B). After 14 days of differentiation, all of the cell lines could rapidly differentiate and produce CD34+ HSCs (Fig. 5C), and the erythroid precursors were detected by CD71 at day 21 (Fig. 5D). These results showed that, compared with the hES-10 cells, the differentiation efficiency of the iPSCs was lower than that of the hES-10 group, and there was no increase in the corrected iPSCs compared with the β-iPS-41/42 cells (Fig. 5E). The differentiated erythrocytes, CD71+ cells, and β-iPS-41/42 cells showed lower differentiation efficiency compared with the hES-10 and the corrected iPSCs (p < 0.05). Compared with the β-iPS-41/42 cells, the differentiation efficiency was improved in the two gene-corrected cells, c-iPS-1 and c-iPS-2 (p < 0.05; Fig. 5F). To determine whether the difference in differentiation efficiency among the cells was caused by the ability of the cells to maintain their pluripotency, we measured the expression levels of pluripotency-related genes in cells that had differentiated for 21 days. The real time PCR results showed that all of the differentiated cells had lost pluripotent gene expression, and the expression of differentiation-associated genes increased dramatically (Fig. 6A).
FIGURE 5.
Differentiation of β-Thal iPSCs and corrected iPSCs. A, schematic view of in vitro hematopoietic differentiation. Sequential stages are mesoderm induction, hematopoietic stem/progenitor cell emergence, and terminal differentiation. B, bright field images of β-iPS-41/42 cells differentiated for 4, 14, and 21 days. C and D, flow cytometry analysis of hES-10, β-iPS-41/42, c-iPS-1, and c-iPS-2 cell surface markers CD34 and CD71 after differentiation for 14 and 21 days. E and F, statistical results of flow cytometric analysis of hES-10, β-iPS-41/42, c-iPS-1, and c-iPS-2 cell surface markers CD34 and CD71 after differentiation for 14 and 21 days. Means ± S.D. (n = 3) are shown. *, p < 0.05 compared with the hES-10 group; #, p < 0.05 compared with the β-iPS-41/42 group. SCF, stem cell factor; EPO, erythropoietin; TPO, thrombopoietin; FL, Flt3 ligand; SSC, side scatter.
FIGURE 6.
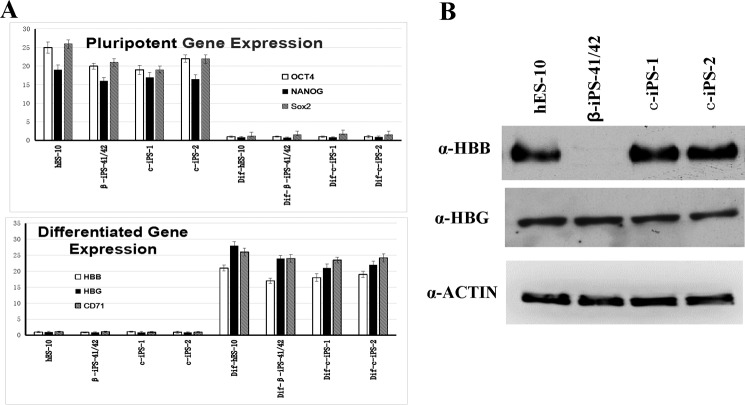
Functional assay of the corrected HBB gene. A, top and bottom, OCT4, NANOG, HBB, HBG, and CD71 gene expression (normalized to actin gene) in pluripotent cells and differentiated cells derived from the indicated iPSCs. Error bars represent S.D. The results showed that before differentiation the cells retained the expression of pluripotent genes, whereas after differentiation the expression of tissue-specific genes increased dramatically. B, Western blotting detection of the HBB and HBG proteins in erythroblasts derived from the indicated cells. The results showed no HBB protein expression in β-iPS-41/42 cells, whereas the gene-corrected iPSC-derived erythroblasts cells restored expression of the full-length HBB protein.
Functional Assay for the Gene-corrected iPSCs
To determine whether the corrected HBB gene restored normal function, first we detected the mRNA expression levels in the differentiated cells. The results showed that the HBB mRNA levels were comparable between the mutated and corrected cells; however, HBB protein expression could only be detected in the gene-corrected cell lines, c-iPS-1 and c-iPS-2 (Fig. 6B). HBG expression at both the mRNA and protein levels showed no difference between the mutated cells and the gene-corrected cells (Fig. 6, A and B). These findings suggested that the CD41/42 mutation in HBB in β-Thal iPSCs was corrected by gene editing, and the function of HBB gene was restored in differentiated erythrocytes.
Discussion
In this study, we demonstrated an efficient method to correct disease-causing mutations by combining CRISPR-mediated gene editing using ssODNs as donor template. We designed gRNAs that recognized the specific CD41/42 mutation sequence of the HBB gene and correction HBB gene mutation by transfection of ssODNs as the donor template and Cas9 gRNA into iPSCs. After antibiotic selection and purification of the cells by applying a PCR-based assay, the corrected clones were selected and sequenced to confirm the mutation correction. After gene editing, the corrected iPSCs retained pluripotency, multidifferentiation ability, and a normal karyotype. After the corrected cells differentiated into erythrocytes, the corrected HBB gene restored the expression of full-length HBB protein. More importantly, we did not detect a high mutation load in the exomes of the corrected iPSCs according to whole-exome sequencing, which is essential for future clinical applications.
In human pluripotent stem cells, homologous recombination gene targeting is particularly difficult (9, 12, 24–26). The recent development of zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and CRISPR technologies provides significant opportunities to enhance the homologous recombination efficiency (14, 17, 28–30). Recently, several studies have been published regarding using the engineered nuclease combined with a donor vector to correct HBB mutations in sickle cell disease and β-Thal iPSCs (14–17, 24–27). Construction of the donor template, selection of gene-corrected clones, and subsequent removal of the selection marker constitute a lengthy process. However, using a recombinase, either Cre or Flp-FRT recombination (FRT), to remove the selection marker unavoidably increases the stress and the risk of generating additional genome instability in human iPSCs (14–17). Although we know that the insertion of a drug selection cassette might interfere with the transcription of a gene-targeted allele, Zou et al. (17) showed that even after Cre-mediated excision of drug selection cassette, the expression of corrected HBB gene was partially repressed. Here, we used ssODNs instead of a donor vector to repair HBB gene mutations and inserted the selection marker with the Cas9 gene, which can avoid the problem encountered when using donor vector as a homology-directed repair template (21). Next, we used antibiotic selection with puromycin to kill the untransfected cells and then refreshed the culture medium, which greatly enhanced the positive rate of corrected clones. Without the long term selection pressure, the chances of exogenous genes integrating into the genome decrease substantially (31). Our results showed that the HBB mRNA expression levels were comparable between the mutated and corrected cells, which suggests that the corrected allele restores complete mRNA expression.
After confirming the gene-corrected clones, we performed whole-exome sequencing to discover new SNVs or indels between the parent cells and the corrected cells. However, all of the discovered mutations were far removed from any predicted CRISPR gRNA1 off-target sites. It is possible that these mutations are located near cryptic off-target sites that are not predicted in silico as similar results associated with zinc finger nuclease and transcription activator-like effector nuclease have been reported (32–34). These results showed that the mutation load to the exome was minimal and that the method was clinically feasible, possibly paving the way for future clinical use. In summary, it seems reasonable to conclude that the mutational load attributable to genome editing technologies in single cell clones can be minimal and should not constitute an inherent obstacle for future applications of single cell clone-based gene editing approaches in human monogenic disease.
Experimental Procedures
Cell Culture
hES-10, which was used as a positive control, was established in the Key Laboratory for Major Obstetric Diseases of Guangdong Province, The Third Affiliated Hospital of Guangzhou Medical University (16). The β-iPS-41/42 cell line was generated using cells from a β-Thal patient with a homozygous CD41/42(−CTTT) HBB mutation and the Sendai virus. hES-10 and β-iPS-41/42 cells were kept in our laboratory and cultured in irradiated mTeSR (STEMCELL Technologies, Vancouver, British Columbia, Canada). The cells were cultured at 37 °C in a humidified chamber with 5% CO2 and passaged at a 1:4 ratio once every week. HEK293T cells (ATCC, Manassas, VA) were cultured in DMEM supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin. The cells were cultured at 37 °C in a humidified chamber with 5% CO2 and passaged at a 1:5 ratio twice every week.
gRNA Assembly and GFP Reporter Vector Construction
Sequences used for cloning gRNA1, gRNA2, gRNA3, gRNA4, and gRNA5 into the lenti-CRISPR V2 vector are provided in supplemental Table 3. To prepare the destination vector, we linearized the lenti-CRISPR V2 cloning vector (Addgene plasmid ID 52961) using BsmbI and isolated the vector by purification. We performed the gRNA assembly reaction (10 μl) using 1 μl of annealed 24-bp fragment, 10 ng of destination backbone, and 1 μl of T4 DNA ligase (New England Biolabs) at 16 °C for 30 min. The reaction can be processed directly for bacterial transformation to colonize individual assemblies. The primers used for GFP reporter vector construction are provided in supplemental Table 3.
Analysis of CRISPR/Cas9-induced Cleavage
The iPSCs were transfected with Cas9 gRNA plasmids for 1 day with 0.5 μg/ml puromycin selection for 2 days. The genomic region surrounding the gRNA targeting site was amplified by PCR, and the products were purified using a gel extraction kit (Thermo). 400 ng of the purified PCR fragments was mixed with 2× PrimeSTAR GXL Buffer (TaKaRa Bio, Shiga, Japan) and distilled water to a final volume of 10 μl and subjected to conditions enabling heteroduplex formation as described previously (18). After annealing, the products were treated with T7 endonuclease 1 for 15 min at 37 °C and analyzed on 10% polyacrylamide gels. The gels were stained with ethidium bromide for 20 min and examined using a Gel DOC imaging system (Bio-Rad).
GFP Reporter Assay
A GFP reporter was constructed by cloning the GFP fragments into the p3xFLAG-CMV-10 vector using primers listed in supplemental Table 3. The target sequence for wild-type HBB was amplified from hES-10 cells, and the mutated CD41/42(−CTTT) variant was amplified from β-iPS-41/42 cells. GFP reporter activation was tested by co-transfecting HEK293T cells with plasmids carrying Cas9, Cas9 gRNA, and GFP reporters. 293T cells were seeded onto 12-well plates the day before transfection, and ∼24 h after seeding, cells were transfected using calcium phosphate. For 12-well plates, we used 0.5 μg of each CRISPR and 0.5 μg of each reporter plasmid per well.
Electroporation and Drug Selection
The procedure for using ssODNs to correct the HBB gene CD41/42(-CTTT) mutation was as follows. Before electroporation, β-iPS-41/42 cells were grown in feeder-free adherent culture in chemically defined mTeSR on plates coated with Matrigel (BD Bioscience). Transfections were done using a P3 Primary Cell 4D Nucleofector X kit (Lonza). Specifically, the cells were pretreated with 10 μm Rho-associated protein kinase (ROCK) inhibitor for 2 h and dissociated into a single cell suspension with 1 mg/ml Accutase (Invitrogen). Subsequently, 8 μg of Cas9 and gRNA plasmid and 2 μg of ssODNs were mixed with 5 × 106 β-iPS-41/42 cells and transferred into a 100-μl Nucleocuvette, and nucleofection was conducted using the CB150 program. Cells were plated on Matrigel-coated plates in mTeSR1 medium supplemented with ROCK inhibitor for the first 24 h. 0.5 μg/ml puromycin was used to select cells for 2 days, and then medium was changed to mTeSR1 medium without puromycin until clones were picked.
PCR Detection of Corrected Clones
PCR was performed using JumpStart Taq (Sigma) according to the manufacturer's instructions. 100 ng of genomic DNA was used in all PCRs. Primers including P1(on HBB locus upstream of ssODN) and P2 (on HBB locus downstream of ssODN) were used to amplify 414-bp products of HBB gene that cross the 5′ and 3′ ends of the ssODN recombination sites (Fig. 2A). Both primers P3 and P4 were located in the ssODNs and annealed to the correction sequence (Fig. 2A). Corrected clones were screened using sequence-specific PCR primer sets P1 and P3 or P2 and P4. Sequencing was confirmed by primers P1 and P2. All primers used are listed in supplemental Table 3.
RNA Extraction and Real Time Quantitative PCR Analysis
To test the expression of HBB and HBG genes, total RNA was isolated using TRIzol (Life Technologies). Total RNA (1 μg) was used for reverse transcription (RT) with the ReScript II RT kit (Qiagen, Hercules, CA), and quantitative PCR was performed using the SYBR PCR kit (Qiagen). All reactions were run at 40 cycles using standard conditions according to the manufacturer's instructions. Actin gene was used to normalize quantitative RT-PCR analysis, and all items were measured in triplicate. The relative quantitative expression of the test gene was described as the ratio of the hematopoietic differentiated hESC/iPSC group and the undifferentiated hESC/iPSC group. The primer sequences used are listed in supplemental Table 3.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 10 min at room temperature for fluorescence staining. After PBS washing, cells were permeabilized with 0.3% Triton X-100 for 45 min at room temperature. After removal of the Triton X-100 solution, cells were washed with PBS and stained at 4 °C overnight with primary antibodies at appropriate dilutions. The primary antibodies used included SSEA-4 (1:100; ab16287, Abcam, Cambridge, MA) and OCT4 (1:100; ab15897, Abcam). Cells were stained at room temperature with the secondary antibody for 2 h at a 1:800 dilution, including FITC-conjugated goat anti-rabbit IgG (Chemicon, Temecula, CA). Nuclei were stained with DAPI (Life Technologies) at a concentration of 1 μg/ml.
Teratoma Formation and Analysis
Cells from a confluent 10-cm plate were harvested by 0.5 mm EDTA digestion, resuspended in Matrigel, and injected subcutaneously into immunodeficient mice. Eight weeks after injection, teratomas were dissected, fixed in 4% paraformaldehyde, and processed for hematoxylin and eosin staining.
Hematopoietic and Erythroblast Differentiation from Human iPSCs
Hematopoietic differentiation was performed according to a protocol described previously (16, 17). Human iPSCs were directly differentiated into hematopoietic progenitor cells by a modified EB formation method as described previously (16). Two weeks after the EB-mediated hematopoietic differentiation, the EB-derived cells were collected and dissociated with Accutase treatment. The dissociated cells were cultured in a serum-free medium containing stem cell factor (SCF) (100 ng/ml), IL-3 (10 ng/ml), insulin-like growth factor II (40 ng/ml), erythropoietin (2 units/ml), and dexamethasone (1 μm) for erythroid cell expansion and maturation (17). Cells were harvested on day 8 of erythroid expansion and differentiation, and the differentiated cells were analyzed by fluorescence-activated cell sorting analysis and quantitative RT-PCR and Western blotting for globin expression.
Hematopoietic and Erythroblast Differentiation Efficiency Assayed by Flow Cytometry
Single cells prepared from EBs were stained with the following fluorochrome-conjugated monoclonal antibodies (mAbs): phycoerythrin-cyanine 7-labeled anti-human CD34 and phycoerythrin-labeled anti-human CD71. These mAbs and their corresponding nonspecific isotype controls were used at 1 mg/ml per 1 × 106 cells. The cells were stained with the mAbs in PBS supplemented with 2% FBS for at least 30 min on ice. The cells were washed twice with 2% FBS in PBS and resuspended with 1 ml of 2% FBS in PBS. The samples were tested using an Aria III flow cytometer (BD Biosciences) and analyzed with FACS Diva software (BD Biosciences).
DNA Isolation
Genomic DNA was extracted with a QIAamp DNA Blood minikit (Qiagen) according to the manufacturer's instructions. RNA erasing was performed at 37 °C for 1 h using RNase (Qiagen). The concentration and quality of each DNA sample were measured using a spectrophotometer (NanoDrop 2000, Thermo Scientific) and 1% agarose gel electrophoresis.
CNV and SNP Genotype Analysis
The Affymetrix Cytoscan HD array (Affymetrix, Santa Clara, CA), which interrogates 2,696,550 copy number markers and >749 thousand SNP probes across the human genome, was used to characterize cell lines of interest to determine whether there were different associations in de novo-generated CNVs among the individual reprogramming methods. Genomic DNA (250 ng) of each tested sample was amplified and labeled according to the manufacturer's instructions. Hybridization of the labeled product using a GeneChip Hybridization Oven 645 (Affymetrix) was followed by washing using the GeneChip Fluidics Station 450 (Affymetrix) and scanning using the GeneChip Scanner 3000 7G (Affymetrix). Data processing was performed using Chromosome Analysis Suite 2.0 (ChAS 2.0) software (Affymetrix) via the standard reference library provided by the manufacturer and based on the hidden Markov model. The segment filter was set to 150 kb and 50 markers for CNVs and 3 Mb and 50 markers for loss of heterozygosity (LOH).
Whole-exome Capture Sequencing and Bioinformatics Analysis
Paired end sequencing was performed using Illumina GAIIx/HiSeq 2000 instruments (Illumina, San Diego, CA), and exon capture was conducted using Agilent Sure Select Technology (Agilent, Santa Clara, CA). For sequence alignment, variant calling, and annotation, the sequences were mapped to their location with the human genome reference sequence (hg19; NCBI Build 37.1) using Burrows-Wheeler Aligner (BWA) (v.0.5.9-r16). Local realignment of the potential insertion/deletion sites was carried out with a genome analysis tool (GATK). SNVs and indel variants were assessed against reference dbSNP 138. All variants were annotated with reference to consensus coding sequences (CCDS) (NCBI release 20090902) and RefSeq (UCSC distribution 20101004). The novel variants were checked using the Integrative Genomics Viewer (IGV).
HBB and HBG Protein Tests by Western Blotting
On day 14, EBs from four groups were collected, washed once with cold PBS, and lysed with cell lysis buffer (catalog number 9803, Cell Signaling Technology) supplemented with 100× phenylmethanesulfonyl fluoride (PMSF; 100 mm) for 5 min. The lysate was centrifuged at 12,000 × g for 10 min at 4 °C to remove the insoluble components. The sample concentrations were determined using the NanoDrop 2000, and 30 mg of protein per group was separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a PVDF membrane. The membrane was blocked with 5% nonfat milk in TBS containing 0.1% Tween 20, incubated overnight with anti-HBB (ab202399, Abgent), anti-HBG1/2 (ab137096, Abcam), and anti-actin (ab8227, Abcam) at 4 °C. The membrane was then incubated with goat anti-rabbit IgG horseradish peroxidase-linked secondary antibody (LP1001a, Abgent) for 1 h at room temperature. The blots were then visualized using an ECL detection kit (23225, Thermo).
Author Contributions
X. N. conducted most of the experiments, analyzed the results, and wrote most of the paper. W. H. conducted experiments on the gene editing. B. S. and D. F. conducted experiments establishing iPSCs. Z. O. and Y. C. conducted experiments on hematopoietic differentiation. Y. F. analyzed the whole-exome sequencing data. X. S. conceived the idea for the project and wrote the paper with X. N.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 31171229, U1132005, 81301184 and 81502445; Natural Science Foundation of Guangdong Province Grant 2014A030312012; International Cooperation Project of Science and Technology Planning Project of Guangdong Province Grant 2013B51000087; and Science and Information Technology of Guangzhou Key Project Grants 201508020258, 201400000003-4, and 201400000004-4. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. 1 and Tables 1–3.
- β-Thal
- β-thalassemia
- HBB
- Homo sapiens hemoglobin β
- CRISPR/Cas9
- clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9)
- gRNA
- guide RNA
- SNV
- single nucleotide variant
- CNV
- copy number variant
- ssODN
- single strand oligodeoxynucleotide
- HSC
- hematopoietic stem cell
- iPSC
- induced pluripotent stem cell
- indel
- insertion and deletion
- EB
- embryoid body
- REP
- reporter
- HBG
- hemoglobin subunit γ
- HBD
- hemoglobin subunit δ.
References
- 1. Finotti A., Breda L., Lederer C. W., Bianchi N., Zuccato C., Kleanthous M., Rivella S., and Gambari R. (2015) Recent trends in the gene therapy of β-thalassemia. J. Blood Med. 6, 69–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Finotti A., and Gambari R. (2014) Recent trends for novel options in experimental biological therapy of β-thalassemia. Expert Opin. Biol. Ther. 14, 1443–1454 [DOI] [PubMed] [Google Scholar]
- 3. Cao A., and Galanello R. (2010) β-Thalassemia. Genet. Med. 12, 61–76 [DOI] [PubMed] [Google Scholar]
- 4. Xu X. M., Zhou Y. Q., Luo G. X., Liao C., Zhou M., Chen P. Y., Lu J. P., Jia S. Q., Xiao G. F., Shen X., Li J., Chen H. P., Xia Y. Y., Wen Y. X., Mo Q. H., Li W. D., Li Y. Y., Zhuo L. W., Wang Z. Q., Chen Y. J., Qin C. H., and Zhong M. (2004) The prevalence and spectrum of α and β thalassaemia in Guangdong Province: implications for the future health burden and population screening. J. Clin. Pathol. 57, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 6. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 7. Park I. H., Zhao R., West J. A., Yabuuchi A., Huo H., Ince T. A., Lerou P. H., Lensch M. W., and Daley G. Q. (2008) Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y., Jiang Y., Liu S., Sun X., and Gao S. (2009) Generation of induced pluripotent stem cells from human β-thalassemia fibroblast cells. Cell Res. 19, 1120–1123 [DOI] [PubMed] [Google Scholar]
- 9. Hanna J., Wernig M., Markoulaki S., Sun C. W., Meissner A., Cassady J. P., Beard C., Brambrink T., Wu L. C., Townes T. M., and Jaenisch R. (2007) Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318, 1920–1923 [DOI] [PubMed] [Google Scholar]
- 10. Ye L., Chang J. C., Lin C., Sun X., Yu J., and Kan Y. W. (2009) Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc. Natl. Acad. Sci. U.S.A. 106, 9826–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu L. C., Sun C. W., Ryan T. M., Pawlik K. M., Ren J., and Townes T. M. (2006) Correction of sickle cell disease by homologous recombination in embryonic stem cells. Blood 108, 1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y., Zheng C. G., Jiang Y., Zhang J., Chen J., Yao C., Zhao Q., Liu S., Chen K., Du J., Yang Z., and Gao S. (2012) Genetic correction of β-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 22, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramalingam S., Annaluru N., Kandavelou K., and Chandrasegaran S. (2014) TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr. Gene Ther. 14, 461–472 [DOI] [PubMed] [Google Scholar]
- 14. Ma N., Liao B., Zhang H., Wang L., Shan Y., Xue Y., Huang K., Chen S., Zhou X., Chen Y., Pei D., and Pan G. (2013) Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J. Biol. Chem. 288, 34671–34679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xie F., Ye L., Chang J. C., Beyer A. I., Wang J., Muench M. O., and Kan Y. W. (2014) Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 24, 1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song B., Fan Y., He W., Zhu D., Niu X., Wang D., Ou Z., Luo M., and Sun X. (2015) Improved hematopoietic differentiation efficiency of gene-corrected β-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 24, 1053–1065 [DOI] [PubMed] [Google Scholar]
- 17. Zou J., Mali P., Huang X., Dowey S. N., and Cheng L. (2011) Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood 118, 4599–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang F., Wen Y., and Guo X. (2014) CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 23, R40–R46 [DOI] [PubMed] [Google Scholar]
- 20. Xu P., Tong Y., Liu X. Z., Wang T. T., Cheng L., Wang B. Y., Lv X., Huang Y., and Liu D. P. (2015) Both TALENs and CRISPR/Cas9 directly target the HBB IVS2–654 (C → T) mutation in β-thalassemia-derived iPSCs. Sci. Rep. 5, 12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bialk P., Rivera-Torres N., Strouse B., and Kmiec E. B. (2015) Regulation of gene editing activity directed by single-stranded oligonucleotides and CRISPR/Cas9 systems. PLoS One 10, e0129308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inui M., Miyado M., Igarashi M., Tamano M., Kubo A., Yamashita S., Asahara H., Fukami M., and Takada S. (2014) Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 4, 5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X., Wang Y., Huang H., Chen B., Chen X., Hu J., Chang T., Lin R. J., and Yee J. K. (2014) Precise gene modification mediated by TALEN and single-stranded oligodeoxynucleotides in human cells. PLoS One 9, e93575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zwaka T. P., and Thomson J. A. (2003) Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 21, 319–321 [DOI] [PubMed] [Google Scholar]
- 25. Leavitt A. D., and Hamlett I. (2011) Homologous recombination in human embryonic stem cells: a tool for advancing cell therapy and understanding and treating human disease. Clin. Transl. Sci. 4, 298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang J. C., Ye L., and Kan Y. W. (2006) Correction of the sickle cell mutation in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Y, Zhang X, Yi L, Hou Z, Chen J, Kou X, Zhao Y, Wang H, Sun XF, Jiang C, Wang Y, Gao S (2016) Naïve induced pluripotent stem cells generated from β-thalassemia fibroblasts allow efficient gene correction with CRISPR/Cas9. Stem Cells Transl. Med. 5, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sebastiano V., Maeder M. L., Angstman J. F., Haddad B., Khayter C., Yeo D. T., Goodwin M. J., Hawkins J. S., Ramirez C. L., Batista L. F., Artandi S. E., Wernig M., and Joung J. K. (2011) In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells 29, 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun N., and Zhao H. (2014) Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol. Bioeng. 111, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 30. Sun N., Liang J., Abil Z., and Zhao H. (2012) Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol. Biosyst. 8, 1255–1263 [DOI] [PubMed] [Google Scholar]
- 31. Papapetrou E. P., Zoumbos N. C., and Athanassiadou A. (2005) Genetic modification of hematopoietic stem cells with nonviral systems: past progress and future prospects. Gene Ther. 12, Suppl. 1, S118–S130 [DOI] [PubMed] [Google Scholar]
- 32. Gabriel R., Lombardo A., Arens A., Miller J. C., Genovese P., Kaeppel C., Nowrouzi A., Bartholomae C. C., Wang J., Friedman G., Holmes M. C., Gregory P. D., Glimm H., Schmidt M., Naldini L., and von Kalle C. (2011) An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 [DOI] [PubMed] [Google Scholar]
- 33. Pattanayak V., Ramirez C. L., Joung J. K., and Liu D. R. (2011) Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suzuki K., Yu C., Qu J., Li M., Yao X., Yuan T., Goebl A., Tang S., Ren R., Aizawa E., Zhang F., Xu X., Soligalla R. D., Chen F., Kim J., Kim N. Y., Liao H. K., Benner C., Esteban C. R., Jin Y., Liu G. H., Li Y., and Izpisua Belmonte J. C. (2014) Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 15, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.