Abstract
β-barrel outer membrane proteins (OMPs) are ubiquitously present in Gram-negative bacteria, mitochondria and chloroplasts, and function in a variety of biological processes. The mechanism by which the hydrophobic nascent β-barrel OMPs are transported through the hydrophilic periplasmic space in bacterial cells remains elusive. Here, mainly via unnatural amino acid-mediated in vivo photo-crosslinking studies, we revealed that the primary periplasmic chaperone SurA interacts with nascent β-barrel OMPs largely via its N-domain but with β-barrel assembly machine protein BamA mainly via its satellite P2 domain, and that the nascent β-barrel OMPs interact with SurA via their N- and C-terminal regions. Additionally, via dual in vivo photo-crosslinking, we demonstrated the formation of a ternary complex involving β-barrel OMP, SurA, and BamA in cells. More importantly, we found that a supercomplex spanning the inner and outer membranes and involving the BamA, BamB, SurA, PpiD, SecY, SecE, and SecA proteins appears to exist in living cells, as revealed by a combined analyses of sucrose-gradient ultra-centrifugation, Blue native PAGE and mass spectrometry. We propose that this supercomplex integrates the translocation, transportation, and membrane insertion events for β-barrel OMP biogenesis.
Keywords: chaperone, complex, membrane biogenesis, membrane protein, protein folding, β-barrel outer membrane protein biogenesis, BamA, SurA, the Sec translocon
Introduction
Biogenesis of membrane-integrated proteins, which have to be folded and inserted into a hydrophobic lipid environment, differs substantially from that of soluble proteins that are folded in a hydrophilic environment (1). In the outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts, the β-barrel outer membrane proteins (β-barrel OMPs)4 primarily comprise β-sheets and adopt a unique cylindrical, barrel-like topology (2). Such β-barrel OMPs are known for being able to maintain their structures under harsh experimental conditions (3). In Gram-negative bacteria, ∼2% of the genome encodes β-barrel OMPs (2), which function in a variety of biological processes, including cell adhesion, envelope organization, nutrient transport, virulence-related protein secretion, and signal transduction (4).
The biogenesis of β-barrel OMPs in Gram-negative bacteria is a complex process, during which the nascent polypeptide chains have to be transported through the hydrophilic periplasmic space before eventually folded and inserted into the hydrophobic outer membrane (5). SurA and BamA have been identified as two key proteins involved in β-barrel OMP biogenesis. Genetic studies have revealed that the deletion of the surA gene significantly decreased the abundance of folded/assembled β-barrel OMPs (6). Indeed, several studies have described SurA as the primary periplasmic chaperone involved in β-barrel OMP biogenesis (7, 8). Biochemical studies have demonstrated that SurA is indispensable for the in vitro refolding/reassembly of β-barrel OMPs (9). Crystallography studies revealed that SurA possesses a core module composed of its N-domain, P1 domain, and C-domain, as well as an additional satellite P2 domain (as shown in panel E in Fig. 1) (10). The β-barrel OMP assembly machine (i.e. the BAM complex) protein BamA, which has homologues in mitochondria (designated as Sam50) and chloroplasts (designated as Toc75), consists of a β-barrel domain that is integrated in the outer membrane and a POTRA (polypeptide translocation-associated) domain that extends into the periplasmic space (11). Genetic and biochemical observations showed that BamA is essential for the folding and membrane insertion of β-barrel OMPs (5, 9, 12, 13). In addition to SurA and BamA, other periplasmic quality control factors such as Skp, FkpA, and DegP in bacteria have been reported to be involved in the biogenesis of β-barrel OMPs (5).
FIGURE 1.
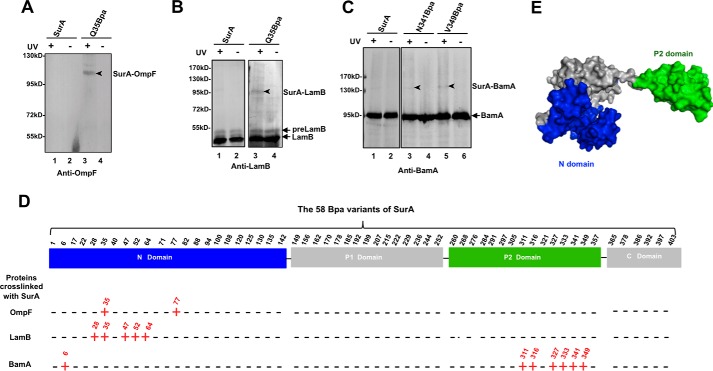
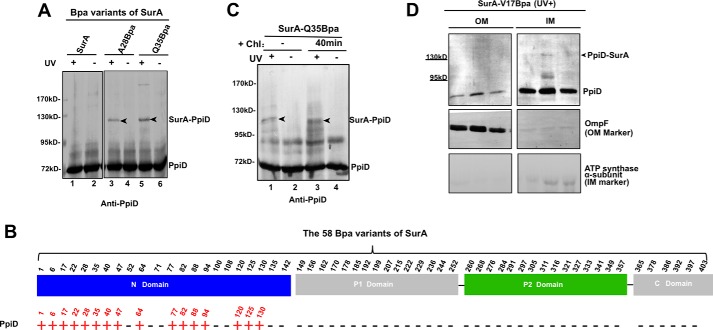
Identification of amino acid residues in SurA that are involved in interacting with β-barrel OMPs and BamA by Bpa-mediated in vivo photo-crosslinking. A–C, representative results of immunoblotting analyses for the in vivo photo-crosslinking products of the 58 Bpa-incorporated variants of SurA, using antibodies against OmpF (A), LamB (B), or BamA (C). The immunoblotting results of photo-crosslinking products for all the 58 Bpa variants of SurA that are displayed in supplemental Figs. S1 (probing OmpF), S2 (probing LamB), S3 (probing BamA), and S4 (probing SurA). It should be noted that the photo-crosslinked SurA-OmpF was assayed using the Ni-NTA affinity-purified SurA-Q35Bpa protein sample, while the photo-crosslinked SurA-LamB and SurA-BamA were directly assayed using the cell lysate preparations (i.e. without purification). D, summary showing the involvement of each of the 58 Bpa variants of SurA in interacting with OmpF, LamB, and BamA, with the localization of each Bpa-substituted position in the four domains (N-domain, P1 domain, P2 domain, and C-domain) of SurA indicated. A detected interaction is indicated by a red “+” sign, while a lack of interaction is indicated by a black “−” sign. E, crystal structure (PDB: 1M5Y) of SurA presented in the “surface” mode, with its four domains colored in blue (for the N-domain), green (for the P2 domain), and gray (for the P1 domain and C-domain).
Although genetic and biochemical studies have revealed that the biogenesis of β-barrel OMPs involves multiple protein factors, its detail molecular mechanism in living cells remains largely undefined. In an attempt to answer this question, we performed systematic studies via in vivo photo-crosslinking as mediated by unnatural amino acids (14–17) to analyze interactions involving the β-barrel OMPs, the primary chaperone SurA, and other factors involved in β-barrel OMP biogenesis. We first observed that, remarkably, SurA interacts with nascent β-barrel OMPs via its N-domain but with its functional partner BamA via its satellite P2 domain. Through systematic analyses of the subcellular localization of the protein factors involved in β-barrel OMP biogenesis, we revealed the presence of a supercomplex that contains the BamA, BamB, SurA, PpiD the Sec translocon, and SecA. This newly identified supercomplex spans the inner and outer membranes, and apparently integrates the translocation, transportation, and membrane insertion events involved in β-barrel OMP biogenesis.
Results
SurA Interacts with β-Barrel OMPs via Its N-Domain and with BamA Mainly via Its Satellite P2 Domain
In an attempt to determine the manner in which the multi-domain primary periplasmic chaperone SurA participates in the biogenesis of β-barrel OMPs, we employed an in vivo photo-crosslinking approach by individually introducing the unnatural amino acid p-benzoyl-l-phenylalanine (Bpa, a photo-activatable crosslinker) into SurA at 58 selected residue positions spreading among its four domains. The photo-crosslinked products for all 58 Bpa variants of His-tagged SurA that were expressed in ΔsurA cells were immunoblotted with antibodies against two representative client β-barrel OMPs (OmpF and LamB) and the potential SurA functional partner BamA. The complete results of these immunoblotting analyses are displayed in supplemental Figs. S1 (probing OmpF), S2 (probing LamB), S3 (probing BamA), and S4 (probing SurA, to gain an overall picture of the photo-crosslinked products). A representative of these immunoblotting results are respectively shown in Fig. 1, A (for OmpF), B (for LamB), and C (for BamA). And all the crosslinking results are summarized in Fig. 1D.
These crosslinking results demonstrate that SurA interacts with its client β-barrel OMPs via its N-domain and with BamA mainly via its satellite P2 domain. It is worth noting that, in addition to the SurA-OmpF and SurA-BamA single-crosslinked products, we also observed photo-crosslinked products with a molecular mass larger than 170 kDa, thus containing multiple (two or three) SurA monomers and one molecule of OmpF (for the SurA-D77Bpa variant; supplemental Fig. S1) or BamA (for the SurA-A311Bpa and SurA-S316Bpa variants; supplemental Fig. S3). We are currently investigating the meaning of these multi-crosslinking results for SurA functioning.
It should be pointed out that complementary expression of most of the Bpa variants of SurA, except SurA-I236Bpa and SurA-I297Bpa, were able to rescue the lethal phenotype of the ΔsurA cells cultured in the presence of EDTA and SDS (supplemental Fig. S5) (18), an envelope-disrupting stress condition, indicating that most of the incorporated Bpa did not significantly affect the function of SurA. Why the substitution of Ile-236 or Ile-297 by Bpa disrupts the function of SurA protein deserves further investigation.
Nascent β-Barrel OMPs Interact with SurA via Their N- and C-terminal Regions
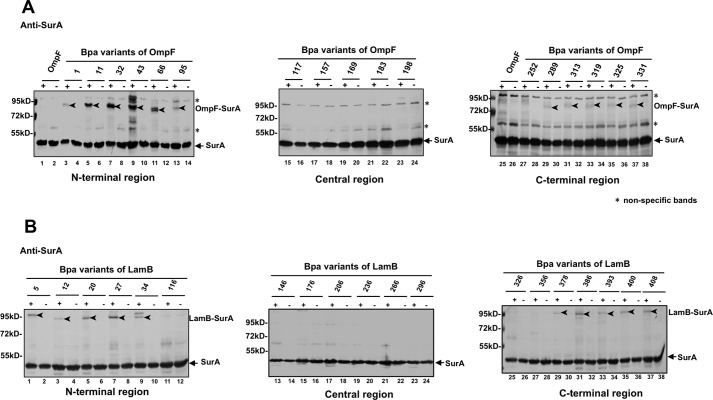
Next, we reciprocally analyzed the direct interactions between SurA and β-barrel OMPs by introducing Bpa into the latter (instead of SurA). For this purpose, Bpa was introduced into OmpF at 17 different residue positions and into LamB at 19 different residue positions (both selected in a largely random manner). We then analyzed the photo-crosslinked products formed between SurA and the Bpa variants of OmpF (Fig. 2A) or LamB (Fig. 2B), probing with the antibody against SurA (instead of OmpF or LamB). Remarkably, the OmpF-SurA and LamB-SurA photo-crosslinked products were both effectively detected, but only when Bpa was placed at the N- or C-terminal region of either OmpF (Fig. 2A) or LamB (Fig. 2B), while almost undetectable when Bpa was placed in the central region of either of these two β-barrel OMPs. We further found that the degree (as reflected by the density of the product bands) of crosslinking between the N-terminal region variants of either OmpF or LamB and SurA was significantly higher than that for their C-terminal region variants (Fig. 2, A and B). The biological meaning of such a unique interaction pattern is being further investigated by us.
FIGURE 2.
Nascent β-barrel OMPs directly interact with SurA via their N- and C-terminal regions. A and B, immunoblotting results of the in vivo photo-crosslinking products for the 17 Bpa variants of OmpF (panel A) and the 19 Bpa variants of LamB (panel B). Immunoblotting was performed using the anti-SurA antibody. Monomeric SurA, photo-crosslinked OmpF-SurA, and photo-crosslinked LamB-SurA are indicated. Cells expressing the wild-type OmpF (with no Bpa incorporation) were analyzed as a negative control (lanes 1 and 2 in panel A). The residue positions of Bpa insertion in both OmpF and LamB are numbered by referring to the mature protein (not the precursor protein).
We also provided evidence to show that it is mainly the nascent (rather than the folded/functional) OmpF that interacts with SurA. This was demonstrated by our observation that the OmpF-SurA photo-crosslinked product was dramatically decreased when cellular protein synthesis was inhibited by adding the antibiotic chloramphenicol (supplemental Fig. S6D). It should be pointed out that during our chase time (of 20 and 40 min), the amount of SurA decreased was little (lanes 2, 4, 6 in supplemental Fig. S6D) and thus the dramatic decrease in the amount of SurA-OmpF photo-crosslinked product was mainly due to the decrease in the amount of nascent β-barrel OMPs.
The expression level as well as the overall picture of photo-crosslinked products for each Bpa variant of the His-tagged OmpF or the His-tagged LamB were examined via immunoblotting analysis using antibodies against the His tag linked to SurA, as respectively displayed in supplemental Fig. S6, A and B. It should be noted that the introduction of Bpa into the β-barrel OMPs had little effect on their biogenesis, in view that the Bpa variants of OmpF was able to fold with an efficiency largely comparable with that of the wild-type OmpF protein (supplemental Fig. S6C).
A Ternary Complex Involving a β-Barrel OMP, SurA, and BamA Is Formed in Living Cells
The respective interaction of the well-separated N-domain and P2 domain of SurA with β-barrel OMPs and BamA suggests the formation of a ternary complex involving β-barrel OMP, SurA, and BamA. To determine whether such a ternary complex is indeed formed in living cells, we initially designed a dual photo-crosslinking strategy in which Bpa was introduced into SurA simultaneously at two positions (one interacting with β-barrel OMPs and the other interacting with BamA; chosen by referring to the crosslinking screening results shown in Fig. 1), but the dual incorporation of Bpa reduced the expression level of SurA protein to one that was insufficient for performing in vivo photo-crosslinking analysis (data not shown).
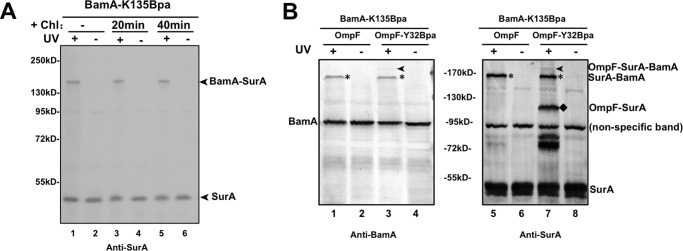
The strategy was therefore modified by simultaneously introducing Bpa into both BamA and a β-barrel OMP (OmpF). For this purpose, we first identified position Lys-135 in the POTRA 2 domain of BamA as one directly interacting with SurA (refer to lane 1 in Fig. 3A) via photo-crosslinking analyses of a series of Bpa variants of BamA (data to be published elsewhere). We further demonstrated that it was the folded BamA rather than nascent BamA that formed this photo-crosslinking product with SurA, because the level of the BamA-SurA product remained almost unaltered when cellular protein synthesis was inhibited by adding the antibiotics chloramphenicol (Fig. 3A). This observation is consistent with results of earlier chemical crosslinking analyses (8) and indicates that the interaction between SurA and functional BamA is independent of β-barrel OMP biogenesis.
FIGURE 3.
An OmpF-SurA-BamA ternary complex was detected by dual photo-crosslinking. A, immunoblotting results of the in vivo photo-crosslinking products for the BamA-K135Bpa variant, using the anti-SurA antibody. Cells were either untreated (lanes 1 and 2) or treated with chloramphenicol (a protein synthesis inhibitor) for 20 (lanes 3 and 4) or 40 (lanes 5 and 6) min before the in vivo photo-crosslinking. B, immunoblotting analysis for the photo-crosslinked OmpF-SurA-BamA ternary complex in cells co-expressing the BamA-K135Bpa and OmpF-Y32Bpa variant proteins. The immunoblotting was performed using antibodies against BamA (left) or SurA (right). The wild-type OmpF (with no Bpa incorporation) was also examined as a negative control (lanes 1, 2, 5, and 6). The positions of the photo-crosslinked OmpF-SurA-BamA, OmpF-SurA, and SurA-BamA complexes are indicated to the right of the gel.
We next performed the dual photo-crosslinking by co-expressing the BamA-K135Bpa and OmpF-Y32Bpa (which was demonstrated to mediate formation of the OmpF-SurA photo-crosslinked product; see lane 7 in Fig. 2A) variant proteins. The immunoblotting results presented in Fig. 3B demonstrate the formation of such a photo-crosslinked OmpF-SurA-BamA ternary complex (lanes 3 and 7 in Fig. 3B), which was detected using antibodies either against BamA (left panel) or SurA (right panel), with a molecular mass of ∼170 kDa (being roughly the sum of the molecular masses of the three constituent proteins). Such an OmpF-SurA-BamA ternary complex was not detectable when BamA-K135Bpa was co-expressed with the wild-type OmpF (i.e. with no Bpa incorporation; lanes 1 and 5 in Fig. 3B). It should be noted that the abundance of such a photo-crosslinked ternary complex is intrinsically low because it relies on the simultaneous crosslinking of a BamA variant and an OmpF variant to one common SurA polypeptide; for example, if the photo-crosslinking efficiency for the formation of either the BamA-SurA or the OmpF-SurA binary complexe was 20%, then the photo-crosslinking efficiency for forming the ternary complex would be lowered to 4% (20% × 20% = 4%).
A Supercomplex That Likely Spans the Inner and Outer Membranes Is Formed, involving BamA, BamB, SurA, SecY, SecE, and SecA
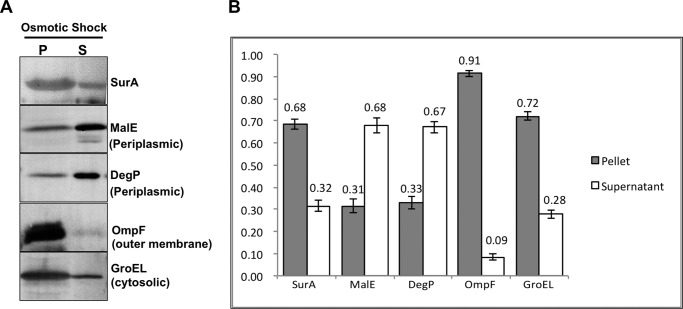
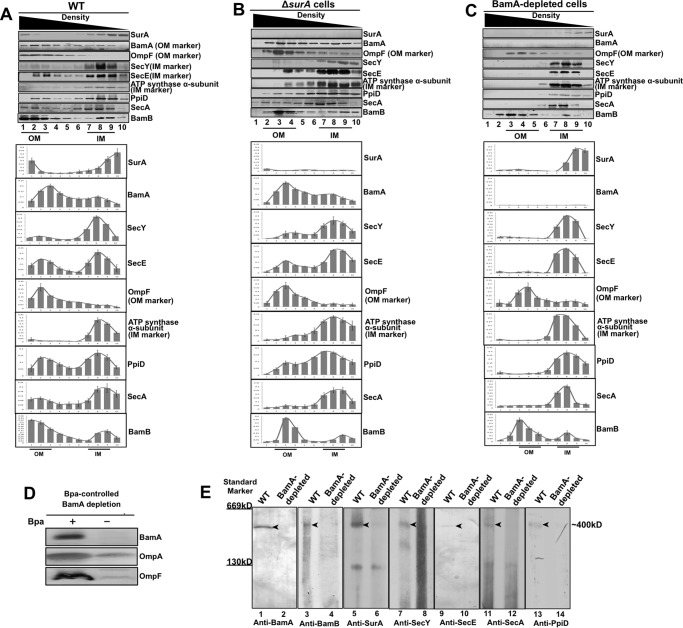
The observations described above (Fig. 3A) demonstrate that SurA interacts with BamA in a somehow permanent manner. In further support of this conclusion, we observed that much of the SurA protein was not released into the soluble part when the cells were subjected to osmotic shock treatment (as commonly applied for releasing freely diffusible periplasmic proteins (19)), in contrast to such typical periplasmic proteins as MalE and DegP, which were effectively released (Fig. 4), demonstrating that SurA largely exists in a non-diffusible manner. Prompted by this result, we measured the abundance of SurA in the inner and outer membrane fractions isolated by sucrose density gradient centrifugation (16). Immunoblotting analyses revealed that, remarkably, SurA was present in both the outer (fractions 1–3 in Fig. 5A) and inner membrane fractions (fractions 7–9 in Fig. 5A). Here BamA and OmpF were initially analyzed as outer membrane marker proteins, with SecY, SecE, and the α-subunit of ATP synthase as inner membrane marker proteins.
FIGURE 4.
SurA is not effectively released from the periplasm by osmotic shock treatment. A, immunoblotting results probing the presence of SurA, MalE, DegP, OmpF, or GroEL in the supernatant (S) and pellet (P) fractions of the wild type cells treated by osmotic shock. The treated cells were centrifuged to separate the two fractions, resolved by SDS-PAGE and probed with antibodies against the indicated proteins. B, relative amounts of the indicated proteins released in the supernatant and remained in the pellet, being quantified (mean ± S.E.; n = 3) from the immunoblotting results shown in panel A.
FIGURE 5.
Distribution of the BamA, BamB, SurA, PpiD, SecY, SecE, or SecA protein in the inner and outer membrane fractions of the wild type, ΔsurA-, or BamA-depleted cells and the detection of a “supercomplex.” A–C, immunoblotting analysis for SurA, BamA, SecY, SecE, PpiD, SecA, or BamB in the inner and outer membrane fractions prepared from the wild-type (A), the ΔsurA (B), or the BamA-depleted (C) cells, with the membrane fractions resolved by sucrose density gradient centrifugation. The relative amounts of the indicated protein in the inner and outer membrane fractions of the three types of cells were quantified (mean ± S.E.; n = 3) from the immunoblotting results and displayed below the corresponding gels in each panel. The α-subunit of ATP synthase and OmpF were analyzed as respective internal protein markers for the inner and outer membranes. D, results of immunoblotting against BamA, OmpA, or OmpF in cells whose chromosome-encoded BamA protein was depleted via a Bpa-controlled strategy (i.e. by subculturing the cells in Bpa-free LB medium, for details see “Experimental Procedures”). E, immunoblotting analysis probing the indicated proteins (BamA, BamB, SurA, PpiD, SecY, SecE, and SecA) in the inner membrane fraction (taken from fraction No. 8, see panel A) prepared from the wild-type or BamA-depleted cells, the membrane sample was resolved by Blue Native PAGE before probed with the indicated antibodies. The molecular mass of the “supercomplex” band was estimated to be ∼400 kDa through regression analysis based on the gel mobility of the “supercomplex” and the molecular size markers.
Surprisingly, we repeatedly detected a significant enrichment of BamA in the inner membrane fractions, in contrast to OmpF that was expectedly enriched only in the outer membrane fractions (Fig. 5A). Equally surprising was our observation that SecY and SecE were significantly enriched in the outer membrane fractions, in contrast to the α-subunit of ATP synthase that was expectedly enriched only in the inner membrane fractions (Fig. 5A).
We next examined whether the unexpected presence of BamA in the inner membrane fractions and SecYE in the outer membrane fractions depends on the presence of SurA. It turned out that such presence of BamA and SecYE were largely unaltered in the ΔsurA cells (Fig. 5B) in comparison with that in the wild-type cells (Fig. 5A). Next, we examined whether the presence of SecYE and SurA in the outer membrane fractions was dependent on the presence of BamA. Because of the essentiality of BamA for cell growth, we adopted a Bpa-controlled BamA depletion strategy (the details are described under “Experimental Procedures”). The immunoblotting analyses revealed that SecY, SecE, and SurA were barely detectable in the outer membrane fractions but remained nearly exclusively in the inner membrane fractions of the BamA-depleted cells (Fig. 5C). The expected decrease in the level of BamA, as well as that of OmpF and OmpA as previously reported (20), in the BamA-depleted cells was confirmed by immunoblotting analysis (Fig. 5D). In further support of the BamA-dependent association of SurA with the outer membrane fraction, we found that the SurA-BamA photo-crosslinked product was also associated with the outer membrane fractions (supplemental Fig. S7).
Taken together, these observations implicate the formation of a supercomplex spanning the inner and outer membranes and at least containing the SecYE, SurA, and BamA proteins. This conclusion was further supported by our unexpected observation that SecA, an inner membrane-associated molecular motor protein that is key for transmembrane protein translocation (21) was also significantly enriched in the outer membrane fractions (Fig. 5A). Additionally, we also observed the enrichment of BamB, a component of the BAM complex (9), in the inner membrane fractions (Fig. 5A).
More importantly, we detected a large protein complex, of ∼400 kDa, in the inner membrane fraction of the wild type cells by immunoblotting with antibodies against BamA, BamB, SurA, SecY, SecE, or SecA after the inner membrane preparation was resolved by Blue-Native PAGE (lanes 1, 3, 5, 7, 9, and 11 in Fig. 5E), but not in that of BamA-depleted cells (lanes 2, 4, 6, 8, 10, and 12 in Fig. 5E). In addition, this 400-kDa band on the Blue Native PAGE was found to contain BamA and SecA as revealed by mass spectrometry analysis (supplemental Table S4), while BamA and SecA were not detectable in the sample taken from the same gel position (i.e. around 400 kDa) when the inner membrane fractions from the BamA-depleted cells were similarly resolved by Blue Native PAGE (supplemental Table S5). This observation, together with that of the BamA-dependent presence of SurA or SecYE in the outer membrane fractions (Fig. 5C), suggest that BamA is the core component required for the formation of the supercomplex that contains BamA, BamB, SurA, SecY, SecE, and SecA.
SurA Directly Interacts with PpiD, a Sec Translocon-associated Inner Membrane Protein
We next tried to find out how SurA associates with the inner membrane. First, our immunoblotting analysis of the photo-crosslinked products of the 58 Bpa variants of SurA, probing with antibodies against SecY and SecE, failed to detect any band that could be rationally assigned as crosslinked SurA-SecY or SurA-SecE (data not shown).
We then repeated the immunoblotting analysis with antibodies against PpiD, an inner membrane protein that was reported to associate with the Sec translocon and to be involved in β-barrel OMP biogenesis (22–24). The photo-crosslinked SurA-PpiD product was clearly detected when Bpa was introduced at multiple positions that are located in the N-domain of SurA (the complete results are shown in supplemental Fig. S8, representative ones in Fig. 6A, summary in Fig. 6B). Notably, we also observed that the interaction between SurA and PpiD, similar to that between SurA and BamA (Fig. 3A), but unlike that between SurA and OmpF (supplemental Fig. S6D), was barely affected when cellular protein synthesis was inhibited by chloramphenicol (Fig. 6C), indicating that SurA interacted with the folded rather than the nascent PpiD.
FIGURE 6.
SurA directly interacts with PpiD as a functional partner. A, immunoblotting results of the in vivo photo-crosslinking products for two representative Bpa variants of SurA, using antibodies against PpiD. The complete immunoblotting results for all the 58 Bpa variants of SurA are displayed in supplemental Fig. S8. B, summary showing the involvement of each of the 58 Bpa substituted positions of SurA in interacting with PpiD, with the localization of each position in the four domains (N-domain, P1 domain, P2 domain, and C-domain) of SurA indicated. A detected interaction is indicated by a red “+” sign, while a lack of interaction is indicated by a black “−” sign. C, immunoblotting analysis of the photo-crosslinked product of the SurA-Q35Bpa variant in cells whose cellular protein synthesis process was suppressed by treating with the antibiotics chloramphenicol for 40 min before the in vivo photo-crosslinking was performed (lanes 3 and 4). D, immunoblotting analysis of the photo-crosslinked SurA-PpiD in the inner and outer membrane fractions. The inner and outer membrane fractions were separated by sucrose density gradient centrifugation and resolved by SDS-PAGE before probed with the indicated antibodies. The α-subunit of ATP synthase and OmpF were analyzed as respective markers of the inner and outer membranes.
We also demonstrated that the photo-crosslinked SurA-PpiD was present in the inner membrane fractions (Fig. 6D). And that PpiD was enriched not only in the inner but also in the outer membrane fractions of wild-type cells (as shown in Fig. 5A). Furthermore, we found that the aforementioned 400 kDa band also contains PpiD, as revealed by immunoblotting (lanes 13 and 14 in Fig. 5E) as well as mass spectrometry analyses (supplemental Table S4). These observations strongly suggest that PpiD is an additional component of the supercomplex.
Discussion
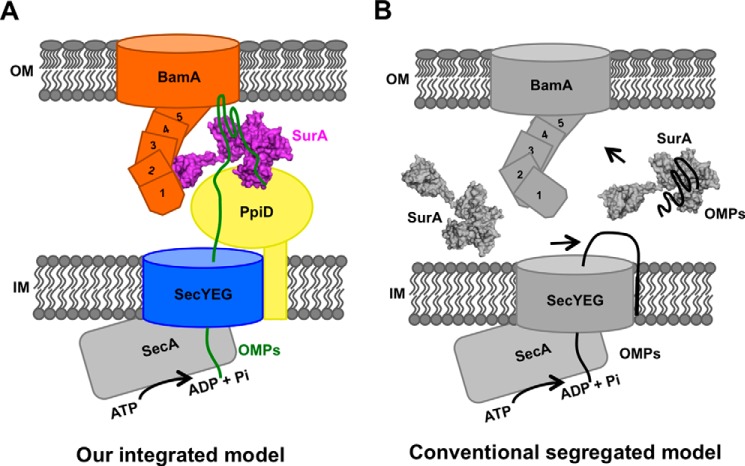
In an attempt to elucidate the molecular mechanism underlying β-barrel OMP biogenesis in living cells, we investigated the manners with which SurA interacts with nascent β-barrel OMPs and with β-barrel assembly machine protein BamA, by systematically performing unnatural amino acid Bpa-mediated in vivo photo-crosslinking analyses. We found that, remarkably, SurA interacts with nascent β-barrel OMPs largely via its N-domain but with the functional partner BamA via its satellite P2 domain (Figs. 1 and supplemental S1, S2, S3). Our dual in vivo photocrosslinking analyses demonstrated the formation of a ternary complex involving β-barrel OMP, SurA, and BamA (Fig. 3B). Additionally and more importantly, we observed the likely formation of a supercomplex spanning the two membranes, involving SecA, SecE, SecY, PpiD, SurA, BamB, and BamA. This was indicated mainly by immunoblotting analyses of membrane fractionations that were resolved by SDS-PAGE (Fig. 5, A–C) or Blue-Native PAGE (Fig. 5E), and by mass spectrometry analysis of a protein band of large molecular mass (supplemental Tables S4 and S5). In light of these observations, we propose that β-barrel OMP biogenesis in Gram-negative bacterial cells occurs via a unique supercomplex, as schematically illustrated in Fig. 7 (panel A).
FIGURE 7.
The supercomplex model for the biogenesis of β-barrel OMPs (A), in comparison with the conventional “segregated” model (B). Our model emphasizes the key role of a supercomplex that spans the inner and outer membranes and integrates the translocation, transportation, and membrane insertion events for the biogenesis of β-barrel OMPs in living cells. For comparison, the conventional model depicts SurA as a chaperone that is freely diffusible in the periplasm.
This supercomplex mediated mechanism can be described as follows. First, the supercomplex is formed mainly around the outer membrane-integrated BamA protein, which extends into the periplasm by its POTRA domains (25–26). Second, the primary periplasmic chaperone SurA anchors to BamA via its satellite P2 domain, whereas interacts with PpiD, an inner membrane protein associated with the SecYEG translocon, via its N-domain. Therefore, SurA exists in an immobilized and non-diffusible fashion when it processes nascent OMPs in the periplasm. Third, other protein factors, such as motor protein SecA, which has been reported to associate with the SecYEG translocon on the inner membrane, and also lipoprotein BamB, a component of the BAM complex, also appear in this supercomplex (27).
This supercomplex evidently integrates multiple events of β-barrel OMP biogenesis in living cells, including the translocation across the inner membrane, transportation through the periplasm, and insertion into the outer membrane. For comparison, in the conventional model (as summarized in panel B, Fig. 7), translocation, transportation, and membrane of the nascent β-barrel OMPs were usually considered as largely segregated events (1, 5, 20, 28), which would result in the undesirable exposure of hydrophobic nascent OMPs in the hydrophilic periplasmic environment. Another evident advantage of forming such a supercomplex is that it allows energy to be directly supplied, via the molecular motor SecA, from the cytoplasm to the β-barrel OMP biogenesis process (as illustrated in Fig. 7), thus bypassing the problem of lacking ATP in the periplasm (1). We believe that the concept of a supercomplex provides a new paradigm for further exploration of the molecular mechanisms underlying β-barrel OMP generation in living cells.
This supercomplex seems to be partially retained even in the lack of nascent β-barrel OMPs, i.e. when cellular protein synthesis was suppressed in the wild type cells by adding chloramphenicol in the culture medium. In such cells, we found that both BamA and SecY were still enriched in not only the inner but also the outer membrane fractions, while SurA, PpiD, SecE, and SecA were no longer enriched in the outer membrane fraction, neither BamB enriched in the inner membrane fraction (supplemental Fig. S9).
Such a supercomplex would have no problem to stretch across the bacterial periplasm, which is about 200 Å in width from the inner to the outer membrane. For example, the POTRA domain of BamA is able to extend about 105 Å in the periplasm (25) and SurA exists as an asymmetric dumbbell with a length of at least 100 Å (PDB: 1M5Y (10)). Furthermore, the periplasmic peptidoglycan, which exists as a mesh network having holes of 15–30 Å in diameter (29), should also allow the supercomplex to pass through. For example, the POTRA domain of BamA and the P2 domain of SurA both have a diameter of about 20 Å (PDB: 3EFC and 1M5Y).
An analogous supercomplex spanning the inner and outer membranes has been proposed to function in the biogenesis of lipopolysaccharides (LPS), which are synthesized at the cytoplasmic leaflet of the inner membrane and transported to the outer leaflet of the outer membrane in the Gram-negative bacteria (30). Furthermore, in the outer membrane of mitochondria, a similar integrated supercomplex, containing the translocase of the outer membrane (TOM) and the sorting and assembly machinery (SAM, a homolog of the BAM complex), was reported to function in mitochondrial β-barrel OMP biogenesis (31). In chloroplasts, β-barrel OMP biogenesis involves the outer membrane translocon TOC, the inner membrane translocon TIC, and the assembly protein Toc-75V, a homolog of bacterial BamA (11). The question of whether a similar integrated supercomplex is also formed in chloroplasts certainly merits further investigation.
Experimental Procedures
Bacteria Strains and Plasmid Construction
All bacterial strains used in this study and their relevant genotypes are listed in supplemental Table S1. The plasmids used for protein expression and chromosome editing are listed in supplemental Table S2. The pBAD-SurA-His6, pBAD-OmpF-His6, and pBAD-LamB-His6 plasmids were constructed by inserting the genes (amplified from the genomic DNA of Escherichia coli cells) encoding SurA, OmpF, or LamB into the pBAD/Myc-His C vector using the restriction enzyme free cloning (RF-cloning) method (32). Site-directed mutagenesis was performed using the Phusion site-directed mutagenesis kit (New England Biolabs).
Expression and Photo-crosslinking of Bpa Variant Proteins
To express the Bpa-incorporated proteins, the pBad-SurA-His6, pBad-OmpF-His6, and pBad-LamB-His6 plasmids were, respectively, transformed into ΔsurA, ΔompF, and ΔlamB mutant E. coli cells, which were all co-transformed with the pSup-BpaRS-6TRN plasmid expressing the orthogonal Bpa-tRNA synthetase/tRNABpa pair for Bpa incorporation (17). The co-transformed cells were cultured in LB broth at 37 °C to an OD600 of ∼0.6, after which Bpa was added to a final concentration of 0.2 mm, while l-arabinose was added to a final concentration of 0.0002% (for expressing OmpF or LamB) or 0.0005% (for expressing SurA) to induce the production of Bpa-incorporated proteins. The cells were cultured for 40 min, after which in vivo photo-crosslinking was performed by exposing the cells to UV irradiation for 10 min in a Hoefer UVC-500 crosslinker.
Protein Purification
The photo-crosslinked SurA-Q35Bpa product was purified via Ni-NTA (GE Healthcare) affinity chromatography in the presence of 8 m urea (a denaturing condition for removing the non-covalently bound proteins) as previously described (14).
Protein Release from Periplasm by Osmotic Shock
The osmotic shock was performed basically according to the method described previously (33). Bacteria cells were cultured in LB medium to an OD600 of ∼1.0 before being harvested by centrifugation at 3,000 × g for 20 min at 4 °C. The pellet was gently resuspended in 0.5 ml TSE buffer (containing 200 mm Tris-HCl, 500 mm sucrose, 1 mm EDTA, and 0.1 mg lysozyme, pH 8) and incubated on ice for 15 min. Then, 0.5 ml ice-cold water was added to such cell suspension before incubating for another 15 min. The supernatant (S, constituting the periplasmic extract) and pellet (P) were separated by centrifugation at 16,000 × g for 30 min at 4 °C.
Separation of the Inner and Outer Membranes
The inner and outer membranes were separated using a two-step sucrose density gradient centrifugation as previously described (8, 16). Briefly, the cells (resuspended in 5 ml of Tris-Buffer containing 10 mm Tris and 1 mm EDTA, pH 7.5) were lysed using a French pressure cell press and centrifuged at 2,000 × g for 10 min. to remove cellular debris and large aggregated proteins. Then 2.5 ml of the resulting supernatant was loaded onto a two-layer sucrose gradient (made of 0.3 ml of 65% and 1 ml of 25% sucrose), centrifuged for 3 h in a Beckman SW55 Ti rotor (4 °C, 220,000 × g). The bottom 0.7-ml fraction (containing the membranes) was subsequently diluted with 1.4 ml of Tris-Buffer and subjected to a multi-layer sucrose gradient (made of 0.5 ml of 65%, 0.5 ml of 55%, 1 ml of 50%, 2 ml of 45%, 2 ml of 40%, 2 ml of 35%, and 1.5 ml of 30% sucrose) centrifugation for 17 h in a Beckman SW41 Ti rotor (4 °C, 220,000 × g).
Bpa-controlled BamA Depletion
The BamA-depletion experiment was performed with cells in which the BamA-K135Bpa protein was expressed from the modified chromosomal DNA. Briefly, the BamA-K135Bpa strain was constructed as follows. First, we modified the wild-type strain BW25113 using a gene doctoring strategy as described earlier (34) to generate the strain LY928, in which the genome encoded the orthogonal aminoacyl-tRNA synthetase and tRNA required for Bpa incorporation. Via the same strategy, the genome of the LY928 strain was further modified to replace the wild type bamA gene with a construct encoding BamA-K135Bpa. The plasmids used to generate the LY928 and BamA-K135Bpa strains are listed in supplemental Table S2.
The BamA depletion assay using the BamA-K135Bpa strain was performed by subculturing the cells in Bpa-free LB medium (200-fold dilution) for 4 h after they were cultured in Bpa-containing LB medium overnight.
Blue Native PAGE
The Blue native gradient gel was generated with a 4–13% gradient of acrylamide. The electrophoresis was first performed at 120 V in Anode Buffer (25 mm imidazole, pH 7.0) and Cathode Buffer A (50 mm Tricine, 7.5 mm imidazole, 0.02% Coomassie Blue G-250, pH 7.0) for 1 h before switching the Cathode Buffer A to Cathode Buffer B (50 mm Tricine, 7.5 mm imidazole, 0.002% Coomassie Blue G-250, pH 7.0) for another 2 h. The proteins on the gel were then transferred onto the PVDF membrane, which was decolored before subject to immunoblotting analysis.
Other methods (semi-native SDS-PAGE, immunoblotting and mass spectrometry analysis) are described in the supplementary information file. The primary and secondary antibodies used in this study are listed in supplemental Table S3.
Author Contributions
Yan Wang and Rui Wang designed and performed the experiments, analyzed the data, and drafted the manuscript. Feng Jin performed some of the experiments. Yang Liu and Jiayu Yu produced some of the bacteria strains used in this study. Prof. Zengyi Chang and Dr. Xinmiao Fu supervised this study.
Supplementary Material
Acknowledgments
We thank Keio Collections for providing us the wild-type, the ΔompF, ΔlamB, and ΔsurA strains. We thank Professor Thomas J. Silhavy (Princeton University) for providing us the anti-LamB serum. We thank Professors Hajime Tokuda (Morioka University) and Kenichi Nishiyama (Iwate University) for providing us the anti-SecE serum. We thank Dr. Wen Zhou, at the Mass Spectrometry Facility of the National Center for Protein Sciences at Peking University, for assistance in performing the mass spectrometry analysis.
This work was supported by Research Grant 2012CB917300 (to Z. Y. C. and X. M. F.) from the National Basic Research Program of China (973 Program) and Research Grants 31470766 (to Z. Y. C.) and 31270804 and 31570778 (to X. M. F.) from the National Natural Science Foundation of China. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S5 and Figs. S1–S9.
- OMP
- outer membrane protein
- LPS
- lipopolysaccharide
- BAM
- β-barrel assembly machine.
References
- 1. Silhavy T. J., Kahne D., and Walker S. (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wimley W. C. (2003) The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 13, 404–411 [DOI] [PubMed] [Google Scholar]
- 3. Haltia T., and Freire E. (1995) Forces and factors that contribute to the structural stability of membrane-proteins. Biochim. Biophys. Acta Bioenergetics 1228, 1–27 [DOI] [PubMed] [Google Scholar]
- 4. Koebnik R., Locher K. P., and Van Gelder P. (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37, 239–253 [DOI] [PubMed] [Google Scholar]
- 5. Bos M. P., Robert V., and Tommassen J. (2007) Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 6. Lazar S. W., and Kolter R. (1996) SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178, 1770–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rouvière P. E., and Gross C. A. (1996) SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10, 3170–3182 [DOI] [PubMed] [Google Scholar]
- 8. Sklar J. G., Wu T., Kahne D., and Silhavy T. J. (2007) Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21, 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hagan C. L., Kim S., and Kahne D. (2010) Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bitto E., and McKay D. B. (2002) Crystallographic structure of SurA, a molecular chaperone that facilitates folding of outer membrane porins. Structure 10, 1489–1498 [DOI] [PubMed] [Google Scholar]
- 11. Kim K. H., Aulakh S., and Paetzel M. (2012) The bacterial outer membrane β-barrel assembly machinery. Protein Sci. 21, 751–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gessmann D., Chung Y. H., Danoff E. J., Plummer A. M., Sandlin C. W., Zaccai N. R., and Fleming K. G. (2014) Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc. Natl. Acad. Sci. U.S.A. 111, 5878–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., and Kahne D. (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 [DOI] [PubMed] [Google Scholar]
- 14. Fu X., Shi X., Yin L., Liu J., Joo K., Lee J., and Chang Z. (2013) Small heat shock protein IbpB acts as a robust chaperone in living cells by hierarchically activating its multi-type substrate-binding residues. J. Biol. Chem. 288, 11897–11906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang M., Lin S., Song X., Liu J., Fu Y., Ge X., Fu X., Chang Z., and Chen P. R. (2011) A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat. Chem. Biol. 7, 671–677 [DOI] [PubMed] [Google Scholar]
- 16. Ge X., Wang R., Ma J., Liu Y., Ezemaduka A. N., Chen P. R., Fu X., and Chang Z. (2014) DegP primarily functions as a protease for the biogenesis of β-barrel outer membrane proteins in the Gram-negative bacterium Escherichia coli. FEBS J. 281, 1226–1240 [DOI] [PubMed] [Google Scholar]
- 17. Chin J. W., Martin A. B., King D. S., Wang L., and Schultz P. G. (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Behrens S., Maier R., de Cock H., Schmid F. X., and Gross C. A. (2001) The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20, 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neu H. C., and Heppel L. A. (1965) The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J. Biol. Chem. 240, 3685–3692 [PubMed] [Google Scholar]
- 20. Hagan C. L., Silhavy T. J., and Kahne D. (2011) β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 [DOI] [PubMed] [Google Scholar]
- 21. Rapoport T. A. (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 [DOI] [PubMed] [Google Scholar]
- 22. Dartigalongue C., and Raina S. (1998) A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17, 3968–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Antonoaea R., Fürst M., Nishiyama K., and Müller M. (2008) The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47, 5649–5656 [DOI] [PubMed] [Google Scholar]
- 24. Sachelaru I., Petriman N. A., Kudva R., and Koch H. G. (2014) Dynamic interaction of the sec translocon with the chaperone PpiD. J. Biol. Chem. 289, 21706–21715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gatzeva-Topalova P. Z., Warner L. R., Pardi A., and Sousa M. C. (2010) Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sinnige T., Weingarth M., Renault M., Baker L., Tommassen J., and Baldus M. (2014) Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. J. Mol. Biol. 426, 2009–2021 [DOI] [PubMed] [Google Scholar]
- 27. Zimmer J., Nam Y., and Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McMorran L. M., Brockwell D. J., and Radford S. E. (2014) Mechanistic studies of the biogenesis and folding of outer membrane proteins in vitro and in vivo: What have we learned to date? Arch. Biochem. Biophys. 564, 265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Demchick P., and Koch A. L. (1996) The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178, 768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chng S. S., Gronenberg L. S., and Kahne D. (2010) Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qiu J., Wenz L. S., Zerbes R. M., Oeljeklaus S., Bohnert M., Stroud D. A., Wirth C., Ellenrieder L., Thornton N., Kutik S., Wiese S., Schulze-Specking A., Zufall N., Chacinska A., Guiard B., et al. (2013) Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 154, 596–608 [DOI] [PubMed] [Google Scholar]
- 32. Unger T., Jacobovitch Y., Dantes A., Bernheim R., and Peleg Y. (2010) Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172, 34–44 [DOI] [PubMed] [Google Scholar]
- 33. Chen Y. C., Chen S. J., Chang M. C., and Chen T. L. (2005) Comparison of various methods for periplasmic release of recombinant creatinase from Escherichia coli. J. Chinese Inst. Chem. Eng. 36, 527–532 [Google Scholar]
- 34. Lee D. J., Bingle L. E., Heurlier K., Pallen M. J., Penn C. W., Busby S. J., and Hobman J. L. (2009) Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains. BMC Microbiol. 9, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.