Abstract
Amaryllidaceae alkaloids are a large group of plant natural products with over 300 documented structures and diverse biological activities. Several groups of Amaryllidaceae alkaloids including the hemanthamine- and crinine-type alkaloids show promise as anticancer agents. Two reduction reactions are required for the production of these compounds: the reduction of norcraugsodine to norbelladine and the reduction of noroxomaritidine to normaritidine, with the enantiomer of noroxomaritidine dictating whether the derivatives will be the crinine-type or hemanthamine-type. It is also possible for the carbon-carbon double bond of noroxomaritidine to be reduced, forming the precursor for maritinamine or elwesine depending on the enantiomer reduced to an oxomaritinamine product. In this study, a short chain alcohol dehydrogenase/reductase that co-expresses with the previously discovered norbelladine 4′-O-methyltransferase from Narcissus sp. and Galanthus spp. was cloned and expressed in Escherichia coli. Biochemical analyses and x-ray crystallography indicates that this protein functions as a noroxomaritidine reductase that forms oxomaritinamine from noroxomaritidine through a carbon-carbon double bond reduction. The enzyme also reduces norcraugsodine to norbelladine with a 400-fold lower specific activity. These studies identify a missing step in the biosynthesis of this pharmacologically important class of plant natural products.
Keywords: crystal structure, enzyme structure, reductase, secondary metabolism, transcriptomics, Amaryllidaceae alkaloids, norbelladine, noroxomaritidine reductase
Introduction
The family of bulbous plants known as the Amaryllidaceae includes ornamental plants such as members of the genera Narcissus (daffodils) and Galanthus (snowdrops). These plants produce a group of alkaloids with a range of potential pharmaceutical uses. Of the Amaryllidaceae alkaloids, galanthamine has been used clinically for treatment of Alzheimer disease symptoms (1, 2). In addition to galanthamine, the chemically diverse Amaryllidaceae alkaloids have many documented pharmacological activities (3). For example, several have cancer fighting potential including hemanthamine, crinine, and lycorine (2, 4, 5). Continued development of Amaryllidaceae alkaloids for clinical and pharmaceutical applications requires the establishment of a cost effective production system for these molecules; however, limited information about the genes and proteins involved in the biosynthesis of Amaryllidaceae alkaloids is available.
For the biosynthesis of all Amaryllidaceae alkaloids, norbelladine, which is made from the condensation of tyramine and 3,4-dihydroxybenzaldehdye followed by imine reduction, is a pathway intermediate (6–8). 3,4-Dihydroxybenzaldehyde is hypothesized to originate from a branch of the phenylpropanoid pathway with demonstrated intermediates trans-cinnamic acid, 4-hydroxycinnamic acid, and either 3,4-dihydroxycinnamic acid or 4-hydroxybenzaldehyde (9). The conversion of 3,4-dihydroxycinnamic acid to 3,4-dihydroxybenzaldehyde could be catalyzed by an enzyme encoded by a paralogue of vanillin synthase. Vanillin synthase converts ferulic acid to vanillin (10). This reaction is similar to the proposed conversion of 3,4-dihydroxycinnamic acid to 3,4-dihydroxybenzaldehyde via a hydratase/lyase-type mechanism. An analogous, non-oxidative pathway is proposed for benzaldehyde biosynthesis. Reactions similar to the β-oxidative and non-oxidative CoA-dependent pathways documented in the biosynthesis of benzaldehyde-type compounds including vanillin and 4-hydroxybenzaldehyde are also a possibility (11). The condensation of tyramine and 3,4-dihydroxybenzaldehyde to the Schiff-base norcraugsodine is analogous to known amine-aldehyde condensing enzymes including norcoclaurine synthase and strictosidine synthase (12, 13).
Considering the simple chemistry required for norcraugsodine formation, it is possible that the condensation reaction occurs either nonenzymatically or in the active site of the enzyme that reduces norcraugsodine to norbelladine. Enzyme superfamilies capable of catalyzing reductions similar to the reduction of norcraugsodine to norbelladine include the aldo-keto reductases and short-chain alcohol dehydrogenase/reductases (SDRs)3 (14, 15). An example of an imine reductase from the alcohol dehydrogenase branch of the SDR superfamily functions as a tetrahydroalstonine synthase in Catharanthus roseus (16). Modification of norbelladine then leads to multiple Amaryllidaceae alkaloids.
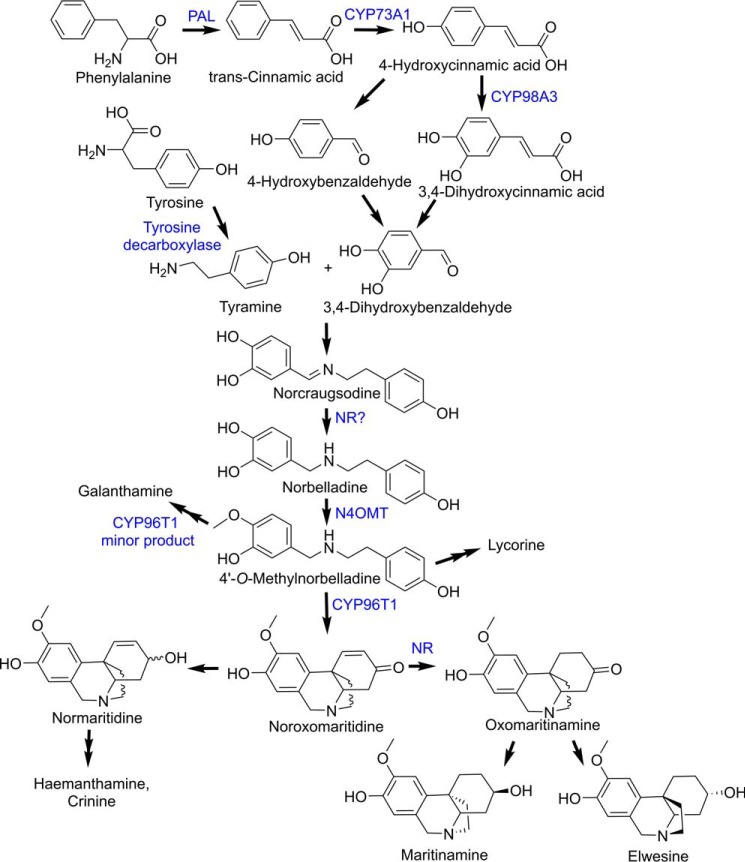
Norbelladine undergoes methylation and phenol-coupling reactions to produce the vast diversity of Amaryllidaceae alkaloids, including galanthamine, lycorine, hemanthamine, and crinine (Fig. 1) (1, 2). Recent studies have identified enzymes catalyzing steps in the core Amaryllidaceae alkaloid biosynthesis pathway, including norbelladine 4′-O-methyltransferase (N4OMT) and the cytochrome P450 CYP96T1 (44). N4OMT catalyzes the 4′-O-methylation of norbelladine to 4′-O-methylnorbelladine (17). CYP96T1 is a para-para′ phenol-phenol coupling cytochrome P450 that generates an enantiomeric mixture of noroxomaritidine (18), which can be further modified into hemanthamine- or crinine-type alkaloids depending on the stereochemistry of noroxomaritidine (19–21). CYP96T1 also converts 4′-O-methylnorbelladine to N-demethylnarwedine, but not as efficiently as it forms noroxomaritidine (18). N-Demethylnarwedine generated by CYP96T1 may contribute to the synthesis of galanthamine-type alkaloids. The enzymes that reduce noroxomaritidine to normaritidine (a precursor of the common hemanthamine-type alkaloids) or oxomaritinamine (a precursor of less prolific alkaloids including maritinamine and elwesine) remain to be identified.
FIGURE 1.
Amaryllidaceae alkaloid biosynthesis pathway. Enzymes involved in the pathway are indicated in blue. PAL, phenylalanine ammonia lyase, N4OMT, and NR are indicated.
In previous work, co-expression analysis of the Narcissus sp. aff. pseudonarcissus, Galanthus sp., and Galanthus elwesii transcriptomes successfully identified N4OMT and CYP96T1 in the core Amaryllidaceae alkaloid biosynthetic pathway (17, 18). For several missing steps in the hemanthamine, crinine, maritinamine, and elwesine pathways, we hypothesized that members of either aldo-keto reductase or SDR enzyme families may catalyze norbelladine synthesis and/or reduction of the noroxomaritidine enantiomers.
Here we identify a N. sp. aff. pseudonarcissus SDR gene that co-expresses with the previously discovered N4OMT (17). Biochemical analysis of the recombinant protein reveals that this N. sp. aff. pseudonarcissus SDR reduces the carbon-carbon double bond of the Amaryllidaceae alkaloid noroxomaritidine to form one of the enantiomeric forms of oxomaritinamine. The enzyme can also produce norbelladine from a solution containing 3,4-dihydroxybenzaldehyde and tyramine with a specific activity above background, but still 400-fold lower than noroxomaritidine conversion. Competition experiments and x-ray crystal structures provide insight on how the N. sp. aff. pseudonarcissus SDR binds tyramine and piperonal, an analogue of 3,4-dihydroxybenzaldehyde, and potentially noroxomaritidine. Given the high specific activity for noroxomaritidine reduction relative to norcraugsodine reduction, we designate this gene/protein as noroxomaritidine reductase (NR).
Results
Identification and Cloning of NR
In previous work, the assembly of a de novo transcriptome for N. sp. aff. pseudonarcissus with 106,450 sequences and a workflow for the identification of Amaryllidaceae alkaloid biosynthetic pathway genes from the 9,505 contigs that co-express with the accumulation of galanthamine was described (17). This resource was used to identify candidate oxidoreductases in the pathway. Because N4OMT catalyzes a reaction in the middle of the pathway, we reasoned that candidate oxidoreductase genes would co-express with the N4OMT gene. When interrogating for N4OMT co-expressing oxidoreductase homologues, 36 transcripts were identified in the N. sp. aff. pseudonarcissus transcriptome.
Of these 36 transcripts, medp_9narc_20101112 58880 was found to co-express with N4OMT in the previously described ABySS and MIRA assemblies for N. sp. aff. pseudonarcissus, G. elwesii, and Galanthus sp. (17, 18). This transcript was also co-expressed with N4OMT in the previously described Trinity-based assembly for Galanthus sp. (18). This made medp_9narc_20101112 58880 distinctive because all the other 36 transcripts were found to co-express with N4OMT in 3 or less of these assemblies. Because the medp_9narc_20101112 58880 was incomplete in the 5′ and 3′ regions, a full-length homolog (medp_9narc_20101112 12438) was used to design primers for PCR amplification. A full-length clone was obtained with the GenBankTM accession number KU295569 that differed from the medp_9narc_20101112 58880 at four nucleic acids (A141G, T183C, T243G, and T300C) with one nonsynonymous change (D81E).
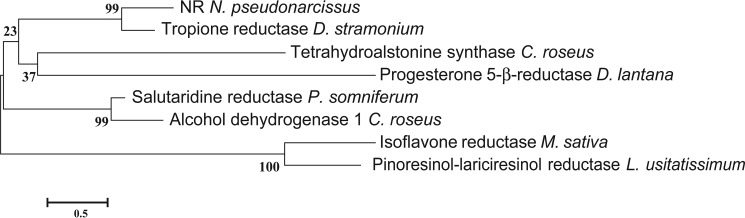
The identified clone from N. sp. aff. pseudonarcissus encodes a 271-amino acid protein (NR) with a predicted molecular mass of 29.0 kDa. Sequence comparison indicates that NR is a member of the SDR enzyme family (Fig. 2). Comparison with representative members of the SDR family shows that NR shares 52% sequence identity to tropinone reductase II from Datura stramonium and less than 20% identity with Papaver somniferum salutaridine reductase, C. roseus alcohol dehydrogenase, Linum usitatissimum pinoresinol/lariciresinol reductase, Medicago sativa isoflavone reductase, C. roseus tetrahydroalstonine synthase, and Digitalis lantana progesterone 5-β-reductase.
FIGURE 2.
Phylogenetic relationship of NR to other plant reductases and dehydrogenases. Amino acid sequences of all of the enzymes were taken from NCBI (PDB 2AE2, Q071NO.1, AJO70763.1, AKF02528.1, AAC48976.1, ABW24501.1, and AAS76634.1). The phylogenetic tree was constructed using MEGA (43), and the amino acid sequences were aligned using MUSCLE. The LG model was used to construct a Maximum Likelihood tree using a bootstrap replicate of 500. The percent identity of the plant reductases and dehydrogenases used in the tree were calculated using the Needleman-Wunsch Global Alignment tool from NCBI with the gap costs set at an existence of 11 and an extension of 2.
Noroxomaritidine Reduction Catalyzed by NR
During the biosynthesis of several prominent alkaloids including hemanthamine in N. sp. aff. pseudonarcissus, the ketone group of noroxomaritidine is reduced to a corresponding alcohol (Fig. 1). The biosynthesis of the Amaryllidaceae alkaloids maritinamine or elwesine requires the reduction of the carbon-carbon double bond in noroxomaritidine, potentially through oxomaritinamine (Fig. 1). Because some members of the SDR enzyme family catalyze similar reduction reactions, for example, the reductions of tropinone or 5β-progesterone (22, 23), we examined the ability of NR to reduce noroxomaritidine. For biochemical and structural studies, the NR protein from N. sp. aff. pseudonarcissus was expressed as an N-terminal His6-tagged protein and purified by affinity and size exclusion chromatography.
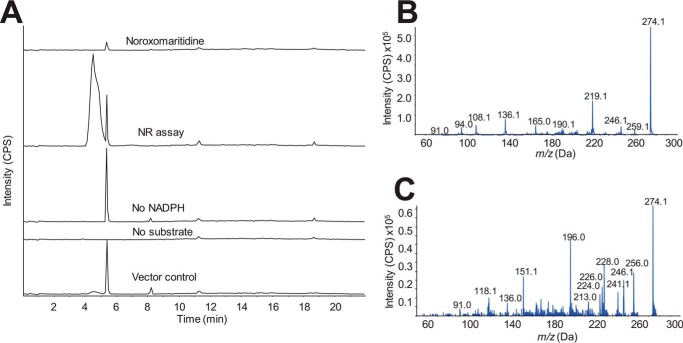
Noroxomaritidine was reduced by NR in the presence of NADPH as indicated by the presence of product +2 m/z from the noroxomaritidine substrate (Fig. 3A). In assays lacking either noroxomaritidine or NADPH, no product formation was observed. Similarly treated TALON resin purified protein extracts from Escherichia coli transformed with empty pET28a vector showed no activity. Incubations using NADH instead of NADPH showed no conversion of noroxomaritidine. Due to limited substrates the enzymatic activities were only quantified by specific activity measurements. The specific activity of NR for reduction of noroxomaritidine was 8600 ± 1250 pmol min−1 mg of protein−1 (Table 1).
FIGURE 3.
Noroxomaritidine reduction by NR. Enzymatic activity of NR for reduction of noroxomaritidine (m/z 274.3) was monitored using LC-MS/MS with the same LC time program found in Ref. 18. A, enzyme assays of NR with 100 mm sodium phosphate buffer, pH 7.0, 100 μm noroxomaritidine, 1 mm NADPH, and 10 μg of pure protein in 100 μl at 30 °C for 2 h. Traces from top to bottom are: noroxomaritidine standard; complete assay with NR, noroxomaritidine, and NADPH; assay without NADPH; assay without noroxomaritidine; and assay with TALON resin purified E. coli empty vector protein extract substituted for NR protein. B, EPI MS/MS of NR product. C, EPI MS/MS of noroxomaritidine reduced with NaBH4.
TABLE 1.
Activity summary of NR
| Substrate | Product monitored | Specific activity |
|---|---|---|
| pmol min−1 mg protein−1 | ||
| Noroxomaritidinea | Oxomaritinamine | 8600 ± 1250 |
| 8,9-O-Didemethyloxomaritidine | 9-O-Demethyloxomaritinamine | −b |
| 3,4-Dihydroxybenzaldehyde and tyraminea | Norbelladine | 21.2 ± 1.2 |
| Isovanillin and tyraminea | 4′-O-Methylnorbelladine | 5.33 ± 0.21 |
| Vanillin and tyramine | 3′-O-Methylnorbelladine | −b |
| Piperonal and tyramine | 4-(2-((1,3-Benzodioxol-5-ylmethyl)amino)ethyl)phenol | −b |
a Measurements are with 500 μm substrate and 1 mm NADPH.
b Product made but not quantified. Values shown are mean ± S.D. for n = 3.
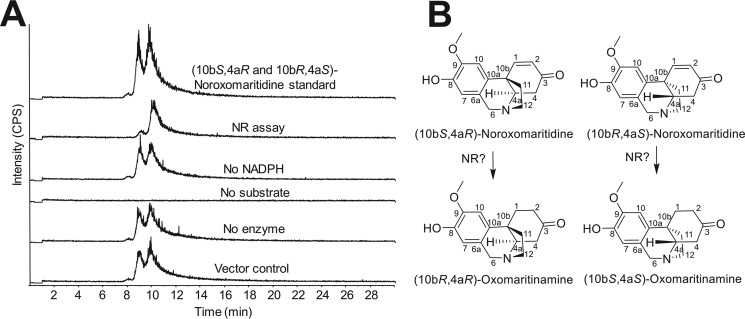
The noroxomaritidine substrate is an enantiomeric mixture. The enantiomers were therefore resolved by chiral chromatography, as previously described for CYP96T1 (18). Following a 2-h incubation, only one of the noroxomaritidine enantiomers was notably consumed and assays lacking NADPH or the NR enzyme did not show this preferential substrate disappearance (Fig. 4A). More than half of the enantiomeric mixture was never consumed, indicating that the enzyme acts preferentially on one of the two enantiomers (Fig. 4B). Optically pure standards of known configuration are lacking, so the absolute configuration of the preferred substrate remains unknown. Comparison of the MS/MS fragmentation patterns of noroxomaritidine reduced by NR (Fig. 3B) with noroxomaritidine reduced by sodium borohydride (Fig. 3C) reveals a clear difference. Aldehyde and ketone double bonds are the typical substrates of sodium borohydride (24). Although carbon-carbon bonds can be reduced by sodium borohydride, the ketone will preferentially be reduced. For this reason, product generated by the sodium borohydride reduction is likely normaritidine. The 1–2 carbon-carbon double bond is, therefore, likely reduced by NR into oxomaritinamine.
FIGURE 4.
NR consumption of 20 μm (10bS,4aR or 10bR,4aS)-noroxomaritidine using the same assay conditions as for specific activity assays incubated 2 h. A, LC-MS using a Chrom Tech, Inc. Chiral-CBH 100 × 4.0-mm, 5-μm column and the same LC setup and time program as for chiral separations in Ref. 18. Samples were monitored at m/z 272.3 with CE (25) and DP (70) on the QTRAP 6500 for the following samples top to bottom: (10bS,4aR and 10bR,4aS)-noroxomaritidine mixed standard; complete assay with NR, (10bS,4aR and 10bR,4aS)-noroxomaritidine and NADPH; assay without NADPH; assay without (10bS,4aR and 10bR,4aS)-noroxomaritidine substrate; assay without NR enzyme; and assay with TALON resin purified E. coli empty vector protein extract substituted for NR protein. B, the two enantiomers of noroxomaritidine and corresponding expected products.
To examine the ability of NR to accept substrates similar to noroxomaritidine, the demethylated form 8,9-O-didemethyloxomaritidine was tested as substrate. 8,9-O-Didemethyloxomaritidine was reduced by NR yielding a product similar to oxomaritinamine in its mass spectrum, indicating that the product is also reduced at the carbon-carbon double bond and therefore is 9-O-demethyloxomaritinamine. Fragments that are identical for these two molecules include m/z 91.1, 94.1, 108.1, and 136.1. Fragments that represent the presence of an additional methyl group in oxomaritinamine compared with 9-O-demethyloxomaritinamine are m/z 246.1 and 232.1, m/z 219.1 and 205.1, m/z 204.1 and 190.1, m/z 190.1 and 176.1, and m/z 165.0 and 151.0.
Identification of Norcraugsodine Reduction Catalyzed by NR
In addition to the reduction of noroxomaritidine, NR was found to catalyze the reduction of the imine in norcraugsodine to form norbelladine, albeit with a substantially lower specific activity. The amine aldehyde condensation of tyramine and 3,4-dihydroxybenzaldehyde to form norcraugsodine can spontaneously occur in solution (Fig. 1). It is not known if a biosynthetic enzyme in this pathway acts on tyramine and 3,4-dihydroxybenzaldehyde as substrates or the pre-condensed substrate norcraugsodine to make norbelladine.
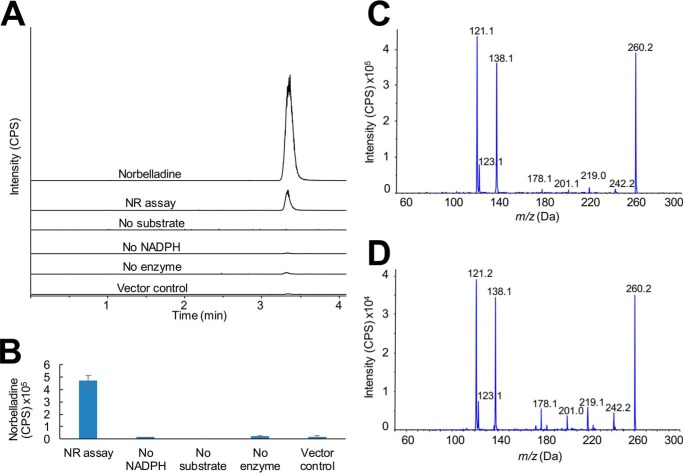
For the assays examining reduction to norbelladine, tyramine and 3,4-dihydroxybenzaldehyde were incubated with NR and NADPH and the resulting product examined by LC-MS/MS. A peak with the same retention time as authentic norbelladine was observed in the complete reaction mixture (Fig. 5A). In the absence of tyramine and 3,4-dihydroxybenzaldehyde, no norbelladine was observed. Assays lacking either NADPH or NR or assays replacing NR with E. coli proteins that co-purify during the TALON protein purification resulted in low levels of norbelladine probably resulting from a background reaction between tyramine and 3,4-dihydroxybenzaldehyde (Fig. 5B); however, these levels were below that observed in the complete assay mixture containing enzyme. Comparison of the MS/MS fragmentation patterns of authentic norbelladine (Fig. 5C) and the product of the NR reaction (Fig. 5D) confirm the identity of the enzymatic product. Given the low abundance of the product formed by NR, assays with varied pH and temperature were performed to maximize enzymatic yields relative to the chemical background reaction. Final assay conditions for norbelladine formation by NR were pH 6.0 at 35 °C with a specific activity of 21.2 ± 1.2 pmol min−1 mg of protein−1 (Table 1).
FIGURE 5.
Norcraugsodine reduction to norbelladine by NR. Enzymatic activity of NR for reduction of norcraugsodine was monitored using the same assay conditions and LC-MS/MS setup as in specific activity assays. A, MRM (m/z 260.1/238.0) of NR assays. Traces from top to bottom are: norbelladine standard; complete assay with NR, tyramine, 3,4-dihydroxybenzaldehyde, and NADPH; assay without NADPH; assay without tyramine and 3,4-dihydroxybenzaldehyde; assay without enzyme; and assay with TALON resin purified E. coli empty vector protein. B, MRM (m/z 260.1/238.0) relative quantification of assays shown in panel A in triplicate. C, EPI MS/MS of norbelladine standard. D, EPI MS/MS of NR product.
Variation of the aldehyde paired with tyramine in the NR-catalyzed reaction was also examined (Table 1). In place of 3,4-dihydroxybenzaldehyde, vanillin, isovanillin, and piperonal were used for incubations with NR, tyramine, and NADPH and led to formation of 3′-O-methylnorbelladine, 4′-O-methylnorbelladine, and 4-(2-((1,3-benzodioxol-5-ylmethyl)amino)ethyl)phenol, respectively (Table 1). The structures of the enzymatic products were confirmed by comparing LC-MS/MS fragmentation patterns to the products produced by chemical condensation and subsequent reduction of vanillin, isovanillin, or piperonal with tyramine in methanol containing sodium cyanoborohydride. The specific activity of the NR-catalyzed reaction with isovanillin and tyramine was determined to be 4-fold lower (5.33 ± 0.21 pmol min−1 mg of protein−1) than the reaction with 3,4-dihydroxybenzaldehyde and tyramine (Table 1). Other rates were not of direct interest to the synthesis of Amaryllidaceae alkaloids and so were not determined. These results indicate that NR can reduce an imine in the conversion of norcraugsodine to norbelladine but the low specific activity could indicate this is a nonspecific reaction.
Inhibition of Noroxomaritidine Reduction
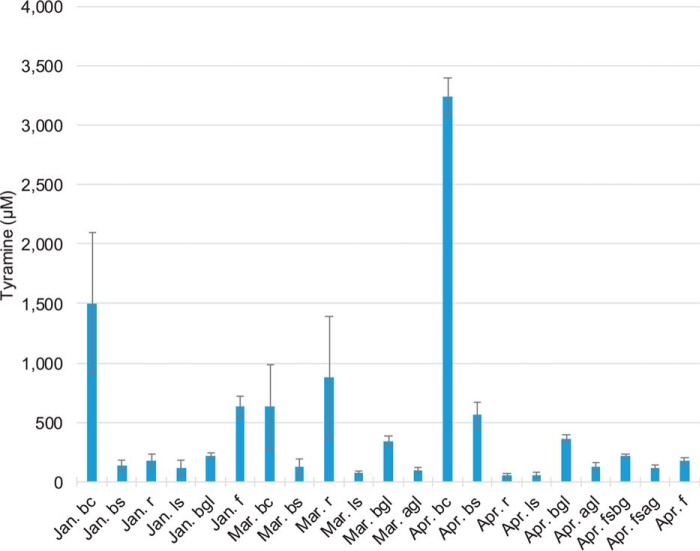
The reduction of noroxomaritidine provided the opportunity to test the occupancy of the active site of NR by 3,4-dihydroxybenzaldehyde and tyramine. The IC50 value of tyramine is 5,230 ± 752 μm. Piperonal has a lower IC50 value (425 ± 62 μm) than 3,4-dihydroxybenzaldehyde (4,960 ± 559 μm). This could result from either the increased stability of piperonal relative to 3,4-dihydroxybenzaldehyde or a tighter binding to the active site. Although these values are high, concentrations of tyramine can be up to 3 mm in N. sp. aff. pseudonarcissus (Fig. 6) and strictosidine synthase, another alkaloid biosynthesis enzyme, has been shown to have a Km = ∼4 mm for tryptamine and secologanin (25). To determine whether these results were likely the result of allosteric interactions or direct competition, several of these compounds were crystallized with NR.
FIGURE 6.
A comparison of the tyramine (CE (15), DP (60), m/z 138.1/121.0 and m/z 138.1/93.0), levels in daffodil organs (above ground leaf (agl), below ground leaf (bgl), leaf scale (ls), bulb scale (bs), bulb core (bc), root (r), flower stalk above ground (fsag), flower stalk below ground (fsbg), and flower (f)). These samples were collected during the months of January (well developed roots with well developed flower primordia), March (all plants form this month lack flower primordia due to sampling limitations), April (plants all flowering), and August (dormant bulbs). The instrument used for the experiment was a QTRAP 6500 with a Phenomenex Luna 5-μm C8(2) 250 × 4.60-mm LC column using the following LC method; A = 0.1% formic acid and B = acetonitrile, start with 10% B for 2 min followed by a linear increase to 40% B for 9.9 min, linear increase to 90% B for 0.1 min, hold for 3 min, linear decrease to 10% B for 0.1 min, and hold 7 min. The following were also monitored by MRM across these conditions but were not detected 3,4-dihydroxybenzaldehyde (CE (15), DP (60), m/z 139.0/111.0 and 139.0/93.0), norbelladine (CE (15), DP (50), m/z 260.1/138.0 and 260.1/121.0), noroxomaritidine (CE (25), DP (70), m/z 272.3/229.0 and 272.1/212.1), and oxomaritinamine (CE (20), DP (60), m/z 274.1/219.1 and 274.1/136.1). Alkaloid samples were prepared by utilizing liquid nitrogen, a mortar and pestle for grinding of frozen tissue, followed by 70% ethanol extraction, PTFE membrane filtration, extract drying, and extract resuspension in mobile phase.
X-ray Crystal Structure of NR
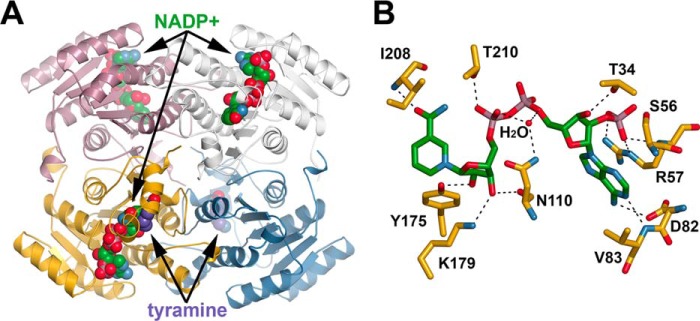
To understand the molecular basis of noroxomaritidine reduction by NR and examine the potential binding of the individual components tyramine and 3,4-dihydroxybenzaldehyde for norbelladine reduction, the x-ray crystal structure of the enzyme was determined. Because of the limited amounts of noroxomaritidine available and instability of 3,4-dihydroxybenzaldehyde, crystallization efforts used tyramine and piperonal as a substrate mimic. No enzymatic activity was observed when these individual molecules were incubated with NR and NADPH. The x-ray crystal structures of N. sp. aff. pseudonarcissus NR complexed with NADP+, NADP+ and tyramine, and NADP+ and piperonal were determined at 1.73-, 1.81-, and 1.50-Å resolution, respectively (Table 2). The NR·NADP+ complex was originally intended to be apoenzyme; however, the cofactor co-purified with the protein. The overall structure of NR reveals a canonical SDR-fold (Fig. 7A). NR forms a tetrameric structure with each 271-amino acid monomer adopting an alternating α/β structure formed by seven parallel β-sheets sandwiched between two layers of three α-helices that shape the NADPH-binding domain (Fig. 7A).
TABLE 2.
Summary of crystallographic data collection and refinement statistics
| NR·NADP+ | NR·NADP+·tyramine | NR·NADP+·piperonal | |
|---|---|---|---|
| Crystal | |||
| Space group | P4222 | P212121 | P322 |
| Cell dimensions (Å) | a = b = 60.38; c = 136.1 | a = 61.80; b = 86.96; c = 186.1 | a = b = 73.39; c = 167.9 |
| Data collection | |||
| Wavelength (Å) | 0.98 | 0.98 | 0.98 |
| Resolution range (Å) (highest shell resolution) | 45.2−1.73 (1.79−1.73) | 39.4−1.81 (1.88−1.81) | 35.9−1.50 (1.55−1.50) |
| Reflections (total/unique) | 366,363/27,021 | 943,975/90,846 | 862,122/84,466 |
| Completeness (highest shell) | 99.7% (98.6%) | 99.2% (91.6%) | 99.9% (98.7%) |
| 〈I/σ〉 (highest shell) | 22.0 (2.4) | 23.7 (2.3) | 17.6 (2.3) |
| Rsym (highest shell) | 10.6% (52.1%) | 9.8% (57.4%) | 7.1% (66.8%) |
| Model and refinement | |||
| RcrystRfree | 18.7%/21.4% | 18.3%/21.3% | 14.9%/16.0% |
| No. of protein atoms | 1870 | 7,650 | 3971 |
| No. of water molecules | 136 | 443 | 644 |
| No. of ligand atoms | 48 | 174 | 107 |
| Root mean square deviation | |||
| Bond lengths (Å) | 0.006 | 0.008 | 0.007 |
| Bond angles (°) | 1.05 | 1.13 | 1.18 |
| Average B-factor (Å2), protein, solvent, ligand | 37.9, 37.7, 41.8 | 41.2, 41.2, 42.4 | 20.2,18.2,33.1 |
| Stereochemistry: most favored, allowed, outliers | 98.8, 1.2, 0% | 97.5, 2.5, 0% | 97.8, 2.2, 0% |
FIGURE 7.
Three-dimensional structure of NR. A, ribbon diagram of the NR tetramer complexed with NADP+ and tyramine is shown. Each monomer is colored differently. NADP+ and tyramine are shown as space-filling models. B, NADP+ binding in NR. Residues interacting with NADP+ are shown as stick figures. Dotted lines represent hydrogen bond and charge-charge interactions.
NR uses NADPH as an electron donor for noroxomaritidine reduction. The crystal structure of NR shows that NADP+ binds the active site of NR through a combination of polar and nonpolar interactions (Fig. 7B). The adenine ring, which is in the anti-conformation, interacts with the side chain carboxylate of Val83 through its exocyclic N6 (3.0 Å) and with an oxygen of Asp82 through hydrogen bonding with N1 (2.8 Å). The 2′-phosphate of the adenine ribose forms charge-charge interactions with the side chain of Arg57 (2.7–2.8 Å) and a hydrogen bond with the backbone amide of Arg57 (2.8 Å). Additionally, the adenine ribose phosphate interacts with the hydroxyl group of Ser56 (2.8 Å). These interactions are key structural features that distinguish the NADPH-specific SDRs from the NADH-specific members of the family (26, 27). The nicotinamide ribose adopts a C3′-endo conformation with the C3 hydroxyl group hydrogen bonding to the side chain hydroxyl group of Thr34 (2.8 Å). The 2′-hydroxyl of the nicotinamide ribose interacts with the catalytic tyrosine (i.e. Tyr175; 2.6 Å) and the 3′-hydroxyl of the nicotinamide ribose interacts with Lys199 (3.0 Å), the second catalytic residue, and the carbonyl of Asn110 (2.7 Å). A water molecule forms hydrogen bonds that bridge two of the nicotinamide ribose phosphates and interacts with the side chain amide nitrogen of Asn110 (2.9 Å). Another hydrogen bond forms between an oxygen on the one nicotinamide ribose phosphate and Thr210 (2.8 Å). The syn conformation of the nicotinamide ring is stabilized by a hydrogen bond involving the carboxamide group to the backbone amide-nitrogen Ile208 (2.8 Å). The A-face of the nicotinamide ring packs against Ile208, Pro225, Gly226, and Ala227 to orient the B-face toward the substrate binding pocket.
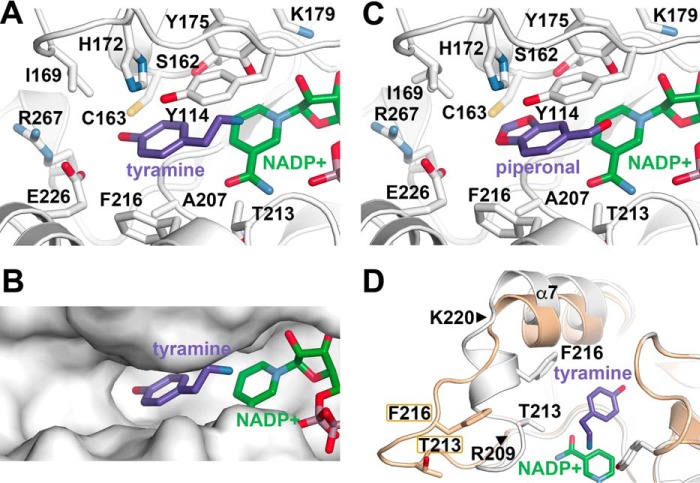
The x-ray crystal structures of NR in complex with NADP+ and either tyramine (Fig. 8, A and B) or piperonal (Fig. 8C) identify how these ligands fit within the active site. In the NR structure with tyramine (Fig. 8A), the amine group of the ligand is positioned roughly equidistant between the hydroxyl group of Tyr175 (3.2 Å) and the C4-position of NADP+ (3.2 Å). The phenolic portion of the ligand binds in a pocket defined by Tyr114 and Phe216 on opposite sides with the tyramine hydroxyl group forming a hydrogen bond with the side chain of Glu226 (3.1 Å). Notably, the ligand binding site of NR is much larger than tyramine (Fig. 8B) with additional space extending toward Ile177 and Arg267. The NR·NADP+·piperonal complex shows a similar mode of binding for the ligand (Fig. 8C). The aldehyde group is bound between Tyr175 and the C4 of NADP+. The benzodioxole ring is sandwiched between Tyr114 and Phe216 with a hydrogen bond interaction made between Ser182 and the O3 of piperonal.
FIGURE 8.
The ligand binding site of NR and loop movement. A, details of the tyramine binding site. B, surface view of the NR ligand binding site showing tyramine and NADP+. C, details of the piperonal binding site. D, comparison of the 209–220 loop movement in the NR·NADP+ (cream) and NR·NADP+·tyramine complexes. The positions of Thr213 and Phe216 are shown in each structure for reference. The positions of the hinge residues Arg209 and Lys220 are noted by triangles.
Experiments on tyramine, piperonal, and 3,4-dihydroxybenzaldehdye leave the substrate of the norbelladine reduction by this NR undetermined. It could be the individual components tyramine and the corresponding aldehyde or preformed norcraugsodine. The uncondensed components bind the NR active site, which could indicate a contribution of the enzyme to condensation through an increased local concentration of the two components but the location of binding is the same. This is not an ideal setup for a condensation on the enzyme because they would be in the same place. Also, none of the NR structures have the enzyme and NADP+ in an orientation that would obviously facilitate the condensation reaction of these substrates. For these reasons, the individual components bind the enzyme active site and can inhibit the noroxomaritidine reduction. It remains unclear if the enzyme actually uses these substrates to make norcraugsodine or takes norcraugsodine from solution.
Comparison of the NR structures reveals conformational flexibility in the NR active site (Fig. 8D). In the “open” active site conformation, the Arg209-Lys220 loop is extended out into solution away from the ligand binding site. In contrast, this region hinges at Arg209 and Lys220 to adopt a “closed” active site conformation in the NR·NADP+·tyramine complex. This shift in structure moves Thr213 and Phe216 10.2 and 12.0 Å, respectively, into the ligand binding site. The Arg209-Lys220 loop also adopts the closed active site conformation in the NR·NADP+·piperonal complex.
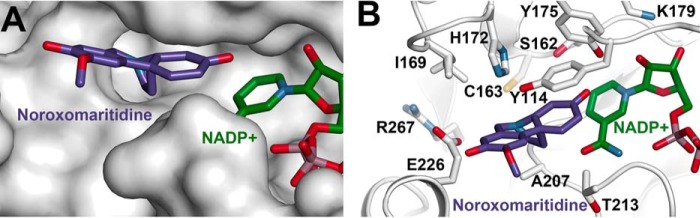
Because of limited availability of noroxomaritidine, computer-aided docking of this ligand into the x-ray structure of NR was performed (Fig. 9). The protein model contained the crystallographically determined NADP+ and the Arg209-Lys220 loop was in the open active site conformation. The closed active site conformation was too constrained to allow for fitting of the ligand in the site. Movement of the Arg209-Lys229 loop provides sufficient space in the NR active site to accommodate noroxomaritidine. The ligand binds with its ketone group and carbon-carbon bond of the A-ring oriented toward the Tyr175-Lys179 catalytic dyad and the C4 position of the cofactor nicotinamide ring to allow for reduction of the substrate.
FIGURE 9.
Structure-based model for noroxomaritidine binding by NR. A, surface representation of noroxomaritidine docked into the open active site conformation of NR. B, residues in the proposed noroxomaritidine binding site.
Discussion
The Amaryllidaceae alkaloids are a large group of plant natural products with over 300 documented structures and diverse biological activities. Moreover, these molecules are of pharmaceutical interest either as potential anti-tumor agents or for the symptomatic treatment of Alzheimer disease (1, 2, 4, 28). Although the general chemical steps in the core Amaryllidaceae alkaloid biosynthetic pathway have been established (Fig. 1), the genes and enzymes of the pathway remain largely unstudied. Recent transcriptome analysis for co-expression of compounds and biosynthetic genes involved in Amaryllidaceae alkaloid biosynthesis have provided molecular insights on this pathway, including the identification and characterization of N4OMT, which catalyzes the methylation of norbelladine, and CYP96T1, which can form noroxomaritidine (17, 18). A similar strategy now identifies a member of the SDR enzyme family from N. sp. aff. pseudonarcissus as a reductase (i.e. NR) that preferentially uses noroxomaritidine as a substrate. 400-Fold lower norcraugsodine reductase activity, which was above background, was also detected.
NR belongs to the SDR superfamily (Fig. 2) (14, 29). Although originally identified as oxidoreductases of ketones and aldehydes, including multiple enzymes involved in the biosynthesis of different types of alkaloids (22, 30, 31). The biochemical repertoire of the SDR superfamily has expanded to include carbon-carbon double bond reductases (23) and imine reductases (16). The co-expression of NR with N4OMT suggested a possible role in the biosynthesis of Amaryllidaceae alkaloids.
Given that SDRs, such as 5β-progesterone reductase (23), can catalyze the reduction of carbon-carbon double bonds, NR was tested using noroxomaritidine as a substrate (Fig. 3 and Table 1). Biochemical analysis of NR indicates that the enzyme functions to reduce noroxomaritidine using NADPH. Although the enantiomeric forms of noroxomaritidine have not been isolated, assays suggest a preference of NR for one of these forms. Depending on the enantiomer being reduced this activity could contribute to the biosynthesis of either maritinamine or elwesine (32, 33). Both of these compounds share a reduced carbon-carbon double bond, but are derived from the (10bR,4aS)-noroxomaritidine and (10bS,4aR)-noroxomaritidine skeletons, respectively. N. sp. aff. pseudonarcissus has not been previously documented to have these compounds and proper standards to determine their presence or absence are lacking. So, the product profiles in N. sp. aff. pseudonarcissus are unknown.
The x-ray crystal structures of NR in complex with NADP+ and either tyramine or piperonal (Figs. 6 and 7) clearly define the common structural features, including the catalytic dyad (i.e. Tyr175 and Lys179) and nucleotide cofactor binding site of the SDR superfamily (14). Moreover, the various structures of NR reveal the conformational flexibility of the Arg209-Lys220 loop in NR. Although the molecular determinants of noroxomaritidine binding in NR remain to be examined in detail, computer-aided docking of the substrate into the open conformation active site (Fig. 9) suggests a possible chemical mechanism for noroxomaritidine reduction.
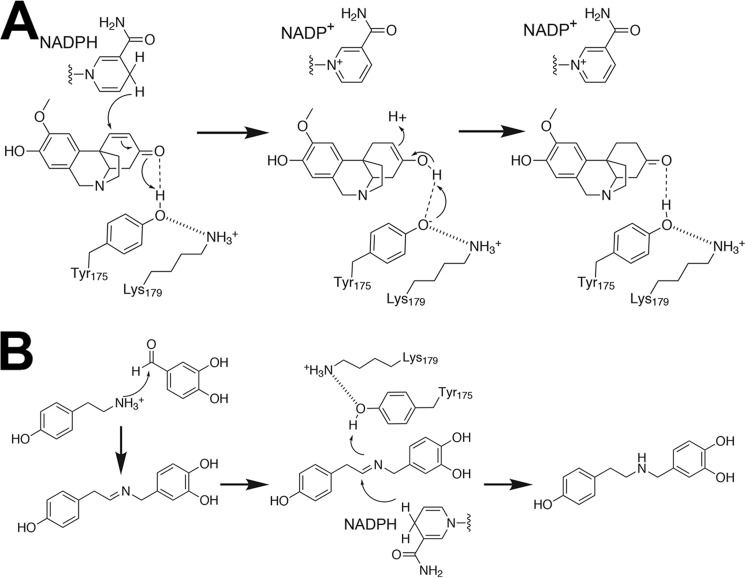
Extensive studies on multiple SDR enzymes support a common reaction mechanism in which the active site tyrosine serves as a general acid to protonate the keto-group of a bound substrate and the lysine in the catalytic dyad lower the pKa of the catalytic tyrosine to promote proton donation (14). The arrangement of Tyr175 and Lys179 in the NR active site (Fig. 8, A and B) suggests a similar chemical mechanism for noroxomaritidine reduction (Fig. 10A). In the reaction catalyzed by NR, binding of noroxomaritidine positions the substrate ketone in proximity to Tyr175 and NADPH. Electrostatic interaction with Lys179 reduces the pKa of Tyr175 to polarize the substrate carbonyl group for protonation and hydride transfer. The resulting enolate-like structure can then tautomerize to yield the ketone product (i.e. oxomaritinamine) with a reduced carbon-carbon double bond. The overall reaction is comparable with those proposed for various steroid carbon-carbon double bond reductases (23, 34, 35). The reduction of noroxomaritidine catalyzed by NR also requires positioning of the carbon-carbon double bond in proximity to the C4 of NADPH to allow for hydride transfer. It is possible that the flexibility of the NR active site loop can aid in the proper orientation of the substrate for catalysis. A similar role for the active site loop as an adaptive feature for allowing the binding of different substrates has been proposed for the SDRs that catalyze reductive terpene cyclization in iridoid alkaloid biosynthesis (36).
FIGURE 10.
Proposed reaction mechanisms for NR catalyzed reduction of noroxomaritidine (A) and norcraugsodine (B).
Reduction of an imine is required in the biosynthesis of norbelladine, a central metabolite in the Amaryllidaceae alkaloid pathway (Fig. 1). Because NR was the oxidoreductase that was co-expressed most consistently with N4OMT, we postulated that NR may also reduce norcraugsodine to norbelladine. Biochemical assays show that incubation of NR with tyramine, 3,4-dihydroxybenzaldehyde, and NADPH leads to norbelladine synthesis at a rate above background, but also with a specific activity substantially lower than that for noroxomaritidine reduction (Fig. 5 and Table 1). Comparable chemistry was also observed when other aldehydes (i.e. vanillin, isovanillin, and piperonal) were used in place of 3,4-dihydroxybenzaldehyde. This substrate flexibility and the low specific activity for this reaction may indicate this is a nonspecific enzymatic reduction.
For NR to catalyze norbelladine synthesis, nucleophilic attack of the tyramine amine group on the aldehyde via a condensation reaction leads to formation of norcraugsodine (Fig. 10B). Whether NR serves to enhance the rate of this reaction relative to the non-enzymatic rate is unclear. The three-dimensional structures of NR and the competition assays demonstrate that tyramine and aldehyde substrates bind at the active site; however, it is not obvious how both molecules would bind for the reaction leading to norcraugsodine. Nonetheless, our data indicates that the reduction of the imine increases in the presence of NR and NADPH. For this reaction, binding of norcraugsodine in the NR active site may position the imine group in proximity to Tyr175 and NADPH. As described above polarization of Tyr175 via interaction with Lys179 would allow the hydroxyl group to serve as a general acid with hydride transfer from NADPH, which results in norbelladine synthesis (Fig. 10B).
The specific activity differences for noroxomaritidine reduction (8600 ± 1250 pmol min−1 mg of protein−1) compared with norcraugsodine reduction (21.2 ± 1.2 pmol min−1 mg of protein−1) (Table 1) suggest that former reaction is preferred and that the latter is a nonspecific reaction. It remains to be determined if the norbelladine formation in planta results from the low side activity of NR or an as yet unidentified enzyme. It should be noted that derivatives of the carbon-carbon double bond reduction of noroxomaritidine are rarely reported in planta. It is possible that NR is normally outcompeted by an unidentified reductase that reduces the ketone group of noroxomaritidine, which is a necessary step for the production of the hemanthamine and crinine alkaloids (20). Ultimately, the identification of NR, along with previous work on N4OMT and CYP96T1 (17, 18), provide biochemical insight on Amaryllidaceae alkaloid biosynthesis and will be useful in future co-expression analysis to search for other genes related to this pathway and to determine the connections between Amaryllidaceae alkaloid biosynthetic gene expression and the accumulation of various alkaloids in different Amaryllidaceae plants.
Experimental Procedures
Plant Tissue and Chemicals
Plant tissues and chemicals used in this study were as previously described (17, 18). In addition, piperonal was purchased from Sigma; 8,9-O-didemethyloxomaritidine and noroxomaritidine were obtained from our natural product collection; dimethyl sulfoxide was from New England BioLabs; NADP+ was from MP Biomedicals; anhydrous citric acid (ACS reagent grade; 99.5%) and sodium borohydride (99%) were from Acros Organics; and sodium cyanoborohydride (reagent grade 95%) was from Aldrich.
Candidate Gene Identification
Candidate genes were selected as in the discovery of CYP96T1 (18), but instead of a list of cytochromes P450, a list of oxidoreductases (supplemental Table S1) were used in the BLASTP query with an e-value cutoff of 1 × 10−4 against ESTScan predicted peptides for each transcriptome. A contig for an SDR was found to co-express with N4OMT in 4 of the 5 available Amaryllidaceae transcriptomes, whereas other BLASTP results showed co-expression with N4OMT in 3 or less of the transcriptomes (18).
Cloning and Recombinant Enzyme Purification
N. sp. aff. pseudonarcissus bulb cDNA was prepared as previously described (18). The outer PCR contained the following components: 25 ng N. sp. aff. pseudonarcissus bulb cDNA, 1× Phusion HF reaction buffer, 1 unit of New England Biolabs Phusion High-Fidelity DNA polymerase, 3% dimethyl sulfoxide, 0.4 mm dNTPs, 0.4 μm NR forward outer (5′-dGGAAAGCCTTCAGAGGAGATT-3′), and 0.4 μm NR reverse outer (5′-dAGATAGCACCGTGGAGAT-3′) primers. The parameters for the PCR were 98 °C 30 s for 1 cycle; 98 °C 10 s, 50 °C 30 s, 72 °C 60 s for 35 cycles; 72 °C 5 min 1 cycle, 4 °C until removed. The inner PCR mixture was the same except 0.2 μm NR forward inner (5′-dAATTGGATCCATGTCGTTGGAGAAGAGATGGT-3′; BamHI site underlined) and 0.2 μm NR reverse inner (5′-dAATTGCGGCCGCTCAACCATTTATGGTCCGTCCT-3′; NotI site underlined) primers were used with the outer PCR product as template instead of cDNA. The inner PCR program had a Tm of 52 °C instead of 50 °C and 25 cycles instead of 35 cycles. The BamHI/NotI-digested PCR fragment was ligated into pET28a to yield the pET28a-NR construct.
The pET28a-NR construct was transformed into E. coli Rosetta II (DE3) cells (EMD Millipore). Cells were cultured in Terrific broth up to A600 nm ∼0.6–0.8. Induction of protein expression used a final concentration of 1 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 16 °C. The enzyme used in enzyme assays was purified with a TALON-cobalt column as previously (17). For protein crystallization, cells were pelleted by centrifugation and resuspended in lysis buffer (50 mm Tris, pH 8.0, 500 mm NaCl, 20 mm imidazole, 10% glycerol, and 1% Tween 20). Following sonication, cell debris were removed by centrifugation, and the resulting lysate was passed over either a Ni2+-nitriloacetic acid (Qiagen) or a TALON-cobalt column equilibrated in lysis buffer. The column was then washed with 50 mm Tris, pH 8.0, 500 mm NaCl, 20 mm imidazole, and 10% glycerol. Bound His-tagged protein was eluted with 50 mm Tris, pH 8.0, 500 mm NaCl, 250 mm imidazole, and 10% glycerol. For protein crystallization, the His-tag was removed by overnight dialysis at 4 °C using thrombin (1:2000 total protein). Dialyzed protein was loaded onto a mixed benzamidine-Sepharose/Ni2+-nitrilotriacetic acid column. The flow-through was loaded onto a Superdex-75 26/60 HiLoad size exclusion column (GE Healthcare) equilibrated with 25 mm HEPES, pH 7.5, and 100 mm NaCl using an Akta Explorer FPLC system. Concentration of all protein purifications was determined by the Bradford method (Protein Assay, Bio-Rad) with bovine serum albumin as a standard.
Enzyme Assays
Assays were performed with 100 mm citrate buffer, pH 6.0, at 35 °C in 40-μl reactions except where noted in figure legends. Compounds screened for enzymatic activity were monitored with the parameters listed in Table 3. Assay-specific conditions are described in appropriate figures and tables. Assays were extracted with ethyl acetate at pH 9.5 as previously described (17). After drying, extracts were re-suspended in mobile phase matching the solvent composition at the beginning of the HPLC program. Samples, except where noted otherwise, were run on a IL-20AC XR prominence liquid autosampler coupled to a 20AD XR prominence liquid chromatography instrument with a Phenomenex Luna 5-μm C8(2) 250 × 4.60-mm column and a QTRAP 4000 as used for CYP96T1 (18). The LC time programs used for assays were as follows: a isocratic time program with 20% acetonitrile and 0.1% formic acid was used for pH optimum, temperature optimum, IC50, and specific activity assays but initial screening of substrates for enzymatic activity used the same time program as for substrate screening of N4OMT (17). The MRM parameters used, with the exception of specific activity measurements, to monitor norbelladine were m/z 260.0/138.0 and 260.0/121.0 with CE (15) and DP (50) and oxomaritinamine was monitored with the following MRMs m/z 274.3/136.1 and 274.3/219.1 with CE (35) and DP (70).
TABLE 3.
Product MS/MS parameters
| Substrate | Product | Parameters (parent ion m/z)(CE)(DP) |
|---|---|---|
| Noroxomaritidine | Oxomaritinamine | (274.3)(35)(70) |
| 8-O-,9-O-Didemethyloxomaritidine | 9-O-Demethyloxomaritinamine | (260.1)(35)(70) |
| 3,4-Dihydroxybenzaldehyde and tyramine | Norbelladine | (260.0)(15)(50) |
| Vanillin and tyramine | 3′-O-Methylnorbelladine | (274.3)(20)(60) |
| Isovanillin and tyramine | 4′-O-Methylnorbelladine | (274.3)(20)(60) |
| Piperonal and tyramine | 4-(2-((1,3-Benzodioxol-5-ylmethyl)amino)ethyl)phenol | (272.1)(20)(60) |
Specific activity measurements required quantification of oxomaritinamine and a new LC-MS/MS setup. Oxomaritinamine was quantified by incubating noroxomaritidine and NR overnight and equating the quantity of noroxomaritidine consumed to the quantity of oxomaritinamine produced using the same HPLC program used to analyze enzyme assays during N4OMT characterization at A288 nm (17). This oxomaritinamine standard was used in specific activity experiments on the LC-MS/MS to determine product quantity. For specific activity measurements, a QTRAP 6500 was used for MRM analysis with an isocratic flow of 20% acetonitrile and 0.1% formic acid in H2O. The MRM parameters for specific activity measurements were norbelladine m/z 260.1/138.0 and 260.1/121.0 CE (10) and DP (60), 4′-O-methylnorbelladine m/z 274.1/137.0 and 274.1/122.0 CE (20) and DP (60), and oxomaritinamine m/z 274.1/136.1 and 274.1/219.1 CE (30) and DP (70).
Norbelladine Analog Synthesis
4-(2-((1,3-Benzodioxol-5-ylmethyl)amino)ethyl)phenol, 3′-O-methylnorbelladine, and 4′-O-methylnorbelladine were prepared by mixing ∼100 mm sodium cyanoborohydride and ∼10 mm tyramine with ∼10 mm piperonal, vanillin, or isovanillin, respectively, in 2.5 ml of anhydrous methanol. After incubation overnight at room temperature, the reactions were evaporated to 0.5 ml with subsequent addition of 2 ml of 1 m sodium carbonate, pH 9.5, and extracted twice with 2 ml of ethyl acetate. After drying, all extracts were dissolved in 1 ml of water, diluted 1/20, and purified by fraction collection with the same HPLC program used to analyze enzyme assays during N4OMT characterization (17).
Protein Crystallography
Purified NR was concentrated to 8 mg ml−1 and crystallized using the hanging-drop vapor-diffusion method with a 2-μl drop (1:1 concentrated protein and crystallization condition). Diffraction quality crystals of the NR·NADP+ complex were obtained at 4 °C with 20% PEG-8000 and 100 mm HEPES buffer, pH 7.5. Crystals of the NR·NADP+·tyramine complex formed at 4 °C in 2 m ammonium sulfate, 100 mm CAPS buffer, pH 10.5, 200 mm lithium sulfate, 1.5 mm NADP+, and 6 mm tyramine. For the NR·NADP+·piperonal complex, 35% 2-methyl-2,4-pentanediol, 100 mm sodium acetate, pH 4.5, and 3 mm piperonal was used for crystallization. Individual crystals were flash-frozen in liquid nitrogen with the mother liquor containing 25% glycerol as a cryoprotectant. Diffraction data (100 K) was collected at the Argonne National Laboratory Advanced Photon Source 19-ID beamline. The data were indexed, scaled, and integrated with HKL3000 (37). Molecular replacement implemented in PHASER (38) used D. stramonium tropinone reductase-II (PDB code 2AE2) (39) as a search model to determine the structure of the NR·NADP+·tyramine complex. Iterative rounds of manual model building and refinement, which included translation-libration-screen models, used COOT (40) and PHENIX (41). The crystal structures of NR·NADP+ and NR·NADP+·piperonal were determined by molecular replacement using PHASER and the NR·tyramine structure with ligands removed as the search model and with building and refinement as described above. Data collection and refinement data are summarized in Table 2. The final model of the NR·NADP+ complex included residues Ser16 to Gly271, NADP+, and 136 waters. The final model of the NR·NADP+·tyramine complex included residues Ser16 to Gly271 for chain A, residues Met15 to Gly271 for chain B, residues Leu17 to Asn270 for chain C, and residues Leu17 to Gly271 for chain D, NADP+ in chains A-C, tyramine in chains B and D, and 443 waters. The final model of the NR·NADP+·piperonal complex included Met15 to Gly271 and NADP+ in chains A and B, piperonal in chain A, and 644 waters. Coordinates and structure factors for NR complexed with NADP+ (PDB 5FEU), NADP+ and tyrosine (PDB 5FF9), and NADP+ and piperonal (PDB 5FFF) have been deposited in the RCSB Protein Data Bank.
Computational Docking of Substrates
Molecular docking of noroxomaritidine into the three-dimensional structure of the NR·NADP+ complex was performed using AutoDock Vina (version 1.1.2) (42). The ligand was generated using ChemDraw 3D and was energy minimized. The ligand was manually placed into the active site by using the position of NADP+ in the active site as a guide with docking using a grid box of 30 × 30 × 30 Å and the level of exhaustiveness set to 8.
Author Contributions
As a part of his Ph.D. thesis M. B. K. conceived the study, performed candidate selection, cloning, and enzymatic experiments, and wrote the majority of the manuscript. C. K. H. performed crystallography and docking experiments and contributed to the writing of the manuscript. J. M. J. contributed to conception of crystallography and docking experiments, revisions of the manuscript, and data analysis. T. M. K. contributed to conception of experiments and revisions of the manuscript.
Supplementary Material
Acknowledgments
Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center of the Advanced Photon Source, a national user facility operated by the University of Chicago for the Department of Energy Office of Biological and Environmental Research under Grant DE-AC02-06CH11357.
This work was supported in part by National Institutes of Health Grant 1RC2GM092561 from the NIGMS and National Science Foundation Grant DBI-0521250 for acquisition of the QTRAP LC-MS/MS. A patent has been filed for the sequence of noroxomaritidine reductase. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
The atomic coordinates and structure factors (codes 5FEU, 5FF9, and 5FFF) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SDR
- short-chain alcohol dehydrogenase/reductase
- N4OMT
- norbelladine 4′-O-methyltransferase
- NR
- noroxomaritidine reductase
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- PDB
- Protein Data Bank
- MRM
- multiple reaction monitoring
- CE
- collision energy
- DP
- declustering potential.
References
- 1. Wilcock G., Howe I., Coles H., Lilienfeld S., Truyen L., Zhu Y., Bullock R., Kershaw P., and Group G.-G.-S. (2003) A long-term comparison of galantamine and donepezil in the treatment of Alzheimer's disease. Drugs Aging 20, 777–789 [DOI] [PubMed] [Google Scholar]
- 2. Havelek R., Seifrtova M., Kralovec K., Bruckova L., Cahlikova L., Dalecka M., Vavrova J., Rezacova M., Opletal L., and Bilkova Z. (2014) The effect of Amaryllidaceae alkaloids haemanthamine and haemanthidine on cell cycle progression and apoptosis in p53-negative human leukemic Jurkat cells. Phytomedicine 21, 479–490 [DOI] [PubMed] [Google Scholar]
- 3. He M., Qu C., Gao O., Hu X., and Hong X. (2015) Biological and pharmacological activities of Amaryllidaceae alkaloids. R.S.C. Adv. 5, 16562–16574 [Google Scholar]
- 4. Lamoral-Theys D., Andolfi A., Van Goietsenoven G., Cimmino A., Le Calvé B., Wauthoz N., Mégalizzi V., Gras T., Bruyère C., Dubois J., Mathieu V., Kornienko A., Kiss R., and Evidente A. (2009) Lycorine, the main phenanthridine Amaryllidaceae alkaloid, exhibits significant antitumor activity in cancer cells that display resistance to proapoptotic stimuli: an investigation of structure-activity relationship and mechanistic insight. J. Med. Chem. 52, 6244–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdel-Halim O. B., Morikawa T., Ando S., Matsuda H., and Yoshikawa M. (2004) New crinine-type alkaloids with inhibitory effect on induction of inducible nitric-oxide synthase from Crinum yemense. J. Nat. Prod. 67, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 6. Battersby A. R., Bink R., and Breuer S. W. (1961) Biosynthesis in the Amaryllidaceae: incorporation of norbelladine into lycorine and norpluvine. Proc. Chem. Soc. 243 [Google Scholar]
- 7. Wildman W. C., Fales H. M., Highet R. J., Breuer S. W., and Battersby A. R. (1962) Biosynthesis in the Amaryllidaceae: evidence for intact incorporation of norbelladine into lycorine, crinamine, and belladine. Pro. Chem. Soc. 180–181 [Google Scholar]
- 8. Battersby A. R., Fales H. M., and Wildman W. C. (1961) Biosynthesis in the Amaryllidaceae: tyrosine and norbelladine as precursors of haemanthamine. J. Am. Chem. Soc. 83, 4098–4099 [Google Scholar]
- 9. Suhadolnik R. J., Fischer A. G., and Zulalian J. (1963) Biogenesis of the Amaryllidaceae alkaloids: II. studies with whole plants, floral primordia and cell free extracts. Biochem. Biophys. Res. Commun. 11, 208–212 [DOI] [PubMed] [Google Scholar]
- 10. Gallage N. J., Hansen E. H., Kannangara R., Olsen C. E., Motawia M. S., Jørgensen K., Holme I., Hebelstrup K., Grisoni M., and Møller B. L. (2014) Vanillin formation from ferulic acid in Vanilla planifolia is catalysed by a single enzyme. Nat. Commun. 5, 4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Widhalm J. R., and Dudareva N. (2015) A familiar ring to it: biosynthesis of plant benzoic acids. Mol. Plant 8, 83–97 [DOI] [PubMed] [Google Scholar]
- 12. Kutchan T. M., Bock A., and Dittrich H. (1994) Heterologous expression of the plant proteins strictosidine synthase and berberine bridge enzyme in insect cell culture. Phytochemistry 35, 353–360 [DOI] [PubMed] [Google Scholar]
- 13. Samanani N., and Facchini P. J. (2002) Purification and characterization of norcoclaurine synthase. The first committed enzyme in benzylisoquinoline alkaloid biosynthesis in plants. J. Biol. Chem. 277, 33878–33883 [DOI] [PubMed] [Google Scholar]
- 14. Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., and Ghosh D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013 [DOI] [PubMed] [Google Scholar]
- 15. Penning T. M. (2015) The aldo-keto reductases (AKRs): overview. Chem. Biol. Interact. 234, 236–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stavrinides A., Tatsis E. C., Foureau E., Caputi L., Kellner F., Courdavault V., and O'Connor S. E. (2015) Unlocking the diversity of alkaloids in Catharanthus roseus: nuclear localization suggests metabolic channeling in secondary metabolism. Chem. Biol. 22, 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kilgore M. B., Augustin M. M., Starks C. M., O'Neil-Johnson M., May G. D., Crow J. A., and Kutchan T. M. (2014) Cloning and characterization of a norbelladine 4′-O-methyltransferase involved in the biosynthesis of the Alzheimer's drug galanthamine in Narcissus sp. aff. pseudonarcissus. PLoS ONE 9, e103223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kilgore M. B., Augustin M. M., May G. D., Crow J. A., and Kutchan T. M. (2016) CYP96T1 of Narcissus sp. aff. pseudonarcissus catalyzes formation of the para-para′ C-C phenol couple in the Amaryllidaceae alkaloids. Front. Plant Sci. 7, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feinstein A. I., and Wildman W. C. (1976) Biosynthetic oxidation and rearrangement of vittatine and its derivatives. J. Org. Chem. 41, 2447–2450 [Google Scholar]
- 20. Fuganti C., and Mazza M. (1972) The absolute configuration of narciclasine: a biosynthetic approach. J. Chem. Soc. Chem. Commun. 1972, 237–239 [Google Scholar]
- 21. Fuganti C. (1973) Evidence for the intermediacy of 11-hydroxyvittatine in the biosynthesis of narciclasine. Gazz. Chim. Ital. 22, 1255–1258 [Google Scholar]
- 22. Reinhardt N., Fischer J., Coppi R., Blum E., Brandt W., and Dräger B. (2014) Substrate flexibility and reaction specificity of tropinone reductase-like short-chain dehydrogenases. Bioorg. Chem. 53, 37–49 [DOI] [PubMed] [Google Scholar]
- 23. Thorn A., Egerer-Sieber C., Jäger C. M., Herl V., Müller-Uri F., Kreis W., and Muller Y. A. (2008) The crystal structure of progesterone 5β-reductase from Digitalis lanata defines a novel class of short chain dehydrogenases/reductases. J. Biol. Chem. 283, 17260–17269 [DOI] [PubMed] [Google Scholar]
- 24. Banfi L., Narisano E., Riva R., Stiasni N., Hiersemann M., Yamada T., and Tsubo T. (2001) Sodium borohydride. in Encyclopedia of Reagents for Organic Synthesis, John Wiley & Sons, Ltd., New York [Google Scholar]
- 25. Hampp N., and Zenk M. H. (1988) Homogeneous strictosidine synthase from cell suspension cultures of Rauvolfia serpentina. Phytochemistry 27, 3811–3815 [Google Scholar]
- 26. Deacon A. M., Ni Y. S., Coleman W. G. Jr., and Ealick S. E. (2000) The crystal structure of ADP-l-glycero-d-mannoheptose 6-epimerase: catalysis with a twist. Structure 8, 453–462 [DOI] [PubMed] [Google Scholar]
- 27. Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., and Oppermann U. (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277, 25677–25684 [DOI] [PubMed] [Google Scholar]
- 28. Evidente A., and Kornienko A. (2009) Anticancer evaluation of structurally diverse Amaryllidaceae alkaloids and their synthetic derivatives. Phytochem. Rev. 8, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moummou H., Kallberg Y., Tonfack L. B., Persson B., and van der Rest B. (2012) The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns. BMC Plant Biol. 12, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brock A., Brandt W., and Dräger B. (2008) The functional divergence of short-chain dehydrogenases involved in tropinone reduction. Plant J. 54, 388–401 [DOI] [PubMed] [Google Scholar]
- 31. Ziegler J., Voigtländer S., Schmidt J., Kramell R., Miersch O., Ammer C., Gesell A., and Kutchan T. M. (2006) Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J. 48, 177–192 [DOI] [PubMed] [Google Scholar]
- 32. Pabuççuoglu V., Richomme P., Gözler T., Kivçak B., Freyer A. J., and Shamma M. (1989) Four new crinine-type alkaloids from Sternbergia species. J. Nat. Prod. 52, 785–791 [Google Scholar]
- 33. Boit H.-G., and Döpke W. (1961) Alkaloide aus Haemanthus-, Zephranthes-, Galanthus- und Crinum-arten. Die Naturwissenschaften 48, 406–407 [Google Scholar]
- 34. Jez J. M., and Penning T. M. (1998) Engineering steroid 5β-reductase activity into rat liver 3α-hydroxysteroid dehydrogenase. Biochemistry 37, 9695–9703 [DOI] [PubMed] [Google Scholar]
- 35. Di Costanzo L., Drury J. E., Penning T. M., and Christianson D. W. (2008) Crystal structure of human liver Δ(4)-3-ketosteroid 5β-reductase (Akr1d1) and implications for substrate binding and catalysis. J. Biol. Chem. 283, 16830–16839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kries H., Caputi L., Stevenson C. E., Kamileen M. O., Sherden N. H., Geu-Flores F., Lawson D. M., and O'Connor S. E. (2016) Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nat. Chem. Biol. 12, 6–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamashita A., Kato H., Wakatsuki S., Tomizaki T., Nakatsu T., Nakajima K., Hashimoto T., Yamada Y., and Oda J. (1999) Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9-Å resolution: implication for stereospecific substrate binding and catalysis. Biochemistry 38, 7630–7637 [DOI] [PubMed] [Google Scholar]
- 40. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hall B. G. (2013) Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235 [DOI] [PubMed] [Google Scholar]
- 44. Kilgore M. B., and Kutchan T. M. (December 23, 2015) U. S. patent WO/2015/196100
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.