Abstract
Periodontitis is a chronic oral inflammatory disease affecting one in five individuals that can lead to tooth loss. CD4+ Th cells activated by a microbial biofilm are thought to contribute to the destruction of alveolar bone surrounding teeth by influencing osteoclastogenesis through IL-17A and receptor activator for NF-κB ligand effects. The relative roles of mucosal Ag presentation cells in directing Th cell immune responses against oral pathogens and their contribution to destruction of alveolar bone remain unknown. We tested the contribution of mucosal Langerhans cells (LCs) to alveolar bone homeostasis in mice following oral colonization with a well-characterized human periodontal pathogen, Porphyromonas gingivalis. We found that oral mucosal LCs did not protect from or exacerbate crestal alveolar bone destruction but were responsible for promoting differentiation of Th17 cells specific to P. gingivalis. In mice lacking LCs the Th17 response was suppressed and a Th1 response predominated. Bypassing LCs with systemic immunization of P. gingivalis resulted in a predominantly P. gingivalis–specific Th1 response regardless of whether LCs were present. Interestingly, we find that in vivo clonal expansion of P. gingivalis–specific Th cells and induced regulatory T cells does not depend on mucosal LCs. Furthermore, destruction of crestal alveolar bone induced by P. gingivalis colonization occurred regardless of the presence of mucosal LCs or P. gingivalis–specific Th17 cells. Our data indicate that both LCs and Th17 cells are redundant in contributing to alveolar bone destruction in a murine model of periodontitis.
Introduction
Periodontitis is a chronic oral disease initiated by microbial biofilms that colonize the tooth surface and drive destructive inflammatory responses involving infiltration of immune cells that secrete proinflammatory cytokines and chemokines into the gingiva (1, 2). Infiltration of these immune cells leads to host-mediated destruction of alveolar bone and loss of connective tissue attachment to the tooth (3–6). In this regard, periodontitis resembles other inflammatory-driven bone destructive diseases where proinflammatory cytokines IL-1, IL-6, IL-17A, receptor activator for NF-κB ligand (RANKL), and TNF-α are important mediators of osteoclast differentiation, activation, and survival (7). At mucosal surfaces the release of proinflammatory cytokines is triggered by the interaction between the surrounding microbial ecosystem and cells responsible for immune surveillance. Dendritic cells (DCs) in the epithelial and subepithelial layers of the oral mucosa survey the microbiota and drive innate and adaptive immune responses through cognate interactions with T cells and bystander release of cytokines (8, 9). DC subsets in the oral mucosa of mice can be divided into four groups based on their anatomical location, migration kinetics, and expression of cell surface markers (10, 11). Of these four, Langerhans cells (LCs) located within the stratified squamous epithelium of the mucosae are ideally placed to be the first immune cells to sample Ags of the periodontal biofilm and elicit regulatory or proinflammatory immune responses (12). However, the exact role played by differentiated CD4+ Th cells Th1, Th2, Th17, or regulatory T cells (Tregs) in the pathogenesis of periodontal bone destruction is unresolved (13–19).
The Gram-negative, anaerobic bacterium Porphyromonas gingivalis is considered a keystone pathogen within the microbial biofilms surrounding the teeth of periodontally diseased subjects (20, 21). In a murine model of periodontitis, P. gingivalis–induced alveolar bone destruction was dependent on MHC class II (MHC-II) Ag presentation, activated Th cells (22, 23), and on the expression of gingipains, which are established virulence factors of P. gingivalis (24, 25). Recent work using this model of P. gingivalis–induced periodontal bone destruction demonstrated that the combined ablation of LCs and CD207 (Langerin+) DCs resulted in significant alveolar bone destruction and higher IFN-γ production but had little to no effect on the expression of IL-17A and IL-10 (26). LCs have well-known immunosuppressive functions, but they can also drive pathogen-specific adaptive Th responses (26–31). Subepithelial Langerin+ DCs in the skin are primarily involved in cross-presentation to CD8+ T cells, but they also sustain Th1-mediated responses during fungal and viral infections. Langerin+ DCs induce retinoic acid–dependent differentiation of IL-10–producing induced Tregs (iTregs) (32, 33) and regulate RANKL expression in osteoclastogenic Th cells (34).
In this study, we test the exclusive contribution of mucosal LCs to alveolar bone destruction in a murine model of periodontitis using the human (hu)Langerin–diphtheria toxin A (DTA) mouse (27). huLangerin-DTA mice express the active subunit of DTA under control of the human langerin promoter, which is uniquely expressed in LCs. When LCs are in the epithelial layer of the skin, expression of DTA leads to early and sustained LC ablation. In other mouse models (35), including murine (mu)Langerin–diphtheria toxin receptor (DTR) mice (36), Langerin+ DCs, LCs, and CD8+ lymphoid-resident DCs that express CD207 are also ablated by the systemic administration of diphtheria toxin (26, 36, 37). Critically for our present study, these non-LC types have been reported to be unchanged in huLangerin-DTA mice when examined in the skin and skin-draining lymph nodes (LNs) (27). Additionally, using a peptide:MHC-II tetramer (38), we track in vivo the clonal expansion and phenotype of CD4+ T cells specific for P. gingivalis and their relative contribution to alveolar bone destruction associated with the exclusive ablation of mucosal LCs.
Materials and Methods
Mice
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Minnesota and performed on age-matched (6–8 wk) mice or littermates. C57BL/6J (H-2b) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). huLangerin-DTA mice have been described elsewhere (27). Foxp3:GFP mice (C57BL/6J background), originally developed in the laboratory of Dr. A. Rudensky (University of Washington, Seattle, WA), were a gift from Dr. S. Way (University of Minnesota). The F1 progeny of Foxp3:GFP and huLangerin-DTA mice were screened to identify LC-deficient and C57BL/6J Foxp3:GFP littermates and used to study the effects of LC ablation on the development of P. gingivalis–specific iTregs. All mice were housed in microisolator cages with food and water ad libitum in a specific pathogen-free animal facility in compliance with the Association for Assessment and Accreditation of Laboratory Animal Care at the University of Minnesota.
Infection of mice with P. gingivalis
P. gingivalis strain ATCC 53977 (A7A1-28) was a gift from Dr. P. Baker (Bates College, Lewiston, ME) (39). P. gingivalis strain ATCC 33277 and its derivatives KDP136 and KDP137 were obtained from Dr. M. Herzberg (University of Minnesota) from stocks sent by Dr. K. Nakayama (Kyushu University, Kyushu, Japan). Generation of the triple rgpA, rgpB, kpg (KDP136) and rgpA, kpg, hagA (KDP137) knockout mutants have been described elsewhere (40). All P. gingivalis strains were grown anaerobically at 37°C for 7 or 14 d in 5% CO2/10% H2/85% N2 in Todd–Hewitt broth and passaged on Todd–Hewitt broth–blood agar plates, both supplemented with 5.0 μg/ml hemin and 0.5 μg/ml menadione. For oral inoculation experiments, mice were pretreated with sulfamethoxazole-trimethoprim antibiotics added to their ad libitum water for 10 d. Mice were orally inoculated with 4 × 109 P. gingivalis CFI/100 μl of prereduced 2% (w/v) carboxymethylcellulose or with 100 μl of prereduced PBS-carboxymethylcellulose (sham inoculated) using a ball-tipped gavage needle every 4 d for the duration of each experiment (22, 38). When required, mice were subjected to s.c. injection at three sites along both flanks with 200 μl of prereduced PBS with or without 1 × 109 CFU of P. gingivalis. Mice were similarly boosted s.c. at day 21 and LNs draining the flanks (inguinal, brachial, and axillary) were harvested for analysis of CD4+ T cells.
Confocal microscopy
Gingival tissues were teased off maxillary jaws as mucoperiosteal flaps using the cutting edge of an 18-gauge needle and placed in OCT embedding medium (Sakura Finetek, Torrance, CA). Cryostat sections (10 μm) were acetone fixed and rehydrated with PBS. Sections were blocked with 5% (v/v) rat serum in PBS and incubated for 30 min at room temperature with 10 μg/ml anti-mouse CD207–Alexa Fluor 647 (918A11; Dendritics, San Diego, CA), CD11b–Alexa Fluor 488 (M1/70; eBioscience, San Diego, CA), CD11c–Alexa Fluor 594 (N418; BioLegend, San Diego, CA), and/or epidermal cell adhesion molecule (EpCAM)–FITC (G8.8; BioLegend) in rat serum–supplemented PBS. Control slides were incubated with the appropriate rat IgG2a, κ isotype controls. After serial washes with PBS–Tween 20 and PBS, DAPI (Life Technologies, Carlsbad, CA)–stained sections were imaged at ×10 and ×40 using an LSM 700 Zeiss confocal laser scanning microscope, and specific signals were quantified using ZEN software (Carl Zeiss Microscopy, Jena, Germany).
Identification of tissue-resident immune cells in oral mucosa by flow cytometry
Groups of 6- to 8-wk-old huLangerin-DTA and C57BL/6J mice were inoculated six times with P. gingivalis by oral gavage at 4-d intervals. Mice were injected retro-orbitally with a nonsaturating amount (1.25 μg) of CD45-FITC mAb (30-F11; eBioscience) 3 min prior to sacrifice. Three minutes is sufficient for the CD45 mAb to circulate in the vascular system and to stain blood-resident immune cells. Addition of this mAb is critical to the unequivocal identification of tissue-resident immune cells (CD45-FITC−) in downstream flow cytometry analysis. The keratinized gingiva of maxillary and mandibular teeth and the entire buccal oral mucosa excluding the anterior two-thirds of the hard palate that overlays the nasal-associated lymphoid tissue were pooled from two mice and placed in 2 ml of complete EHAA (Life Technologies). Tissue was incubated in a shaking 37°C incubator for 60 min in the presence of 2 mg/ml collagenase D (Roche Diagnostics, Indianapolis, IN) and 1 mg/ml DNase I (Sigma-Aldrich). EDTA was added for the last 10 min to a final concentration of 5 mM. Tissue was minced and single-cell suspensions were stained with cell viability dye Zombie Aqua (BioLegend) according to the manufacturer’s protocol. Cells were then stained with anti-mouse CD45, CD11b, CD11c, I-Ab (M5/114.15.2; BioLegend), EpCAM, F4/80 (BM8; BioLegend), and Ly-6G (IA8; BioLegend) flurochrome-conjugated mAbs. To detect langerin, cells were permeabilized and stained intracellularly with anti-CD207 mAb conjugated to Alexa Fluor 647. Cell fluorescence emissions were acquired on an LSR II flow cytometer (BD Biosciences, San Jose, CA) and analyzed with FlowJo software (Tree Star, Ashland, OR).
FITC painting of gingival tissue
FITC (Sigma-Aldrich, St. Louis, MO) was dissolved in a 1:1 (v/v) acetone/dibutyl phthalate (DBP) (Sigma-Aldrich) solution at 10 mg/ml. Mice anesthetized i.p. with a standard ketamine/xylazine regimen (100 mg/10 mg per kilogram body weight) were laid on their backs and their mouths held open by a micro cheek retractor that moved the buccal mucosa away from the maxillary molars. Vaseline was liberally applied to the lips to prevent delivery of FITC/DBP to the peri-oral skin. Left and right maxillary molars and gingival surfaces were dried with a micro cotton swab and 10 μl of FITC/DBP solution was applied to the left and right palatal gingiva using a flexible fine gel-loading micropipette tip. Mice were kept on their backs (5 min) to allow the FITC solution to dry before they were returned to their cage for recovery.
Identification of DC subsets in cervical LNs
Cervical LNs (CLNs) were harvested from mice at 0, 24, and 96 h following palatal painting with FITC. For optimal DC isolation, CLNs were incubated for 30 min at 37°C in Dulbecco’s PBS with Ca2+ and Mg2+, supplemented with 2 mg/ml collagenase D and 1 mg/ml DNase I. CD11c+ cells were enriched from single-cell suspensions by immunomagnetic positive selection (CD11c MicroBeads; Miltenyi Biotec, Auburn, CA). Cells were stained with the following fluorochrome-conjugated anti-mouse mAbs: CD11c, I-Ab, B220 (RA3-6B2; BioLegend), CD3 (145-2C11), CD8α (53-6.7), CD103 (2E7; BioLegend), and EpCAM. Cells were permeabilized and stained intracellularly with anti-CD207 mAbs to detect langerin. Cell fluorescence emissions were acquired on an LSR II flow cytometer and analyzed with FlowJo software.
Assessment of alveolar bone levels
Maxillae were harvested from mice at day 48 and processed for alveolar bone loss assessment as previously described (41). Jaws were photographed in a standardized position and were measured from the cementum–enamel junction of each tooth to the alveolar bone crest at 10 landmark points along the buccal surface of the three molar teeth using Adobe Photoshop Elements (Adobe, San Jose, CA). Measurements were normalized to the mesiodistal size of molar no. 2 when measured at the cementum–enamel junction.
ELISPOT analysis
ELISPOT was used to assess the phenotype of CD4+ T cell populations in C57BL/6J mice to determine the total P. gingivalis–specific CD4+ T cell response. Single-cell preparations from CLNs of P. gingivalis– or sham-colonized C57BL/6J mice were prepared. CD4+ T cells were purified to >94% by immunomagnetic negative selection (CD4+ T cell isolation kit, Miltenyi Biotec) and incubated in equal number with 5 × 105 irradiated naive splenocytes in 96-well filter plates (EMD Millipore, Billerica, MA) coated with cytokine capture Abs (eBioscience) specific for mouse IFN-γ (AN-18), IL-4 (11B11) or IL-17A (eBio17CK15A5) as previously described (38). Cytokine-releasing clones were identified as spot-forming units (SFU) following incubation with biotinylated anti–IFN-γ (R4-6A2), IL-4 (BVD6-24G2), or IL-17A (eBio17B7) detection Abs (eBioscience), streptavidin-conjugated HRP (eBioscience), and 3-amino-9-ethylcarbazole substrate solution (BD Biosciences). SFU were automatically counted using the standardized threshold algorithm (SmartCount) in the ImmunoSpot software coupled to an ELISPOT plate reader (CTL-ImmunoSpot S6; Cellular Technology, Cleveland, OH).
Quantification of P. gingivalis–specific CD4+ T cells or iTregs
CLNs were harvested on days 0, 21, 28, 35, and 48 and single-cell suspensions were prepared using standard methods. A two-step process was used to quantify the total number of P. gingivalis–specific iTregs or CD4+ T cells residing in each CLN sample. First, we added PKH26 reference microbeads (Sigma-Aldrich) to a 200-μl sample aliquot and counted this sample on a flow cytometer to determine the total number of lymphocytes present. Second, we determined the percentage of activated P. gingivalis–specific CD4+ T cells or iTregs in the lymphocyte population. To do this, we stained an aliquot of 5 × 106 lymphocytes with 5–25 nM PE-conjugated pR/Kgp::I-Ab tetramer at room temperature followed by a panel of anti-mouse mAbs to identify Ag-experienced CD44hiCD4+ T cells as previously described (22, 38). The epitope (YASSTGNDA) recognized by the pR/Kgp::I-Ab tetramer occurs once in RgpA, one to two times in Kgp, and four to five times in the HagA protein. Epitope copy number is P. gingivalis strain–dependent. For iTreg assessment, Foxp3:GFP mice were used and anti-mouse CD25 (3C7; eBioscience) mAb was added to the staining panel. Cells were counted on an LSR II flow cytometer and analyzed with FlowJo software. Absolute numbers of each population were calculated by multiplying the number of lymphocytes determined in each CLN/mouse sample (step 1) by the percentage of each P. gingivalis–specific CD44hiCD4+ T cell or iTreg population (step 2).
Cytokine expression in P. gingivalis–specific CD4+ T cells or iTregs
CD4+ T cells were enriched from single-cell preparations prepared from CLNs or flank-draining LNs by immunomagnetic negative selection (Miltenyi Biotec). CD4+ T cells were cultured in complete EHAA (Life Technologies) and incubated with PMA (50 ng/ml, Sigma-Aldrich), ionomycin (500 ng/ml, Sigma-Aldrich), and brefeldin A (10 μg/ml, BD Biosciences) for 6 h to induce expression and intracellular retention of cytokines. Cells were washed twice with FACS buffer, then stained with PE-conjugated pR/Kgp::I-Ab tetramer followed by cell-surface staining with anti-mouse CD3 and CD4 with or without CD25 (iTreg assessment only) and a cell-exclusion dump comprising anti-mouse CD8α, B220, F4/80, and CD11b/c to unambiguously identify P. gingivalis–specific CD4+ T cells or Tregs. Cells were fixed, made permeable with Cytofix/Cytoperm buffer (BD Biosciences), and incubated with anti-mouse IFN-γ (XMG1.2), IL-4 (11B11), and IL-17A (TC11-18H10.1) mAbs to determine Th1, Th2, or Th17 phenotypes, respectively, or anti-mouse IL-10 (JES5-16E3) and TGF-β (LAP; TW7-16B4) mAbs to examine Tregs. All anti-mouse cytokine mAbs were purchased from BioLegend.
Statistical analysis
CD4+ T cell frequency and bone level data were analyzed and plotted using Prism 6 software (GraphPad Software, San Diego, CA) and expressed as mean ± SEM. Treatment and control groups as well as data from huLangerin-DTA versus C57BL/6J mice were compared using one- or two-tailed Student t tests. A p value < 0.05 was considered significant.
Results
Mucosal LCs are ablated in the gingiva of huLangerin-DTA mice
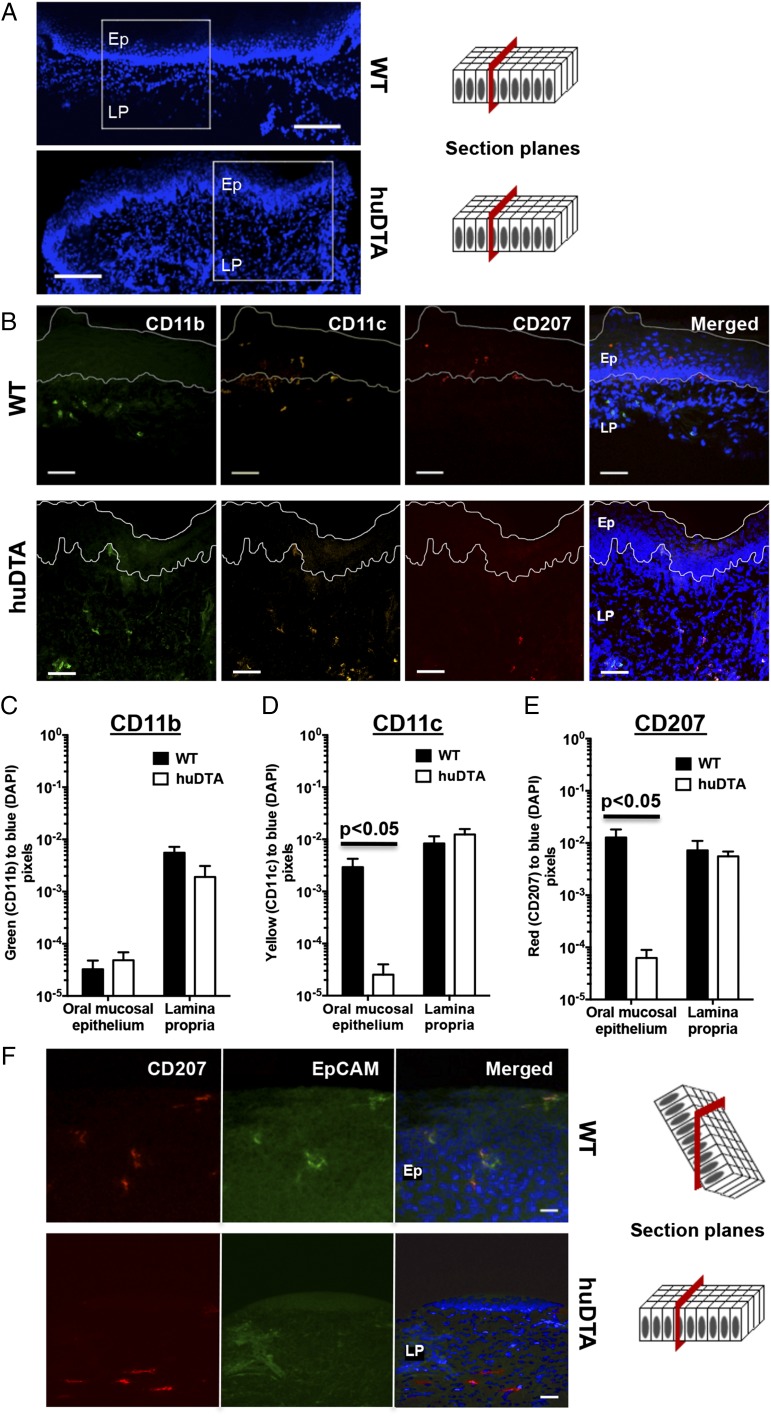
The effects of DTA expression on skin LCs and dermal Langerin+ DC populations of huLangerin-DTA mice have been well described (35), but they have not been examined in oral mucosae that surround teeth similar to the keratinized gingiva for example. To validate this model to study mucosal LCs, we first examined frozen gingival sections of huLangerin-DTA and wild-type C57BL/6J mice for expression of CD207, CD11c, CD11b, and EpCAM, a marker of LCs (Fig. 1A, 1B). We found colocalization of CD207 and CD11c expression on cells within the stratified squamous epithelial layer of C57BL/6J mice marking these cells as resident mucosal LCs (Fig. 1B). These LCs were absent in the equivalent gingival layer of huLangerin-DTA mice. Additionally, we established no evidence of compensatory intraepithelial migration of myeloid CD11c+ or CD11b+ into these gingival tissues (Fig. 1C, 1D). The lamina propria of both mouse strains harbored cells coexpressing CD207 and CD11c, and these likely represent Langerin+ DCs, rather than transiting LCs, as they do not express EpCAM (Fig. 1B, 1F). We found that CD11b-, CD11c-, and CD207-specific signals in the subepithelial lamina propria were similar in the two mouse strains (Fig. 1C–E). CD11c and CD207 signals were significantly decreased in the squamous epithelial layer of huLangerin-DTA mice (Fig. 1D, 1E) consistent with ablation of mucosal LCs. In skin at least, EpCAM expression has been used to differentiate LCs from Langerin+ DCs (42, 43). In double-stained frozen gingival sections, we found that EpCAM and CD207 colocalized only in the epithelial layer of C57BL/6J mice (Fig. 1F). In the lamina propria, CD207 and EpCAM did not colocalize marking these CD207-stained cells as Langerin+ DCs. Taken together, these immunohistology results demonstrated that under steady-state conditions LCs are ablated without a compensatory influx of other APCs into the gingiva of huLangerin-DTA mice, and that the population of Langerin+ DCs remains unchanged.
FIGURE 1.
huLangerin-DTA mouse gingiva lack mucosal LCs at steady-state. Frozen maxillary gingival sections from C57BL/6J (wild-type [WT]) and huLangerin-DTA (huDTA) mice were stained with DAPI and probed with flurochrome-conjugated anti-mouse mAbs to detect CD207, CD11b, CD11c, and EpCAM. Sections were examined by confocal scanning laser microscopy. (A) DAPI signal on stained gingival sections taken at original magnification ×10 showing regions examined at higher magnification in (B). Scale bars, 100 μm. (B) Separate and merged images showing DAPI-, CD11b-, CD11c-, and CD207-specific staining of representative gingival sections (original magnification, ×40). Scale bars, 40 μm. Stratified squamous epithelium (Ep) and lamina propria (LP) regions are labeled and delineated by a dashed white line to facilitate the signal quantification summarized in (C)–(E). (C–E) Quantification of (C) CD11b-, (D) CD11c-, and (E) CD207-specific signals in Ep and LP regions as the ratio of green, yellow, or red to blue (DAPI) pixels, respectively. Pooled pixel ratios obtained from 11 separate sections from each mouse strain were compared using two-tailed Student t test and presented as mean ± SEM. (F) Representative separate and merged images showing DAPI-, CD207-, and EpCAM-specific staining in Ep or LP gingival regions taken from WT or huDTA mice, respectively (original magnification, ×40). Scale bars, 20 μm.
LCs do not repopulate the oral mucosa of huLangerin-DTA mice under inflammatory conditions
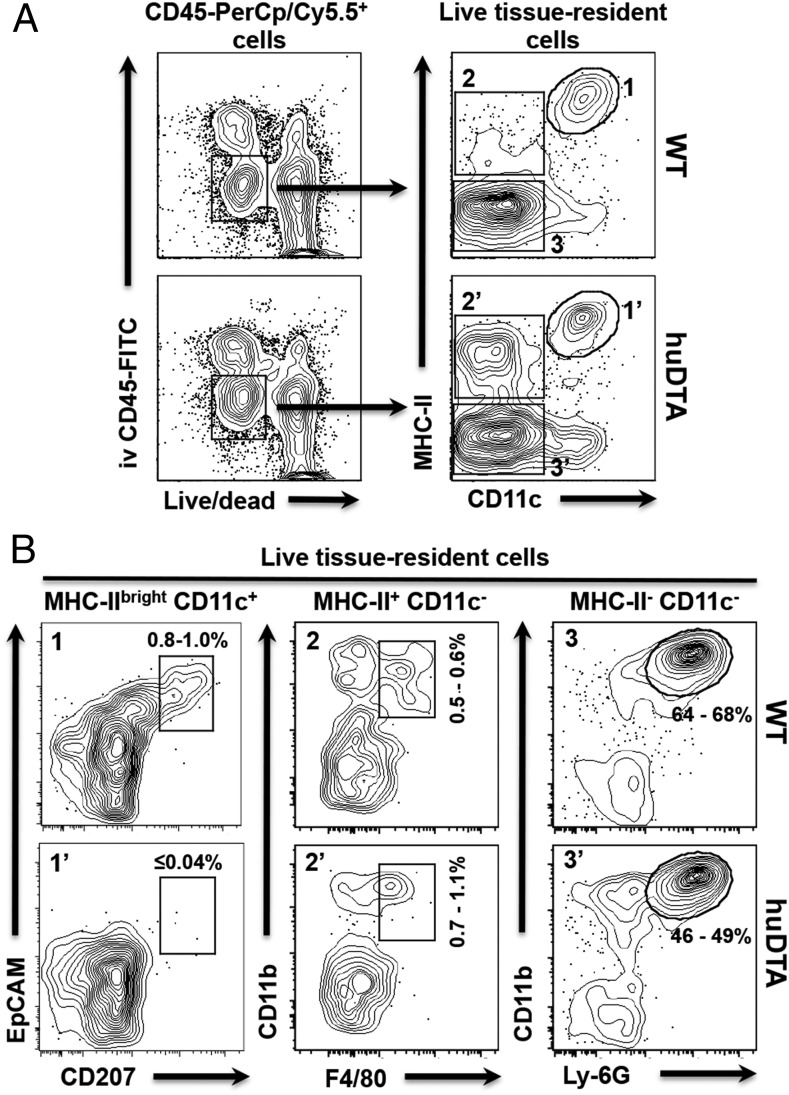
Capucha et al. (44) recently demonstrated that oral mucosal LCs do not share the same ontology as skin-resident LCs of mice. LCs located within the keratinized stratified epithelium of the gingiva are derived from circulating pre-DCs and monocytic progenitors and are constantly being replenished during steady-state (44). It is unknown how inflammation affects recruitment of mucosal LCs from circulating precursors and when, and if, these de novo–differentiated LCs are ablated in the huLangerin-DTA mouse model. To address this issue, we compared oral mucosal tissue from huLangerin-DTA and C57BL/6J mice that had been chronically exposed to P. gingivalis. We used a simple in vivo assay to stain immune cells circulating within the vasculature, allowing us to differentiate these cells from those that have extravasated and reside within tissue (Fig. 2A). We routinely found that 20–35% of cells stained with CD45 mAbs actually reside within the vasculature of the oral mucosa (data not shown). Gating on live, tissue-resident (CD45-FITC−) immune cells we found no evidence that LCs (MHC-IIbrightCD11c+EpCAM+CD207+) stably repopulated the oral mucosa of huLangerin-DTA mice even after sustained exposure to P. gingivalis (Fig. 2B). In C57BL/6J mouse oral mucosa, LCs ranged from 0.8 to 1.0% of the live tissue-resident immune cells, whereas these cells were absent in huLangerin-DTA mice (Fig. 2B, panel 1 versus 1′). Interestingly, we found trends of higher tissue-resident macrophage (MHC-II+CD11c−CD11b+ F4/80+) frequencies in huLangerin-DTA mice (Fig. 2B, panel 2 versus 2′) and higher neutrophil (MHC-II−CD11c−CD11b+Ly-6G+) frequencies in C57BL/6J mice (Fig. 2B, panel 3 versus 3′). Similarly to what we observed in the gingiva at steady-state, P. gingivalis–induced inflammation in huLangerin-DTA mice does not lead to accumulation of mucosal LCs in the oral mucosa. We hypothesize that the human langerin promoter controlling DTA expression is activated when circulating pre-DCs and monocytic progenitors enter the epithelial layer and commit to becoming mucosal LCs.
FIGURE 2.
LCs do not repopulate the oral mucosa of huLangerin-DTA (huDTA) mice following sustained oral colonization with P. gingivalis. Groups of 6- to 8-wk-old huDTA and C57BL/6J (wild-type [WT]) mice were inoculated six times with P. gingivalis strain 53977 by oral gavage at 4-d intervals. Mice were injected retro-orbitally with a nonsaturating amount of FITC-conjugated anti-mouse CD45 mAb (1.25 μg) 3 min prior to sacrifice to allow FITC-stained blood-resident immune cells to be excluded from downstream flow cytometry analysis. Single-cell suspensions prepared from pooled (two mice) oral mucosal tissue were stained with cell Live/Dead discriminating dye followed by anti-mouse CD45, CD11b, CD11c, I-Ab (MHC-II), EpCAM, CD207, F4/80, and Ly-6G flurochrome-conjugated mAbs and analyzed by flow cytometry. (A) Flow cytometry gating strategy used to identify live tissue-resident immune cells. Representative dot plots for pooled WT and huDTA mouse samples are shown. Initial gates were drawn around CD45-PerCP/Cy5.5+ cells that were alive (low fluorescence dye staining) and residing within the tissue (i.v. CD45-FITC−). Cells residing in the vasculature are CD45-FITC+ and are not included in these gates. The live, tissue-resident immune cells were divided into three distinct populations based on expression of MHC-II and CD11c (MHC-IIbrightCD11c+, gates 1 and 1′; MHC-II+CD11c−, gates 2 and 2′; and MHC-II−CD11c−, gates 3 and 3′). (B) Representative dot plots for WT and huDTA mouse samples are shown with gates drawn around LCs (EpCAM+CD207+; panels 1 and 1′), macrophages (CD11b+F4/80+; panels 2 and 2′), and neutrophils (CD11b+Ly-6G+; panels 3 and 3′) derived from their respective parental gates. The number of cells within each of the drawn gates is expressed as a percentage of the total live tissue-resident immune cell population and indicates the range found among all samples analyzed.
Mucosal LCs do not affect migration of other DC subsets from oral mucosa under steady-state or inflammatory conditions
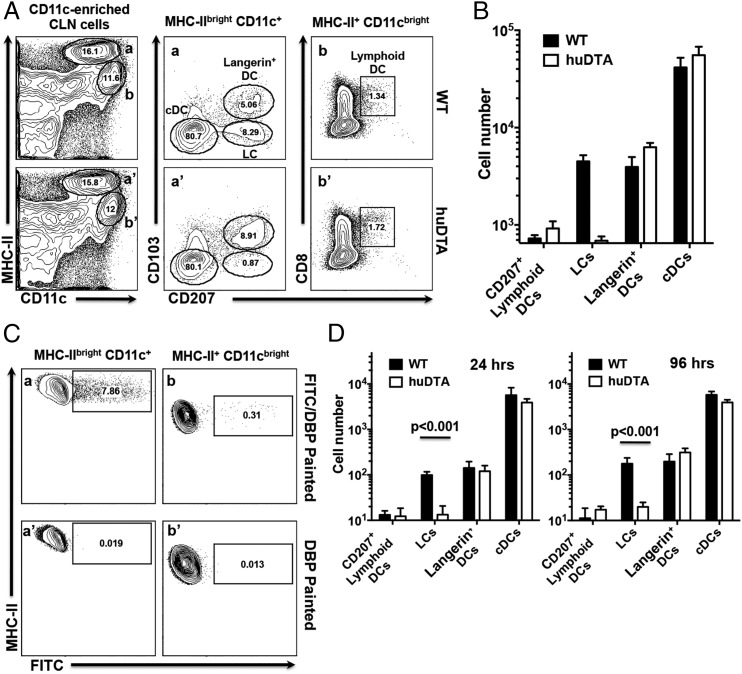
We next asked whether LC deficiency in huLangerin-DTA mice leads to qualitative or quantitative differences in DC migration that might impact Ag presentation in CLNs that drain the oral mucosae. Enriched CD11c+ cells from CLNs were stained with mAbs against MHC-II, CD11c, CD11b, CD207, CD103, and CD8α and analyzed by flow cytometry. Mature migratory DCs increase expression of MHC-II and can be readily identified by their MHC-IIbrightCD11c+ phenotype (Fig. 3A, gates a and a′) that separates them from MHC-II+CD11cbright lymphoid-resident DC populations (Fig. 3A, gates b and b′) (45). We further divided the migratory DCs into three groups based on their expression of CD207 and CD103 (Fig. 3A): LCs (CD207+CD103−), Langerin+ DCs (CD207+CD103+), and conventional DCs (cDCs) (CD207−CD103−CD11b+ [data not shown]). A small population of lymphoid-resident DCs (MHC-II+CD11cbrightCD8α+) also expresses CD207 (Fig. 3A, panels b and b′ panel). C57BL/6J and huLangerin-DTA mice have similar number of cDCs, Langerin+ DCs, and CD207+ lymphoid-resident DCs in CLNs (Fig. 3B). Consistent with our immunohistology data, only LCs were absent in the CLNs of huLangerin-DTA mice (Fig. 3B). The small numbers of CD207+ CD103− cells detected in the MHC-IIbrightCD11c+ gated population in huLangerin-DTA mice (Fig. 3A, panel a′) may represent CD103−Langerin+ DCs (43).
FIGURE 3.
Migration of DC subsets to CLNs under steady-state and inflammatory conditions is not inhibited in LC-deficient huLangerin-DTA (huDTA) mice. Single-cell suspensions prepared from CLNs harvested from C57BL/6J (wild-type [WT]) and huDTA mice were enriched for CD11c+ cells, stained with anti-mouse CD11b, CD11c, I-Ab (MHC-II), CD103, EpCAM, CD8α, CD207, CD3, and B220 flurochrome-conjugated mAbs, and analyzed by flow cytometry to identify LCs and DC subsets. (A) Flow cytometry gating strategy used to identify DC subsets within CLNs. Representative dot plots for a WT and huDTA mouse are shown. Migratory (gates a and a′) and lymphoid-resident (gates b and b′) DCs were identified as MHC-IIbrightCD11c+CD3−B220− or MHC-II+CD11cbrightCD3−B220− populations, respectively. Migratory DCs were further divided into populations of cDCs (CD103−CD207−), LCs (CD103−CD207+), and Langerin+ DCs (CD103+CD207+) based on the differential expression of CD207 and CD103 (panels a and a′). Lymphoid-resident DCs expressing CD207 were identified based on expression of CD8α and CD207 (panels b and b′). Numbers within individual gates refer to percentage of cells in the parent gate. (B) Total number of cDCs, LCs, Langerin+ (CD207+) and CD207+ lymphoid-resident DCs found in CLNs of WT and huDTA mice. Total numbers of each subpopulation were determined from pre-enrichment sample aliquots stained to identify MHC-IIbrightCD11c+CD3−B220− and MHC-II+CD11cbrightCD3−B220− cells coupled with percentage of cells found in each subpopulation gate after enrichment. Data pooled from two independent experiments (WT, n = 5; huDTA, n = 6) were compared using a two-tailed Student t test and are presented as mean ± SEM. (C) Representative flow cytometry dot plots showing FITC signal on MHC-IIbrightCD11c+CD3−B220− (panels a and a′) and MHC-II+CD11cbrightCD3−B220− (panels b and b′) CLN cell populations following application of FITC/DBP or DBP to mouse gingiva 24 h earlier. (D) Summary of FITC+ DC populations found in CLNs of WT and huDTA mice painted with FITC 24 or 96 h earlier. CLNs were harvested and single-cell suspensions were enriched, stained, and analyzed by flow cytometry to identify specific DC subpopulations as described in (A). Data from two independent experiments of at least five mice in each group were compared using a two-tailed Student t test and presented as mean ± SEM.
Next we examined migration of mucosal APCs to CLNs following application of a local inflammatory stimulus. Mice gingivae were treated the irritant DBP and FITC. APCs residing in the stratified squamous epithelial layer and lamina propria are stained locally by the penetrating FITC and subsequently migrate to draining CLNs as FITC+ cells (Fig. 3C). We used the same staining and gating strategy described earlier to identify LCs, Langerin+ DCs, cDCs, and CD207+ lymphoid-resident DCs among the MHC-IIbrightCD11c+ (Fig. 3C, panels a and a′) and MHC-II+CD11cbright (Fig. 3C, panels b and b′) populations, respectively. CLNs of C57BL/6J and huLangerin-DTA contained similar numbers of FITC+Langerin+ DCs and FITC+ cDCs at 24 and 96 h, indicating that their migration from the oral mucosa is not impacted by LCs (Fig. 3D). Consistent with the immunohistology and flow cytometry analyses presented earlier, FITC+ mucosal LCs were found only in the CLNs of C57BL/6J mice (Fig. 3D). Interestingly, the total number of recently migrated cDCs exceeds that of Langerin+ DCs and LCs in C57BL/6J mice by at least a factor of 30, reflecting greater numbers of cDCs in the tissue and/or faster migration kinetics. Because FITC is taken up locally in the gingiva/oral mucosae, it is entirely consistent that FITC-stained cells would not be a significant component of the CD207+ lymphoid-resident DC population in either C57BL/6J or huLangerin-DTA mice (Fig. 3D).
Mucosal LCs are not required to drive alveolar bone destruction in mice orally colonized with P. gingivalis
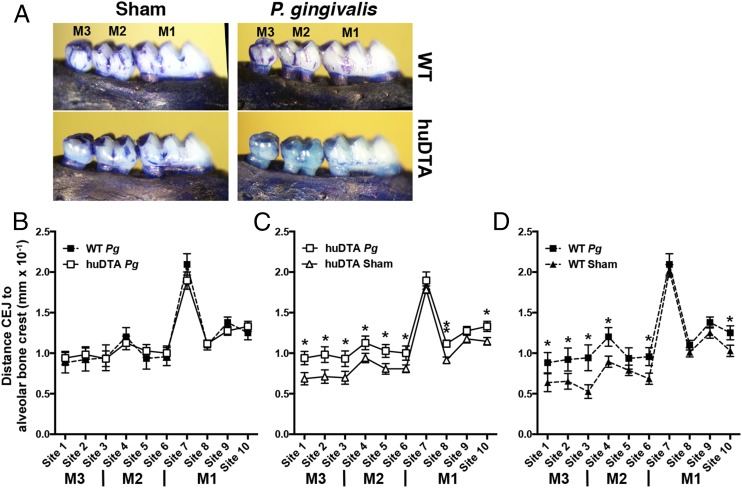
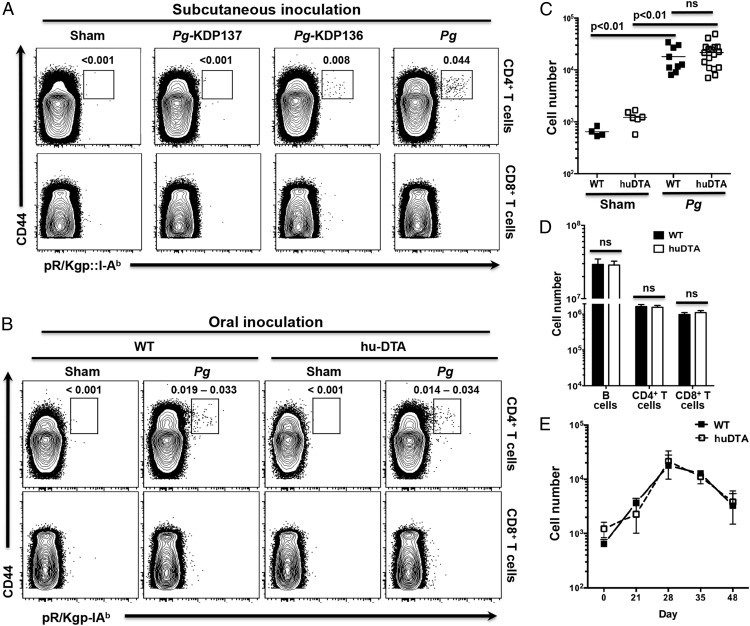
Recent studies demonstrated that the combined ablation of LCs and Langerin+ DCs resulted in significant alveolar bone destruction and higher IFN-γ production in P. gingivalis–colonized muLangerin-DTR mice (26). We have demonstrated that Langerin+ DCs in the oral mucosa of huLangerin-DTA mice behave like those in wild-type C57BL/6J mice. Importantly, we showed that other DCs do not occupy the gingival niche vacated by mucosal LCs in huLangerin-DTA mice. These results prompted us to reexamine the conclusions of Arizon et al. (26) by using huLangerin-DTA mice to test what effect the absence of mucosal LCs alone has on alveolar bone destruction mediated by P. gingivalis. In direct contrast to these authors, we found no significant differences in crestal alveolar bone levels (p > 0.05) across all 10 measured maxillary sites in huLangerin-DTA and C57BL/6J mice 48 d after initial oral colonization with P. gingivalis strain 53977 (Fig. 4A, 4B). Both mice strains experienced similar levels of alveolar bone loss at each measured site irrespective of the presence of mucosal LCs. Even though the C57BL/6J strain is considered less susceptible to P. gingivalis–induced periodontitis than are BALB/c mice, all mice orally colonized with P. gingivalis developed significantly more alveolar bone destruction than did their respective sham-colonized controls (p < 0.05) (Fig. 4C, 4D). Collectively, these data clearly indicate that mucosal LCs alone do not protect against or drive alveolar bone destruction. This finding prompted us to investigate the kinetics of CD4+ T cell activation in huLangerin-DTA and C57BL/6J mice.
FIGURE 4.
LC-deficient mice are not protected from crestal alveolar bone destruction induced by oral colonization with P. gingivalis. Groups of 6- to 8-wk-old LC-deficient (huLangerin-DTA [huDTA]) and C57BL/6J (wild-type [WT]) mice were orally inoculated with bone loss–inducing P. gingivalis strain 53977 by oral gavage at 4-d intervals. Six weeks after the first feeding, the maxillae were de-fleshed and examined for alveolar bone changes at 10 points spanning the buccal surface of all three maxillary molar teeth by measuring the distance between the cementum–enamel junction (CEJ) and the alveolar bone crest (ABC). (A) Representative images of de-fleshed and methylene blue–stained maxillae from sham- or P. gingivalis (Pg)-infected mice. (B) Alveolar bone destruction across 10 measurement sites of P. gingivalis–inoculated WT and huDTA mice was similar when each measurement was compared with a two-tailed Student t test. (C) Alveolar bone destruction in P. gingivalis–inoculated huDTA mice was compared with sham-inoculated huDTA mice at the 10 measurement sites. (D) Alveolar bone destruction in P. gingivalis–inoculated WT mice was compared to sham-inoculated WT mice at the 10 measurement sites. Measurements were normalized across mice based on the mesiodistal size of the second maxillary molar. Data expressed as mean distance from CEJ to alveolar bone crest ± SEM resulted from three independent experiments totaling 10 to 13 mice per group. An increase in mean bone destruction in the P. gingivalis– versus sham-inoculated groups was assessed by a one-tailed Student t test at each of the 10 measurement sites. *p < 0.05, **p < 0.01.
Mucosal LCs are dispensable for expansion of P. gingivalis–specific CD4+ T cells
LCs have been attributed both immunogenic and immunosuppressive functions. To examine the contribution of mucosal LCs to the adaptive immune response against P. gingivalis, we compared clonal expansion of CD4+ T cells in huLangerin-DTA and C57BL/6J mice using a peptide:MHC-II tetramer (pR/Kgp::I-Ab) that identifies cells responding to unique and specific P. gingivalis Ags (38). C57BL/6J mice were s.c. inoculated with P. gingivalis strains 33277, KDP136, or KDP137, and the fidelity of the pR/Kgp::I-Ab tetramer for specific Ag-experienced (CD44hi) CD4+ T cells was examined under inflammatory conditions. The wild-type P. gingivalis strain 33277 expresses all three (RgpA, Kgp, HagA) proteins that share the YASSTGNDA epitope, and the KDP136 mutant expresses only HagA, whereas KDP137 lacks all three (40). Consistent with the prevailing epitope copy number, expansion of tetramer+CD44hiCD4+ T cells was highest in mice inoculated with 33277 or 53977 and absent in sham- or KDP137-inoculated mice (Fig. 5A). Interestingly, KDP136 with its reduced epitope copy number induced a smaller expansion of tetramer+CD44hiCD4+ T cells. In all mice, nonspecific background staining of the tetramer reagent was low as indicated by low signal on CD8+ T cells (Fig. 5A). Next, we compared the expansion of tetramer+CD44hiCD4+ T cells found in the CLNs of huLangerin-DTA and C57BL/6J mice following sustained oral inoculation with P. gingivalis strain 53977 during 28 d (Fig. 5B). The number of tetramer+CD44hiCD4+ T cells increased significantly in both P. gingivalis–colonized huLangerin-DTA and C57BL/6J mice when compared with their respective sham-colonized controls (Fig. 5C). Moreover, we saw no significant difference in the magnitude of the adaptive CD4+ T cell response in the absence of mucosal LCs. Owing to technical limitations, we were unable to reliably quantify tetramer+CD4+ T cells in the gingiva (data not shown). We also found that P. gingivalis–colonized huLangerin-DTA and C57BL/6J mice had similar numbers of CD44hiCD4+ cells, CD44hiCD8+ T cells, and B cells in their CLNs (Fig. 5D). Clonal expansion and contraction of P. gingivalis–specific CD4+ T cells assessed during a 48-d infection cycle was also similar in both mouse strains, ruling out any effect that mucosal LCs may have on the timing of the CD4+ T cell response against P. gingivalis (Fig. 5E). Decrease in cell numbers seen after 28 d is likely due to emigration of newly differentiated P. gingivalis–specific CD4+ T cells from CLNs to sites of P. gingivalis persistence in the oral mucosa (Fig. 5E). Collectively, these experiments demonstrate that clonal expansion of P. gingivalis–specific CD4+ T cells is due to DCs other than mucosal LCs and that they do not make a significant direct contribution to the magnitude of this adaptive immune response.
FIGURE 5.
Clonal expansion of Ag-experienced P. gingivalis (Pg)-specific CD4+ T cells is normal in LC-deficient mice. C57BL/6J (wild-type [WT]) and huLangerin-DTA (huDTA) mice were orally or s.c. inoculated with P. gingivalis, and at defined time points single-cell suspensions from harvested LNs were stained with anti-mouse CD3, B220, CD8α, CD4, and CD44 flurochrome-conjugated mAbs and pR/Kgp::I-Ab tetramer to identify Ag-experienced tetramer+CD4+ T cells by flow cytometry. (A) Flow cytometry plots that demonstrate the utility and fidelity of the pR/Kgp::I-Ab tetramer to identify specific CD4+ T cells that recognize an epitope unique to P. gingivalis proteins RgpA, Kgp, and HagA. Plots display gated CD4+ T cells (CD3+CD4+CD8−B220−) or CD8+ T cells (CD3+CD4−CD8+B220−) from C57BL/6J mice that were s.c. injected with PBS (sham), P. gingivalis, or mutants KDP136 (Δ RgpA and Kgp) and KDP137 (Δ RgpA, Kgp, and HagA). Percentages of cells within the tetramer+CD44high CD4+ T cell gate are reported. The experiment was repeated twice with similar results. (B) Representative flow cytometry plots of CD4+ or CD8+ T cells isolated from the CLNs of orally inoculated (sham or P. gingivalis strain 53977) WT and huDTA mice at day 28. Numbers indicate the range of cells within the tetramer+ gate as a percentage of the displayed CD4+ T cell population. (C and D) Absolute number of tetramer-specific CD44hi (Ag experienced) CD4+ T cells (C) or Ag-experienced CD4+ and CD8+ T cells, and B cells (D) found in CLNs of orally inoculated WT or huDTA mice at day 28. Cell numbers were calculated from the total CLN lymphocyte population and from the percentage of lymphocytes found in the tetramer-specific Ag-experienced CD4+ T cell gate as described in Materials and Methods. Data are pooled from at least three independent experiments involving at least nine mice in each P. gingivalis treated group. Means ± SEM were compared using a two-tailed Student t test. (E) Absolute number of Ag-experienced gingipain-specific CD4+ T cells found in CLNs of P. gingivalis–inoculated WT or huDTA mice at day 0, 21, 28, 35, or 48. Data for day 28 are from (C); otherwise, data are pooled from at least two independent experiments involving at least eight mice (mean ± SEM). ns, not significant.
Mucosal LCs are not required for the expansion of P. gingivalis–specific iTregs
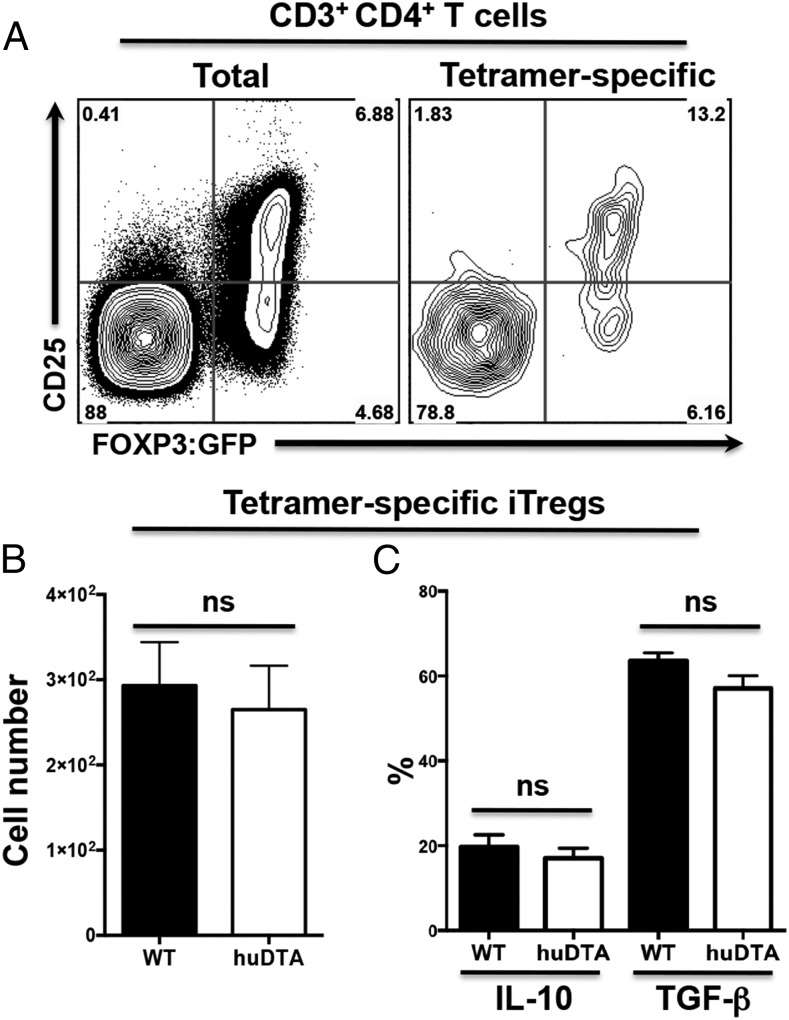
Arizon et al. (26) reported increased clonal expansion of CD4+ T cells attributed to P. gingivalis and decreased iTreg numbers in muLangerin-DTR mice. However, given that we did not find increased tetramer+CD44hiCD4+ T cell numbers in huLangerin-DTA mice (Fig. 5), we examined the effect of mucosal LCs on the differentiation of iTregs in a newly developed LC-deficient mouse (huLangerin-DTA) that expresses GFP under the control of the endogenous Foxp3 promoter. LC-deficient and LC-competent Foxp3:GFP littermates were tested for clonal expansion of Foxp3:GFP+CD25hiCD4+ iTregs following oral inoculation with P. gingivalis (Fig. 6A). The number of tetramer+Foxp3:GFP+CD25hiCD4+ iTregs in CLNs was similar regardless of the presence or absence of LCs (Fig. 6B). Although the number of iTregs was similar, it is possible that expression of TGF-β and IL-10, two key cytokines produced by iTregs, could differ in the absence of mucosal LCs. However, we found that the frequency of P. gingivalis–specific iTregs expressing TGF-β or IL-10 was similar in LC-deficient and LC-competent Foxp3:GFP littermates (Fig. 6C).
FIGURE 6.
The iTreg response to oral inoculation with P. gingivalis is unaltered in LC-deficient mice. huLangerin-DTA (huDTA) Foxp3:GFP and C57BL/6J [wild-type (WT)] Foxp3:GFP mice were orally infected with P. gingivalis strain 53977, and iTregs present in CLNs were analyzed by flow cytometry at day 28 as describe in Fig. 5. (A) Representative flow cytometry dot plots obtained from analysis of CD3+CD4+ lymphocytes (gated as CD3+CD4+B220−CD8−) from a huLangerin-DTA Foxp3:GFP mouse. iTregs were identified as the CD25hi GFP+ (Foxp3) population in both the CD3+CD4+ lymphocyte population (total iTregs, left panel) and CD3+CD4+ pR/Kgp-IAb+ lymphocyte population (tetramer-specific iTregs, right panel). Percentage of cells within each of the four quadrants is indicated. (B) Summary data of CD3+CD4+pR/Kgp-I-Ab+ iTreg population. Data were pooled from three independent experiments totaling at least 14 mice per group. Data expressed as mean ± SEM (two-tailed Student t test). (C) Percentage of iTregs found expressing intracellular IL-10 or TGF-β was determined in tetramer-specific iTreg populations by flow cytometry following PMA/ionomycin treatment of enriched CD4+ T cells obtained from CLNs of P. gingivalis colonized mice at day 28. Data pooled from three independent experiments totaling at least 13 mice per group were compared using two-tailed Student t test and presented as mean ± SEM. ns, not significant.
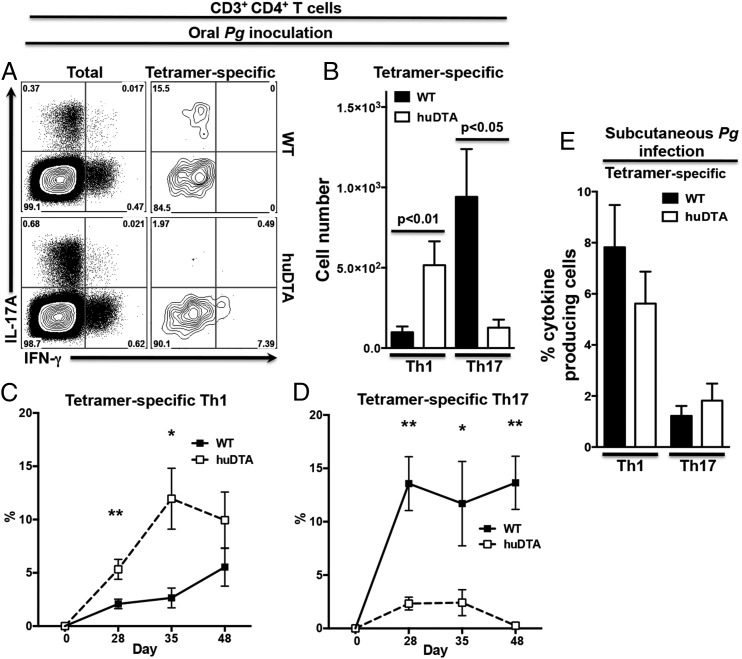
Mucosal LCs drive primarily a P. gingivalis–specific Th17 response and dampen the Th1 response in mice orally colonized with P. gingivalis
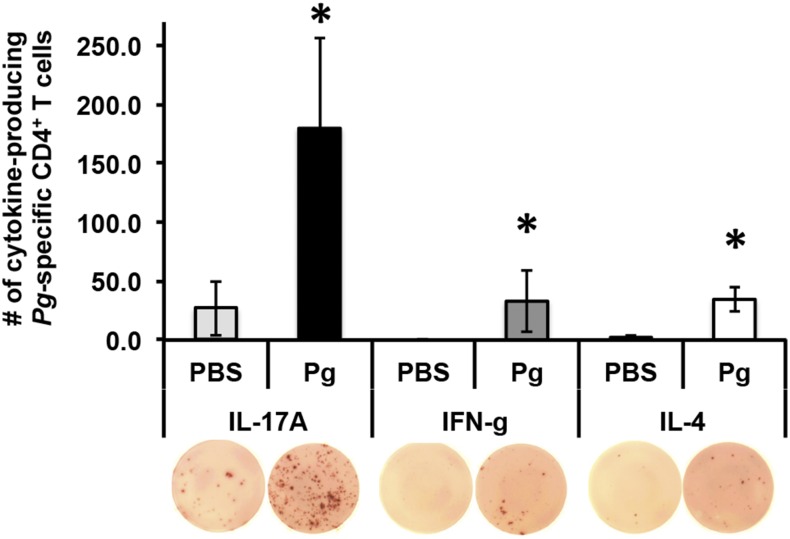
Initial ELISPOT experiments demonstrated that oral colonization with P. gingivalis induced differentiation of IL-17A–expressing P. gingivalis–specific CD4+ T cells in C57BL/6J mice with smaller numbers expressing IFN-γ (Fig. 7). In ELISPOT, IL-4–producing cells could be related to cells other than CD4+ T cells because irradiated naive splenocytes from C57BL/6J mice reveal high IL-4 background production when restimulated with P. gingivalis (data not shown). To avoid this issue, we examined cytokine expression in P. gingivalis–specific CD4+ Th cells identified by our pR/Kgp::I-Ab tetramer (38). We found that P. gingivalis–specific CD4+ Th cells from C57BL/6J mice primarily differentiated into Th17 cells with a minor Th1 component (Fig. 8A) and few or no Th2 cells (data not shown). C57BL/6J mice had on average 8-fold more P. gingivalis–specific Th17 cells compared with P. gingivalis–specific Th1 cells at day 28 (Fig. 8B). In contrast, P. gingivalis–specific CD4+ T cells differentiated into significantly greater numbers of Th1 cells in huLangerin-DTA mice compared with C57BL/6J mice, indicating that mucosal LCs are essential to drive differentiation of P. gingivalis–specific Th17 cells and may act to dampen a Th1-type response (Fig. 8B). This sustained P. gingivalis–specific Th1 response in LC-deficient mice peaks at day 35 (Fig. 8C) and the weak Th17 response completely disappears by day 48 (Fig. 8D). We can also rule out a defect in the Th17 differentiation pathway in huLangerin-DTA mice, as CD3+CD4+ T cells clearly have the ability to differentiate into Th17 cells, presumably under different pathogen stimuli (Fig. 8A). Moreover, using s.c. P. gingivalis immunization to bypass mucosal surfaces revealed that the number of P. gingivalis–specific Th cells expressing IL-17A was similar in huLangerin-DTA and C57BL/6J mice (Fig. 8E). Interestingly, this s.c. route of P. gingivalis infection favored a Th1-type response over Th17 in both mouse strains, supporting the idea that mucosal LCs are responsible for directing a Th17 response against P. gingivalis.
FIGURE 7.
Oral P. gingivalis (Pg) primarily induces differentiation of IL-17A–expressing CD4+ T cells in C57BL/6J mice. CD4+ T cells isolated from CLNs of C57BL/6J mice orally inoculated with P. gingivalis strain 53977 every 4 or 24 d were incubated with irradiated naive splenocytes and restimulated with O2-killed whole P. gingivalis or PBS control for 72 h in an ELISPOT assay. Bars display the net average number of SFU ± SEM identifying IL-17A–, IFN-γ–, or IL-4–producing CD4+ T cells after subtraction of SFU generated by irradiated splenocytes. Representative ELISPOT well images are shown (original magnification ×2). One-tailed Student t test was used to determine whether recall sample means were significantly larger than the restimulation without Ag (PBS). *p < 0.05.
FIGURE 8.
LCs resident in the oral mucosa drive differentiation of Th17 cells following oral infection with P. gingivalis (Pg). LC-deficient (huDTA) and C57BL/6J (wild-type [WT]) mice were inoculated with P. gingivalis strain 53977 by oral gavage or s.c. injection and enriched CD4+ T cells from CLN were stimulated with PMA/ionomycin in the presence of brefeldin A, stained for CD3+CD4+ T cells, and analyzed to identify IL-17A– and IFN-γ–expressing Th lymphocytes by flow cytomtery. (A) Representative flow cytometry dot plots showing expression of IL-17A and IFN-γ at day 28 in the total (gated as CD3+CD4+B220−CD11c/b−CD8−F4/80−) or tetramer-specific (gated as CD3+CD4+pR/Kgp-IAb+B220−CD11c/b−CD8−F4/80−) CD4+ T cell populations. Percentage of cells within each of the four quadrants is indicated. (B) Total number of tetramer-specific CD3+CD4+ T cells expressing IL-17A or IFN-γ at day 28 in CLNs of mice orally inoculated with P. gingivalis. Data pooled from three independent experiments totaling at least 13 mice per group were compared using a two-tailed Student t test and presented as mean ± SEM. (C and D) Percentage of tetramer-specific CD3+CD4+ T cells expressing IL-17A (C) or IFN-γ (D) at days 0, 28, 35, or 48. Data are presented as mean ± SEM and compared using a two-tailed Student t test. *p < 0.05, **p < 0.01. (E) huDTA and WT mice were inoculated with P. gingivalis s.c. into their flanks, boosted at day 21, and enriched CD4+ T cells from flank-draining LNs (inguinal, brachial, axillary) were analyzed for the frequency of tetramer-specific CD4+ T cells expressing IL-17A or IFN-γ 12 d later as described above. Data were pooled from two independent experiments totaling at least five mice per group and means were compared using a two-tailed Student t test. ns, not significant.
Discussion
Skin-resident LCs have been well characterized (35), and there is emerging evidence that suggests mucosal LCs have a different ontology to skin-resident LCs (44). However, how mucosal LCs direct adaptive immune responses against oral pathogens is understudied. In this study, we demonstrate that mucosal LCs are dispensable for directing alveolar bone destruction in mice triggered by oral infection with P. gingivalis. We observed similar levels of alveolar bone destruction in LC-deficient and wild-type mice, yet the P. gingivalis–specific adaptive immune response switched from Th17 in wild-type mice to a predominantly Th1-type response in LC-deficient mice. This result was somewhat unexpected given the accumulating evidence that links Th17 cells and IL-17A, in particular, to bone destructive pathologies such as arthritis, osteoporosis, and inflammatory-driven and age-related periodontal diseases (16, 18, 46, 47). Both IL-17A and IFN-γ are considered proinflammatory cytokines even though they activate different arms of innate immunity. Although typically viewed as being a negative regulator of osteoclastogenesis (48), IFN-γ can also promote osteoclastogenesis under certain circumstances (15, 49, 50). In LC-deficient mice we propose that IFN-γ expression by enhanced Th1 cell numbers compensates for the defect in Th17-mediated expression of IL-17A, thereby inducing similar alveolar bone destruction in the two mouse strains we tested. Consistent with this idea is that Arizon et al. (26) saw increased IFN-γ expression and enhanced alveolar bone destruction in their muLangerin-DTR periodontitis mouse model.
Inflammation in the oral cavity is a relatively rare occurrence, and resident DCs would appear tasked with maintaining immunological homeostasis even in the face of a considerable bacterial burden. In human periodontal lesions, diminished iTregs are associated with lower levels of IL-10 and increased numbers of CD4+ T cells expressing the osteoclastogenic factor RANKL (34). Arizon et al. (26) have suggested that LCs play a regulatory role in experimental periodontitis by dampening the magnitude of the activated CD4+ T cell response through a mechanism involving iTregs. These findings should be interpreted with caution, however, as LCs, Langerin+ DCs, and CD8+ lymphoid-resident DCs that express CD207 can be affected in the muLangerin-DTR experimental system. iTreg and Th cell numbers are often inversely correlated owing to feedback mechanisms that control cell number via production and consumption of IL-2 (51, 52). Consistent with this model, as well as our iTreg data, we demonstrate that Foxp3+CD25hi P. gingivalis–specific CD4+ T cells did not increase in LC-deficient mice compared with wild-type mice, suggesting that mucosal LCs do not influence iTreg numbers. Moreover, we did not observe a significant decrease in the frequency of P. gingivalis–specific iTregs expressing anti-inflammatory cytokines IL-10 or TGF-β. We conclude that other mucosal DCs, possibly Langerin+ (CD103+) DCs or ALDH+Langerin−CD11b+CD24int migratory DCs, are involved in iTreg induction. Resident CD103+ DCs in the gut and liver (32) and ALDH+Langerin−CD103−CD11b+CD24int DCs in the skin (53) have been shown to actively induce iTreg cells in regional draining LNs via the retinoic acid–dependent mechanism (54).
We present evidence from immunohistological and flow cytometry data that indicates the presence of Langerin+ DCs in the gingiva of mice (Figs. 1, 3), contrary to what has been reported by others (11, 26). These DCs are likely equivalent to dermal-resident Langerin+CD103+ migratory DCs and are distinguishable from skin-resident LCs based on three lines of evidence: 1) ontogeny, because they are continuously replenished by bone marrow precursors (55); 2) anatomical location, because they reside in the subepithelial layer (56); and 3) function, because they are required for optimal production of β-galactosidase–specific IgG2a/c and IgG2b in the acute phase (42, 57). The discrepancy between our work and that reported by Arizon et al. (26) could be partly attributed to their gating strategy that would have counted Langerin+CD103− DCs as LCs due to the acquisition of an EpCAM+ surface phenotype upon migration to LNs (42, 43), a phenotype we have also observed on migratory DCs resident in CLNs (data not shown). Our evidence suggests that mucosal Langerin+ DCs, similar to mucosal LCs, are well positioned to act as APCs to acquire Ag from microorganisms that potentially invade the periodontal pocket and could be involved in immunogenic as well as tolerogenic responses.
We demonstrated that LC-deficient huLangerin-DTA mice are substantially similar to C57BL/6 control mice in terms of cDC and Langerin+ DC populations found in CLNs under both steady-state and inflammatory conditions. By 24 h, >95% of FITC+ cells arriving in the CLNs were conventional lamina propria DCs (CD11b+CD103−CD207−) and this number only dropped slightly by 96 h. In CLNs analyzed by 96 h, the numbers of FITC+ LCs and Langerin+ DCs had doubled in number but still were a relatively minor component (2 and 3%, respectively). This imbalance is likely due to a delayed or a slower rate of migration of LCs and Langerin+ DCs combined with a difference in numbers residing in the oral mucosa (10, 42).
Consistent with what has been reported by others for cutaneous DCs (43), we detected EpCAM at some levels on all FITC+ migratory DCs in the CLNs (data not shown) whereas the expression of EpCAM in the gingiva is confined to be of mucosal LCs (Fig. 1F). The functional significance of an increase and de novo expression of EpCAM by these DC populations remains unknown. EpCAM is involved in homophilic cell-to-cell interactions in the epithelia, is widely expressed on cancer cells that have an epithelial origin, and is thought to have a role in their migration (58, 59). Thus, it is tempting to speculate that EpCAM may also play a role in migration of DCs to draining LNs as has been recently described for LCs (60).
In LC-deficient huLangerin-DTA mice we observed a significant decrease in the number of P. gingivalis–specific Th17 cells and a concomitant increase in Th1 cells. Importantly, even with a dampened Th17 response, LC deficiency did not impact the total clonal expansion of P. gingivalis–specific CD4+ T cells. Interestingly, in LC-deficient mice we observed reduced recruitment of neutrophils to the oral mucosa compared with wild-type mice, which might be a direct consequence of the dampened Th17 response. Driving the differentiation of Th17 cells specific for P. gingivalis appears to be a unique function of mucosal-resident LCs. This conclusion appears at odds with their poor ability to process and present bacterial Ags (61), but it does not exclude an indirect effect of mucosal LCs on subepithelial Langerin+ DCs and/or cDCs before they migrate to regional LNs. Using the same LC-deficient huLangerin-DTA mouse model, it has been shown that skin-resident LCs preferentially promote Ag-specific Th17 responses against Candida albicans in a cutaneous candidiasis model (30, 62). Recent work reported that mucosal LCs are not required for Th17 differentiation against C. albicans (63). However, this research relied on the Langerin-DTR model (36) that is known to not only deplete LCs temporarily but also to deplete Langerin+ DCs and lymphoid-resident CD8+ DCs. In a series of elegant experiments, Igyártó et al. (30) demonstrated that skin-resident LCs suppress the Th1 polarizing ability of dermal Langerin+ DCs. We hypothesize that gingiva-resident Langerin+ DCs may also be responsible for the predominantly Th1 response we see in huLangerin-DTA mice against P. gingivalis, although this function may also be performed in conjunction with, or solely by, cDCs. This idea is consistent with the increase in IFN-γ expression reported in the muLangerin-DTR periodontitis model (26). In experiments where P. gingivalis was delivered s.c., thereby circumventing skin-resident LCs, we demonstrated a pronounced Th1 response in both wild-type and LC-deficient mice.
Finally, our results reveal that mucosal LCs have a disproportionate influence over shaping the type of adaptive immune response mounted against infection by oral pathogens such as P. gingivalis. Shklovskaya et al. (31) found a 1:3 ratio of migratory skin LCs to migratory dermal DCs in cutaneous LNs. In our analysis, migratory mucosal LCs were at best 2–3% of the total number of FITC+CD11c+MHC-IIbright cells seen in CLNs and, significantly, these cells migrated more slowly and in fewer numbers than did the cDCs migrating out of the underlying lamina propria. Additionally, it has been reported that mucosal LCs are unable to present P. gingivalis Ags to CD4+ T cells in vitro (26). Given these findings, we hypothesize that mucosal LCs, following engagement with P. gingivalis, “educate” underlying lamina propria–resident DCs through a modulatory bystander effect to drive polarization of naive CD4+ T cells to a Th17 phenotype. In the absence of LCs or through bypassing LCs via s.c. immunization, the intrinsic polarizing program of lamina propria–resident DCs takes precedence and drives naive CD4+ T cell to a Th1 phenotype. Our findings have implications for vaccine strategies that seek to specifically target DC subsets in the oral mucosa to achieve protection. Our proposed model of bystander activation of lamina propria–resident DCs by mucosal LCs is novel and warrants further investigation.
Acknowledgments
We thank Dr. Marc C. Herzberg for graciously providing the P. gingivalis mutant strains, Drs. Seshagiri Nandula and Yuping Wei for help with anesthetizing mice, and Talia Just for technical support.
This work was supported in part by National Institutes of Health/National Institute of Dental and Craniofacial Research Grant R21 DE022858 (to M.C.) and by the Schaffer Chair for Periodontal Research at the University of Minnesota.
- cDC
- conventional dendritic cell
- CLN
- cervical lymph node
- DBP
- dibutyl phthalate
- DC
- dendritic cell
- DTA
- diphtheria toxin A
- DTR
- diphtheria toxin receptor
- EpCAM
- epithelial cell adhesion molecule
- hu
- human
- iTreg
- induced Treg
- LC
- Langerhans cell
- LN
- lymph node
- MHC-II
- MHC class II
- mu
- murine
- RANKL
- receptor activator for NF-κB ligand
- SFU
- spot-forming unit
- Treg
- regulatory T cell.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Costalonga M., Herzberg M. C. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 162(2 Pt A): 22–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffen A. L., Beall C. J., Campbell J. H., Firestone N. D., Kumar P. S., Yang Z. K., Podar M., Leys E. J. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6: 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gamonal J., Acevedo A., Bascones A., Jorge O., Silva A. 2000. Levels of interleukin-1β, -8, and -10 and RANTES in gingival crevicular fluid and cell populations in adult periodontitis patients and the effect of periodontal treatment. J. Periodontol. 71: 1535–1545. [DOI] [PubMed] [Google Scholar]
- 4.Berglundh T., Donati M. 2005. Aspects of adaptive host response in periodontitis. J. Clin. Periodontol. 32(Suppl. 6): 87–107. [DOI] [PubMed] [Google Scholar]
- 5.Teng Y. T. 2003. The role of acquired immunity and periodontal disease progression. Crit. Rev. Oral Biol. Med. 14: 237–252. [DOI] [PubMed] [Google Scholar]
- 6.Dixon D. R., Bainbridge B. W., Darveau R. P. 2004. Modulation of the innate immune response within the periodontium. Periodontol. 2000 35: 53–74. [DOI] [PubMed] [Google Scholar]
- 7.Braun T., Zwerina J. 2011. Positive regulators of osteoclastogenesis and bone resorption in rheumatoid arthritis. Arthritis Res. Ther. 13: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banchereau J., Steinman R. M. 1998. Dendritic cells and the control of immunity. Nature 392: 245–252. [DOI] [PubMed] [Google Scholar]
- 9.Randolph G. J., Jakubzick C., Qu C. 2008. Antigen presentation by monocytes and monocyte-derived cells. Curr. Opin. Immunol. 20: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aramaki O., Chalermsarp N., Otsuki M., Tagami J., Azuma M. 2011. Differential expression of co-signal molecules and migratory properties in four distinct subsets of migratory dendritic cells from the oral mucosa. Biochem. Biophys. Res. Commun. 413: 407–413. [DOI] [PubMed] [Google Scholar]
- 11.Hovav A. H. 2014. Dendritic cells of the oral mucosa. Mucosal Immunol. 7: 27–37. [DOI] [PubMed] [Google Scholar]
- 12.Malissen B., Tamoutounour S., Henri S. 2014. The origins and functions of dendritic cells and macrophages in the skin. Nat. Rev. Immunol. 14: 417–428. [DOI] [PubMed] [Google Scholar]
- 13.Teng Y. T., Nguyen H., Hassanloo A., Ellen R. P., Hozumi N., Gorczynski R. M. 1999. Periodontal immune responses of human lymphocytes in Actinobacillus actinomycetemcomitans-inoculated NOD/SCID mice engrafted with peripheral blood leukocytes of periodontitis patients. J. Periodontal Res. 34: 54–61. [DOI] [PubMed] [Google Scholar]
- 14.Gao L., Faibish D., Fredman G., Herrera B. S., Chiang N., Serhan C. N., Van Dyke T. E., Gyurko R. 2013. Resolvin E1 and chemokine-like receptor 1 mediate bone preservation. J. Immunol. 190: 689–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stashenko P., Gonçalves R. B., Lipkin B., Ficarelli A., Sasaki H., Campos-Neto A. 2007. Th1 immune response promotes severe bone resorption caused by Porphyromonas gingivalis. Am. J. Pathol. 170: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardoso C. R., Garlet G. P., Crippa G. E., Rosa A. L., Júnior W. M., Rossi M. A., Silva J. S. 2009. Evidence of the presence of T helper type 17 cells in chronic lesions of human periodontal disease. Oral Microbiol. Immunol. 24: 1–6. [DOI] [PubMed] [Google Scholar]
- 17.Eskan M. A., Jotwani R., Abe T., Chmelar J., Lim J. H., Liang S., Ciero P. A., Krauss J. L., Li F., Rauner M., et al. 2012. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat. Immunol. 13: 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moutsopoulos N. M., Kling H. M., Angelov N., Jin W., Palmer R. J., Nares S., Osorio M., Wahl S. M. 2012. Porphyromonas gingivalis promotes Th17 inducing pathways in chronic periodontitis. J. Autoimmun. 39: 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y., Wang L., Liu D., Lin X. 2014. Tamibarotene modulates the local immune response in experimental periodontitis. Int. Immunopharmacol. 23: 537–545. [DOI] [PubMed] [Google Scholar]
- 20.Hajishengallis G. 2014. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darveau R. P., Hajishengallis G., Curtis M. A. 2012. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91: 816–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker P. J., Dixon M., Evans R. T., Dufour L., Johnson E., Roopenian D. C. 1999. CD4+ T cells and the proinflammatory cytokines γ interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect. Immun. 67: 2804–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker P. J., Howe L., Garneau J., Roopenian D. C. 2002. T cell knockout mice have diminished alveolar bone loss after oral infection with Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 34: 45–50. [DOI] [PubMed] [Google Scholar]
- 24.Wilensky A., Polak D., Houri-Haddad Y., Shapira L. 2013. The role of RgpA in the pathogenicity of Porphyromonas gingivalis in the murine periodontitis model. J. Clin. Periodontol. 40: 924–932. [DOI] [PubMed] [Google Scholar]
- 25.Pathirana R. D., O’Brien-Simpson N. M., Brammar G. C., Slakeski N., Reynolds E. C. 2007. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect. Immun. 75: 1436–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arizon M., Nudel I., Segev H., Mizraji G., Elnekave M., Furmanov K., Eli-Berchoer L., Clausen B. E., Shapira L., Wilensky A., Hovav A. H. 2012. Langerhans cells down-regulate inflammation-driven alveolar bone loss. Proc. Natl. Acad. Sci. USA 109: 7043–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan D. H., Jenison M. C., Saeland S., Shlomchik W. D., Shlomchik M. J. 2005. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity 23: 611–620. [DOI] [PubMed] [Google Scholar]
- 28.Igyarto B. Z., Jenison M. C., Dudda J. C., Roers A., Müller W., Koni P. A., Campbell D. J., Shlomchik M. J., Kaplan D. H. 2009. Langerhans cells suppress contact hypersensitivity responses via cognate CD4 interaction and langerhans cell-derived IL-10. J. Immunol. 183: 5085–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bobr A., Olvera-Gomez I., Igyarto B. Z., Haley K. M., Hogquist K. A., Kaplan D. H. 2010. Acute ablation of Langerhans cells enhances skin immune responses. J. Immunol. 185: 4724–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igyártó B. Z., Haley K., Ortner D., Bobr A., Gerami-Nejad M., Edelson B. T., Zurawski S. M., Malissen B., Zurawski G., Berman J., Kaplan D. H. 2011. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity 35: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shklovskaya E., O’Sullivan B. J., Ng L. G., Roediger B., Thomas R., Weninger W., Fazekas de St Groth B. 2011. Langerhans cells are precommitted to immune tolerance induction. Proc. Natl. Acad. Sci. USA 108: 18049–18054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coombes J. L., Siddiqui K. R., Arancibia-Cárcamo C. V., Hall J., Sun C. M., Belkaid Y., Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J. Exp. Med. 204: 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwata M., Hirakiyama A., Eshima Y., Kagechika H., Kato C., Song S. Y. 2004. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21: 527–538. [DOI] [PubMed] [Google Scholar]
- 34.Ernst C. W., Lee J. E., Nakanishi T., Karimbux N. Y., Rezende T. M., Stashenko P., Seki M., Taubman M. A., Kawai T. 2007. Diminished forkhead box P3/CD25 double-positive T regulatory cells are associated with the increased nuclear factor-κB ligand (RANKL+) T cells in bone resorption lesion of periodontal disease. Clin. Exp. Immunol. 148: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan D. H. 2010. In vivo function of Langerhans cells and dermal dendritic cells. Trends Immunol. 31: 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett C. L., van Rijn E., Jung S., Inaba K., Steinman R. M., Kapsenberg M. L., Clausen B. E. 2005. Inducible ablation of mouse Langerhans cells diminishes but fails to abrogate contact hypersensitivity. J. Cell Biol. 169: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kissenpfennig A., Henri S., Dubois B., Laplace-Builhé C., Perrin P., Romani N., Tripp C. H., Douillard P., Leserman L., Kaiserlian D., et al. 2005. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity 22: 643–654. [DOI] [PubMed] [Google Scholar]
- 38.Bittner-Eddy P. D., Fischer L. A., Costalonga M. 2013. Identification of gingipain-specific I-Ab-restricted CD4+ T cells following mucosal colonization with Porphyromonas gingivalis in C57BL/6 mice. Mol. Oral Microbiol. 28: 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker P. J., Evans R. T., Roopenian D. C. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39: 1035–1040. [DOI] [PubMed] [Google Scholar]
- 40.Shi Y., Ratnayake D. B., Okamoto K., Abe N., Yamamoto K., Nakayama K. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274: 17955–17960. [DOI] [PubMed] [Google Scholar]
- 41.Costalonga M., Batas L., Reich B. J. 2009. Effects of Toll-like receptor 4 on Porphyromonas gingivalis-induced bone loss in mice. J. Periodontal Res. 44: 537–542. [DOI] [PubMed] [Google Scholar]
- 42.Nagao K., Ginhoux F., Leitner W. W., Motegi S., Bennett C. L., Clausen B. E., Merad M., Udey M. C. 2009. Murine epidermal Langerhans cells and langerin-expressing dermal dendritic cells are unrelated and exhibit distinct functions. Proc. Natl. Acad. Sci. USA 106: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henri S., Poulin L. F., Tamoutounour S., Ardouin L., Guilliams M., de Bovis B., Devilard E., Viret C., Azukizawa H., Kissenpfennig A., Malissen B. 2010. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med. 207: 189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capucha T., Mizraji G., Segev H., Blecher-Gonen R., Winter D., Khalaileh A., Tabib Y., Attal T., Nassar M., Zelentsova K., et al. 2015. Distinct murine mucosal Langerhans cell subsets develop from pre-dendritic cells and monocytes. Immunity 43: 369–381. [DOI] [PubMed] [Google Scholar]
- 45.Dakic A., Shao Q. X., D’Amico A., O’Keeffe M., Chen W. F., Shortman K., Wu L. 2004. Development of the dendritic cell system during mouse ontogeny. J. Immunol. 172: 1018–1027. [DOI] [PubMed] [Google Scholar]
- 46.Pöllinger B., Junt T., Metzler B., Walker U. A., Tyndall A., Allard C., Bay S., Keller R., Raulf F., Di Padova F., et al. 2011. Th17 cells, not IL-17+ γδ T cells, drive arthritic bone destruction in mice and humans. J. Immunol. 186: 2602–2612. [DOI] [PubMed] [Google Scholar]
- 47.Won H. Y., Lee J. A., Park Z. S., Song J. S., Kim H. Y., Jang S. M., Yoo S. E., Rhee Y., Hwang E. S., Bae M. A. 2011. Prominent bone loss mediated by RANKL and IL-17 produced by CD4+ T cells in TallyHo/JngJ mice. PLoS One 6: e18168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B., Ivashkiv L. B. 2011. Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res. Ther. 13: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teng Y. T., Mahamed D., Singh B. 2005. Gamma interferon positively modulates Actinobacillus actinomycetemcomitans-specific RANKL+ CD4+ Th-cell-mediated alveolar bone destruction in vivo. Infect. Immun. 73: 3453–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Y., Grassi F., Ryan M. R., Terauchi M., Page K., Yang X., Weitzmann M. N., Pacifici R. 2007. IFN-γ stimulates osteoclast formation and bone loss in vivo via antigen-driven T cell activation. J. Clin. Invest. 117: 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson A., Murray S., Divakar P., Burnaevskiy N., Pifer R., Forman J., Yarovinsky F. 2012. Microbial infection-induced expansion of effector T cells overcomes the suppressive effects of regulatory T cells via an IL-2 deprivation mechanism. J. Immunol. 188: 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liston A., Gray D. H. 2014. Homeostatic control of regulatory T cell diversity. Nat. Rev. Immunol. 14: 154–165. [DOI] [PubMed] [Google Scholar]
- 53.Guilliams M., Crozat K., Henri S., Tamoutounour S., Grenot P., Devilard E., de Bovis B., Alexopoulou L., Dalod M., Malissen B. 2010. Skin-draining lymph nodes contain dermis-derived CD103− dendritic cells that constitutively produce retinoic acid and induce Foxp3+ regulatory T cells. Blood 115: 1958–1968. [DOI] [PubMed] [Google Scholar]
- 54.Sun C. M., Hall J. A., Blank R. B., Bouladoux N., Oukka M., Mora J. R., Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ginhoux F., Collin M. P., Bogunovic M., Abel M., Leboeuf M., Helft J., Ochando J., Kissenpfennig A., Malissen B., Grisotto M., et al. 2007. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J. Exp. Med. 204: 3133–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bursch L. S., Wang L., Igyarto B., Kissenpfennig A., Malissen B., Kaplan D. H., Hogquist K. A. 2007. Identification of a novel population of Langerin+ dendritic cells. J. Exp. Med. 204: 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poulin L. F., Henri S., de Bovis B., Devilard E., Kissenpfennig A., Malissen B. 2007. The dermis contains langerin+ dendritic cells that develop and function independently of epidermal Langerhans cells. J. Exp. Med. 204: 3119–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Litvinov S. V., Velders M. P., Bakker H. A., Fleuren G. J., Warnaar S. O. 1994. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 125: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baeuerle P. A., Gires O. 2007. EpCAM (CD326) finding its role in cancer. Br. J. Cancer 96: 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaiser M. R., Lämmermann T., Feng X., Igyarto B. Z., Kaplan D. H., Tessarollo L., Germain R. N., Udey M. C. 2012. Cancer-associated epithelial cell adhesion molecule (EpCAM; CD326) enables epidermal Langerhans cell motility and migration in vivo. Proc. Natl. Acad. Sci. USA 109: E889–E897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Aar A. M., Picavet D. I., Muller F. J., de Boer L., van Capel T. M., Zaat S. A., Bos J. D., Janssen H., George T. C., Kapsenberg M. L., et al. 2013. Langerhans cells favor skin flora tolerance through limited presentation of bacterial antigens and induction of regulatory T cells. J. Invest. Dermatol. 133: 1240–1249. [DOI] [PubMed] [Google Scholar]
- 62.Haley K., Igyártó B. Z., Ortner D., Bobr A., Kashem S., Schenten D., Kaplan D. H. 2012. Langerhans cells require MyD88-dependent signals for Candida albicans response but not for contact hypersensitivity or migration. J. Immunol. 188: 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trautwein-Weidner K., Gladiator A., Kirchner F. R., Becattini S., Rülicke T., Sallusto F., LeibundGut-Landmann S. 2015. Antigen-specific Th17 cells are primed by distinct and complementary dendritic cell subsets in oropharyngeal candidiasis. PLoS Pathog. 11: e1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]