Abstract
Watermelon (Citrullus lanatus) is one xerophyte that has relative higher tolerance to drought and salt stresses as well as more sensitivity to cold stress, compared with most model plants. These characteristics facilitate it a potential model crop for researches on salt, drought or cold tolerance. In this study, a genome-wide comprehensive analysis of the ClNAC transcription factor (TF) family was carried out for the first time, to investigate their transcriptional profiles and potential functions in response to these abiotic stresses. The expression profiling analysis reveals that several NAC TFs are highly responsive to abiotic stresses and development, for instance, subfamily IV NACs may play roles in maintaining water status under drought or salt conditions, as well as water and metabolites conduction and translocation toward fruit. In contrast, rapid and negative responses of most of the ClNACs to low-temperature adversity may be related to the sensitivity to cold stress. Crosstalks among these abiotic stresses and hormone (abscisic acid and jasmonic acid) pathways were also discussed based on the expression of ClNAC genes. Our results will provide useful insights for the functional mining of NAC family in watermelon, as well as into the mechanisms underlying abiotic tolerance in other cash crops.
The NAC [no apical meristem (NAM), Arabidopsis thaliana transcription activation factor (ATAF1/2) and cup-shaped cotyledon (CUC2)] gene family is one of the largest plant-specific transcription factor (TF) families. NAC proteins play key roles in regulating gene expression at the transcription level by binding to specific cis-acting elements in the promoters of target genes. Commonly, NAC proteins possess a conserved NAM domain at the N-terminus and a divergent transcription regulation domain at the C-terminus, forming the typical protein model of NAC transcription factors1,2. NAC domains are usually composed of nearly 150 amino acid residues and divided into five subdomains A–E1,3. Among them, subdomains C and D are conserved and bind to DNA. Subdomain A plays an important role in NAC dimeric proteins. Subdomains B and E are highly divergent and might confer functional diversity to NAC TFs4,5. The NAC domain’s crystal structure in ANAC019 from Arabidopsis and in stress-response NAC1 from rice were similar to the structure of WRKY4,6. Another study showed a high similarity between the protein domain structures of NAC and GLIA CELL MISSING (GCM)7. Therefore, NAC proteins are classified as members of the WRKY-GCM1 super family.
Increasing evidences indicate roles for NAC proteins in biological processes and transcriptional regulatory networks8. For example, ATAF1/2, CUC2, and ANAC036 are involved in cell division9,10,11. SECONDARY WALL NAC DOMAIN PROTEIN1 in rice and NAC SECONDARY WALL THICKENING PROMOTING FACTOR 2 (NST2) in Arabidopsis are concerned with the secondary growth12,13. CUC2 is involved in shoot apical meristem development14, AtNAM plays a role in embryo development15, ANAC029 (also known as AtNAP) and EPHEMERAL1 are involved in plant senescence16,17, AtNAC2 and TaNAC1 are implicated in lateral root development18,19, and some other NAC TFs play roles in nutrition transportation20, flowering time21, and cell death22. A tomato NAC gene is a positive regulator of carotenoid accumulation and fruit ripening23 and PpNAC1 activates the biosynthesis of anthocyanin in peach24, implying roles for NAC TFs in plant fruit development. Increasing amounts of evidence indicate that NAC is involved in xylem development25. The essential roles of the NAC family in both water-contributing and supporting cells indicated the contribution of this family to plants adaptation to land26.
NAC domain-containing proteins are also involved in plant abiotic and biotic responses. In Arabidopsis, ANAC019, ANAC055 and ANAC072 were markedly up-regulated by drought, salt, and abscisic acid (ABA) treatments, and consequently improve plant drought resistance27. Moreover, ANAC072 and ANAC019 also have the ability to positively regulate ABA signaling27,28,29. ANAC019 and ANAC055 can promote the expression of VEGETATIVE STORAGE PROTEIN1 (VSP1) and LIPOXYGENASE2 (LOX2), which are involved in jasmonic acid (JA) signaling30. In addition, the overexpression of a Lepidium latifolium NAC gene in tobacco enhanced its cold tolerance31. The Ataf1-1 mutant showed decreased resistance to Blumeria graminis f.sp. hordei, suggesting a positive role for ATAF1 in pathogen tolerance32, while ATAF2 exhibited a positive response to JA and salicylic acid (SA)33.
Citrullus lanatus is one xerophyte that has relative higher tolerance to drought and salt stresses as well as more sensitivity to cold stress, compared with most other crops. However, a systematic analysis on ClNAC family genes and their responsive patterns to diverse abiotic stresses is lacking. Here, we identified 80 ClNAC TFs and predicted their induced patterns and functions through a genome-wide bioinformatics analysis. Furthermore, a global landscape of NAC expression patterns in response to abiotic stresses (drought, salt and cold) and phytohormones (ABA and JA) was investigated. This study will lay the basis of functional characterization of NAC TFs, as well as the advancement of research on abiotic tolerance in cash crops.
Results and Discussion
Identification of NAC TFs
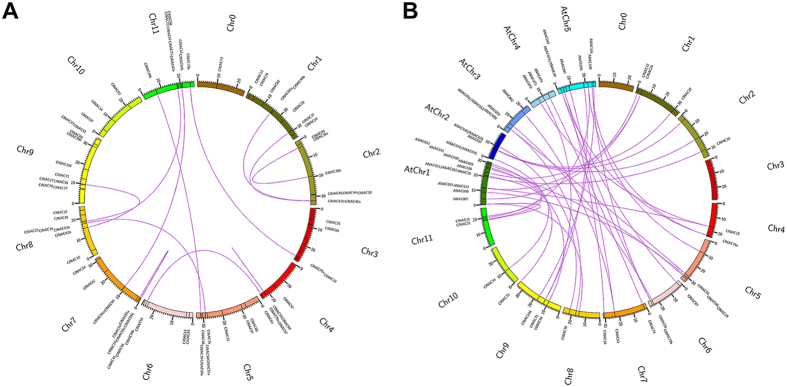
To identify ClNAC proteins, searches of the Citrullus lanatus genome using the BLASTp algorithm were performed with Arabidopsis and rice NAC proteins sequences as the query. In total, 80 putative NAC TFs with conserved NAM domain were identified (Table 1), which is in agreement with the watermelon NAC gene family in the Plant Transcription Factor Database (PlantTFDB; http://planttfdb.cbi.pku.edu.cn). The number of NAC TFs in watermelon is less (80) than in Arabidopsis (138) and rice (140). Owing to the lack of a designated standard annotation for the 80 NAC genes in watermelon, we named them ClNAC1-ClNAC104 based on their homology to the Arabidopsis NAC proteins (highest to lowest sequence similarity level) and some numbers were omitted due to the lack of ANAC homologies in watermelon. The NAC TF genes identified in watermelon encoded proteins ranging from 153 to 642 amino acid (aa) residues in length, with an average of 346 aa (Table 1). Seventy-nine of the ClNACs were distributed across the 11 watermelon chromosomes, with ClNAC73 putatively being located on the Chromosome 0 (Table 1, Fig. 1A). In an neighbor-joining (NJ) phylogenetic analysis, 12 pairs of duplicate/triplicate genes were identified, including two pairs of tandem duplicate genes (ClNAC59 and ClNAC60 on chromosome 4, and ClNAC55b and ClNAC55c on chromosome 7) (Fig. 1A; Supplementary Fig. S1). Most of the ClNAC duplicate genes had similar N-myristoylation motifs (Supplementary Fig. S2). These duplicate genes contributed significantly to the expansion of the watermelon NAC TF gene family. Simultaneously, 30 pairs of putative orthologs of NAC TFs, between watermelon and Arabidopsis, were found (Fig. 1B, Supplementary Figs S1 and S3).
Table 1. NAC transcription factor gene family in watermelon.
| Gene symbol | Gene locus | Length(aa) | Gene Location | Putative Arabidopsis orthologs | The closest genes | E-value |
|---|---|---|---|---|---|---|
| ClNAC01 | Cla007853 | 289 | Chr2:1955638..1956791 | ANAC002/ATAF1 | 1.00E-103 | |
| ClNAC02a | Cla023182 | 299 | Chr11:18231295..18232770 | ANAC002/ATAF1 | 1.00E-135 | |
| ClNAC02b | Cla013922 | 296 | Chr8:14908050..14909118 | ANAC002/ATAF1 | 1.00E-140 | |
| ClNAC05 | Cla006268 | 334 | Chr5:7417554..7418938 | ANAC007/VND4 | 1.00E-114 | |
| ClNAC06 | Cla004626 | 379 | Chr9:31582624..31584970 | ANAC008 | 5.00E-76 | |
| ClNAC07 | Cla005677 | 363 | Chr10:3535524..3538561 | ANAC007/VND4 | 1.00E-105 | |
| ClNAC08 | Cla020366 | 410 | Chr5:30465691..30470090 | ANAC008 | 1.00E-142 | |
| ClNAC09a | Cla010181 | 388 | Chr5:31397122..31398806 | ANAC009 | 1.00E-104 | |
| ClNAC09b | Cla003347 | 327 | Chr11:7128468..7130130 | ANAC009 | 4.00E-82 | |
| ClNAC10 | Cla009648 | 212 | Chr1:31813698..31814520 | ANAC010/SND3 | 2.00E-79 | |
| ClNAC12 | Cla011325 | 389 | Chr1:1270767..1272154 | ANAC012/SND1/NST3 | 1.00E-107 | |
| ClNAC15 | Cla012377 | 358 | Chr8:2676356..2682444 | ANAC070, ANAC015 | E-112, 2E-90 | |
| ClNAC16 | Cla013643 | 538 | Chr8:18071416..18074448 | ANAC016 | 1.00E-126 | |
| ClNAC17 | Cla016331 | 562 | Chr9:9847279..9851129 | ANAC017 | 1.00E-125 | |
| ClNAC18 | Cla011315 | 487 | Chr3:27577243..27579904 | ANAC018/NTL9 | 1.00E-52 | |
| ClNAC20 | Cla013445 | 361 | Chr2:29297528..29301836 | ANAC20 | 1.00E-102 | |
| ClNAC21 | Cla023219 | 317 | Chr11:18613972..18618963 | ANAC021/ANAC022/AtNAC1 | 1.00E-108 | |
| ClNAC23 | Cla021917 | 336 | Chr8:18558531..18559867 | ANAC031/CUC3 | 1.00E-76 | |
| ClNAC24 | Cla002713 | 625 | Chr7:279898..281775 | ANAC030/VND7 | 5.00E-14 | |
| ClNAC25 | Cla019475 | 353 | Chr3:5597680..5600578 | ANAC025 | 4.00E-93 | |
| ClNAC26 | Cla011554 | 329 | Chr1:3745458..3747358 | ANAC007/VND4, ANAC026 | E-112, E-104 | |
| ClNAC27 | Cla022514 | 433 | Chr8:24287182..24289453 | ANAC028 | 9.00E-09 | |
| ClNAC28 | Cla009127 | 642 | Chr1:22943282..22947465 | ANAC028 | 1.00E-152 | |
| ClNAC29 | Cla010201 | 283 | Chr5:31276248..31277300 | ANAC029/ATNAP/NAP | 1.00E-111 | |
| ClNAC30 | Cla016349 | 153 | Chr9:9685786..9687158 | ANAC030/VND7 | 2.00E-88 | |
| ClNAC31 | Cla023471 | 411 | Chr11:20978854..20981134 | ANAC031/CUC3 | 4.00E-98 | |
| ClNAC32 | Cla002170 | 240 | Chr5:20029237..20030302 | ANAC083 | 2.00E-40 | |
| ClNAC33 | Cla005472 | 320 | Chr9:34952827..34954767 | ANAC033 | 1.00E-103 | |
| ClNAC34 | Cla004555 | 416 | Chr10:10290284..10291953 | ANAC034/ANAC035 | 1.00E-110 | |
| ClNAC36a | Cla015772 | 279 | Chr2:3373610..3374942 | ANAC036 | 1.00E-109 | |
| ClNAC36b | Cla006906 | 296 | Chr2:19551544..19553134 | ANAC036 | 1.00E-97 | |
| ClNAC37 | Cla014269 | 303 | Chr1:29585085..29587217 | ANAC037/VND1 | 6.00E-97 | |
| ClNAC38 | Cla022231 | 351 | Chr8:21813445..21817442 | ANAC038/ANAC039 | 1.00E-108 | |
| ClNAC40a | Cla008629 | 341 | Chr2:32424448..32427343 | ANAC040/NTL8 | 3.00E-84 | |
| ClNAC40b | Cla011058 | 367 | Chr1:16131062..16138877 | ANAC040/NTL8 | 7.00E-89 | |
| ClNAC41 | Cla019304 | 244 | Chr6:26797719..26798537 | ANAC083 | 1.00E-29 | |
| ClNAC42 | Cla005508 | 325 | Chr9:34507930..34509090 | ANAC042 | 4.00E-81 | |
| ClNAC43 | Cla006697 | 400 | Chr6:3348856..3350249 | ANAC043/NST1, ANAC066 | 8E-94, 5E-80 | |
| ClNAC44 | Cla013474 | 319 | Chr2:28979835..28981165 | ANAC042 | 3.00E-81 | |
| ClNAC45 | Cla013475 | 326 | Chr2:28970371..28972299 | ANAC042 | 9.00E-78 | |
| ClNAC47 | Cla023239 | 176 | Chr11:18821525..18822788 | ANAC042 | 6.00E-76 | |
| ClNAC50 | Cla020528 | 467 | Chr5:29010431..29015195 | ANAC050, ANAC051/ANAC052 | 3E-93, 5E-91 | |
| ClNAC53a | Cla020527 | 563 | Chr5:29018616..29021862 | ANAC053, NAC2 | 1E-145,1E-117 | |
| ClNAC53b | Cla013731 | 563 | Chr8:17220446..17223318 | ANAC053, NAC2 | 1E-134,1E-111 | |
| ClNAC54 | Cla010881 | 235 | Chr7:30741029..30741978 | ANAC083 | 2.00E-95 | |
| ClNAC55a | Cla002217 | 310 | Chr7:786430..787518 | ANAC056/AtNAC2 | 1.00E-09 | |
| ClNAC55b | Cla002680 | 294 | Chr7:5608..6648 | ANAC056/AtNAC2 | 6.00E-10 | |
| ClNAC55c | Cla002681 | 294 | Chr7:10329..11369 | ANAC056/AtNAC2 | 6.00E-10 | |
| ClNAC56a | Cla011760 | 340 | Chr7:10715001..10716327 | ANAC056/AtNAC2 | 1.00E-108 | |
| ClNAC56b | Cla023408 | 320 | Chr11:20484013..20485439 | ANAC056/AtNAC2 | 1.00E-101 | |
| ClNAC57 | Cla018634 | 248 | Chr4:23877496..23880341 | ANAC057 | 1.00E-136 | |
| ClNAC58 | Cla018973 | 345 | Chr6:23992729..23994725 | ANAC058 | 2.00E-91 | |
| ClNAC59 | Cla018410 | 262 | Chr4:21728169..21729673 | ANAC090 | 3.00E-69 | |
| ClNAC60 | Cla018411 | 279 | Chr4:21736304..21738286 | ANAC090 | 1.00E-72 | |
| ClNAC61a | Cla003039 | 240 | Chr1:15383814..15384842 | ANAC090, ANAC061 | 5E-63, 4E-61 | |
| ClNAC61b | Cla008633 | 203 | Chr2:32391575..32392368 | ANAC090, ANAC061 | 8E-64, 4E-60 | |
| ClNAC62 | Cla002400 | 576 | Chr7:23188084..23191054 | ANAC091, ANAC062 | 4E-83,4E-82 | |
| ClNAC63 | Cla021063 | 448 | Chr5:85305..87840 | ANAC062 | 1.00E-30 | |
| ClNAC68 | Cla019693 | 336 | Chr3:8593061..8596774 | ANAC073 | 1.00E-101 | |
| ClNAC69 | Cla011761 | 302 | Chr7:10570728..10571833 | ANAC072/RD26 | 1.00E-115 | |
| ClNAC71 | Cla016169 | 341 | Chr9:12586253..12587805 | ANAC096, ANAC071 | 2E-87, 2E-82 | |
| ClNAC72 | Cla023407 | 321 | Chr11:20462289..20463440 | ANAC072/RD26 | 1.00E-125 | |
| ClNAC73 | Cla000378 | 298 | Chr0:10012381..10015299 | ANAC073 | 1.00E-112 | |
| ClNAC74 | Cla005970 | 289 | Chr7:1814933..1818320 | ANAC074 | 2.00E-78 | |
| ClNAC75a | Cla011248 | 490 | Chr3:26766834..26770238 | ANAC075 | 1.00E-138 | |
| ClNAC75b | Cla016810 | 467 | Chr11:25081349..25086384 | ANAC075 | 1.00E-139 | |
| ClNAC76 | Cla020655 | 299 | Chr5:27964577..27965999 | ANAC074 | 2.00E-14 | |
| ClNAC77 | Cla014880 | 154 | Chr9:7121234..7124037 | ANAC074 | 5.00E-29 | |
| ClNAC78 | Cla014910 | 201 | Chr9:7520978..7522919 | ANAC074 | 7.00E-55 | |
| ClNAC79a | Cla018596 | 332 | Chr4:23607586..23608786 | ANAC100/ATNAC5 | E-103, 3E-97 | |
| ClNAC79b | Cla019099 | 362 | Chr6:25092288..25093541 | ANAC100/ATNAC5 | E-115, E-106 | |
| ClNAC82 | Cla008434 | 480 | Chr1:9372652..9375621 | ANAC082, ANAC103 | 2E-67, 2E-67 | |
| ClNAC83 | Cla001495 | 256 | Chr6:1853182..1854287 | ANAC083 | 1.00E-107 | |
| ClNAC87 | Cla012144 | 323 | Chr4:15660560..15662087 | ANAC087, ANAC046 | 3E-78, 7E-73 | |
| ClNAC92 | Cla016990 | 265 | Chr10:21307912..21309583 | ANAC092/ATNAC2/ATNAC6 | 7.00E-81 | |
| ClNAC96 | Cla019229 | 367 | Chr6:26229431..26230741 | ANAC096 | 2.00E-13 | |
| ClNAC97 | Cla004290 | 289 | Chr5:9565473..9568819 | ANAC098/CUC2 | 8.00E-93 | |
| ClNAC98 | Cla023357 | 368 | Chr11:19954210..19956573 | ANAC098/CUC2 | 1.00E-107 | |
| ClNAC100 | Cla010317 | 331 | Chr9:30221670..30222842 | ANAC100/ATNAC5 | 2.00E-96 | |
| ClNAC104 | Cla009439 | 202 | Chr9:17570927..17572481 | ANAC104/XND1 | 5.00E-59 |
Figure 1. Visualization of the NAC TF linkage groups.
(A) Chromosomal distributions of NAC TFs in the watermelon genome. The lines represent duplicate pairs of watermelon NAC genes. (B) Putative orthologs of NAC TFs in watermelon and Arabidopsis.
Phylogenetic analysis
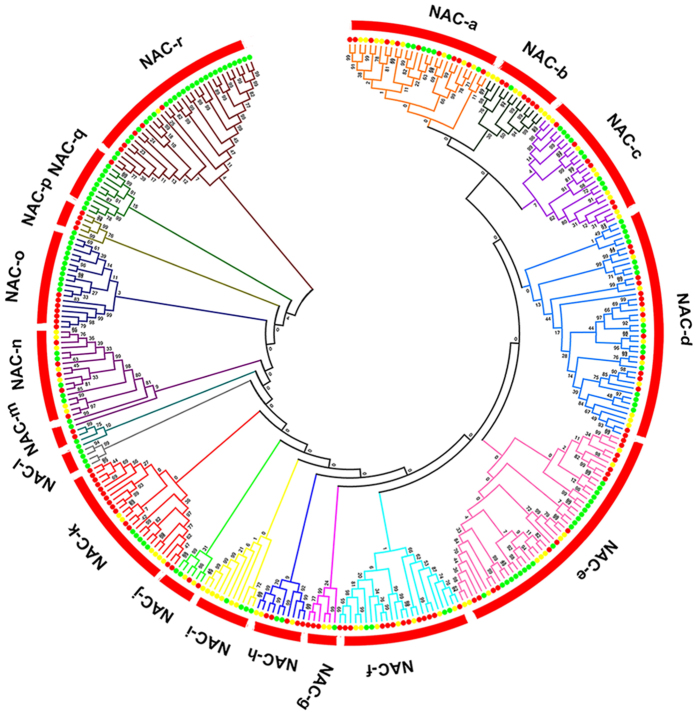
To investigate the evolutionary relationships among the NAC TFs, 329 NAC domain sequences were predicted from Arabidopsis, rice, and watermelon using alignments of the full-length NAC sequences. These NAC proteins were classified into 18 groups (namely NAC-a to NAC-r; Fig. 2, Supplementary Fig. S1), which is in strong agreement with the results found in Populus34. NAC TFs in same group are likely to possess similar functions. For example, group NAC-a includes NAC proteins such as RD26, ANAC019, and ANAC055 and are involved in stress responses28,30, while group NAC-b possesses all of the NAC proteins, such as CUC1 and CUC2, that function in the delimitation of the shoot organ boundary14,35. The 80 ClNAC TFs are distributed throughout most of the groups, indicating multiple and various functions of NAC TFs in watermelon. Interestingly, ClNAC TF is absent in the NAC-m, NAC-o and NAC-p groups, which implies that these groups might be lost in watermelon during evolution. This finding may explain why watermelon contains fewer NAC TFs than Arabidopsis, even though these two plants have similar numbers of protein-coding genes. Similarly, group NAC-i did not contain any Arabidopsis members (Fig. 2, Supplementary Fig. S1). Additionally, group NAC-l and group NAC-q contain only rice members, suggesting that these groups were either acquired after the divergence of monocots and dicots, or were lost in watermelon and Arabidopsis.
Figure 2. Phylogenetic tree of NAC proteins from watermelon, Arabidopsis, and rice.
The phylogenetic tree is based on a sequence alignment of 329 NAC protein sequences from watermelon, Arabidopsis, and rice. The unrooted tree was generated with MEGA5.0 using the NJ method. Bootstrap values are indicated at each node. The NAC proteins are grouped into 18 distinct clades (a–r). The yellow, red, and green dots represent watermelon, Arabidopsis, and rice NACs, respectively.
Gene structure and conserved motifs
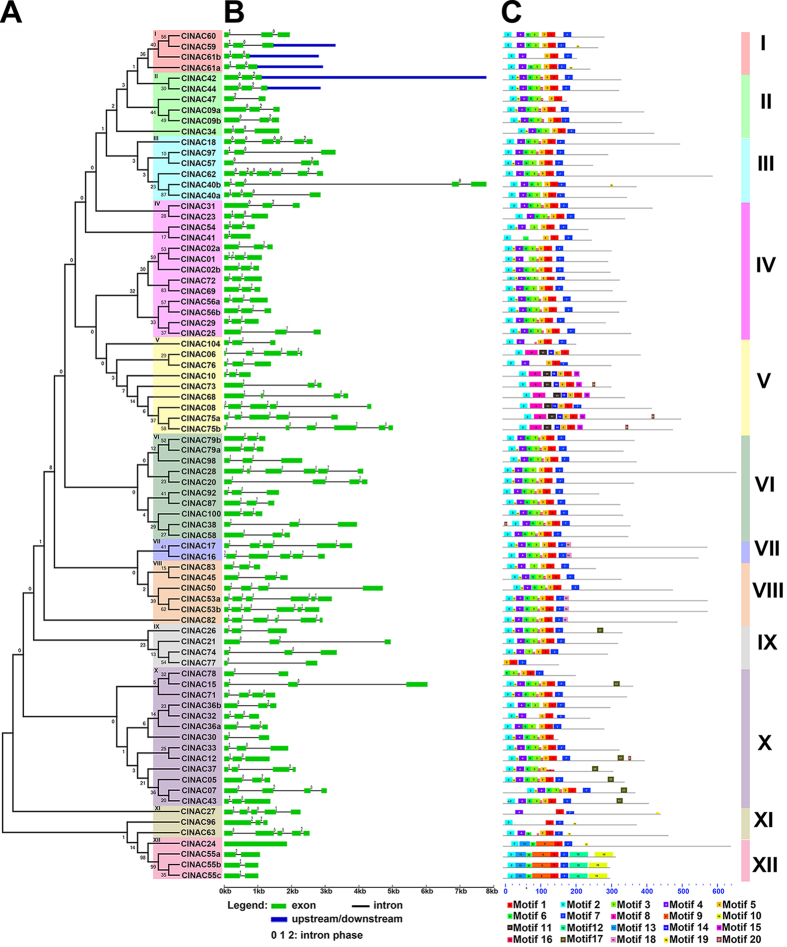
To get a better understanding of the structural diversity of ClNAC TFs, we compared the exon/intron organization in their coding sequences. The 80 ClNAC TFs were divided into 12 subfamilies in the NJ phylogenetic tree. Among them, subfamily IV and X with 13 members were the highest in numbers and subfamily VII was the lowest with only two members (Fig. 3A). Members in the same subfamily shared similar exon/intron structures in terms of intron phase, intron number, and exon length. For instance, the NAC genes in subfamily V and XI harbored two to four introns, while those in subfamily XII possessed only one intron, with the exception of ClNAC24 which had no intron. By contrast, subfamily VII had the largest number of 4 to 5 introns. Interestingly, the intron number varied significantly, while the intron phase and exon length were highly conserved in subfamilies III, VI, and VIII (Fig. 3B).
Figure 3. Phylogenetic relationships, gene structures and protein structures of the ClNAC TFs.
(A) The phylogenetic tree was constructed with MEGA 5.0 using the NJ method with 1,000 bootstrap replicates based on a multiple alignment of 80 NAC amino acid sequences from watermelon. The 12 major subfamilies are indicated (I–XII) and are marked with different colored backgrounds. (B) Exon/intron structures of NAC genes from watermelon. Exons and introns are represented by green boxes and black lines, respectively. The sizes of the exons and introns are estimated using the scale at the bottom. (C) Schematic of the conserved motifs in the NAC proteins from watermelon elucidated by MEME. Every motif is represented by one colored box with a number. The black lines represent the non-conserved sequences. Refer to Supplementary Table S1 for individual motif details.
To reveal the diversity of ClNAC TFs, the MEME program was used to predict putative motifs. Ultimately, 20 distinct motifs were identified (Supplementary Table S1). Most of the NAC TF proteins contained A to E motifs in the N-termini, which conferred DNA-binding activity1. Here, motif 2, 4, 3, 1 and 7 specified the NAM subdomains A to E, respectively. Most of the ClNAC proteins contain all of these five motifs, except for subfamily XII, which had no motif B, and subfamily V, which had neither subdomain A nor B. However, these two subfamilies had their specific motifs, such as motifs 9, 10, 12, and 13 in subfamily XII, and motifs 8, 11, 14, and 15 in subfamily V. Even if the divergence level in C-terminal regions of the NAC TF proteins was relatively high, some conserved motifs were also identified in these regions in some specific subfamilies, for example, motif 17 in subfamily X and motif 18 in subfamilies VII and VIII (Fig. 3C). These results suggested that the specific functions of different subfamilies might be owing to specific motifs.
NAC gene response, localization and function predictions
Gene expression responses are largely related to their promoters; therefore, we investigated the putative stimulus-responsive cis-elements in the promoter regions of all of the ClNAC genes (Supplementary Table S2). Nine types of cis-elements were detected, including cis-acting regulatory elements (AREs) that are essential for anaerobic induction; two cis-acting regulatory elements (TGACG-motif and CGTCA-motif) that are involved in MeJA responsiveness; MYB-binding sites (MBS) associated with drought inducibility; low-temperature-responsive elements (LTRs); ABA-responsive elements (ABREs); SA-responsive elements (TCA-elements); heat shock-responsive elements (HSEs) and ET-responsive elements (EREs)36,37,38,39,40,41. Every NAC gene contains at least one cis-element type in their promoter sequences (Supplementary Table S3), suggesting that these ClNACs are involved in watermelon response to different abiotic stresses and/or hormone signaling. Surprisingly, differences in the types and numbers of cis-elements were observed in some duplicate gene pairs. Two ERE elements exist in the promoter of ClNAC09a, while none could be found in its duplicate gene, ClNAC09b (Supplementary Table S3). A comparison of the promoter regions of all the duplicate gene pairs showed their divergence, although conserved regions were also observed (Fig. 4). Additionally, the protein’s function is related to its localization in some way42. Based on the subcellular localization predictions, most ClNACs probably function in the nucleus, while others were located in different organelles or the cytoplasm. For instance, ClNAC06, ClNAC62, ClNAC50, ClNAC53a and ClNAC74 might be located in chloroplasts; ClNAC07 and ClNAC30 might be located in mitochondria; and ClNAC01, ClNAC02b, ClNAC77, ClNAC78, and ClNAC104 might be located in the cytoplasm. Moreover, of the 80 ClNACs, only ClNAC06 contains a signal peptide, indicating that it has an important role in protein subcellular localization (Supplementary Fig. S4). Moreover, phosphorylation could adjust the cellular localization of TFs, and change their activities43. Each ClNAC protein sequence contains these three types of phosphorylation sites, with S phosphorylation being the most common (Supplementary Table S5, Supplementary Fig. S5). These phosphorylation sites might be involved in the regulation of protein activities when plants are subject to stresses.
Figure 4. Comparative analysis of the promoter regions in ClNAC duplications.
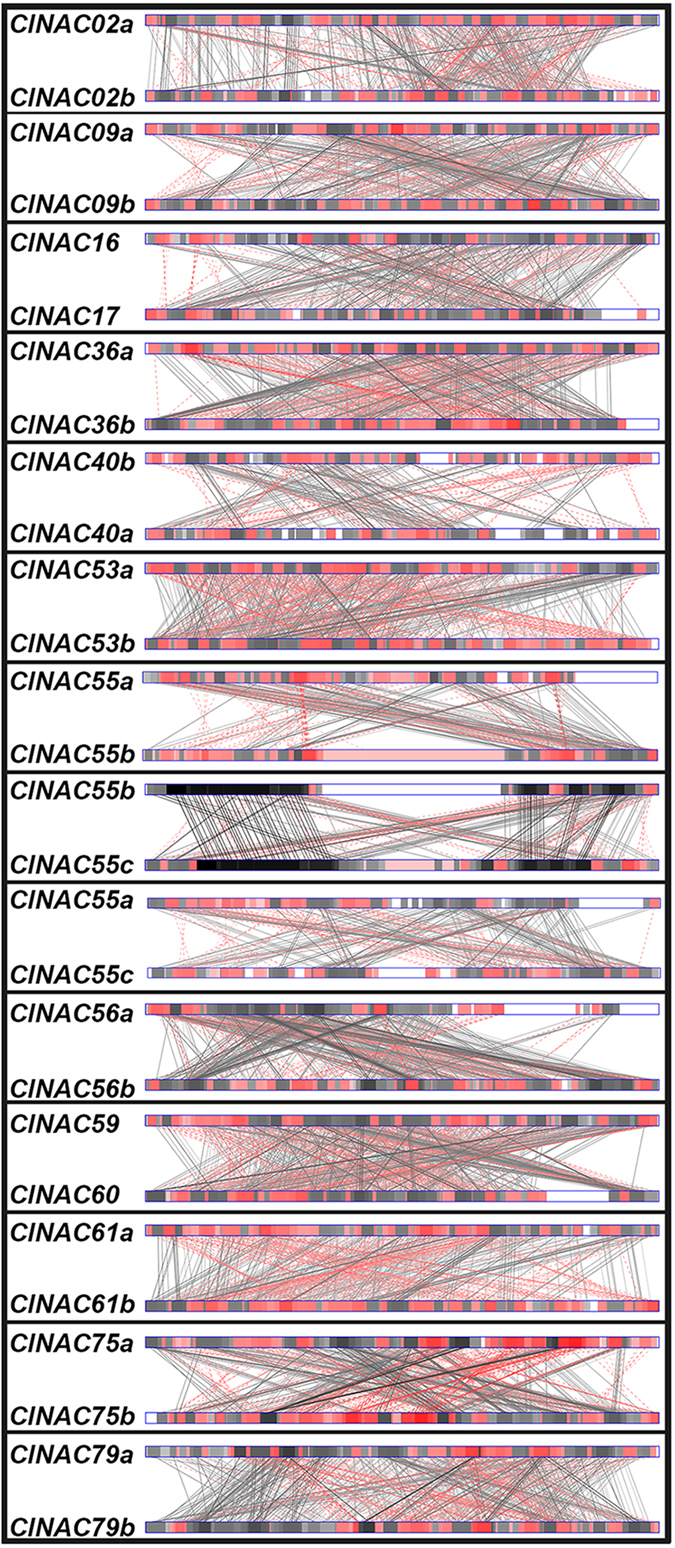
Black and red boxes with connecting lines between duplicate genes represent similar regions in their promoters. The depths of the different colors represent the similarities of conserved regions. Solid dark lines connect similar regions and red broken lines connect matched regions in the reversed orientation. White boxes without connecting lines represent divergent regions.
Expression profiles of ClNACs in tissues and fruit developmental stages
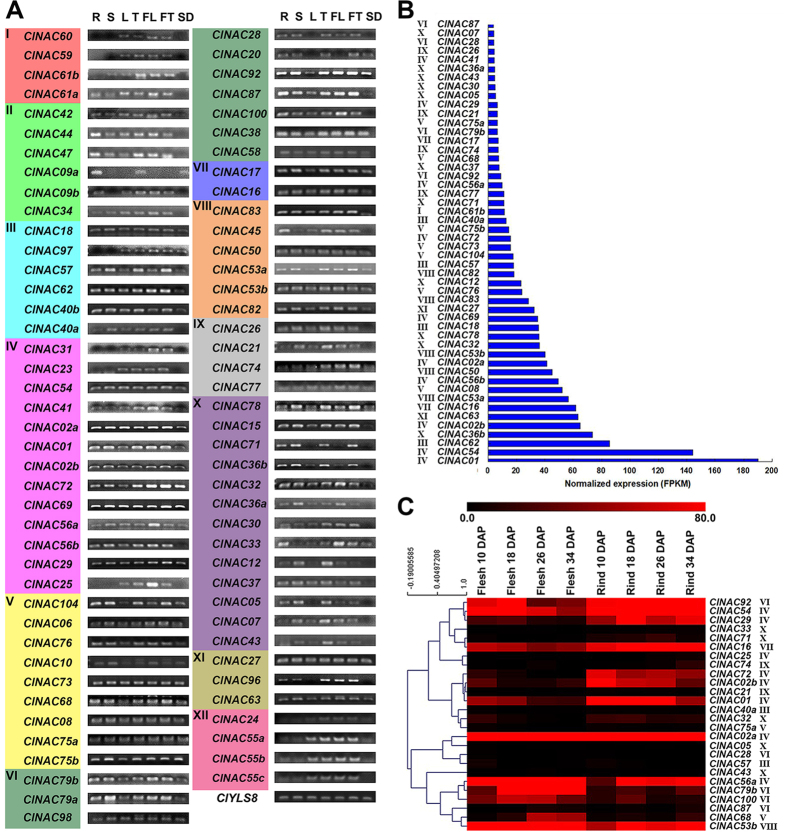
In total, 45 NAC TFs could be detected in all of the tissues, suggesting that they may have various regulatory roles in multiple tissues at multiple developmental stages. Besides, the expression of all ClNACs can be detected in young fruit, except for ClNAC09a. While subfamily XII exhibit the most uniform expression pattern, and all of the members could be detected in tissues of young leaf, tendril, flower, and young fruit. Furthermore, most duplicated gene pairs shared similar expression patterns (Fig. 5A).
Figure 5. Expression profiles of watermelon NAC TFs in different tissues and at different fruit developmental stages.
(A) RT-PCR analyses of NAC TFs in seven watermelon tissues. R: root; S: stem apex; L: young leaf; T: tendril; FL: flower; F: young fruit; SD: seed. CIYLS8 was used as the control. (B) The transcript levels of NAC TFs in watermelon vascular tissues. Numbers on the vertical axis represent the normalized expression (FPKM) of 50 NAC TFs in watermelon vascular tissues. (C) Expression profiles of watermelon NAC TFs across different fruit developmental stages. The scale representing the relative signal intensity values is shown above. DAP: Days After Pollination.
As a drought-tolerant crop with a high water demand, a powerful vascular system is essential for watermelon to maintain its water status to keep homeostasis under water-deficit conditions. Moreover, increasing evidence indicates that NAC TFs play important roles in the development of vascular tissues2,25,44,46, as well as in the adaptation of plants to land26. To determine the functions of ClNAC TFs in the development of vascular system, we analyzed the normalized expression of ClNAC TFs using published transcriptome sequencing data45. ClNAC54 and ClNAC01, which belong to the subfamily IV, show extremely higher expression levels. The expression levels of six ClNAC genes (ClNAC07, ClNAC05, ClNAC26, ClNAC30, ClNAC24, and ClNAC37), homologous to Arabidopsis VASCULAR-RELATED NAC-DOMAIN (VND) genes25,44,46, were relatively lower. The expression of ClNAC43 and ClNAC18 were also detected, and their putative homologs, NST1 and SECONDARY WALL-ASSOCIATED NAC DOMAIN 1 (SND1), play crucial roles in secondary wall thickening47,48. Interestingly, 10 out of 13 subfamily IV members had detectable expression levels that were mostly relatively higher (Fig. 5B), indicating that subfamily IV may be involved in the vascular system development. The occurrence of the plant vascular system is a striking innovation that enabled its colonization of land, and NAC proteins played essential roles in the adaptation of plants to land26. The putative functions of subfamily IV ClNACs in vascular development suggested that subfamily IV is likely involved in the evolutionary process of water conduction in watermelon.
Given the expression of almost all ClNACs in young fruit (Fig. 5A), we analyzed the involvement of NAC TFs in different parts of the fruit during different fruit stages (Fig. 5C). The expression levels of ClNAC16, ClNAC92, ClNAC54, and ClNAC29 were relatively higher in the rind at all of the stages, while their expression in the flesh was higher in the early stages and decreased from 26 days after pollination. Moreover, the transcript levels of ClNAC32, ClNAC72, ClNAC02b, and ClNAC01 were higher in the rind than in the flesh, and their expression levels were relatively higher in the earlier stages of each tissue development, which suggested that these genes might play more important roles in the early stages of rind development. However, some ClNACs, such as ClNAC56a, ClNAC79b, ClNAC100, and ClNAC53b, showed relatively higher levels in the later stages (Fig. 5C). These results indicated that different NAC TFs play roles in different fruit ripening stages. Most (10 of 12) of the highly expressed genes detected in this analysis belonged to subfamily IV or VI, indicating that these two subfamilies might be important for fruit development. The vascular system is essential for water and sugar transportation during fruit development. Here, 21 common ClNACs were detected in both vascular tissues and fruit, with 10 of them belonging to subfamily IV or VI (Fig. 5B,C). This suggested that these two subfamilies were important in correlating the development of vascular tissues and fruit in watermelon. In particular, ClNAC01, ClNAC02a and ClNAC02b, which presented quite high expression levels in both vascular and fruit (Fig. 5B,C), were similar to SlNAC4 in protein sequence and expression profiles. This tomato NAC gene is a positive regulator of carotenoid accumulation and fruit ripening23. Additionally, TtNAM-B1, which had a sequence similarity with ClNAC56a and ClNAC56b, increases nutrient remobilization in wheat20. All of these ClNACs belong to subfamily IV, implying that this subfamily is important for the transport of nutrients and metabolites to watermelon fruit via the vascular system.
Expression profiles of the ClNACs under abiotic stress
Given that Citrullus lanatus is tolerant to salt and drought stresses, but sensitive to low temperatures; and NAC TFs are likely to be involved in physiological adaptations in response to these stresses18,49,50. We examined the expression levels of some ClNACs under salt, drought and low-temperature treatments. Salt stress caused quick and significant responses of 10 ClNACs (ClNAC74, ClNAC59, ClNAC60, ClNAC23, ClNAC31, ClNAC36b, ClNAC56a, ClNAC56b, ClNAC72, and ClNAC69) in roots. It also caused a quick but transient increase in the expression level of nine ClNACs (ClNAC78, ClNAC24, ClNAC07, ClNAC61b, ClNAC25, ClNAC77, ClNAC09a, ClNAC96, and ClNAC09b) (Fig. 6A). In Arabidopsis, there are three closely related stress-response NAC genes (ANAC019, ANAC055 and ANAC072), which were induced by drought, salinity, and the hormones ABA and JA27,28,29,30. Here, their watermelon orthologs (ClNAC72 and ClNAC69) also showed positive responses to the NaCl treatment. Notably, all of these members of subfamily IV were extremely sensitive to NaCl treatment, which is in strong agreement with their functions in vascular development. Furthermore, 3 quarters members of subfamily I also showed rapid and positive responses after the NaCl treatment. The high response of subfamilies I and IV NACs to salt treatment provided primary evidence for their possible participation in plant salt stress tolerance.
Figure 6. Expression analyses of NAC TFs in the roots of watermelon exposed to NaCl, PEG and low temperature.
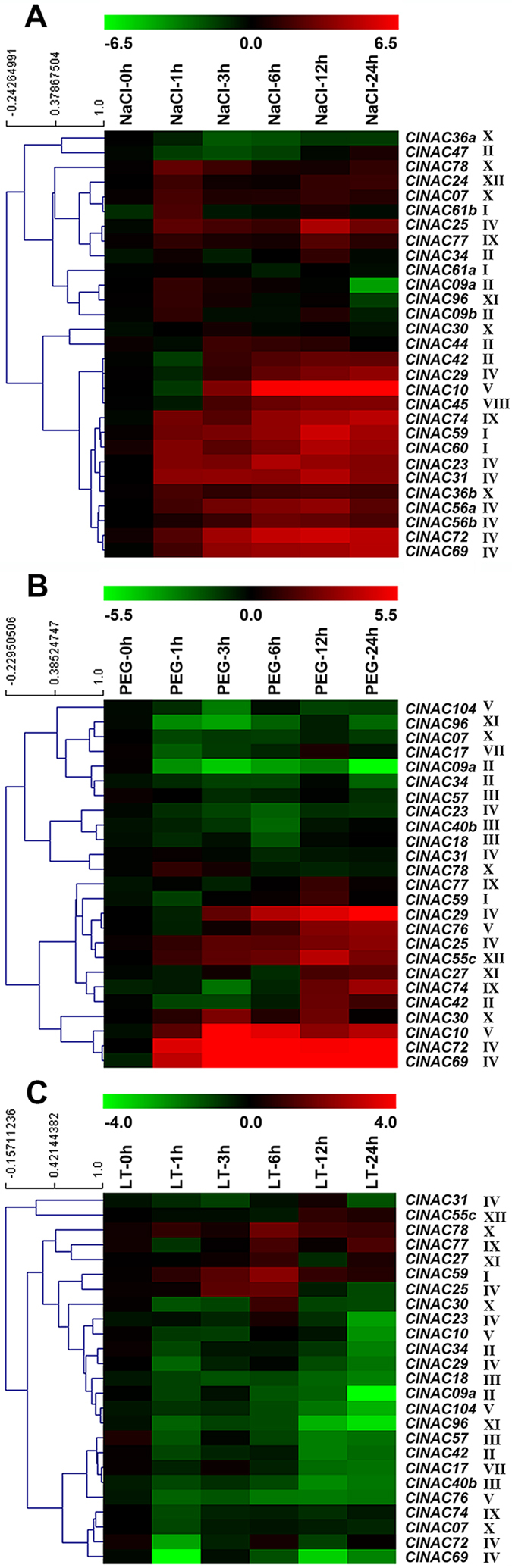
Expression analysis of NAC TFs in the roots of watermelon exposed to 200 mM NaCl. (B) Expression analysis of 23 NAC TFs in the roots of watermelon exposed to 20% PEG. (C) Expression analysis of 23 NAC TFs in the roots of watermelon exposed to 8 °C (low-temperature, LT). The scale representing the relative signal intensity values is shown above. Hierarchical clustering was used in the data analysis.
After PEG treatment, several genes, including ClNAC29, ClNAC25, ClNAC55c, ClNAC30, ClNAC10, ClNAC72 and ClNAC69, showed rapid and positive responses (Fig. 6B). Among them, ClNAC72 and ClNAC69 were the most outstanding responsers. In contrast, there were about half of the detected ClNAC genes were quickly and markedly down-regulated. Among them, ClNAC96 and ClNAC09a showed the most significant decrease, suggesting their potential involvement in drought tolerance in a negative manner (Fig. 6B). Interestingly, four highly expressed ClNACs (ClNAC29, ClNAC25, ClNAC72 and ClNAC69) belonged to subfamily IV, which also participates in vascular development and salt response (Figs 5B and 6A). As the function of ANAC019, ANAC055 and ANAC072, homologs of ClNAC72 and ClNAC69, in drought tolerance have been demonstrated in transgenic plant27, and proteins with similar structure have the same kinds of function, we hypothesized that subfamily IV ClNACs may play similar roles for plant responses to water stresses.
Under low-temperature stress, most of the detected ClNACs showed negative responses (Fig. 6C), which was assumed to be attributed to the sensitivity of watermelon to this stress. There were also few ClNAC genes that were induced by the low-temperature. Among them, ClNAC25, ClNAC78 and ClNAC59, exhibited quicker responses to the low-temperature and higher fold changes in expression levels than the others. Almost all of positive-responding genes showed their expressional peak at 6h after treatment, suggesting their earlier responses to low-temperature stress (Fig. 6C). Additionally, the LTR element, which is responsible for low-temperature inducibility, could only be found in the promoters of some ClNAC genes, such as ClNAC30, ClNAC31, ClNAC55c and ClNAC77 (Supplementary Table S3), and all of these ClNACs were found to be up-regulated under low temperature. Notably, there were four ClNACs (ClNAC25, ClNAC77, ClNAC78 and ClNAC59), exhibited positive response to drought, salt and low-temperature stresses, implying their involvement in the crosstalk of abiotic stress signal pathways.
Expression profiles of the ClNACs in response to exogenous ABA and JA
Given that ABA plays crucial roles in response to environmental stresses51,52,53, the response of several selected NAC TFs to exogenous ABA were examined (Fig. 7A). There are five ClNACs (ClNAC42, ClNAC72, ClNAC34, ClNAC30 and ClNAC69) that showed positive responses quickly and persistently. Whereas, the expression of some ClNACs (ClNAC74, ClNAC25, ClNAC56a, ClNAC09a, ClNAC09b, ClNAC59, ClNAC23, and ClNAC60) was significantly enhanced after a transient inhibition. Not surprisingly, ABRE elements were observed in most of their promoters (Fig. 7A, Supplementary Table S3). In contrast, some negative responding ClNACs (ClNAC45, ClNAC44, ClNAC36a, ClNAC01, ClNAC02b, ClNAC61b, ClNAC36b, and ClNAC78) were also found. Interestingly, ClNAC56a, ClNAC59, and ClNAC60 positively responded to both NaCl and ABA treatments, and ClNAC72, ClNAC69, ClNAC42 and ClNAC10 were up-regulated by PEG, NaCl and ABA treatments. Moreover, ClNAC25 was induced by salt, drought, low-temperature, and ABA treatments (Figs 6 and 7A). Thus, these ClNACs might confer abiotic stress responses through the ABA pathway. Additionally, there are some ClNAC genes, such as ClNAC07, ClNAC56b, ClNAC31, ClNAC36b and ClNAC74, that were highly up-regulated by abiotic stress, but not enhanced by ABA treatment (Figs 6A,B and 7A), implying that they may participate in responses to abiotic stresses via an ABA-independent pathway.
Figure 7. Expression analyses of NAC TFs in the roots of watermelon exposed to ABA and JA.
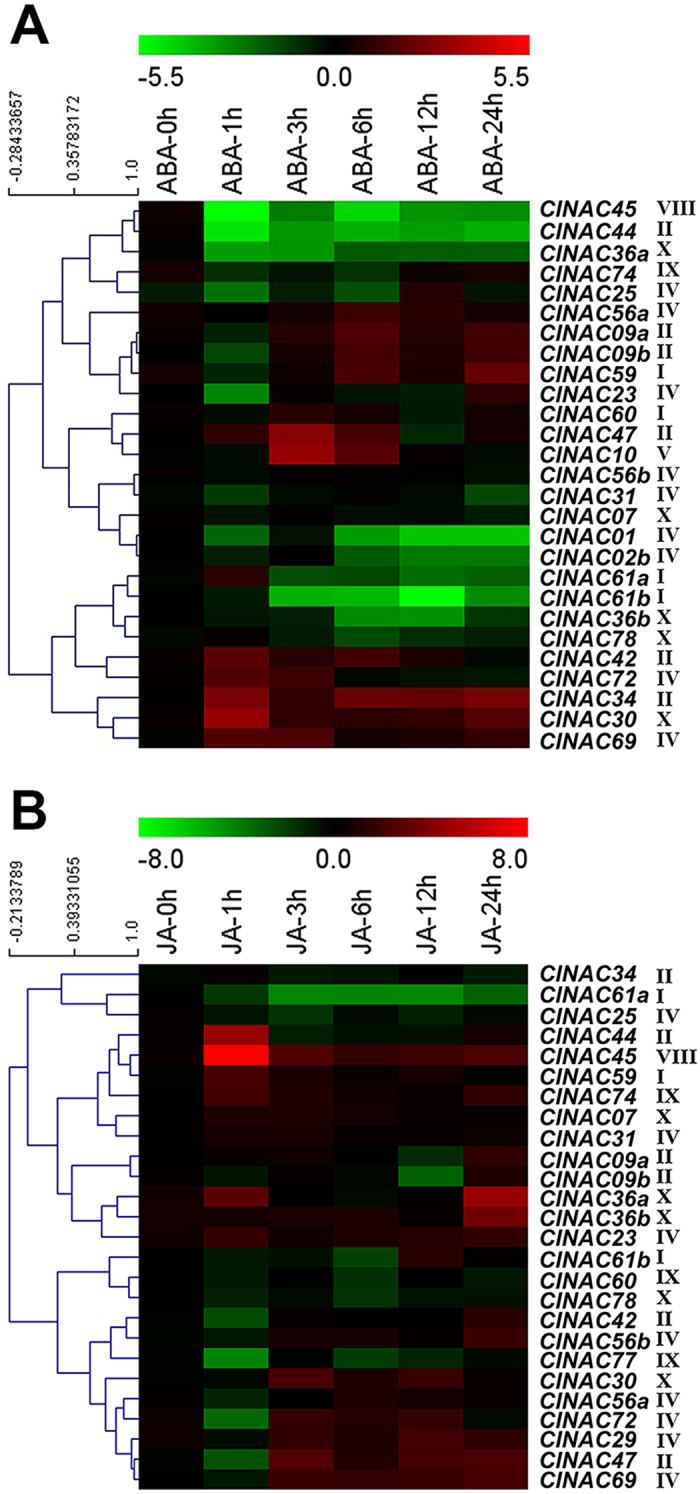
(A) Expression analysis of NAC TFs in the roots of watermelon exposed to ABA. (B) Expression analysis of 24 NAC TFs in the roots of watermelon exposed to JA. The scale representing the relative signal intensity values is shown above. Hierarchical clustering was used in the data analysis.
JA is an important hormone that regulates plant defense responses against biotic stresses, as well as a moderator of abiotic tolerance54,55. Thus, we analyzed the expression of ClNACs in response to JA. Some ClNACs showed positive responses to the exogenous JA treatment, which might result from the MeJA-responsiveness cis-acting regulatory elements (T GACG-motif and CGTCA-motif) present in most of the ClNAC promoters (Fig. 7B; Supplementary Table S3). Several ClNACs (ClNAC29, ClNAC23, ClNAC31, ClNAC56b, ClNAC44, ClNAC45, ClNAC36b, ClNAC72, ClNAC69, and ClNAC74) positively responded to both NaCl and JA treatments (Figs 6A and 7B), implying that they participate in salt stress responding via the JA pathway. Interestingly, ClNAC77 and ClNAC78 may participate in responding to all abiotic stresses above through ABA- and JA-independent pathways, as they showed no or negative responses to ABA or JA. While some other ClNACs (ClNAC59, ClNAC47, ClNAC30, ClNAC72 and ClNAC69) were induced by both JA and ABA treatments (Fig. 7A,B), suggesting that they may be the common targets downstream of the ABA- and JA-mediated stress responses. In Arabidopsis, ANAC072 and ANAC019 have the ability to positively regulate ABA signaling28,29. Moreover, ANAC019 and ANAC055 function as activators of JA-signaled defense responses30. Here, ClNAC72 and ClNAC69 exhibited similar expression patterns as those of their putative homologs (ANAC072, ANAC019 and ANAC055) under NaCl, PEG, ABA and JA treatments (Fig. 7)27. This suggests that ClNAC72 and ClNAC69 may also act as positive regulators of ABA and JA signaling in salt and drought responses. Notably, all of the subfamily IV NACs, involved in abiotic stress responses, were mediated by ABA and/or JA treatment (Figs 6 and 7). This implies that subfamily IV may be important downstream regulators of ABA- and/or JA- signal-induced stress defenses.
In conclusion, we selected 80 NAC genes and classified them into subfamilies based on their amino acid sequences for the first time in watermelon. Here we showed a global expression landscape of NAC TFs in response to various abiotic stresses. The watermelon ClNACs from different subfamilies exhibited diverse responsive patterns to environmental adversity. However, some subfamilies are highly responsive to abiotic stresses, such as salinity, cold and water deficiency, as well as involved in some distinctive vascular tissue and fruit development. The results also uncovered that the sensitivity of watermelon to cold stress might be related to the rapid and negative response of NAC TFs to low-temperature exposure. Given further studies are still needed to unravel the roles of ClNACs in the regulation of plant abiotic tolerance, our findings provide valuable clues for further functional research on NAC TF family in crop and its adaptation improvement to abiotic stresses via molecular approaches.
Methods
Plant materials, growth conditions and stress treatments
Watermelon of Citrullus lanatus cv. IVSM9 seedlings were used in this study. For the abiotic stress conditions, watermelon seedlings three true-leaves stage were grown in Hoagland solution containing 200 mM NaCl, 20% PEG6000 (w/v), 100 μΜ ABA, and 50 μΜ JA, respectively, under a photoperiod of 16 h at 27 °C (day) and 8 h at 24 °C (night) in a phytotron. The low-temperature treatment was carried out at 8 °C under the same photoperiod.
Sequence database searches
To identify the watermelon NAC TF gene family, Arabidopsis (https://www.arabidopsis.org/) and rice (http://rapdb.dna.affrc.go.jp) NAC TF protein sequences were used to search the watermelon genome database (version 1; http://www.icugi.org/) using BLASTP, and then, a self-BLAST of the sequences was performed to remove redundancy. All of the putative candidates were manually verified using NCBI (http://www.ncbi.nlm.nih.gov/) to confirm the presence of the protein NAM conserved domain. They were then further examined to obtain all of the protein sequences using SMART (http://smart.embl-heidelberg.de/) and Pfam (http://pfam.sanger.ac.uk). Finally, all of the obtained protein sequences were compared with the watermelon NAC TF sequences downloaded from the PlantTFDB (http://planttfdb.cbi.pku.edu.cn/).
Phylogenetic analysis
Multiple sequence alignments of the full-length amino acid sequences were aligned using Clustal W. The unrooted phylogenetic trees were constructed according to the NJ method using MEGA 5.0, and the bootstrap test was carried out with 1,000 iterations.
Gene homologs and chromosomal location
The duplicate genes and the homologous genes between watermelon and Arabidopsis, based on the NAC protein phylogenetic tree from watermelon, Arabidopsis, and rice, were identified using the protocol of Kong et al.56. The tandem duplicated genes were identified and are defined as an array of two or more genes that were in the same phylogenetic group and found within a 100-kb chromosomal fragment57. All of the NAC genes chromosomal locations were found in the Cucurbit Genomics Database and then were visualized in a Circos map using CIRCOS software (http://circos.ca).
Genomic structure and conserved motifs
The Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/) program was used to elucidate the exon/intron organization of NAC genes. The Multiple Expectation Maximization for Motif Elicitation (MEME; http://meme-suite.org/) program was used to illustrate the motifs in 80 putative ClNAC protein sequences.
Prediction of promoter cis-elements, subcellular localizations, phosphorylation sites, and signal peptides
The putative cis-acting regulatory DNA elements (cis-elements) in the promoter regions of NAC genes were identified using the PlantCARE (http://bioinformatics.psb.ugent. be/webtools/plantcare/html/) program. Cis-elements were identified within the 1000-bp genomic DNA sequence upstream of the initiation codon (ATG)58. The GATA program was used to perform a comparative analysis of the promoter regions59. WoLF PSORT (http://wolfpsort.seq.cbrc.jp) was used to predict the subcellular localization, while phosphorylation sites and signal peptides were identified using NetPhos2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/) and SignalP (http://www.cbs.dtu.dk/services/SignalP), respectively.
Expression patterns analyses by RT-PCR and qRT-PCR
Total RNA was extracted from all of the tissue samples using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. First-strand cDNAs were synthesized using the Transcriptor First Strand cDNA Synthesis kit (Roche, Switzerland). To detect PCR products, 2% agarose gel electrophoresis was used. qRT-PCR reactions were performed in the ABI PRISM 7900HT (Applied Biosystems, USA) using FastStart Universal SYBR Green Master (Roche, Switzerland) according to the manufacturer’s instructions. The relative expression levels of NAC genes were calculated according to the method of Livak and Schmittgen60. The primers used in this analysis are described in Supplementary Table S6.
Transcriptome sequencing data analysis
The transcriptome sequencing data for vascular and fruit developmental stages were obtained from a published paper45 using the identified ClNAC ID. The expression profiles were analyzed and visualized by MeV4.9.0 software (The Institute for Genomic Research, USA).
Additional Information
How to cite this article: Lv, X. et al. Global Expressions Landscape of NAC Transcription Factor Family and Their Responses to Abiotic Stresses in Citrullus lanatus. Sci. Rep. 6, 30574; doi: 10.1038/srep30574 (2016).
Supplementary Material
Acknowledgments
This work was supported by the earmarked fund for Modern Agro-Industry Technology Research System of China (CARS-26-17), National Natural Science Foundation of China (31372077; 31501782) and Key Science and Technology Program of Zhejiang Province (2012C129031-2-11).
Footnotes
Author Contributions Z.H., J.Y. and M.Z. conceived and designed the study. X.L. and S.L. performed the experiments. X.L. and K.M.G. analyzed the data, and X.L. and Z.H. wrote the paper. All authors reviewed the manuscript.
References
- Ooka H., Satoh K. & Doi K. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247 (2003). [DOI] [PubMed] [Google Scholar]
- Shen H., Yin Y., Chen F., Xu Y. & Dixon R. A. A Bioinformatic Analysis of NAC Genes for Plant Cell Wall Development in Relation to Lignocellulosic Bioenergy Production. Bioenerg. Res. 2, 217–232 (2009). [Google Scholar]
- Aida M., Ishida T., Fukaki H., Fujisawa H. & Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst H. A., Olsen A. N., Larsen S. & Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K. et al. Transcriptional regulation by an NAC (NAM–ATAF1,2–CUC2) transcription factor attenuates aba signalling for efficient basal defence towards blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 56, 867–880 (2008). [DOI] [PubMed] [Google Scholar]
- Chen Q., Wang Q., Xiong L. & Lou Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2, 55–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. X. et al. Structure of the GCM domain-DNA complex: a DNA-binding domain with a novel fold and mode of target site recognition. EMBO J. 22, 1835–1845 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K. & Skriver K. NAC transcription factor gene regulatory and protein-protein interaction networks in plant stress responses and senescence. IUBMB Life 66, 156–166 (2014). [DOI] [PubMed] [Google Scholar]
- Kim Y. S. et al. Amembrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V. et al. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 15, 913–922 (2008). [DOI] [PubMed] [Google Scholar]
- Kato H., Motomura T., Komeda Y., Saito T. & Kato A. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167, 571–577 (2010). [DOI] [PubMed] [Google Scholar]
- Chai M. et al. The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 58, 44–51 (2015). [Google Scholar]
- Sakamoto S. & Mitsuda N. Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant Cell Physiol. 56, 299–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K. et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M., Hsieh T. F., Kim S. Y. & Thomas T. L. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248 (2002). [DOI] [PubMed] [Google Scholar]
- Shibuya K., Shimizu K., Niki T. & Ichimura K. Identification of a NAC transcription factor, EPHEMERAL1, that controls petal senescence in Japanese morning glory. Plant J. 79, 1044–1051 (2014). [DOI] [PubMed] [Google Scholar]
- Kim H. J. et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. J. et al. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916 (2005). [DOI] [PubMed] [Google Scholar]
- Wang F. et al. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front. Plant. Sci. 6, 1–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A. & Dubcovsky J. A. NAC Gene regulating senescence improves grain protein, Zn, and Fe content in wheat. Science 314, 1298–1301 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y. Q. et al. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic. Acids. Res. 43, 1469–1484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu F. et al. Canola (Brassica napus L.) NAC103 transcription factor gene is a novel player inducing reactive oxygen species accumulation and cell death in plants. Biochem. Biophys. Res. Commun. 454, 30–35 (2014). [DOI] [PubMed] [Google Scholar]
- Zhu M. et al. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant & Cell Physiol. 55, 119–135 (2014). [DOI] [PubMed] [Google Scholar]
- Zhou H. et al. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121 (2015). [DOI] [PubMed] [Google Scholar]
- Endo H. et al. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254 (2015). [DOI] [PubMed] [Google Scholar]
- Xu B. et al. Contribution of NAC transcription factors to plant adaptation to land. Science 343, 1505–1508 (2014). [DOI] [PubMed] [Google Scholar]
- Tran L. S. et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876 (2004). [DOI] [PubMed] [Google Scholar]
- Jensen M. K. et al. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196 (2010). [DOI] [PubMed] [Google Scholar]
- Bu Q. Y. et al. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 18, 756–767 (2008). [DOI] [PubMed] [Google Scholar]
- Grover A. et al. Overexpression of NAC gene from Lepidium latifolium L. enhances biomass, shortens life cycle and induces cold stress tolerance in tobacco: potential for engineering fourth generation biofuel crops. Mol. Biol. Rep. 41, 7479–7489 (2014). [DOI] [PubMed] [Google Scholar]
- Jensen M. K. et al. The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 65, 137–150 (2007). [DOI] [PubMed] [Google Scholar]
- Delessert C. et al. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 43, 745–757 (2005). [DOI] [PubMed] [Google Scholar]
- Hu R., Qi G. & Kong Y. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10, 1–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K., Takada S. & Tasaka M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36, 687–696 (2003). [DOI] [PubMed] [Google Scholar]
- Holmgren R., Corces V., Morimoto R., Blackman R. & Meselson M. Sequence homologies in the 5′regions of four Drosophila heatshock genes. Proc. Natl. Acad. Sci. USA 78, 3775–3778 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. B. A regulatory upstream promoter element in the Drosophila Hsp70 heat-shock gene. Cell 30, 517–528 (1982). [DOI] [PubMed] [Google Scholar]
- Pla M. J. et al. The cis-regulatory element CCACGTGG is involved in ABA and water-stress responses of the maize gene rab28. Plant Mol. Biol. 21, 259–266 (1993). [DOI] [PubMed] [Google Scholar]
- Roderick E. M., Wu-Peng X. S., Yen P. M. et al. Interactions of Estrogen- and Thyroid Hormone Receptors on a Progesterone Receptor Estrogen Response Element (ERE) Sequence: a Comparison with the Vitellogenin A2 Consensus ERE. Mol. Endo. 11, 1581–1591 (1997). [DOI] [PubMed] [Google Scholar]
- Dunn M. A., White A. J., Vural S. & Hughes M. A. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol. Biol. 38, 551–564 (1998). [DOI] [PubMed] [Google Scholar]
- Narusaka Y. et al. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148 (2003). [DOI] [PubMed] [Google Scholar]
- Wallbach M. et al. Distinct functions of the dual leucine zipper kinase depending on its subcellular localization. Cellular Signalling. 28, 272–283 (2016). [DOI] [PubMed] [Google Scholar]
- Kumer S. C. & Vrana K. E. Intricate regulation of tyrosine hydroxylase activity and gene expression. J. Neurochem. 67, 443–462 (1996). [DOI] [PubMed] [Google Scholar]
- Yamaguchi M. et al. Arabidopsis NAC domain proteins VND-INTERACTING1 and ANAC103 interact with multiple NAC domain proteins. Plant Biotechnology 32, 119–123 (2015). [Google Scholar]
- Guo S. et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat.Genet. 45, 51–58 (2013). [DOI] [PubMed] [Google Scholar]
- Kubo M. et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev.,19, 1855–1860 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K. & Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R. Q., Demura T. & Ye Z. H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290 (2009). [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K. & Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochimica. Et. Biophysica. Acta. 1819, 97–103 (2012). [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K. S. & Zhu J. K. Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. T. C. & Hiron R. W. P. (+)-abscisic acid, the growth inhibitor induced in detached wheat leaves by a period of wilting. Nature 224, 719–720 (1969). [Google Scholar]
- Savouré A., Hua X. J., Bertauche N., Van Montagu M. & Verbruggen N. Abscisic acid-independent and abscisic acid-dependent regulation of proline biosynthesis following cold and osmotic stresses in Arabidopsis thaliana. Mol. Gen. Genet. 254, 104–109 (1997). [DOI] [PubMed] [Google Scholar]
- Wasternack C. & Parthier B. Jasmonate-signalled plant gene expression. Trends Plant Sci. 2, 302–307 (1997). [Google Scholar]
- Wasternack C. & Hause B. Jasmonates and octadecanoids: signals in plant stress responses and development. Prog. Nucleic. Acid. Res. Mol. Biol. 72, 165–221 (2002). [DOI] [PubMed] [Google Scholar]
- Kong X. et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 433, 1471–2164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. et al. The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 148, 1189–1200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombauts S., Dehais P., Van Montagu M. & Rouze P. Plant-CARE, a plant cis-acting regulatory element database. Nucleic. Acids. Res. 27, 295–296 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix D. A. & Eisen M. B. GATA: a graphic alignment tool for comparative sequence analysis. BMC Bioinformatics 6, 285–299 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆ CT Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.