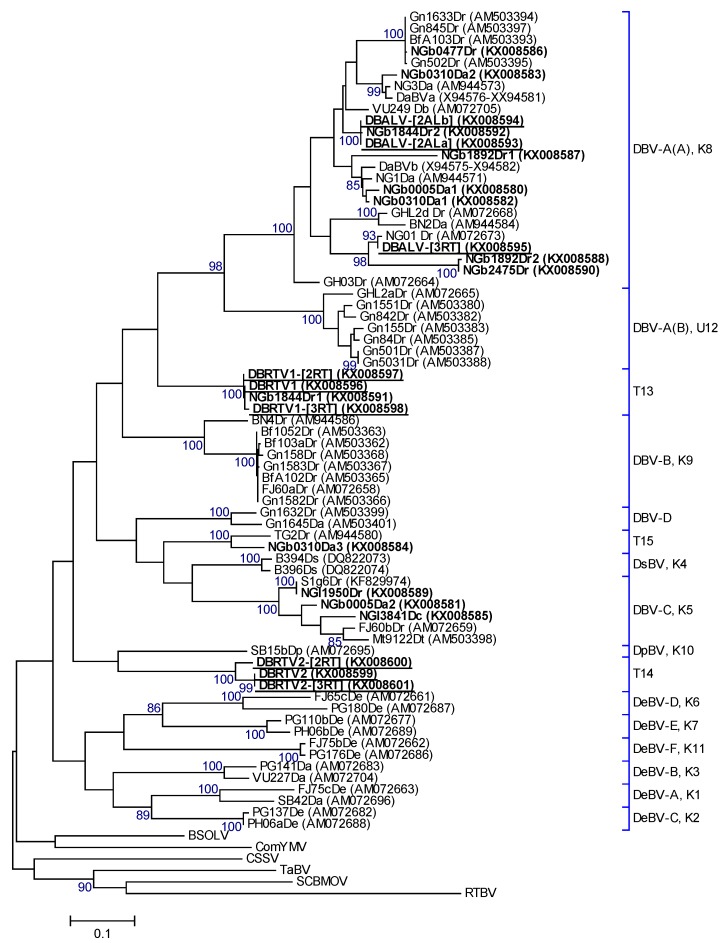
Figure 2.
Bootstrap consensus phylogenetic tree using Maximum Likelihood method built from badnavirus 528 bp long partial RT-RNaseH nucleotide sequences of 22 yam badnavirus sequences determined in this study representing two already described and three newly proposed distinct monophyletic species groups (T13, T14, T15). Included in the analysis are partial RT-RNaseH sequences from GenBank of previously analysed yam samples by Seal et al. [3] and Eni et al. [8]. Equivalent sequences from CSSV (AJ781003), BSOLV (AJ002234), ComYMV (NC001343), SCBMOV (M89923), TaBV (AF357836) and outgroup RTBV (X57924) were added, as well as representative sequences of all monophyletic groups described by Bousalem et al. [9], where DBV-A = Dioscorea bacilliform virus A (A and B subgroups); DBV-B = Dioscorea bacilliform virus B; DBV-C = Dioscorea bacilliform virus C; DBV-D = Dioscorea bacilliform virus D; DeBV-A = Dioscorea esculenta bacilliform virus A; DeBV-B = Dioscorea esculenta bacilliform virus B; DeBV-C = Dioscorea esculenta bacilliform virus C; DeBV-D = Dioscorea esculenta bacilliform virus D; DeBV-E = Dioscorea esculenta bacilliform virus E; DeBV-F = Dioscorea esculenta bacilliform virus F; and DpBV = Dioscorea pentaphylla bacilliform virus. One monophyletic group denoted by Umber et al. [30] and 11 corresponding Kenyon et al. [2] groupings to these monophyletic groups are also given and denoted by U12 and K1–K11 respectively. Sequence names: bold = partial RT-RNaseH sequences from RCA clones, bold and underlined represent partial RT-RNaseH sequences and their GenBank accessions of the characterised full-length genomes obtained in this study. The bootstrap analysis of the sequences was 1000 replicates and the cut-off value was 85%.