Abstract
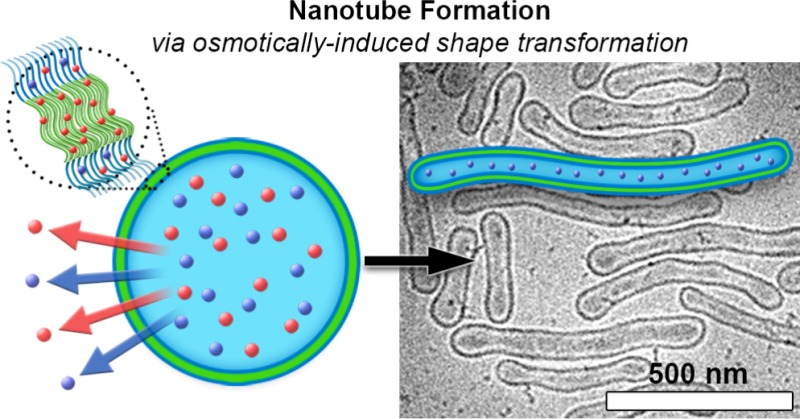
Polymersomes are robust, versatile nanostructures that can be tailored by varying the chemical structure of copolymeric building blocks, giving control over their size, shape, surface chemistry, and membrane permeability. In particular, the generation of nonspherical nanostructures has attracted much attention recently, as it has been demonstrated that shape affects function in a biomedical context. Until now, nonspherical polymersomes have only been constructed from nondegradable building blocks, hampering a detailed investigation of shape effects in nanomedicine for this category of nanostructures. Herein, we demonstrate the spontaneous elongation of spherical polymersomes comprising the biodegradable copolymer poly(ethylene glycol)-b-poly(d,l-lactide) into well-defined nanotubes. The size of these tubes is osmotically controlled using dialysis, which makes them very easy to prepare. To confirm their utility for biomedical applications, we have demonstrated that, alongside drug loading, functional proteins can be tethered to the surface utilizing bio-orthogonal “click” chemistry. In this way the present findings establish a novel platform for the creation of biocompatible, high-aspect ratio nanoparticles for biomedical research.
Polymersomes, nanoscopic polymer vesicles self-assembled from synthetic block copolymers, have shown great promise in biomedical applications from drug delivery to artificial organelles.1 Polymersomes display enhanced stability and membrane integrity under a wide range of conditions when compared to liposomes, frequently used in nanomedical systems, due to their thicker membranes.2 Polymersome morphology can be controlled by copolymer composition and subsequently re-engineered in response to chemical or physical stimuli such as pH, osmotic pressure, temperature, or magnetic fields.3 The importance of gaining control over the size and shape of nanoparticles is essential to directing their function in vivo.4 While spherical polymersomes have shown to be successful candidates for some applications in a biological context,5 the ability to transform their morphology can lead to new biomedical opportunities such as targeted drug delivery.4f
For example, it has been shown that high-aspect ratio structures lead to enhanced specific accumulation and reduced nonspecific adhesion to cells.6 Elongated nanoparticles interact with cells via distinct mechanisms, leading to enhanced uptake, in a similar fashion to the uptake of rod-like bacteria by nonphagocytotic cells.7 Moreover, the unique ability of tubular morphologies, in contrast to spheres, to interact with the immune system through multivalency is an exciting prospect for the development of artificial antigen-presenting cells.4g However, up to this point the bulk of research toward engineering elongated polymeric vesicles has focused on nonbiocompatible polymers. For example, tubular nanostructures have been reported, with shape transformation accomplished by using chemical stimuli such as the covalent introduction of a redox cleavable cross-linker8 or by swelling the membrane with lipids or cholesterol.9 However, the utility of such methodologies is limited by their nonbiodegradable components or lack of structural control. It is therefore of critical importance to develop new methodologies for the generation of well-defined, elongated polymersomes utilizing biodegradable polymers. Biodegradable copolymers have shown excellent promise in the development of biocompatible nanomaterials for medical applications.4f,10 Biodegradable nanotubes can therefore be directly implemented in nanomedical research due to their inherent biocompatibility.11
Here, we present a methodology for the formation of functional, polymeric nanotubes from biodegradable polymersomes comprising poly(ethylene glycol)-b-poly(d,l-lactide) (PEG-PDLLA). Poly(lactides) have established utility in biomedical engineering due to their biocompatibility and degradation behavior, which has been shown to vary between days or weeks, depending on the conditions employed.12 Furthermore, the formation of polymersomes from PEG-PDLLA block copolymers has previously been reported.12a Herein, we demonstrate that spherical polymersomes comprising PEG-PDLLA block copolymers can be transformed into well-defined nanotubes by exposure to hypertonic conditions, and thereby osmotic pressure, at low temperature. In this way, nanotubes of different lengths are controllably obtained by varying the electrolyte concentration. Moreover, such nanotubes can readily be functionalized through incorporation of hydrophobic drugs or via surface modification with proteins utilizing bio-orthogonal “click” chemistry.
Previously we reported the osmotically induced shape transformation of poly(ethylene glycol)-b-poly(styrene) (PEG-PS) polymersomes into bowl-shaped stomatocytes through deflation of the inner volume and membrane indentation.13 This shape was maintained because PS in the membrane has a high glass transition temperature, which provides the necessary membrane rigidity at room temperature to prevent structural relaxation toward a spherical morphology. In order to understand in more detail the nature of the shape transformation that occurs when spherical polymersomes are exposed to hypertonic conditions, it is necessary to consider the bending energy (Eb) that can be described by eq 1:14,15
| 1 |
Eq 1 allows us to describe the energetic state of a polymersomal membrane as a function of 3 parameters: the bending rigidity (k), the mean surface curvature (C), and the spontaneous curvature (C0). The bending rigidity (k) is dictated by the chemical properties of the membrane (polymer composition and length) alongside environmental factors (temperature and solvent composition). The mean surface curvature (C) describes the degree of curvature at different positions on the membrane and is contingent upon the shape. In contrast to C, the spontaneous curvature (C0) is not a consequence of the shape; rather, it arises from asymmetry in copolymer conformation between the inner and outer surfaces and is therefore sensitive to the membrane microenvironment. From eq 1 we understand that the effect of C0 is to either reduce or increase Eb for a given morphology and, as such, can be used to gain control over the shape transformation process.14 Under the influence of osmotic pressure, which can be orders of magnitude greater than Eb, as a spherical polymersome undergoes reduction in volume there are 2 energetic pathways that can be taken. Such deflation can proceed via prolates (toward tubes) or oblates (toward discs and stomatocytes) as a result of the minimization of Eb, with the determining factor being C0. Without significant contribution from the effect of C0, the prolate pathway has a lower energetic profile, however, a positive or negative contribution of C0 would favor either prolates or oblates, respectively.15 With this in mind, it is clear that finding a suitable biodegradable replacement for poly(styrene), which is sufficiently rigid while tending toward prolates and tubes rather than discoidal structures, is a challenging task.
Shape transformations of self-assembled structures using most common poly(esters) and poly(carbonates), even with careful tuning of conditions, are extremely challenging due to their amorphous (low Tg) or crystalline (high Tm) character. However, atactic polylactide, synthesized from a mixture of lactide isomers, possesses a Tg above 20 °C (as opposed to the isotactic form that is highly crystalline) and is therefore an excellent candidate for the formation of biodegradable polymersomes and their shape transformed counterparts.16 PEG-PDLLA copolymers were synthesized by ring-opening polymerization (ROP) with the organic base 1,8-diazabicycloundec-7-ene (DBU) as a catalyst, according to previous literature (Figure 1a).17 The reaction progress and the composition of the final product were calculated from the NMR spectra (Figures S1 and S2) with the resulting copolymers having PDI values of <1.1 (Figure S3). Two copolymers were synthesized, PEG22-b-PDLLA45 (Mw ≈ 7.5 kDa) and PEG44-b-PDLLA90 (Mw ≈ 14.9 kDa), both comprising ∼13 wt % PEG. Differential scanning calorimetry (DSC) data confirmed that the Tg values for PEG22-PDLLA45 and PEG44-PDLLA90 were ∼26 and 30 °C, respectively (Figure S4).
Figure 1.
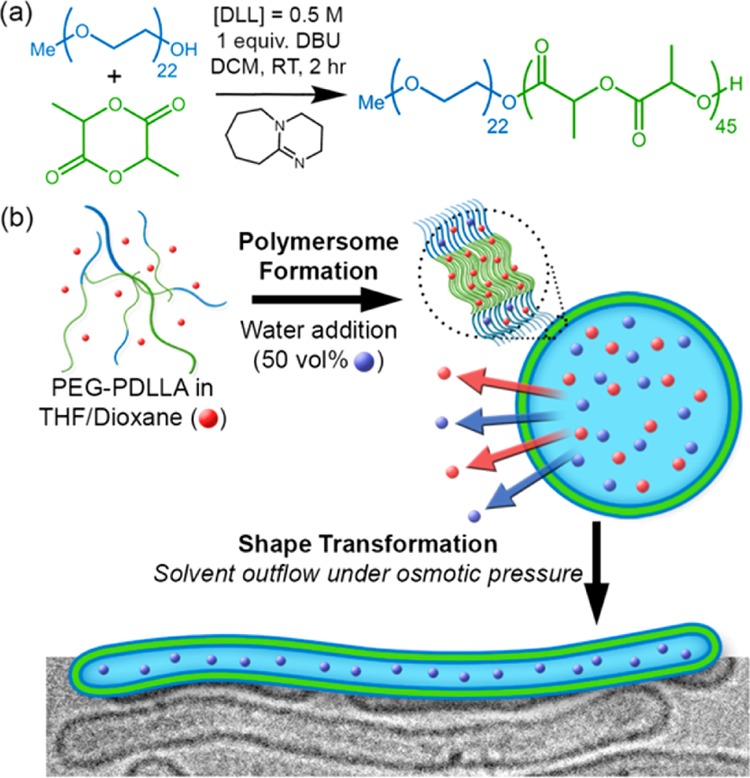
Schematic outlining (a) the organobase-catalyzed synthesis of PEG-PDLLA and (b) the osmotically induced shape transformation of spherical polymersomes into nanotubes.
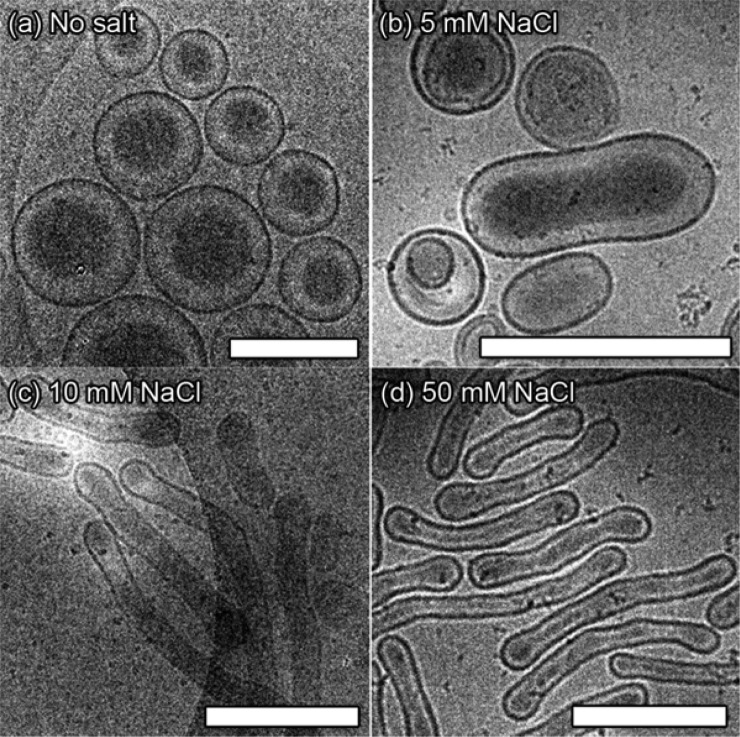
Assembly tests using PEG-PDLLA were conducted using an established method for polymersome formation via solvent switch, whereby a solution of block copolymer in organic solvent (4:1, THF:dioxane) was diluted by water (up to 50% v/v at 1 mL/h, Figure 1b).18 Interestingly, the longer copolymer PEG44-PDLLA90 yielded micelles (spherical and rods, Figure S5a), whereas PEG22-PDLLA45 yielded polymersomes with a hydrodynamic radius (Rh) ≈ 165 nm and a 15 nm thick membrane (Figures 2a and S5b–d). In order to transform the spherical polymersomes into nanotubes, the plasticized dispersion was dialyzed against aqueous solutions with varying salt concentration (5, 10, and 50 mM) at 4 °C so that the glassiness of the membrane was maintained after the main part of organic solvent was removed. It was observed that after overnight dialysis against 5 mM NaCl the polymersomes had partially transformed into prolate structures, and a mixture of morphologies was observed (Figures 2b and S6a). By comparison, dialysis against higher [NaCl] (10 and 50 mM) resulted in the predominant formation of elongated (prolate) nanotubes of increasing length (Figures 2c,d and S6b,c). Interestingly, polymersome dialysis against 100 mM NaCl resulted in the formation of elongated ribbons, identified as collapsed nanotubes (Figure S6d). Conversely, when dialyzing the spherical polymersomes at 25 °C, no shape transformation was observed while at 30 °C collapsed structures were obtained (Figures S6e-f), highlighting the importance of polymer rigidity for establishing and maintaining the shape transformation.
Figure 2.
Cryo-TEM images following the shape transformation of PEG22-PDLLA45 from (a) spherical polymersomes into elongated nanotubes under the influence of osmotic pressure by dialysis, as a function of [NaCl]; (b) 5, (c) 10, and (d) 50 mM. All scale bars = 500 nm.
Nanotubes formed using dialysis against 10 mM NaCl were on average shorter, with average dimensions (length × width) of 110 × 800 nm, whereas dialysis against 50 mM NaCl yielded longer tubes of 90 × 1040 nm (Figures 2c,d, S6b,c, and S7). Crucially, these dimensions correspond to average surface areas of ∼3 × 106 nm2, similar to that of the original polymersomes, however, the internal volume significantly decreased by 39 and 51% for the samples dialyzed against 10 and 50 mM NaCl, respectively (Table S1). This highlights that increased osmotic pressure results in nanotube elongation due to the geometrical constraint of surface area during osmosis and concomitant reduction of internal volume. Asymmetric flow field-flow fractionation (AF4, settings in Table S2), in combination with static and dynamic light scattering, were used in order to gain a quantitative understanding of the shape transformation from vesicles to tubes. Comparing the radius of gyration (Rg) to the hydrodynamic radius (Rh) gives information on particle shape where values of Rg/Rh for spheres are ≤1 and for prolates (rod-like shapes) are >1.36.19 Elution profiles, with corresponding values for Rg, clearly showed that the spherical PEG-PDLLA polymersomes have a uniform ratio value near to 1 (hollow spheres), whereas the nanotubes have an average value of 1.5 with all values >1.3, which quantitatively demonstrates that the predominant morphology is that of tubes (Figure S8). Light scattering and cryo-TEM were used to confirm that nanotubes, once formed, are stable under increased temperature and salt concentration, as would be encountered under physiological conditions, which is vital for application toward biomedical research. Indeed, tubular morphology was sustained after dialysis against PBS, with both the Rh and scattering intensity remaining constant during heating to 40 °C, due to lack of osmotic pressure (Figure S9).
Under hypertonic conditions, polymersomes comprising PEG22-PDLLA45 deflate into nanotubes rather than stomatosomal or nested structures, which are more commonly encountered.13,20 As has already been discussed, the contribution of C0 is a deciding factor for the direction of shape transformations. In our previous work using PEG-PS polymersomes, a less permeable PS membrane and longer PEG chains at the surface led to a negative C0, which induced the formation of oblates, stomatocytes and nested vesicles.13,14b In comparison, for the shape transformation of PEG-PDLLA polymersomes, increasing the [NaCl] during dialysis not only results in greater volume reduction but also influences the transformation pathway. Dialyzing polymersomes against 5 mM NaCl resulted in the formation of a population of heterogeneous morphologies, dominated by prolates, which is unsuitable for quantification (Figures 2b and S6a). However, increasing the [NaCl] to 10 and 50 mM gave a significant improvement in the quantity (and quality) of prolate structures, concurrently lowering the percentage of oblate morphologies to ∼10 and 4%, respectively (Figures 2c,d and S6b). Interestingly, dialysis under increased osmotic pressure (100 mM NaCl) led to the formation of nanotubes that underwent total collapse due to outflow of all internal solvent molecules during the shape transformation process. These results suggest that not only does NaCl contribute toward solution osmolarity, which drives volume reduction and elongation, but it also contributes toward a positive C0, promoting the formation of prolates and tubes. Although the physical origins of C0 are not well documented, it is evident that it arises from a mismatch between the external and internal membrane surfaces. Such a mismatch is accentuated by chemical differences between external and internal environments, which is the case in the current situation. Although it is beyond the scope of the present work, these findings warrant further investigation into the unique properties of PEG-PDLLA in order to develop experimental insight into the contribution of C0 in shape transformations.
In order to establish the functional capacity of PEG-PDLLA nanotubes, experiments were conducted to demonstrate biomolecular labeling (Figure 3a–c) and drug loading (Figure 3d–f). Biomolecular labeling of nanoparticles is an important procedure by which biological activity can be introduced to the surface through covalent modification with antibodies or antigens in order to provide site-selective specificity in cellular uptake or to stimulate specific immune responses.21 It is therefore important to demonstrate that the nanotubes can be modified with surface-bound biomolecules, which retain their native function after attachment. For this purpose, enhanced green fluorescent protein (eGFP) containing the clickable unnatural amino acid bicyclo[6.1.0]nonyne-l-lysine (BCN-lys) was expressed and purified (eGFPBCN, Figures 3a and S10). Azido-modified copolymer (N3-PEG75-PDLLA45) was introduced into the nanotubes at different amounts (2 and 5 wt %) to demonstrate that control over the density of biomolecular labeling can be accomplished through simple formulation. Flow cytometry and confocal microscopy were used to confirm successful labeling of eGFP onto the nanotube, with a positive signal indicating that the tethered eGFP maintains its native structure (Figure 3).22 The stability of eGFP modified N3-nanotubes was confirmed by cryo-TEM (Figure S11a). With flow cytometry the N3-nanotubes showed increases in their fluorescent signal upon increasing the concentration of eGFPBCN in the reaction mixtures from an equimolar amount to a 3-fold excess (Figures 3b and S11c) as well as by increasing the total N3-copolymer content from 2 to 5 wt % (Figure S11d). It was furthermore investigated if hydrophobic cargo could be loaded in the polymer membrane without undesired effects on nanotube formation, at levels comparable to literature.23 To this end, PEG-PDLLA nanotubes were prepared in the presence of 2 and 5 wt % of hydrophobic doxorubicin (DOX), which was dissolved into the organic solvent phase and then became incorporated into the copolymer membrane. As was confirmed by cryo-TEM, DOX did not impede the subsequent shape transformation (Figure S11b). Flow cytometry was used to confirm nanotube fluorescence, which increased with greater amounts of DOX, although it is likely that 5 wt % is above the capacity for this system as the signal did not greatly increase in comparison with 2 wt % loading (Figure 3e). Specific association of the drug with the nanotubes was confirmed using confocal microscopy (Figure 3f).
Figure 3.
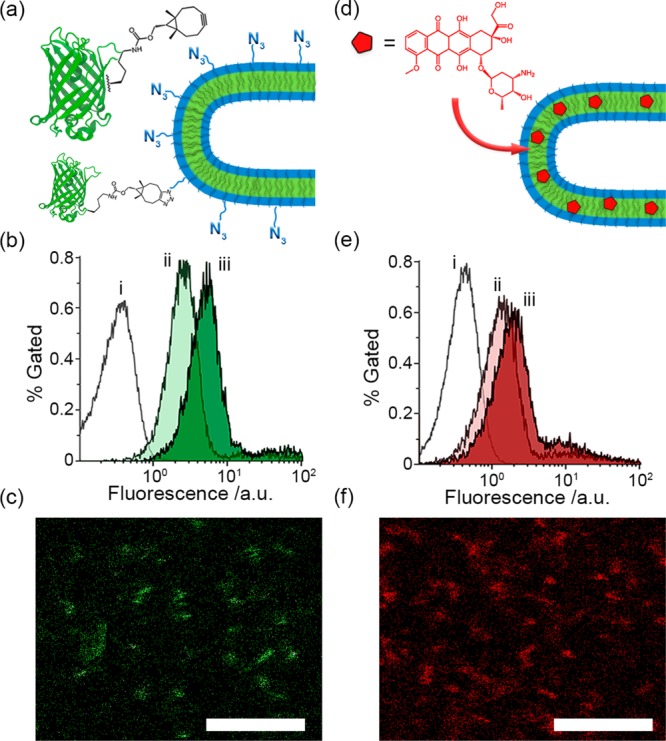
(a) Schematic outlining the covalent modification of azide-modified nanotubes using eGFPBCN, (b) flow cytometry data for eGFP modification of 5% azido nanotubes using an equimolar (ii) or a 3-fold excess (iii) of eGFPBCN compared to unmodified tubes (i) and (c) confocal visualization of green-fluorescent nanotubes. (d) Schematic outlining the loading of nanotubes with fluorescent DOX, (e) flow cytometry data of nanotubes loaded with 2 (ii) and 5 (iii) wt % DOX compared to unloaded tubes (i) and (f) confocal visualization of DOX-loaded nanotubes (5 wt % preparation). All scale bars = 5 μm.
In summary, we have presented the synthesis of biodegradable PEG-PDLLA copolymers that spontaneously assemble into spherical polymersomes. These biodegradable polymersomes can then undergo shape transformation into nanotubes upon dialysis under hypertonic conditions, with increasing [NaCl] leading to both structural enrichment and elongation. Such well-defined nanoparticles, which can be easily functionalized, are of great interest for biomedical research where nanoscopic control over size and shape is highly valuable.
Acknowledgments
Authors acknowledge the support from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-20012)/ERC-StG 307679 “StomaMotors” and the support from the Ministry of Education, Culture and Science (Gravitation program 024.001.035). The authors thank Geert-Jan Janssen and Liesbeth Pierson (General Instrumentation, Radboud University) and Rob Woestenenk (Flow Cytometry Facility, Radboud University Medical Centre) for their practical assistance.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b03984.
Experimental details and characterization data (PDF)
Author Contributions
‡ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- a LoPresti C.; Lomas H.; Massignani M.; Battaglia G. J. Mater. Chem. 2009, 19, 3576. 10.1039/b818869f. [DOI] [Google Scholar]; b Peters R. J. R. W.; Louzao I.; van Hest J. C. M. Chem. Sci. 2012, 3, 335. 10.1039/C2SC00803C. [DOI] [Google Scholar]; c Palivan C. G.; Goers R.; Najer A.; Zhang X.; Car A.; Meier W. Chem. Soc. Rev. 2016, 45, 377. 10.1039/C5CS00569H. [DOI] [PubMed] [Google Scholar]; d Li Y.; Liu G.; Wang X.; Hu J.; Liu S. Angew. Chem., Int. Ed. 2016, 55, 1760. 10.1002/anie.201509401. [DOI] [PubMed] [Google Scholar]
- Discher B. M.; Won Y.-Y.; Ege D. S.; Lee J. C. -M.; Bates F. S.; Discher D. E.; Hammer D. A. Science 1999, 284, 1143. 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- a Chang H. Y.; Sheng Y. J.; Tsao H. K. Soft Matter 2014, 10, 6373. 10.1039/C4SM01092B. [DOI] [PubMed] [Google Scholar]; b Blanazs A.; Madsen J.; Battaglia G.; Ryan A. J.; Armes S. P. J. Am. Chem. Soc. 2011, 133, 16581. 10.1021/ja206301a. [DOI] [PubMed] [Google Scholar]; c Hu X.; Hu J.; Tian J.; Ge Z.; Zhang G.; Luo K.; Liu S. J. Am. Chem. Soc. 2013, 135, 17617. 10.1021/ja409686x. [DOI] [PubMed] [Google Scholar]
- a Petros R. A.; DeSimone J. M. Nat. Rev. Drug Discovery 2010, 9, 615. 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]; b Albanese A.; Tang P. S.; Chan W. C. W. Annu. Rev. Biomed. Eng. 2012, 14, 1. 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]; c Perry J. L.; Herlihy K. P.; Napier M.; DeSimone J. M. Acc. Chem. Res. 2011, 44, 990. 10.1021/ar2000315. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Decuzzi P.; Pasqualini R.; Arap W.; Ferrari M. Pharm. Res. 2009, 26, 235. 10.1007/s11095-008-9697-x. [DOI] [PubMed] [Google Scholar]; e Geng Y.; Dalhaimer P.; Cai S.; Tsai R.; Tewari M.; Minko T.; Discher D. E. Nat. Nanotechnol. 2007, 2, 249. 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Sancho Oltra N.; Swift J.; Mahmud A.; Rajagopal K.; Loverde S. M.; Discher D. E. J. Mater. Chem. B 2013, 1, 5177. 10.1039/c3tb20431f. [DOI] [PubMed] [Google Scholar]; g van der Weijden J.; Paulis L. E.; Verdoes M.; van Hest J. C. M.; Figdor C. Chem. Sci. 2014, 5, 3355. 10.1039/C4SC01112K. [DOI] [Google Scholar]
- Wang L.; Liu G.; Wang X.; Hu J.; Zhang G.; Liu S. Macromolecules 2015, 48, 7262. 10.1021/acs.macromol.5b01709. [DOI] [Google Scholar]
- Kolhar P.; Anselmo A. C.; Gupta V.; Pant K.; Prabharkarpandian B.; Ruoslahti E.; Mitragotri S. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 10753. 10.1073/pnas.1308345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gratton S. E. A.; Ropp P. A.; Pohlhaus P. D.; Luft J. C.; Madden V. J.; Napier M. E.; DeSimone J. M. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 11613. 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Park J.-H.; von Maltzahn G.; Zhang L.; Derfus A. M.; Simberg D.; Harris T. J.; Ruoslahti E.; Bhatia S. N.; Sailor M. J. Small 2009, 5, 694. 10.1002/smll.200801789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers M. C. M.; Rutjes F. P. J. T.; van Hest J. C. M. J. Am. Chem. Soc. 2013, 135, 16308. 10.1021/ja408754z. [DOI] [PubMed] [Google Scholar]
- Robertson J. D.; Yealland G.; Avila-Olias M.; Chierico L.; Bandmann O.; Renshaw S. A.; Battaglia G. ACS Nano 2014, 8, 4650. 10.1021/nn5004088. [DOI] [PubMed] [Google Scholar]
- Liu G.-Y.; Chen C.-J.; Jian J. Soft Matter 2012, 8, 8811. 10.1039/c2sm25721a. [DOI] [Google Scholar]
- Tian H.; Tang Z.; Zhuang X.; Chen X.; Jing X. Prog. Polym. Sci. 2012, 37, 237. 10.1016/j.progpolymsci.2011.06.004. [DOI] [Google Scholar]
- a Meng F.; Hiemstra C.; Engbers G. H. M.; Feijen J. Macromolecules 2003, 36, 3004–3006. 10.1021/ma034040+. [DOI] [Google Scholar]; b Samarajeewa S.; Shrestha R.; Li Y.; Wooley K. L. J. Am. Chem. Soc. 2012, 134, 1235. 10.1021/ja2095602. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Fukushima K.; Feijoo J. L.; Yang M.-C. Eur. Polym. J. 2013, 49, 706. 10.1016/j.eurpolymj.2012.12.011. [DOI] [Google Scholar]
- a Kim K. T.; Zhu J.; Meeuwissen S. A.; Cornelissen J. J. L. M.; Pochan D. J.; Nolte R. J. M.; van Hest J. C. M. J. Am. Chem. Soc. 2010, 132, 12522. 10.1021/ja104154t. [DOI] [PubMed] [Google Scholar]; b Wilson D. A.; Nolte R. J. M.; van Hest J. C. M. Nat. Chem. 2012, 4, 268. 10.1038/nchem.1281. [DOI] [PubMed] [Google Scholar]; c Rikken R. S. M.; Kerkenaar H. H. M.; Nolte R. J. M.; Maan J. C.; van Hest J. C. M.; Christianen P. C. M.; Wilson D. A. Chem. Commun. 2014, 50, 5394. 10.1039/C3CC47483F. [DOI] [PubMed] [Google Scholar]; d Abdelmohsen L. K. E. A.; Nijemeisland M.; Pawar G. M.; Janssen G. J. A.; Nolte R. J. M.; van Hest J. C. M.; Wilson D. A. ACS Nano 2016, 10, 2652. 10.1021/acsnano.5b07689. [DOI] [PubMed] [Google Scholar]
- a Góźdź W. T. Langmuir 2004, 20, 7385. 10.1021/la049776u. [DOI] [PubMed] [Google Scholar]; b Rikken R. S. M.; Engelkamp H.; Nolte R. J. M.; Maan J. C.; van Hest J. C. M.; Wilson D. A.; Christianen P. C. M.. Nat. Comm. 2016, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert U.; Berndl K.; Lipowsky R. Phys. Rev. A: At., Mol., Opt. Phys. 1991, 44, 1182. 10.1103/PhysRevA.44.1182. [DOI] [PubMed] [Google Scholar]
- Slomkowski S.; Penczek S.; Duda A. Polym. Adv. Technol. 2014, 25, 436. 10.1002/pat.3281. [DOI] [Google Scholar]
- Lohmeijer B. G. G.; Pratt R. C.; Leibfarth F.; Logan J. W.; Long D. A.; Dove A. P.; Nederberg F.; Choi J.; Wade C.; Waymouth R. M.; Hedrick J. L. Macromolecules 2006, 39, 8574. 10.1021/ma0619381. [DOI] [Google Scholar]
- Peng F.; Tu Y.; van Hest J. C. M.; Wilson D. A. Angew. Chem., Int. Ed. 2015, 54, 11662. 10.1002/anie.201504186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer A.; Striegel A. Analyst 2011, 136, 515. 10.1039/C0AN00738B. [DOI] [PubMed] [Google Scholar]
- Salva R.; Le Meins J.-F.; Sandre O.; Brûlet A.; Schmutz M.; Guenoun P.; Lecommandeux S. ACS Nano 2013, 7, 9298. 10.1021/nn4039589. [DOI] [PubMed] [Google Scholar]
- a Debets M. F.; Leenders W. P. J.; Verrijp K.; Zonjee M.; Meeuwissen S. a.; Otte-Höller I.; van Hest J. C. M. Macromol. Biosci. 2013, 13, 938. 10.1002/mabi.201300039. [DOI] [PubMed] [Google Scholar]; b Zou T.; Dembele F.; Beugnet A.; Sengmanivong L.; Trepout S.; Marco S.; Marco A. d.; Li M.-H. J. Biotechnol. 2015, 214, 147. 10.1016/j.jbiotec.2015.09.034. [DOI] [PubMed] [Google Scholar]
- Nallani M.; Woestenenk R.; de Hoog H.-P. M.; van Dongen S. F. M.; Boezeman J.; Cornelissen J. J. L. M.; Nolte R. J. M.; van Hest J. C. M. Small 2009, 5, 1138. 10.1002/smll.200990048. [DOI] [PubMed] [Google Scholar]
- Xu J.; Zhao Q.; Jin Y.; Qiu L. Nanomedicine 2014, 10, 349. 10.1016/j.nano.2013.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.