Abstract
The free radical nitric oxide (NO•) regulates diverse physiological processes from vasodilation in humans to gas exchange in plants. S-nitrosoglutathione (GSNO) is considered a principal nitroso reservoir due to its chemical stability. GSNO accumulation is attenuated by GSNO reductase (GSNOR), a cysteine-rich cytosolic enzyme. Regulation of protein nitrosation is not well understood since NO•-dependent events proceed without discernible changes in GSNOR expression. Because GSNORs contain evolutionarily-conserved cysteines that could serve as nitrosation sites, we examined the effects of treating plant (Arabidopsis thaliana), mammalian (human), and yeast (Saccharomyces cerevisiae) GSNORs with nitrosating agents in vitro. Enzyme activity was sensitive to nitroso donors, while the reducing agent dithiothreitol (DTT) restored activity, suggesting catalytic impairment was due to S-nitrosation. Protein nitrosation was confirmed by mass spectrometry, by which mono-, di-, and tri-nitrosation were observed, and these signals were sensitive to DTT. GSNOR mutants in specific non-zinc coordinating cysteines were less sensitive to catalytic inhibition by nitroso donors and exhibited reduced nitrosation signals by mass spectrometry. Nitrosation also coincided with decreased tryptophan fluorescence, increased thermal aggregation propensity, and increased polydispersity—properties reflected by differential solvent accessibility of amino acids important for dimerization and the shape of the substrate and coenzyme binding pockets as assessed by hydrogen-deuterium exchange mass spectrometry. Collectively, these data suggest a mechanism for NO• signal transduction in which GSNOR nitrosation and inhibition transiently permit GSNO accumulation.
Nitric oxide (NO•) influences myriad developmental and pathological processes via post-translational modification of cysteines, tyrosines, and organometallic centers1, 2. In animals, NO• is synthesized from L-arginine by constitutive and inducible nitric oxide synthase (NOS) enzymes3. Plants utilize both nitrite reductase and an arginine-dependent NOS-like activity to generate NO•4. NO• stability is inversely proportional to concentration, and oxygen (O2)-mediated NO• oxidation to nitrous anhydride (N2O3) predominates in vivo5, 6.
O2 and NO• are both present at similar (nanomolar to micromolar) concentrations in vivo7, so NO•-derived N2O3 favors cysteine S-nitrosothiol (RSNO) synthesis6. Owing to high concentrations of glutathione (GSH), the most abundant intracellular RSNO is S-nitrosoglutathione (GSNO)8. GSNO is formed both by nucleophilic attack of the GS− anion on N2O3 and by radical coupling between NO• and GS• thiyl radicals6. GSNO S-nitrosates protein cysteines by nitroso group transfer (transnitrosation)8, yielding GSH and a nitrosated protein. Reversible by thioredoxin enzymes, S-nitrosation is theorized to protect cysteines from irreversible oxidation and tune the activity of target proteins9, 10. Biotin switch and related indirect detection technologies have been developed to address the lability and low abundance of S-nitrosation in vivo. While these advances have informed the predictive power of in silico algorithms (e.g., GPSSNO) 11, 12, methods are needed to directly interrogate the effects of S-nitrosation on protein structure and function.
If GSNO is a capacitor of bioactive NO•, then the resistor that attenuates S-nitrosation is GSNO reductase (GSNOR), a dimeric, 2-zinc alcohol dehydrogenase (ADH) found in all eukaryotes and most bacteria13-17. With electrons from NADH, GSNOR reduces GSNO to an intermediate that, under physiological conditions, decomposes to glutathione disulfide (GSSG) and hydroxylamine. The same protein also catalyzes a parallel NAD+-dependent oxidation reaction proposed to prevent accumulation of endogenous formaldehyde18. Both activities may operate simultaneously in living cells via nicotinamide recycling19, a tempting hypothesis given the interdependence of reactive oxygen and nitrogen biochemistry. As expected for an enzyme of antioxidant metabolism, the consequences of GSNOR ablation are pleiotropic: mice exhibit enhanced hepatocarcinogenesis and cardiomyocyte regeneration, while reduced fertility, thermotolerance, and herbicide sensitivity are observed in the plant Arabidopsis thaliana 16, 20-22. In contrast, over-active GSNOR correlates with asthma in humans23.
The regulation of GSNOR is poorly understood. Some proteins contain cysteines whose oxidation state is remarkably sensitive to changes in intracellular redox poise (termed “redox switches”24). We previously showed that plant GSNORs contain positionally-conserved cysteines that do not chelate zinc, a subset of which (Cys10, Cys271, and Cys370 in Arabidopsis GSNOR) are predicted to be nitrosation targets by GPS-SNO11, 13. GSNORs from both Arabidopsis and poplars were found to be nitrosated in vivo upon genetic and abiotic stresses that increase endogenous NO• levels, and GSNOR activity decreased upon addition of nitroso donors to Arabidopsis leaf extracts25, 26. While demonstrating in vivo GSNOR nitrosation, these studies stopped short of investigating the mechanism or significance of these phenomena.
We tested the hypothesis that conserved GSNOR cysteines are redox switches. Our findings confirmed that GSNORs from plant, mammalian, and yeast lineages are all enzymatically inhibited via nitrosation of non-zinc coordinating cysteines and that these modifications engender subtle conformational changes that alter the shape of the substrate binding cavity and the interaction of GSNOR monomers within the same dimer. We propose a model by which constitutive GSNOR activity is negatively regulated by high levels of nitrosating equivalents, thus allowing NO• signaling to proceed during NO• bursts.
Experimental Procedures
Chemicals and reagents
All reagents were purchased from Sigma-Aldrich unless otherwise stated. S-nitroso-N-acetylpenicillamine (SNAP) was furnished by Santa Cruz Biotechnology. S-nitrosoglutathione (GSNO) was made in-house from a solution of 666mM L-glutathione via acid-catalyzed nitrosation as described previously27 and stored at −20 °C with dessicant. S-nitrosocysteine (CysNO) was freshly prepared from a solution of 200mM L-cysteine by acid-catalyzed nitrosation28 and used the same day. Acidified cysteine (CysSH) lacking sodium nitrite was prepared identically to CysNO and used as a control.
DNA sequences and constructs
GSNORs from Arabidopsis thaliana (AtGSNOR, Genbank ID AED95034), Homo sapiens (HsGSNOR, Genbank ID NP_000662), and Saccharomyces cerevisiae (ScSFA1, Genbank ID CAA98742) were expressed as N-terminal polyhistidine fusion proteins via pET expression plasmids (Novagen). AtGSNOR was amplified from 4-week Arabidopsis leaf cDNA13 with primers containing 5′ BamHI and 3′ EcoRI restriction sites, respectively. The PCR product and pET28 vector were cut, the vector treated with Antarctic phosphatase (New England Biolabs), and the PCR product purified (Qiagen), after which the two were ligated via Quick Ligase (New England Biolabs). HsGSNOR was amplified from a Hep2G cDNA library with primers containing 5′ XhoI and 3′ XhoI sites, respectively, after which it was cloned into pET16b. The pET28a_ScSFA1 clone, flanked 5′ and 3′ with BamHI and XhoI sites, respectively, was ordered from DNA2.0 (Menlo Park, CA). Cysteine-to-alanine mutants were developed via the Stratagene quickchange method (Agilent) and substitution confirmed by sequencing and mass spectrometry of purified proteins (see below). Sequence alignments were made with Jalview29.
Protein expression and purification
Wild-type and most mutant constructs were transformed into BL21 Escherichia coli and grown to a 600nm light attenuance (D600) of 0.4-0.6, after which isopropyl thiogalactoside (ITPG; Gold Biotechnology) was added to 0.5mM and cells were grown at 16 °C for 16 hours. For AtGSNORC10/271/370A (C3→A3), HsGSNORC195A, and HsGSNORC268A, IPTG was added to 0.2M at D600 of 1.2. Cells were collected by centrifugation at 10,000 × g at 4 °C, resuspended in lysis buffer ([20mM Tris + 300mM KCl + 30mM imidazole + 10% (v/v) glycerol] pH 7.5 + 1mM dithiothreitol (DTT) + 0.25x EDTA-free Complete Protease Inhibitors (Roche)), lysed with a microfluidizer (Microfluidics), and centrifuged at 18,000 × g, after which soluble material was combined with lysis buffer-equilibrated nickel sepharose resin (GE Healthcare) at a ratio of 1ml resin per 1 L of cells, and incubated in batch at 4 °C with mild agitation for 30 minutes. Resin was collected by centrifugation at 1,000 × g, washed in batch with 10 column volumes of lysis buffer, and protein eluted with 2 column volumes lysis buffer + 300mM imidazole. Eluates were buffer exchanged with 20mM Tris + 300mM KCl pH 7.5 using Amicon ultracell 10,000 Da MWCO centrifugal concentrators (EMD Millipore) at 4 °C, after which protein solutions were aliquoted, flash-frozen in liquid nitrogen, and stored at −80 °C. AtGSNORC3→A3, HsGSNORC195A and HsGSNORC268A were additionally purified on a HiPrep 26/60 S200-HR sephacryl gel filtration column (GE Healthcare) run at 1 ml/min in 20mM Tris + 300mM KCl pH 7.5 using an AKTA Prime FPLC system. Protein concentration was determined by the Bradford method using the Biorad reagent, and purity assessed by SDS-PAGE and Coomassie Blue R-250 protein stain, which indicated ~95% purity by densitometry.
Treatment of GSNOR proteins with reactive nitrogen donors and scavengers
For studies with SNAP, AtGSNOR was brought to 20μM in 20mM Tris pH 8.0, 1mM EDTA, 300μM DTT, with or without 1mM SNAP in 200μl and incubated in the dark at room temperature for 30 minutes, after which samples were buffer exchanged with 20mM Tris pH 8.0 at 4 °C using Amicon 0.5ml concentrators (EMD Millipore) until ≥99.9% of the initial solution was displaced (5-6 centrifuge spins). Alternatively, modified and mock-treated samples were buffer exchanged with 20mM ammonium acetate or 25mM HEPES , 5mM MgCl2, 150mM KCl pH 7.5 (HMK buffer) for mass spectrometry or thermal and chemical stability measurements, respectively. AtGSNOR, HsGSNOR, and ScSFA1 were similarly treated with 2mM CysNO (or 2mM CysSH as a control), except EDTA was replaced with DTPA. For GSNO modification in Fig. 1A, the buffer exchange step was omitted. For experiments to assess reversibility by reducing agents, DTT was added at 5mM either directly before, or 30 minutes following, introduction of the nitroso donor, after which samples were incubated on ice for 30 minutes prior to buffer exchange.
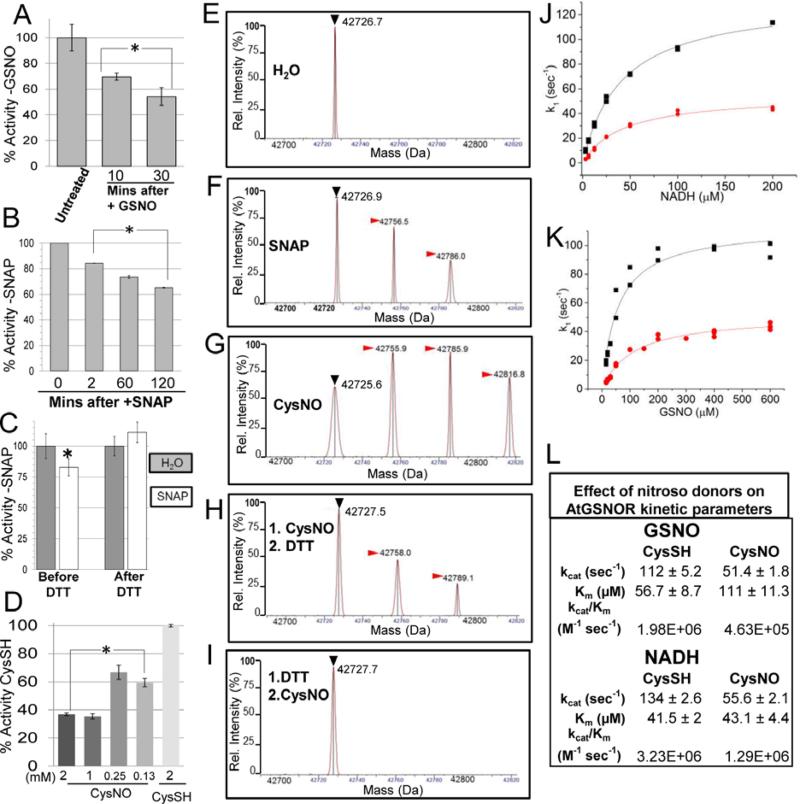
Figure 1. Modification of Arabidopsis GSNOR with reactive nitrogen donors inhibits enzyme activity and is reversed by DTT in vitro.
A. AtGSNOR activity decreases when incubated with the substrate and nitrosating agent GSNO. B. AtGSNOR activity decreases when incubated with the nitroso donor SNAP. C. Inhibition of AtGSNOR by SNAP is reversed by the reducing agent DTT. Before DTT: AtGSNOR was incubated with SNAP or water for 30 minutes prior to assessing enzyme activity. After DTT: SNAP- or water-treated protein solutions were brought to 5mM DTT and incubated for 5 minutes prior to assessing enzyme activity. D. Concentration dependence of CysNO-induced inhibition of AtGSNOR. Enzymes were assayed in triplicate (A-C) or quadruplicate (D). Error bars signify two times standard error. Asterisks indicatestatistically significant differences for a treatment relative to an initial time point or condition. E-I: Deconvolved mass spectra of AtGSNOR treated with water (D), SNAP (E), the NO donor CysNO alone (F), followed by DTT (G), or preceded by DTT (H). Black (▶) and red ( ) arrowheads designate unmodified GSNOR and nitrosated GSNOR adducts (+29 Da), respectively. J-L: CysNO treatment reduces the enzymatic efficiency of AtGSNOR. CysSH- (black) and CysNO- (red) treated AtGSNOR were assayed at saturating concentrations of NADH (J) or GSNO (K) over a range of concentrations of the corresponding substrate. L. Summary of kinetic parameters, plus or minus twice the standard error.
) arrowheads designate unmodified GSNOR and nitrosated GSNOR adducts (+29 Da), respectively. J-L: CysNO treatment reduces the enzymatic efficiency of AtGSNOR. CysSH- (black) and CysNO- (red) treated AtGSNOR were assayed at saturating concentrations of NADH (J) or GSNO (K) over a range of concentrations of the corresponding substrate. L. Summary of kinetic parameters, plus or minus twice the standard error.
Enzyme assays
Assays were conducted in triplicate or quadruplicate in 96-well plates with a SpectraMax M5 spectrophotometer (Molecular Devices) using Softmax Pro software in kinetic mode. AtGSNOR and ScSFA1 activities were assessed at 25 °C by adding enzyme to 1-10nM in 20mM Tris pH 8.0, 200μM NADH, 400μM GSNO, after which the decrease in absorbance at 340nm (dA340/dt) was measured at 10 second intervals for 120 seconds. HsGSNOR was assayed at 100μM NADH + 200μM GSNO. Initial velocities were estimated from slopes (dA340/dt) using a combined extinction coefficient of 7.06mM−1cm−1 to account for NADH and GSNO absorbance maxima overlap, as previously reported15. To derive kinetic parameters, [GSNO] was varied from 15-600μM (from 1-200μM for HsGSNOR) at a fixed [NADH] of 200μM (100μM for HsGSNOR), or [NADH] was varied from 3-200μM at a fixed [GSNO] of 400μM, after which data were fit to a Michaelis-Menten model using Origin Pro software. % activity of control (i.e., Fig. 4) refers to the specific activity (first-order rate constant) of a particular wild-type or mutant GSNOR treated with CysNO divided by the specific activity of the same enzyme treated with CysSH, and this quotient multiplied by 100.
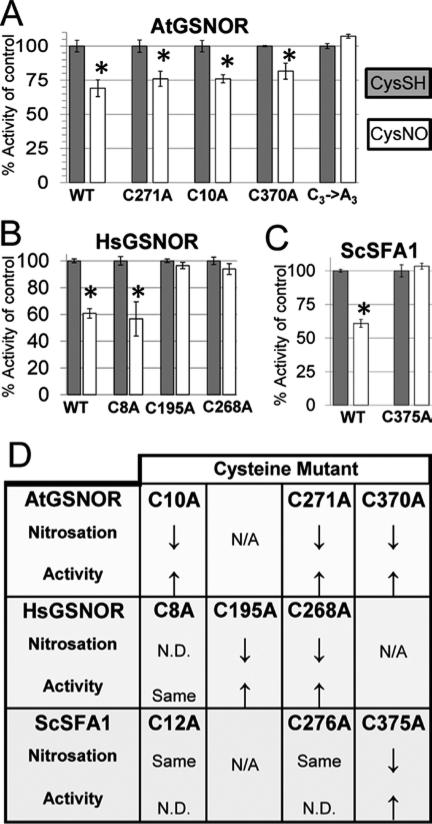
Figure 4. Nitrosation-induced inhibition of GSNOR enzyme activity is blocked by mutation of conserved cysteine residues.
Arabidopsis (A), human (B), and yeast (C) wild-type and cysteine mutant GSNOR isoforms were treated with CysNO or CysSH as a control, after which proteins were buffer exchanged with 20mM Tris pH 8 and assayed as described in triplicate. Error bars signify two times standard error. Asterisks indicate statistically significant differences in catalysis upon CysNO treatment of each protein relative to its CysSH control. D: Summary of the effect of replacement of single conserved cysteines on nitrosation and nitrosation-induced enzyme activity inhibition. Up- and down-arrows are relative to wild-type. N/A: Not applicable (there is no homologous cysteine at this position ). N.D.: Not determined.
Thermal stability assays
Following protein aggregation methods30, 31, CysSH- or CysNO-treated proteins were exchanged into HMK buffer, brought to 5-50μM in 50μl, and incubated at 45 °C for 30 minutes, after which samples were placed on ice for 2 minutes, centrifuged at 16,000 × g for 15 minutes at 4 °C, and the soluble fraction aspirated from the insoluble protein pellet. Pellets were resuspended in a volume of HMK buffer equal to the aspirated soluble fraction (45-50μl), Laemmli buffer was added to both fractions, and samples were separated by SDS-PAGE on 10% (w/v) acrylamide gels. Gels were stained with Coomassie Blue G-250 and densitometry of protein band intensities were quantified with a LiCoR Odyssey flatbed scanner via Image Studio 4.0 software using a 700nm excitation wavelength. Relative soluble protein was calculated by normalizing the ratio of soluble protein signal to insoluble protein signal.
Intrinsic fluorescence and chemical denaturation studies
20μM protein in HMK buffer was excited at 280nm, and fluorescence emission was collected at either discrete wavelengths or from 315-355nm using a PTI fluorometer (Photon Technology International) with 1nm excitation and emission slit widths. Percent control fluorescence was calculated from the ratio of fluorescence counts for either control or nitrosated protein to the fluorescence counts at the maximum emission wavelength of the control protein. To assess protein stability in denaturants, control and nitrosated AtGSNOR were diluted to 2μM in 150μl HMK buffer containing 0-8M urea or 0-4M guanidine hydrochloride (GdnHCl), and samples were incubated at 25 °C for 3 hours prior to fluorometry. Fluorescence at 335nm was normalized for each sample, and ΔGfoldapp. and m values were derived with Origin Pro 2015 using a two-state model 32.
Hydrogen-deuterium exchange measurement and nitrosated peptide discovery
Hydrogen deuterium exchange (HDX) assays33 were performed to assess the solvent accessibility of Arabidopsis and human GSNOR. Protein solutions were buffer exchanged with 10mM potassium phosphate (in H2O) pH 7.0 (buffer E), diluted ≥10-fold into 10mM potassium phosphate (in D2O, Cambridge Isotope Laboratories) pD 7.4 (buffer L) to 5-10μM in 50μl, and incubated at room temperature for 10-300 seconds, or at a fixed time point in triplicate depending on the experiment, after which HDX was quenched with 50μl ice-cold 100mM potassium phosphate (in H2O) pH 2.5 (buffer Q), and samples were promptly flash-frozen in liquid nitrogen. Control “mapping” reactions, (buffer E substituted for buffer L) were performed simultaneously in triplicate to obtain consensus maps of peptides produced by pepsin digestion of each GSNOR. D2O and mock-treated proteins were processed with an M-class nanoACQUITY UPLC-coupled Synapt G2Si-HDMS (ESI-Q-TOF) mass spectrometer using the MassLynx software (Waters). All UPLC columns were maintained at 10 °C in the HDX manager chamber to minimize hydrogen-deuterium back-exchange. Upon thawing, 80μl of quenched sample was loaded by Hamilton syringe via the HDX manager injection port onto a 5μm BEH 2.1 × 30mm Enzymate immobilized pepsin column (Waters) and eluted at 25μl/min with 0.1% formic acid pH 2.5 for 5 minutes (average pressure 1200psi). Peptides were collected with an in-line 1.7μm BEH C18 2.1 × 5mm VanGuard pre-column (Waters). Following digestion, peptides were separated with a 1.8μm HSS T3 C18 2.1 × 30mm ACQUITY UPLC column (Waters) eluted at 40μl/min with a 0.1% formic acid pH 2.5 (solvent A) /acetonitrile (solvent B) gradient as follows: 0-7 mins, 5-35% B; 7-8mins, 35-85% B; 8-10 mins, 85% B; 10-11mins, 85-5% B; 11-12mins, 5% B (average column pressure 9000 psi at 5% B). Eluted peptides, along with a lockspray comprising 2pmol/μl Leu-enkephalin (556.27 ± 0.25 m/z), then entered the mass spectrometer from separate capillaries at 2μl/min by a Z-spray ionization source at 3,000V with these specifications: Source temperature = 70 °C, sampling cone = 10, source offset = 10, desolvation temperature = 150 °C, cone gas = 0, desolvation gas = 600, nebulizer gas = 6, source offset = 10, trap and collision energy = off, gas controls = automatic, resolving quadrupole = 4.9. Peptides from mapping runs were also subjected to MSE fragmentation34 to validate peptide maps. Data from mapping and experimental runs were analyzed using the automated PLGS 3.0 and Dynamx programs (Waters), respectively, to obtain peptide maps and assess deuterium incorporation. Quality thresholds of MS1 signal intensity ≥ 5,000; sequence length 4-20 amino acids; maximum mass error 1ppm; and ≥0.3 products/amino acid were applied. Automated results were then manually refined. Peptides with overlapping isotopic distributions were excluded prior to assembly of HDX heat maps, as was any peptide with a standard error >±20% of the average percent deuterium incorporation. Cartoons of x-ray crystal structures were drawn with PyMol 35. Nitrosated peptides were identified in PLGS 3.0 by searching MS1 datasets for 29 Da adducts with the aforementioned MS2 quality criteria.
Native and denaturing intact protein mass spectrometry
AtGSNOR, HsGSNOR, and ScSFA1 were buffer exchanged with 20mM ammonium acetate made with Fisher Optima-grade water, denatured in 47% (w/v) methanol + 4% acetic acid + 49% water to 5-10μM, and infused onto an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher) at a spray voltage of 3500 V. Data were collected at 1 microscan per second from 1000-3000 m/z at a resolution of 60,000 with the orbitrap detector in intact protein positive ion mode following these specifications: Capillary temperature = 275 °C, sheath gas = 5, auxiliary gas = 5, sweep gas = 0, AGC Target = 4*105, maximum injection time ~20 milliseconds, S lens = 60%. To offset ion suppression of HsGSNORC195A, HsGSNORC268A, or AtGSNORC3→A3, sheath gas and auxiliary gas parameters were increased to 20 and 15, respectively, and data were collected from 1150-3000 m/z. To calculate protein masses, spectral RAW files were processed with Protein Deconvolution 3.0 (Thermo) in isotopically unresolved mode (Manual ReSpect). Ion chromatograms were extracted from total ion intensities for relevant m/z ranges, charge states of +10-+100 were considered for species 40,000-50,000 Da in mass (30,000-40,000 Da for ADH1), assuming the proton as the charge carrier. Three iterations were performed during deconvolution, and filters of ±20ppm mass tolerance, 95% confidence noise reduction, and 5% relative species abundance thresholds were applied. The standard deviations for measured accurate masses of protein species (nitrosated and unmodified) were less than or equal to ±1.5 Da for AtGSNOR, ±3.6 Da for HsGSNOR, and ±1.8 Da for ScSFA1 between successive runs.
Mass spectrometry of native nitrosated and mock-treated AtGSNOR was carried out with the Synapt G2Si in TOF mode. Samples were buffer exchanged into 20mM ammonium acetate, loaded into gold-painted borosilicate emitters with orifice widths trimmed in house with a needle puller to ~5-10μm (kindly provided by Tyler Marcinko), installed into a nanospray ESI source block, and sprayed at 1,600 V. Settings of Offset = 3-10, Trap Gas = 2-10, Nanoflow Gas = 0 were used, and protein deconvolution was performed manually after smoothing raw spectra in Masslynx (Waters).
Results
Reactive nitrogen donors inhibit Arabidopsis GSNOR enzyme activity in a thiol reducing agent-sensitive manner
The aim of our study was to determine how nitrosation affects the structure and function of eukaryotic S-nitrosoglutatione reductases (GSNORs). Initial experiments focused on Arabidopsis GSNOR (AtGSNOR) due to our previous work on its role in acquired thermotolerance and redox homeostasis in plants13, 16, the availability of crystal structures (PDB code: 3UKO), and a recent report that it is nitrosated in vivo.25
Incubation with GSNO caused a time-dependent decrease in the specific activity of unmodified AtGSNOR (Fig. 1A), suggesting that GSNOR is sensitive to its substrate. The molar ratio of protein (20μM) to nitroso donor (1mM) was chosen to approximate physiological conditions, assuming low micromolar [GSNO] and tens of nanomolar [AtGSNOR]16. Treatment with another nitroso donor, S-nitroso-N-acetyl-penicillamine (SNAP), produced similar results (Fig. 1B). We hypothesized enzymatic inactivation was caused by nitrosation of cysteine thiols, which is reversible by the thiol reducing agent dithiothreitol (DTT) 36. Indeed, SNAP-treated GSNOR brought to a concentration of DTT sufficient to quench all RSNO (5mM) reversed the inhibitory effect of SNAP on GSNOR activity (Fig. 1C). The relative specific activity of AtGSNOR was also sensitive to the transnitrosation agent S-nitrosocysteine (CysNO), decreasing by ~40% in the presence of 125μM CysNO after one hour (Fig. 1D). Protein modification (Supp. Fig. S1) increased concomitantly with nitroso donors (discussed below). These findings strongly suggested that AtGSNOR is sensitive to inhibition by nitrosation.
Reactive nitrogen donors nitrosate Arabidopsis GSNOR
We assessed the purity and nitrosation of AtGSNOR by mass spectrometry. Extracted ion chromatograms (Supp. Fig. S2A-B) revealed two species corresponding to the masses of His-tagged AtGSNOR with and without gluconate (+178 Da) (Fig. 1E and data not shown), a sugar acid commonly observed to modify His-tagged proteins37. In the SNAP-treated sample two adducts (+29 Da and +58 Da) were detected that were absent from the water control (compare Fig. 1E and 1F, red arrowheads). Application of CysNO produced a third species (Supp. Fig. S2C-D), and adducts differed in mass by multiples of 29 Da (Fig. 1G). Mass increases of 29 Da are consistent with nitroso moiety addition38. Similar patterns were observed for gluconated AtGSNOR, and no adducts >87 Da (i.e., 3* 29 Da larger than the precursor mass) were detected (data not shown). Moreover, DTT markedly decreased the abundance of nitrosated adducts (Fig. 1H), and prior application of DTT blocked AtGSNOR nitrosation completely (Fig. 1I). Differences in adduct accumulation between donors presumably reflect different rates of transnitrosation39. These data substantiate the conclusion that AtGSNOR can be nitrosated by SNAP and CysNO at no more than three sites per protomer.
The GSNOR mechanism is random sequential with respect to NADH and GSNO40. To explore the modality of nitrosation-induced inhibition, CysSH- and CysNO-treated AtGSNOR were assayed with a concentrations range of one substrate while holding the other constant. One hour incubation with CysNO reduced kcat by ~55% with both substrates relative to CysSH-treated controls (Fig. 1J-L), but substrate affinity was differentially affected; while KmGSNO increased ~2-fold, KmNADH was virtually unchanged (Fig. 1L). These data suggest nitrosation alters the affinity of GSNOR for GSNO, but not NADH, and that concomitant changes to the tertiary structure may impede catalysis even when both substrates are bound.
Substitution of conserved cysteines with alanine decreases sensitivity of AtGSNOR to nitrosation
Three AtGSNOR cysteines that do not chelate zinc (Cys10, Cys271, and Cys370) are conserved among plant GSNORs and are putative nitrosation sites11, 13. To test for nitrosation, these cysteines and catalytic zinc-coordinating Cys177 (not a predicted nitrosation target) were substituted with alanine. While the C177A mutation abolished activity, substitution of other cysteines decreased kcat by no more than 40% (Supp. Table S1 and Supp. Fig. S3A).
Wild-type and mutant AtGSNORs were then incubated with CysNO or SNAP (or CysSH or water as controls) and analyzed by mass spectrometry. Wild-type AtGSNOR and AtGSNORC177A were nitrosated similarly (Fig. 2, B and D). However, only two adducts were observed for single cysteine mutants (Fig. 2F, H, J). AtGSNOR271/370A and AtGSNORC10/271A exhibited mono-nitrosation (Fig. 2N, P), and nitrosation was blocked by substitution of all three cysteines (AtGSNORC3→A3; Fig. 2L). Collectively, these results confirm that AtGSNOR is nitrosated at conserved cysteines that do not chelate zinc.
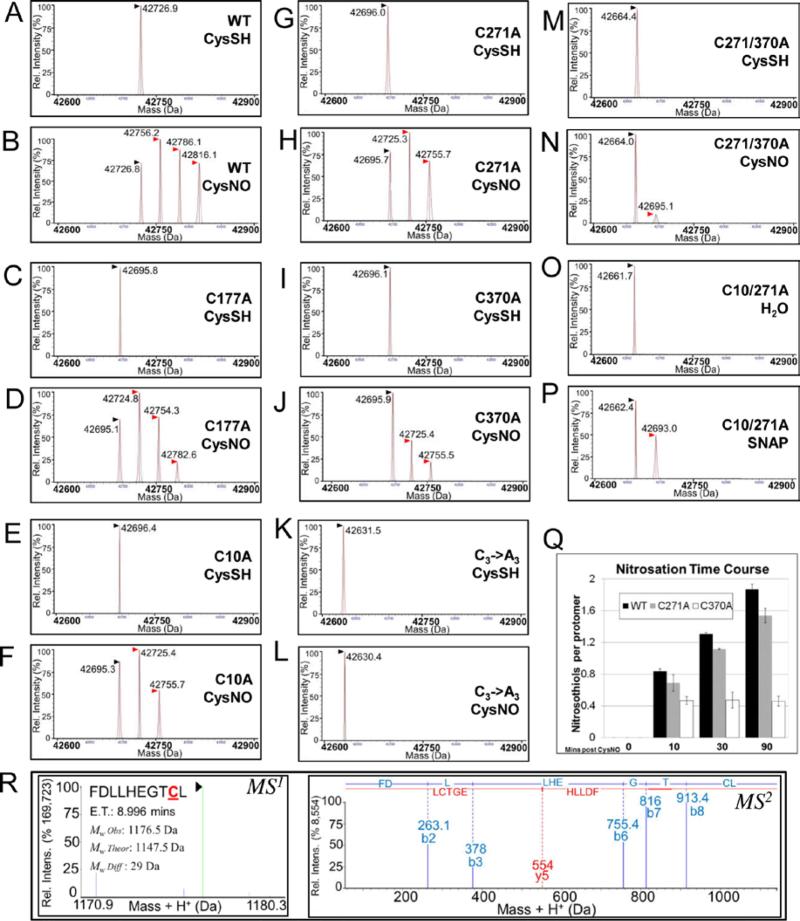
Figure 2. AtGSNOR nitrosation is decreased by replacement of conserved cysteines with.
A-P: Deconvolved mass spectra of wild-type and AtGSNOR cysteine mutants treated with the NO donors CysNO (B,D,F,H,J,L,N) or SNAP (P) or with CysSH (A,C,E,G,I,K,M) or water (O) as controls. Black (▶) and red ( ) arrowheads designate unmodified GSNOR and nitrosated GSNOR adducts (+29 Da), respectively. CysSH: Cysteine. CysNO: S-nitrosocysteine. Q: Time course of CysNO-induced nitrosation of wild-type and mutant AtGSNORs. Nitrosothiols per protomer were calculated from relative distributions of deconvolved mass adducts in two biological replicates, and error bars indicate one standard deviation. R: Representative MS1 (left) and MS2 (right) spectra of a nitrosated peptide from CysNO-treated AtGSNOR. ET: Elution time.
) arrowheads designate unmodified GSNOR and nitrosated GSNOR adducts (+29 Da), respectively. CysSH: Cysteine. CysNO: S-nitrosocysteine. Q: Time course of CysNO-induced nitrosation of wild-type and mutant AtGSNORs. Nitrosothiols per protomer were calculated from relative distributions of deconvolved mass adducts in two biological replicates, and error bars indicate one standard deviation. R: Representative MS1 (left) and MS2 (right) spectra of a nitrosated peptide from CysNO-treated AtGSNOR. ET: Elution time.
AtGSNORC370A and AtGSNORC271/370A were less nitrosated than AtGSNORC10A, AtGSNORC271A, or AtGSNORC10/271A, suggesting Cys370 modification potentiates nitrosation of other cysteines. To test this, nitrosated adduct accumulation was measured over time for CysNO-treated wild-type AtGSNOR, AtGSNORC271A, and AtGSNORC370A. While AtGSNORC271A was less nitrosated than wild-type at any time point, their rates of nitrosation were comparable (Fig. 2Q and Supp. Fig. S3C-D). In contrast, AtGSNOR370A nitrosation was immeasurably slow after marginal di-nitrosation was reached early in the time course. To assess nitrosation at amino acid resolution, UPLC-tandem mass spectrometry after pepsin proteolysis was performed on nitrosated and unmodified wild-type AtGSNOR. In three separate experiments, 2 to 6 peptides were observed with mass shifts consistent with Cys370 nitrosation (Fig. 2R). However, additional cysteines were not observed to be nitrosated (data not shown), probably due to the lability of this S-N bond during proteolysis and ionization, and the search algorithm did not identify Cys370 nitrosation in unmodified samples. Overall, these findings suggest Cys370 is the preferred AtGSNOR nitrosation site. It is possible that Cys370 modification reduces kinetic and/or thermodynamic barriers to nitrosation of Cys10 and Cys271 by inducing a conformational change that alters the solvent accessibility and electrostatic environment of these residues.
Human and yeast GSNOR nitrosation sites include conserved cysteines
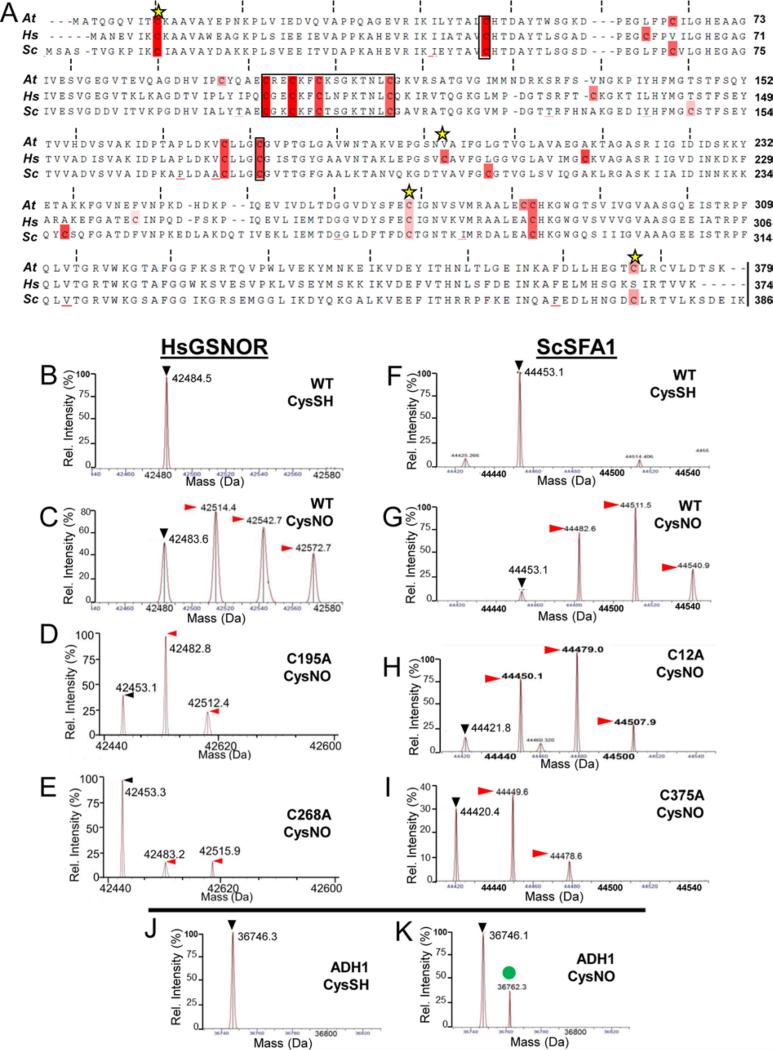
Like AtGSNOR, GSNORs from Homo sapiens (HsGSNOR) and Saccharomyces cerevisiae (ScSFA1, Sensitive to Formaldehyde 141) contain nine cysteines that do not chelate zinc (Fig. 3A), of which three (Cys8, Cys195, and Cys268 in HsGSNOR; Cys12, Cys276, and Cys375 in ScSFA1) are predicted nitrosation sites according to GPS-SNO. AtGSNOR kinetic parameters were observed to be 5-10 times greater than those of HsGSNOR (Supp. Table S1), as previously observed42. As with AtGSNOR, HsGSNOR and ScSFA1 were mono-, di-, and tri-nitrosated by CysNO (Fig. 3, panels C and G). HsGSNORC195A, HsGSNORC268A, and ScSFA1C375A were less nitrosated than corresponding wild-type proteins (Fig. 3, compare D,E,I with C,G), thus confirming that nitrosatable cysteines that do not chelate zinc are a common feature of eukaryotic GSNORs (e.g., Cys370 in Arabidopsis and yeast and Cys271 in Arabidopsis and human). However, ScSFA112A and ScSFA1276A were modified indistinguishably from wild-type ScSFA1, and nitrosated HsGNSORC8A was inhibited similarly to wild-type HsGSNOR (Fig. 3H and data not shown). Therefore, while some conserved cysteines are nitrosated in GSNORs across taxa, others are not, thus underscoring the limitation of nitrosation prediction algorithms such as GPS-SNO.
Figure 3. Nitrosation of human and yeast GSNORs also depends on conserved cysteine residues.
A. Multiple sequence alignment of GSNORs examined in this study. Black boxes denote zinc-coordinating cysteines, while yellow stars indicate putative nitrosation sites according to GPS SNO [34]. B-I: Deconvolved mass spectra of wild-type and cysteine mutant human GSNOR (B-E) and the yeast ortholog SFA1 (F-I) treated with the NO donor CysNO. Black (▶) and red ( ) arrowheads designate unmodified GSNOR and nitrosated GSNOR adducts (+29 Da), respectively. J-K: Yeast alcohol dehydrogenase 1 (ADH1) is not nitrosated under the same conditions. The green circle (
) arrowheads designate unmodified GSNOR and nitrosated GSNOR adducts (+29 Da), respectively. J-K: Yeast alcohol dehydrogenase 1 (ADH1) is not nitrosated under the same conditions. The green circle ( ) demarcates an unknown 16 Da adduct in the CysNO-treated sample.
) demarcates an unknown 16 Da adduct in the CysNO-treated sample.
As an additional test of the specificity of this modification for GSNOR, nitrosation was examined for yeast ADH1, which shares six zinc-chelating cysteines, a nicotinamide binding domain, and the same basic protein fold in common with GSNORs14. No nitrosated ADH1 adducts were detected under our experimental conditions (Fig. 3J-K). This result strongly suggests GSNOR non-zinc chelating cysteines in particular, and not ADH cysteines in general, can be nitrosated.
Nitrosation-induced inhibition of GSNOR enzyme activity is blocked by mutation of conserved cysteine residues
To further test the role of conserved cysteines, we measured the specific activities of CysSH- and CysNO-treated wild-type and mutant AtGSNOR, HsGSNOR, and ScSFA1 as in Fig. 1D. As expected, most Cys→Ala mutants were less sensitive to CysNO than corresponding wild-types in a manner that mirrored nitrosation profiles. For example, AtGSNOR single mutants and AtGSNORC3→A3 were marginally and completely insensitive to CysNO, respectively (Fig. 4A). HsGSNORC195A, HsGSNORC268A, and ScSFA1C375A—all of which exhibited reduced nitrosation (Fig. 3D-E, I)—were also nearly insensitive to CysNO (Fig. 4B-C). However, ScSFA1C12A and HsGSNORC8A were as sensitive to nitrosation as their corresponding wild-types (summarized in Fig. 4D). Taken together, these results indicate that nitrosation of some, but not all, conserved cysteines decreases enzyme activity of eukaryotic GSNORs.
Similar regions of Arabidopsis and human GSNOR exhibit differential hydrogen-deuterium exchange due to nitrosation
The reversible inhibition of enzyme activity by nitrosation (Fig. 1A-D) suggested it effects local conformational changes rather than global deformation of the protein fold. To test this hypothesis, hydrogen-deuterium exchange (HDX), a metric for solvent accessibility, was examined for both nitrosated and unmodified AtGSNOR and HsGSNOR. Because deuterium incorporation increased linearly over an interval of 10-300 seconds (data not shown), proteins were subsequently exposed to deuterium for 210 seconds in triplicate experiments.
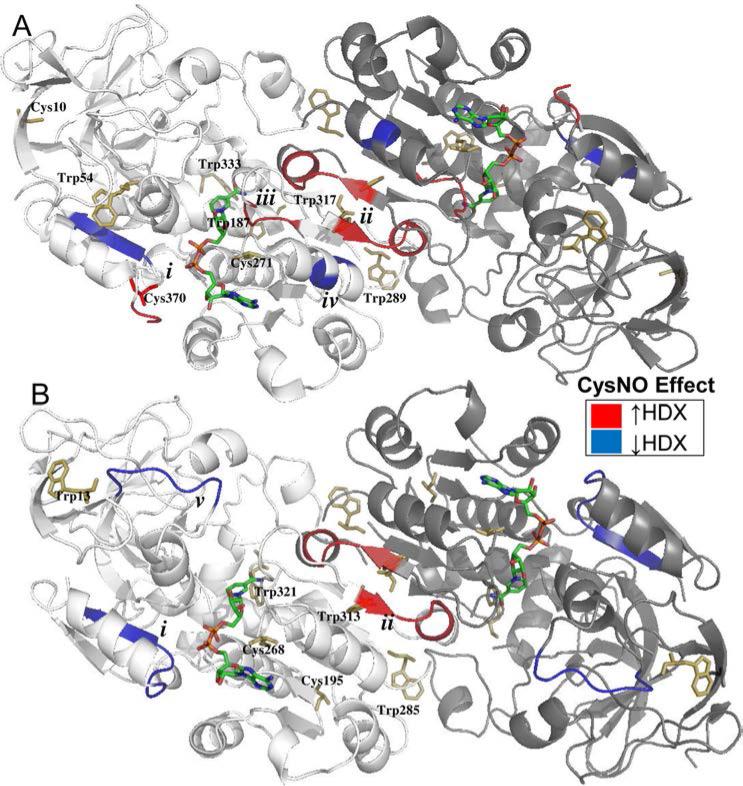
Heat maps of deuterium uptake, based on >95% sequence coverage and 5.1× average redundancy for both CysSH- and CysNO-treated AtGSNOR (Supp. Fig. S4), were applied to a known AtGSNOR crystal structure (PDB: 3UKO) and revealed nearly identical patterns of HDX between nitrosated and unmodified AtGSNOR (Supp. Fig. S4A). Noteworthy exceptions (summarized in Supp. Table S2) included differential HDX at the C-terminus (i), increased HDX at the interface between monomers (ii), increased HDX for NADH-binding amino acids (iii), and decreased HDX for an alpha helix adjacent to the dimer interface (iv) (Fig. 6A). Val295-Val297 in (iii.) make contacts with NADH phosphates 43 and are located 5-6 Å from the putative nitrosation site Cys271 and 6-8Å from (iv.). Moreover, (i.) contains the putative nitrosation site 370, which is 5Å from Gly204 and Thr205 that contact the NADH ribose moiety. While some regions of HsGSNOR (PDB: 1M6H) exhibited similar behavior, others were distinct (Fig. 5B). Like AtGSNOR, homologous amino acids at the HsGSNOR C-terminus (Gly365-Thr371; (i)) and dimer interface (Ala301-Gln306; (ii)) exhibited differential HDX upon nitrosation (Fig. 5B and Supp. Fig. S5). In addition, CysNO treatment correlated with reduced HDX for Pro57-Val63 (v.), which forms part of the roof of the substrate-binding cavity. In summary, nitrosation subtly changed the solvent accessibility of amino acids that either form the substrate binding cavity or comprise the dimer interface of HsGSNOR and AtGSNOR.
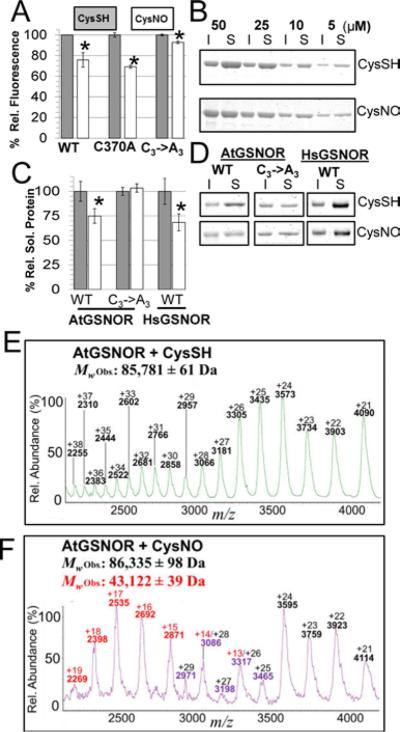
Figure 6. Intrinsic fluorescence, thermal stability, and polydispersity are affected by GSNOR nitrosation.
A. Normalized maximum fluorescence emission intensities (Ex: 280nm, Em: 335-340nm) of wild-type and mutant AtGSNORs treated with CysSH (gray) or CysNO (white). Error bars represent 2 times standard error from three or more experiments. B. AtGSNOR treated with CysSH or CysNO was incubated at 45 °C for 30 mins at the indicated concentrations, after which insoluble (I) and soluble (S) material were separated by SDS-PAGE and visualized by Coomassie G-250 staining. C. Experiments were carried out as in B with the indicated proteins at 20 μM, after which protein band intensities were quantified with a LiCoR Odyssey flatbad scanner. D. Representative Coomassie-stained gels for quantification in C. E-F: Distribution of native AtGSNOR charge states under control (E) and nitrosating (F) conditions.
Figure 5. Nitrosation affects the hydrogen-deuterium exchange of similar conserved regions of Arabidopsis and Human GSNOR.
A-B: X-ray structure cartoons of Arabidopsis (A, PDB 3UKO) and Human (B, PDB 1MP0) GSNOR co-crystalized with NADH (multicolored sticks). One monomer in each dimer is shaded more darkly for clarity. Tryptophans and predicted nitrosated cysteines are depicted with yellow sticks. Relative to the CysSH-treated control, CysNO induced similar regions of both proteins to exhibit decreased (blue) or increased (red) hydrogen-deuterium exchange. A. AtGSNOR. i. Glu367-Leu375, contains cysteine 370 and located 5Å from residues that bind NADH. ii. Ser305-Gln310, comprises part of the dimer interface. iii. Ile294-Ala298, contains NADH-binding residues and located 5Å from cysteine 271. iv. Ala280-Leu282, located 6 Å from (iii). B. HsGSNOR. i. Gly365-Val372, located 5Å from residues that bind NADH, homologous to (i) in AtGSNOR. ii. Ala301-Gln306, comprises part of the dimer interface, homologous to (ii) in AtGSNOR. v. Pro57-Val63, constitutes part of the roof of the substrate-binding cavity.
Nitrosation also alters GSNOR polydispersity, intrinsic fluorescence, and thermostability
The impact of nitrosation on enzyme activity and local protein conformation led us to test whether this modification affected other physicochemical properties. It was hypothesized that nitrosation may perturb protein thermodynamic stability (ΔGf), the free energy difference between the folded and unfolded state. Protein aggregation upon heating precluded measurement of ΔGf by thermal denaturation (Fig. 6B-C and data not shown). As an alternative approach, thermodynamic parameters were estimated from the effect of urea and guanidinium hydrochloride (GdnHCl)-induced denaturation on tryptophan fluorescence. Re-folding chaotrope dilution assays indicated AtGSNOR denaturation is not completely reversible (data not shown). Therefore, all derived thermodynamic parameters were “apparent.” While the apparent folding energy (ΔGfap) increased, and folding cooperativity (m-value) decreased upon nitrosation using both denaturants (Supp. Fig. S6A-B), the magnitude of change was not appreciable.
Nitrosation decreased folded AtGSNOR fluorescence (Supp. Fig. S6C-D), and time-dependent loss of AtGSNOR fluorescence was detectable down to 100μM CysNO (Supp. Fig. S1E). Parallel phenomena were observed for HsGSNOR and ScSFA1 (Supp. Fig. S6E-F). As expected, fluorescence of AtGSNORC3→A3 was almost unaffected by CysNO (Fig. 6A, Supp. Fig. S6D). However, single point mutants of GSNORs from all three species were only exhibited similar fluorescence loss to corresponding wild-types (Supp. Fig. S6G-H). RSNOs, several amino acid sidechains, and the peptide backbone can quench fluorescence44,45. Mere mono-nitrosation therefore seems sufficient to alter the conformation of one or more buried tryptophans.
Since thermal melt experiments were foiled by irreversible protein aggregation, it was hypothesized that nitrosation may affect thermal aggregation propensity. Following incubation at 45 °C, protein solutions were separated into soluble and insoluble fractions by centrifugation and resolved by SDS-PAGE. CysNO treatment promoted the formation of thermally-catalyzed protein aggregates (defined as protein species pelleting at 16,000 × g) over a range of AtGSNOR concentrations (Fig. 6B). While, nitrosation also enhanced aggregation of HsGSNOR, the solubility of non-nitrosatable AtGSNORC3→A3 was apparently unaffected by CysNO relative to CysSH treatment (Fig. 6C-D). Collectively, these data indicate nitrosation induces conformational changes that increase the propensity of GSNOR to aggregate upon heating.
Given that CysNO treatment facilitated increased HDX of residues comprising the dimer interface (Fig. 5, Supp. Table S2), it was speculated that nitrosation could alter the GSNOR monomer-dimer equilibrium. To explore this possibility, CysSH and CysNO-treated AtGSNOR samples were analyzed by native mass spectrometry. While the control sample ion series comprised charge states of an AtGSNOR dimer, the nitrosated protein produced overlapping ion distributions whose deconvolution was consistent with a mixture of both monomeric and dimeric AtGSNOR (Fig. 6E-F). Taken together, these data support the hypothesis that nitrosation decreases the self-affinity of GSNOR protomers.
Discussion
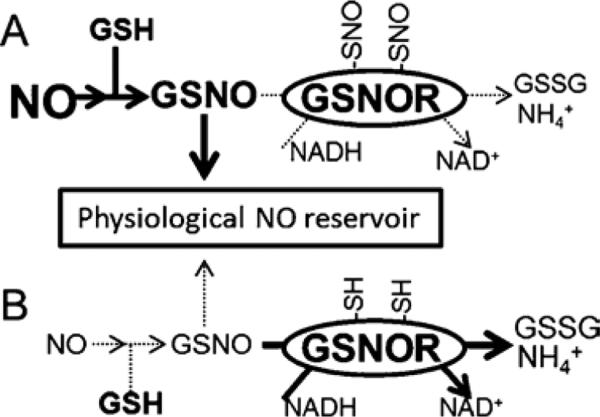
It may seem paradoxical that GSNOR, a negative regulator of NO• responses, would itself be downregulated by nitrosation. Constituting ~0.015% of soluble Arabidopsis protein by mass and constitutively expressed at moderately-high levels in animals and plants, GSNOR is poised to attenuate NO•-dependent biological processes16, 46, 47. To be effective, a sufficient quantity of bioactive NO• must reach target proteins. We therefore offer a testable model for the role of GSNOR in NO• homeostasis (Fig. 7): At low NO• concentrations, GSNO is efficiently catabolized by constitutive GSNOR activity. Induced NO• bursts reversibly inhibit GSNOR, allowing for GSNO accumulation and nitrosation of intended targets. GSNOR is reactivated by the cytosolic reducing environment, and GSNO is decreased to basal concentrations after the NO• burst ceases. This model could explain how NO• signaling proceeds in the presence of GSNOR. Indeed, GSNOR activity decreases and GSNOR nitrosation increases in Arabidopsis that over-express NO•25.
Figure 7. A model for the function of reversible GSNOR nitrosation.
A: During a nitric oxide burst, GSNOR is nitrosated on conserved cysteines, leading to a decrease in enzymatic activity. Lower GSNOR activity facilitates accumulation of GSNO, itself an agent of protein nitrosation. By allowing GSNO to accumulate, down-regulation of GSNOR permits robust nitric oxide signaling. B: Eventually GSNOR is re-reduced by cellular antioxidant machinery, whereupon GSNO concentrations are returned to basal levels by GSNOR.
We have shown here that cysteine nitrosation decreases the catalytic activity of Arabidopsis, human, and budding yeast GSNORs (Figs. 1-4). Frungillo et al. reported that GSNOR activity observable in Arabidopsis cell lysates decreases upon NO• fumigation25. Providing direct evidence of nitrosation by intact protein and peptide tandem mass spectrometry (Figs. 1F, 2R), our data confirm these findings and suggest a fraction of GSNOR cysteines are evolutionarily-conserved redox switches. This was validated by DTT-sensitivity of nitrosated adducts (Fig. 1HI) and decreased modification and enzymatic inhibition of cysteine to alanine mutants (Fig. 2F-P; Fig. 3D-E, H-I; Fig. 4A-C). Cys8 of HsGSNOR and Cys12 and Cys276 of ScSFA1 do not appear to be nitrosated (Fig. 3H and data not shown), but all three wild-type proteins exhibited tri-nitrosation. While this indicates moderate divergence in nitrosation preference among the three eukaryotic lineages, all modification sites are likely to be non-zinc chelating cysteines. AtGSNOR active site Cys177 is not nitrosated (Fig. 2D), and the corresponding residue in other GSNORs is also an unlikely target since it is shared by ADH1, which is not nitrosated (Fig. 3K).
Conserved cysteine nitrosation is known to negatively regulate many metabolic enzymes including cytosolic glyceraldehyde-3-phosphate dehydrogenase36, glutathione transferase P148, and protein disulfide isomerase49. In these cases, modified cysteines are either active site residues or are located in catalytic domains. In contrast, AtGSNOR cysteines 10, 271, and 370 and HsGSNOR cysteines 195 and 268 are on the solvent-accessible surface, distal from catalytic sites. Thus, nitrosation apparently decreases GSNOR activity by allostery rather than direct inactivation of the active site. The ND3 subunit of mitochondrial Complex I similarly attenuates NADH dehydrogenase activity upon nitrosation50, suggesting nitrosation-induced allosteric inactivation may be a general mechanism of protein regulation.
Occasionally, substitution of one cysteine disproportionately decreased nitrosation of remaining cysteines (e.g., AtGSNORC370A and HsGSNORC268A; Figs. 2J,N and 3E). This could reflect a cooperative mechanism whereby mono-nitrosation at a favored site thermodynamically poises the protein for additional nitrosation, a feature reminiscent of hierarchical phosphorylation51. The detection of peptides with MSE fragmentation indicative of AtGSNOR Cys370 nitrosation (Fig. 2R) supports this notion. While it cannot be completely ruled out that cysteine to alanine mutations occluded the accessibility of nitrosatable cysteines, it is unlikely since single cysteine substitution negligibly affected enzyme kinetics (Supp. Table S2 and Crotty42), and nitrosation (Fig. 2D).
Loss of protein fluorescence (Fig. 6A; Supp. Figs. S1E, S6C-H) corresponds to local changes in buried amino acid solvent accessibility, particularly given the increased propensity of nitrosated GSNOR to aggregate upon heating (Fig. 6B-D). Fluorescence loss after tri-nitrosation of wild-type GSNOR is comparable to that of di-nitrosated single cysteine mutants of all three lineages (Fig. 6A; Supp. Fig. S6G-H), so nitrosation per se probably contributes little to quenching. Two of the four tryptophans shared by HsGSNOR and AtGSNOR are located at the dimer interface, which incidentally exhibits differential HDX upon nitrosation (Fig. 5A-B). Weakened interaction between the two GSNOR protomers, as indicated by native mass spectrometry (Fig. 6E-F), could explain the decreased fluorescence as well as the increased aggregation propensity, possibly due to the exposure of buried hydrophobic regions in the dimer interface. C-terminal amino acids that contact an NADH-binding glycine in both AtGSNOR and HsGSNOR exhibited diminished HDX upon nitrosation (Supp. Table S2), which may partially explain concomitant decreases in enzyme activity. Since the AtGSNOR Km NADH did not change upon nitrosation (Fig. 1L), these displacements may be balanced by conformational changes that preserve the binding affinity of NADH for the enzyme, albeit in a manner counter-productive to catalysis.
Despite the correlation between nitrosation and aggregation propensity (Fig. 6B-D), nitrosation minimally affected protein thermodynamic stability (Supp. Fig. S6A-B). ΔGf subsumes myriad local rearrangements, and aggregation propensity can be uncoupled from thermodynamic destabilization52. The extent to which individual nitrosation events affect GSNOR structure is not completely clear because experiments were performed on ensembles of unmodified, mono-, di-, and tri-nitrosated proteins. To our knowledge, separation of differentially nitrosated GSNORs in their native conformations is presently not feasible. It may prove fruitful in future studies to optically trap and denature individual proteins to estimate with greater precision how each cysteine modification affects enzyme activity and the integrity of individual domains.
The coincidence of asthma and diminished nitrosothiol accumulation in human patients23, 53 serve to highlight that NO• homeostasis is delicately balanced between excess and deficiency. We propose that reversible inhibition of GSNOR during NO• bursts could allow a cell to clear reactive nitrogen following perception of an NO• signal, and this work provides evidence that conserved cysteine residues may enable such dynamic regulation of GSNOR. An improved understanding of this process will aid the development of therapeutics that reversibly and specifically target GSNOR activity.
Supplementary Material
Acknowledgments
The authors thank Thomas Sawyer, Tereza Tichá, Sara Kimiko Suzuki, Sam Del'Olio, and Shellyhan Gordon for assisting in the cloning, purification, and/or kinetic assays of several Cys→Ala mutants. We are grateful to Dr. Lila Gierasch for furnishing a spectrofluorometer, and Karan Hingorani, Dr. Indu Santhanagopalan, Dr. Abhay Thakur, and Joe Tilitsky for interpretation of fluorescence data. We are particularly indebted to Dr. Jeanne Hardy for discussions on in vitro nitrosation experiments. The authors also thank Dr. Steve Eyles for access to the UMass mass spectrometry center, Dr. Eugenia Clerico and Tyler Marcinko for instruction and troubleshooting on the Orbitrap and Synapt mass spectrometers, and Dr. Daniel J. Guerra for careful reading and critique of this manuscript prior to submission.
Funding Sources
This work was supported by a Massachusetts Life Sciences Center new faculty research award (E.V.) and grants from the National Science Foundation (MCB1517046 to E.V., which funded D.G. and I.T.) and National Institutes of Health (T32GM008515 to K.B.).
Abbreviations
- PTM
post-translational modification
- NO•
nitric oxide
- GSNO
S-nitrosoglutathione
- GSNOR
S-nitrosoglutathione reductase
- HsGSNOR
GSNOR from Homo sapiens
- AtGSNOR
GSNOR from Arabidopsis thaliana
- ScSFA1
GSNOR from Saccharomyces cerevisiae
- ADH
alcohol dehydrogenase
- RNS
reactive nitrogen species
- GSH
glutathione
- SNAP
S-nitroso-N-acetyl- penicillamine
- CysNO
S-nitrosocysteine
- cPTIO
2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- EDTA
ethylenediaminetetraaceticacid
- DTPA
diethylene triamine pentaacetic acid
- ITPG
isopropyl β-D-1-thiogalactopyranoside
- DTT
dithiothreitol
- TCEP
Tris(2-carboxyethyl)phosphine
- GdnHCl
guanidine hydrochloride
- HDX
hydrogen-deuterium exchange
Footnotes
Supporting Information
Supporting information for the manuscript can be found online at http://pubs.acs.org.
References
- 1.Mur LAJ, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, Hall MA, Harren FJM, Hebelstrup KH, Gupta KJ. Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants. 2013;5 doi: 10.1093/aobpla/pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choudhari SK, Chaudhary M, Bagde S, Gadbail AR, Joshi V. Nitric oxide and cancer: a review. World J. Surg. Oncol. 2013;11:118. doi: 10.1186/1477-7819-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur. Heart J. 2012;33:829–837. 837a–837d. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corpas FJ, Barroso JB. Peroxisomal plant nitric oxide synthase (NOS) protein is imported by peroxisomal targeting signal type 2 (PTS2) in a process that depends on the cytosolic receptor PEX7 and calmodulin. FEBS Lett. 2014;588:2049–2054. doi: 10.1016/j.febslet.2014.04.034. [DOI] [PubMed] [Google Scholar]
- 5.Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. The American journal of physiology. 1996;271:C1424–1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 6.Keszler A, Zhang Y, Hogg N. The Reaction between Nitric Oxide, Glutathione and Oxygen in the Presence and Absence of Protein: How are S Nitrosothiols Formed? Free radical biology & medicine. 2010;48:55–64. doi: 10.1016/j.freeradbiomed.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohlen HG. Is the Real In Vivo Nitric Oxide Concentration Pico or Nano Molar? Influence of Electrode Size on Unstirred Layers and NO Consumption. Microcirculation. 2013;20:30–41. doi: 10.1111/micc.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broniowska KA, Diers AR, Hogg N. S-nitrosoglutathione. BBA-Gen. Subjects. 2013;1830:3173–3181. doi: 10.1016/j.bbagen.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cejudo FJ, Meyer AJ, Reichheld J-P, Rouhier N, Traverso JA. Thiol-based redox homeostasis and signaling. Front. Plant Sci. 2014;5:266. doi: 10.3389/fpls.2014.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 11.Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J. GPS-SNO: Computational Prediction of Protein <italic>S</italic>-Nitrosylation Sites with a Modified GPS Algorithm. PLoS ONE. 2010;5:e11290. doi: 10.1371/journal.pone.0011290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaki M, Kovacs I, Spannagl M, Lindermayr C. Computational prediction of candidate proteins for S-nitrosylation in Arabidopsis thaliana. PLoS One. 2014;9:e110232. doi: 10.1371/journal.pone.0110232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S, Guerra D, Lee U, Vierling E. S-nitrosoglutathione reductases are low-copy number, cysteine-rich proteins in plants that control multiple developmental and defense responses in Arabidopsis. Front. Plant Sci. 2013;4:430. doi: 10.3389/fpls.2013.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 15.Jensen DE, Belka GK, Du Bois GC. S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem J. 1998;331:659–668. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell. 2008;20:786–802. doi: 10.1105/tpc.107.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubienova L, Kopecny D, Tylichova M, Briozzo P, Skopalova J, Sebela M, Navratil M, Tache R, Luhova L, Barroso JB, Petrivalsky M. Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum. Biochimie. 2013;95:889–902. doi: 10.1016/j.biochi.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ, Chandrasekaran G, Broecker V, Wei W, Liu L, Swenberg JA, Crossan GP, Patel KJ. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Mol Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staab CA, Ålander J, Brandt M, Lengqvist J, Morgenstern R, Grafström RC, Höög J-O. Reduction of S-nitrosoglutathione by alcohol dehydrogenase 3 is facilitated by substrate alcohols via direct cofactor recycling and leads to GSH-controlled formation of glutathione transferase inhibitors. Biochem. J. 2008;413:493–504. doi: 10.1042/BJ20071666. [DOI] [PubMed] [Google Scholar]
- 20.Tang C-H, Wei W, Hanes MA, Liu L. Increased Hepatocarcinogenesis from GSNOR Deficiency in mice is Prevented by Pharmacological Inhibition of iNOS. Cancer Res. 2013;73:2897–2904. doi: 10.1158/0008-5472.CAN-12-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatzistergos KE, Paulino EC, Dulce RA, Takeuchi LM, Bellio MA, Kulandavelu S, Cao Y, Balkan W, Kanashiro-Takeuchi RM, Hare JM. S-Nitrosoglutathione Reductase Deficiency Enhances the Proliferative Expansion of Adult Heart Progenitors and Myocytes Post Myocardial Infarction. J. Am. Heart Assoc. 2015;4:e001974. doi: 10.1161/JAHA.115.001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, Zuo J. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19:1377–1387. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- 23.Green LS, Chun LE, Patton AK, Sun X, Rosenthal GJ, Richards JP. Mechanism of inhibition for N6022, a first-in-class drug targeting S- nitrosoglutathione reductase. Biochemistry. 2012;51:2157–2168. doi: 10.1021/bi201785u. [DOI] [PubMed] [Google Scholar]
- 24.Luebke JL, Giedroc DP. Cysteine sulfur chemistry in transcriptional regulators at the host-bacterial pathogen interface. Biochemistry. 2015;54:3235–3249. doi: 10.1021/acs.biochem.5b00085. [DOI] [PubMed] [Google Scholar]
- 25.Frungillo L, Skelly MJ, Loake GJ, Spoel SH, Salgado I. S- nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat. Commun. 2014;5:5401. doi: 10.1038/ncomms6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng T, Chen J, Ef AA, Wang P, Wang G, Hu X, Shi J. Quantitative proteomics analysis reveals that S-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta. 2015;242:1361–1390. doi: 10.1007/s00425-015-2374-5. [DOI] [PubMed] [Google Scholar]
- 27.Duong HT, Kamarudin ZM, Erlich RB, Li Y, Jones MW, Kavallaris M, Boyer C, Davis TP. Intracellular nitric oxide delivery from stable NO-polymeric nanoparticle carriers. Chem. Commun. (Camb.) 2013;49:4190–4192. doi: 10.1039/c2cc37181b. [DOI] [PubMed] [Google Scholar]
- 28.Kumar V, Martin F, Hahn MG, Schaefer M, Stamler JS, Stasch J-P, van den Akker F. Insights into BAY 60-2770 Activation and S-Nitrosylation- Dependent Desensitization of Soluble Guanylyl Cyclase via Crystal Structures of Homologous Nostoc H-NOX Domain Complexes. Biochemistry. 2013;52:3601–3608. doi: 10.1021/bi301657w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mogk A, Schlieker C, Friedrich KL, Schonfeld HJ, Vierling E, Bukau B. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 2003;278:31033–31042. doi: 10.1074/jbc.M303587200. [DOI] [PubMed] [Google Scholar]
- 31.Basha E, Jones C, Wysocki V, Vierling E. Mechanistic differences between two conserved classes of small heat shock proteins found in the plant cytosol. J. Biol. Chem. 2010;285:11489–11497. doi: 10.1074/jbc.M109.074088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl .alpha.-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 33.Yan X, Maier CS. Hydrogen/deuterium exchange mass spectrometry. Methods Molec. Biol. 2009;492:255–271. doi: 10.1007/978-1-59745-493-3_15. [DOI] [PubMed] [Google Scholar]
- 34.Blackburn K, Mbeunkui F, Mitra SK, Mentzel T, Goshe MB. Improving Protein and Proteome Coverage through Data-Independent Multiplexed Peptide Fragmentation. J. Proteome Res. 2010;9:3621–3637. doi: 10.1021/pr100144z. [DOI] [PubMed] [Google Scholar]
- 35.PyMOL. The PyMol Molecular Graphics System. Version 1.6 Schrodinger, LLC.; [Google Scholar]
- 36.Zaffagnini M, Morisse S, Bedhomme M, Marchand CH, Festa M, Rouhier N, Lemaire SD, Trost P. Mechanisms of nitrosylation and denitrosylation of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. J. Biol. Chem. 2013;288:22777–22789. doi: 10.1074/jbc.M113.475467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geoghegan KF, Dixon HB, Rosner PJ, Hoth LR, Lanzetti AJ, Borzilleri KA, Marr ES, Pezzullo LH, Martin LB, LeMotte PK, McColl AS, Kamath AV, Stroh JG. Spontaneous alpha-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: the cause of extra mass of 258 or 178 Da in fusion proteins. Anal Biochem. 1999;267:169–184. doi: 10.1006/abio.1998.2990. [DOI] [PubMed] [Google Scholar]
- 38.Zech B, Wilm M, van Eldik R, Brune B. Mass spectrometric analysis of nitric oxide-modified caspase-3. J. Biol. Chem. 1999;274:20931–20936. doi: 10.1074/jbc.274.30.20931. [DOI] [PubMed] [Google Scholar]
- 39.Wang K, Wen Z, Zhang W, Xian M, Cheng JP, Wang PG. Equilibrium and kinetics studies of transnitrosation between S-nitrosothiols and thiols. Bioorganic & medicinal chemistry letters. 2001;11:433–436. doi: 10.1016/s0960-894x(00)00688-0. [DOI] [PubMed] [Google Scholar]
- 40.Sanghani PC, Davis WI, Fears SL, Green S-L, Zhai L, Tang Y, Martin E, Bryan NS, Sanghani SP. Kinetic and Cellular Characterization of Novel Inhibitors of S-Nitrosoglutathione Reductase. J. Biol. Chem. 2009;284:24354–24362. doi: 10.1074/jbc.M109.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wehner EP, Rao E, Brendel M. Molecular structure and genetic regulation of SFA, a gene responsible for resistance to formaldehyde in Saccharomyces cerevisiae, and characterization of its protein product. Molecular & general genetics : MGG. 1993;237:351–358. doi: 10.1007/BF00279438. [DOI] [PubMed] [Google Scholar]
- 42.Crotty J. PhD Dissertation. 2009. Crystal Structures and Kinetics of S-Nitrosoglutathione Reductase from Arabidopsis thaliana and Human. [Google Scholar]
- 43.Crotty J, Greving M, Brettschneider S, Weichsel A, Wildner GF, Vierling E, Montfort WR. Crystal structure and kinetic behavior of alcohol dehydrogenase III/S-nitrosoglutathione reductase from Arabidopsis thaliana. 2013 Unpublished. [Google Scholar]
- 44.Sillen A, Hennecke J, Roethlisberger D, Glockshuber R, Engelborghs Y. Fluorescence quenching in the DsbA protein from Escherichia coli: Complete picture of the excited-state energy pathway and evidence for the reshuffling dynamics of the microstates of tryptophan. Proteins: Struc. Func. Bioinform. 1999;37:253–263. doi: 10.1002/(sici)1097-0134(19991101)37:2<253::aid-prot10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 45.Akhter S, Vignini A, Wen Z, English A, Wang PG, Mutus B. Evidence for S-nitrosothiol-dependent changes in fibrinogen that do not involve transnitrosation or thiolation. Proc. Natl. Acad. Sci. 2002;99:9172–9177. doi: 10.1073/pnas.142136499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estonius M, Danielsson O, Karlsson C, Persson H, Jornvall H, Hoog JO. Distribution of alcohol and sorbitol dehydrogenases. Assessment of mRNA species in mammalian tissues. Eur. J. Biochem. 1993;215:497–503. doi: 10.1111/j.1432-1033.1993.tb18059.x. [DOI] [PubMed] [Google Scholar]
- 47.Leterrier M, Chaki M, Airaki M, Valderrama R, Palma JM, Barroso JB, Corpas FJ. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Sig. Behav. 2011;6:789–793. doi: 10.4161/psb.6.6.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balchin D, Wallace L, Dirr HW. S-nitrosation of glutathione transferase p1-1 is controlled by the conformation of a dynamic active site helix. J. Biol. Chem. 2013;288:14973–14984. doi: 10.1074/jbc.M113.462671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 50.Chouchani ET, Methner C, Nadtochiy SM, Logan A, Pell VR, Ding S, James AM, Cocheme HM, Reinhold J, Lilley KS, Partridge L, Fearnley IM, Robinson AJ, Hartley RC, Smith RAJ, Krieg T, Brookes PS, Murphy MP. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat Med. 2013;19:753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J. Biol. Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 52.Brubaker WD, Freites JA, Golchert KJ, Shapiro RA, Morikis V, Tobias, Douglas J, Martin RW. Separating Instability from Aggregation Propensity in γS-Crystallin Variants. Biophys. J. 2011;100:498–506. doi: 10.1016/j.bpj.2010.12.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H, Romieu I, Sienra-Monge J-J, Rio-Navarro B. E. d., Anderson DM, Jenchura CA, Li H, Ramirez-Aguilar M, Lara-Sanchez I. d. C., London SJ. Genetic Variation in S-nitrosoglutathione Reductase (GSNOR) and Childhood Asthma. J. Allerg. Clincal. Immunol. 2007;120:322–328. doi: 10.1016/j.jaci.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.