Abstract
STUDY QUESTION
Can spontaneous premature ovarian failure (POF) patients derived from population-based biobanks reveal the association between copy number variations (CNVs) and POF?
SUMMARY ANSWER
CNVs can hamper the functional capacity of ovaries by disrupting key genes and pathways essential for proper ovarian function.
WHAT IS KNOWN ALREADY
POF is defined as the cessation of ovarian function before the age of 40 years. POF is a major reason for female infertility, although its cause remains largely unknown.
STUDY DESIGN, SIZE, DURATION
The current retrospective CNV study included 301 spontaneous POF patients and 3188 control individuals registered between 2003 and 2014 at Estonian Genome Center at the University of Tartu (EGCUT) biobank.
PARTICIPANTS/MATERIALS, SETTING, METHODS
DNA samples from 301 spontaneous POF patients were genotyped by Illumina HumanCoreExome (258 samples) and HumanOmniExpress (43 samples) BeadChip arrays. Genotype and phenotype information was drawn from the EGCUT for the 3188 control population samples, previously genotyped with HumanCNV370 and HumanOmniExpress BeadChip arrays. All identified CNVs were subjected to functional enrichment studies for highlighting the POF pathogenesis. Real-time quantitative PCR was used to validate a subset of CNVs. Whole-exome sequencing was performed on six patients carrying hemizygous deletions that encompass genes essential for meiosis or folliculogenesis.
MAIN RESULTS AND THE ROLE OF CHANCE
Eleven novel microdeletions and microduplications that encompass genes relevant to POF were identified. For example, FMN2 (1q43) and SGOL2 (2q33.1) are essential for meiotic progression, while TBP (6q27), SCARB1 (12q24.31), BNC1 (15q25) and ARFGAP3 (22q13.2) are involved in follicular growth and oocyte maturation. The importance of recently discovered hemizygous microdeletions of meiotic genes SYCE1 (10q26.3) and CPEB1 (15q25.2) in POF patients was also corroborated.
LIMITATIONS, REASONS FOR CAUTION
This is a descriptive analysis and no functional studies were performed. Anamnestic data obtained from population-based biobank lacked clinical, biological (hormone levels) or ultrasonographical data, and spontaneous POF was predicted retrospectively by excluding known extraovarian causes for premature menopause.
WIDER IMPLICATIONS OF THE FINDINGS
The present study, with high number of spontaneous POF cases, provides novel data on associations between the genomic aberrations and premature menopause of ovarian cause and demonstrates that population-based biobanks are powerful source of biological samples and clinical data to reveal novel genetic lesions associated with human reproductive health and disease, including POF.
STUDY FUNDING/COMPETING INTEREST
This study was supported by the Estonian Ministry of Education and Research (IUT20-43, IUT20-60, IUT34-16, SF0180027s10 and 9205), Enterprise Estonia (EU30020 and EU48695), Eureka's EUROSTARS programme (NOTED, EU41564), grants from European Union's FP7 Marie Curie Industry-Academia Partnerships and Pathways (IAPP, SARM, |EU324509) and Horizon 2020 innovation programme (WIDENLIFE, 692065), Academy of Finland and the Sigrid Juselius Foundation.
Keywords: copy number variation, CNV, menopause, premature ovarian failure, POF, SNP genotyping, population-based biobank
Introduction
Menopause is a normal part of the female aging process. The median age of natural menopause is approximately 51 years, although variance in age is strongly determined by genetic (de Bruin et al., 2001) and environmental factors such as smoking, parity and diet (Lund, 2008). However, ∼1% of women experience premature menopause with cessation of ovarian function before the age of 40 (Haller-Kikkatalo et al., 2015), a condition commonly known as premature ovarian failure (POF). Owing to estrogen deficiency, POF patients suffer from symptoms similar to menopause, but also present a higher risk of developing osteoporosis, cardiovascular diseases and neurodegenerative disorders (Goswami and Conway, 2005). However, the most widely recognized effects of POF concern the reproductive system. The first symptoms that usually precede POF are irregular menstrual cycles (Haller-Kikkatalo et al., 2015) and female infertility or poor outcome of infertility treatment (de Boer et al., 2003).
The designation of POF has been under discussion for a long time, but it is still preferred among many alternatives, including ovarian insufficiency, dysfunction or insult (Shelling, 2010). POF is highly heterogeneous and can be associated with iatrogenic factors such as oophorectomy, ovarian resection or cystectomy reducing ovarian reserve (Busacca et al., 2006) or chemotherapy, and systemic diseases (Group, 2002). Furthermore, to minimize misinterpretation (Shah and Nagarajan, 2014) we use the term ‘spontaneous POF’ to define retrospectively ovarian quiescence in patients younger than 40 who do not present with any other known extraovarian causes for premature menopause. Therefore, spontaneous POF can arise as a result of noniatrogenic conditions affecting primarily ovaries, and include autoimmune reactions (Monnier-Barbarino et al., 2005) and genetic factors (Pouresmaeili and Fazeli, 2014).
Studies of familial spontaneous POF cases showed that genetic background strongly influences the onset of POF with familial incidences ranging from around 4% up to 30% of cases (Cramer et al., 1995; Conway et al., 1996; Vegetti et al., 1998). Disorders of the X chromosome are considered to be one of the main causes of POF, and conventional karyotyping has identified several X chromosome regions containing genes required for normal ovarian function (Beke et al., 2013). Mutations in X-linked genes have been described and include FMR1 (MIM*309550), BMP15 (MIM*300247) and DACH2 (MIM*300608). In addition, some autosomal genes are associated with POF, such as GDF9 (MIM*601918), FSHR (MIM*136435), LHR (MIM*152790) and NR5A1 (MIM*184757) (reviewed in Persani et al., 2010). The association between mutations in meiotic genes and ovarian decline has also been shown in POF patients (Lacombe et al., 2006; Caburet et al., 2014) and women with 46,XX gonadal dysgenesis (Zangen et al., 2011), highlighting the importance of meiotic genes in the regulation of ovarian function. Nevertheless, the cause of premature menopause remains unknown in many cases.
Advanced DNA microarray technologies have highlighted the importance of submicroscopic copy number variations (CNVs) in the pathogenesis of rare and complex diseases (Vulto-van Silfhout et al., 2013). CNVs represent deletions, duplications and insertions that are >1 kb in size and are present in variable copy number compared with the reference human genome (Feuk et al., 2006). Associations between CNVs and a certain disease may reveal novel candidate regions and genes involved in the pathogenesis of the disease. A limited number of studies have already demonstrated a possible association between the POF phenotype and CNVs. However, most of these studies used low-resolution array comparative genome hybridization (aCGH) platforms with a mean spatial resolution of 0.7 Mb or average probe spacing of 21.7 kb (Aboura et al., 2009; Ledig et al., 2010), or targeted the X chromosome only (Quilter et al., 2010; Knauff et al., 2011).
Until now, only two CNV studies in POF patients used high-resolution array platforms, either whole-genome SNP array (McGuire et al., 2011) or a custom-designed aCGH with average probe spacing of 2.2 kb that targeted genes implicated in gender development (Norling et al., 2014). Using high-resolution arrays, these studies were able to identify novel microdeletions and microduplications previously not associated with POF. However, the cohort sizes of these studies remained relatively modest, and involved 89 and 26 patients, respectively. Thus, in the current study we used high-resolution SNP genotyping of 301 spontaneous POF patients derived from Estonian Genome Center at the University of Tartu (EGCUT) biobank to reveal novel CNV regions and candidate genes related to POF phenotype. In addition, whole-exome sequencing (WES) was performed on genomic DNA of six women with spontaneous POF carrying hemizygous deletions to determine whether these deletions can result in unmasking of recessive mutations in POF candidate genes in undeleted regions of the homologous chromosomes, or reveal other POF-associated variants in the exome. The data reported here reveal new associations between rare CNVs and susceptibility to spontaneous POF. Furthermore, our study indicates that using samples from population-based biobanks may be a useful and efficient way to recruit patients for large-scale retrospective CNV and disease association studies.
Materials and Methods
Ethical approval
The Ethics Review Committee on Human Research of the University of Tartu and Scientific Committee of the EGCUT approved this study.
Population-depictive cohort samples from EGCUT
EGCUT is the population-based biobank (www.geenivaramu.ee, 6 June 2016, date last accessed) formed according to the Estonian Gene Research Act and all participants have signed the broad informed consent. The cohort of EGCUT currently includes >51 000 gene donors over 18 years of age. EGCUT represents 5% of Estonian population and closely reflects the age, gender and geographical distribution of the Estonian population. All subjects were recruited randomly and voluntarily by general practitioners and physicians in hospitals. A Computer Assisted Personal Interview (CAPI) was completed at a doctor's office and included personal, genealogical, educational, occupational history and lifestyle data. Anthropometric measurements, blood pressure and resting heart rate were measured, and venous blood was taken during the visit. Medical history, including parameters of reproductive health, was collected retrospectively during the recruitment. All past and current medical conditions were recorded according to the International Statistical Classification of Diseases (ICD-10) codes.
Women with spontaneous POF
Participants of the study were selected from 34 041 women at the EGCUT according to phenotype. The ovarian cause for spontaneous premature menopause was considered if secondary amenorrhea occurred before the age of 40 years, but no exclusive criteria were reported by participants or recorded by family physicians. The exclusive criteria were: iatrogenic manipulation (surgical hysterectomy, bilateral oophorectomy or unilateral oophorectomy diminishing ovarian reserve, use of oral contraceptives or other medications causing amenorrhea), comorbidities affecting menstrual cycle (including obesity with body mass index (BMI) ≥30 kg/m2, hyperprolactinemia, renal failure and HIV infection), sex chromosome abnormalities (including Turner syndrome, 46,XY or 47,XXX), developmental disorders (uterine agenesis), as well as primary amenorrhea. As a result of the selection, this study included a total of 301 women with spontaneous POF (Table I). The average age at the time of the study was 57.8 ± 12.5 years (mean ± SD) and on average, 20.7 years had passed since the onset of menopause, which started at an average age of 37.2 years. The average BMI value by the time of study was 28.9 kg/m2, indicating overweight among participants (BMI 25.1–30 kg/m2 indicates overweight and BMI ≥ 30 kg/m2 stands for obesity). Also, the waist-to-hip ratio of study participants exceeded 0.8, indicating a moderately increased risk for being overweight (0.8–0.85 indicates moderate risk, while >0.85 indicates substantial risk).
Table I.
Anamnestic data of 301 women with spontaneous premature ovarian failure (POF).
| Age at study (years) | 57.8 ± 12.5 |
| Duration of amenorrhea (years) | 20.7 ± 11.6 |
| Smoking status (n) | |
| Never smoked | 206 (68.4%, 62.8–73.6) |
| Former smoker | 27 (9.0%, 6.1–12.9) |
| Current smoker | 68 (22.6%, 18.1–27.8) |
| Height (cm) | 163.0 ± 6.5 |
| BMI (kg/m2) | 28.9 ± 6.7 |
| Waist-to-hip ratio | 0.84 ± 0.08 |
| Age at menarche (years) | 13.8 ± 1.7 |
| Length of menstrual cycle at age 25–35 | |
| 25–29 days | 174 (57.8%, 52.0–63.4) |
| ≤20 days | 8 (2.7%, 1.2–5.4) |
| 21–24 days | 57 (18.9%, 14.8–23.9) |
| 30–35 days | 12 (4.0%, 2.2–7.0) |
| >35 days | 1 (0.3%, 0–2.1) |
| Irregular | 25 (8.3%, 5.6–12.2) |
| Do not know | 12 (4.0%, 2.2–7.0) |
| Duration of fertility (years) | 23.6 ± 5.1 |
| Age of 1st pregnancy (years) | 22.5 ± 3.6 |
| No. of pregnancies | 3.6 ± 2.0 |
| No. of live births | 2.0 ± 1.1 |
| Age at menopause (years) | 37.2 ± 4.4 |
Data are means ± SD or count (percentage, 95% CI of percentage).
Since the EGCUT cohort is a volunteer-based longitudinal population biobank, additional kinship analysis using the web-based PLINK v1.07 toolset (http://pngu.mgh.harvard.edu/purcell/plink/, 6 June 2016, date last accessed) was performed (Purcell et al., 2007) to identify possible familial cases among the POF patients.
Reference data
The Database of Genomic Variants (DGV; http://dgv.tcag.ca/, 6 June 2016, date last accessed) was used as a reference dataset to exclude benign polymorphisms. DGV holds information on common CNVs found in more than 20 000 healthy control samples and serves as a catalog of control data for correlating genomic variation with phenotypic data (MacDonald et al., 2013). In addition, genotype and phenotype information was drawn for the Estonian general population samples (n = 3188) from the EGCUT, previously subjected to SNP genotyping with HumanCNV370 (n = 489) and HumanOmniExpress (n = 2699) BeadChip arrays (Illumina, Inc., San Diego, CA, USA) to determine and exclude benign population-specific CNV regions. The derived EGCUT dataset represented control group of women with normal age at menopause, as the group included >41 years old pre- and post-menopausal women, but excluded potential POF cases.
SNP genotyping and CNV calling
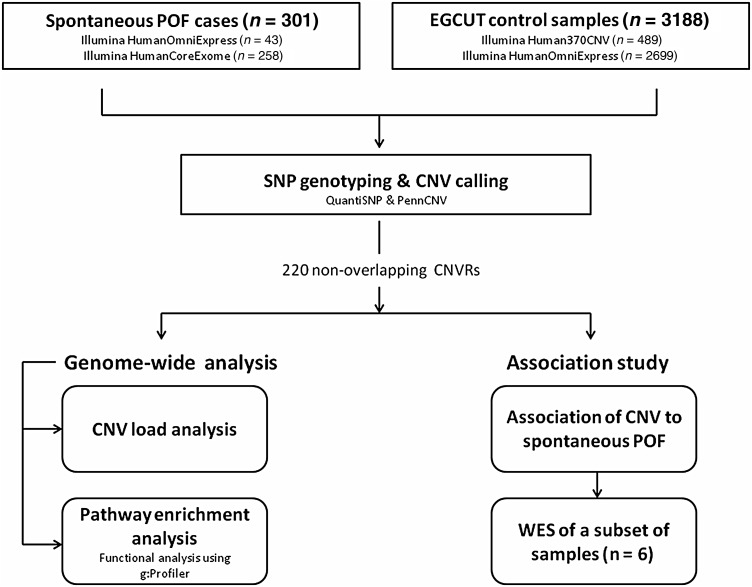
The study design is represented schematically in Fig. 1. Infinium® II whole-genome genotyping assay with HumanOmniExpress (n = 43) or HumanCoreExome (n = 258) BeadChip arrays (Illumina, Inc.) were used for whole-genome SNP genotyping. The HumanOmniExpress BeadChip contains >715 000 SNP markers with median marker spacing of 2.1 kb, while HumanCoreExome BeadChip has 547 644 markers, with 265 919 of them covering exonic regions, and a median marker spacing of 1.9 kb. SNP genotyping wet-lab experiments were performed according to the manufacturer's protocols.
Figure 1.
Schematic representation of study design. A total of 301 spontaneous premature ovarian failure (POF) patients were enrolled in the study. After single nucleotide polymorphism (SNP) genotyping and copy number variation (CNV) calling, genome-wide CNV load analysis was performed per individual to estimate the effect of CNV load in the genome on time of menopause. Subsequently, all the detected CNVs were compared against the EGCUT control population (n = 3188) to identify nonoverlapping CNV regions (CNVRs) (n = 220), for which functional enrichment and association studies were performed. After the following interpretation of each CNVR, six individuals with hemizygous deletions were also selected for whole-exome sequencing (WES) to determine whether identified deletions can result in unmasking recessive mutations or other POF-associated variants in the exome.
Genotypes were called by GenomeStudio software Genotyping Module v.3.1 (Illumina, Inc.). A call rate of >98% was accepted as the primary quality control for each sample on both arrays. Log R Ratio and B Allele Frequency values generated by the GenomeStudio software were applied in further CNV calling using two independent Hidden Markov Model-based algorithms: QuantiSNP v.2.1 (Colella et al., 2007) and PennCNV (Wang et al., 2007). Parameters suggested by the authors were applied to each algorithm. The genomic size threshold for CNVs was set to 50 kb for the HumanOmniExpress array and to 100 kb for the HumanCoreExome array, and CNV contained at least 10 consecutive SNP markers. To minimize the number of false-positive findings, initial CNV calls from the two algorithms were merged and only CNVs called by both algorithms and visually confirmed in GenomeStudio Genome Viewer were considered for further data interpretation. All CNVs identified in this study were submitted to ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/, 6 June 2016, date last accessed; accession numbers SCV000212280 to SCV000212499).
For the EGCUT control group, the HumanCNV370 and HumanOmniExpress microarray data were processed in the same manner by performing independent CNV calling using QuantiSNP and PennCNV and by considering only CNV regions that were called by both algorithms.
Interpretation of CNV results
The genomic context of aberrant regions was studied using the UCSC Genome Browser database (http://genome.ucsc.edu/) along with the OMIM database (www.omim.org, 6 June 2016, date last accessed) to evaluate the potential clinical relevance of a particular CNV. The interpretation of CNVs was performed according to the ACMG Guidelines (Kearney et al., 2011). Briefly, chromosomal aberrations were deemed clinically significant if they overlapped with a genomic region associated with a well-established syndrome or contained a gene or a part of a gene implicated in a known disorder. Findings were considered likely benign if they were present in healthy individuals (e.g. >1% of Estonian population or DGV) and/or they were gene deserts or gene-poor regions and/or did not encompass any previously known disease-associated genes. All remaining findings were categorized as variants of unknown clinical significance.
Functional analysis of results
Functional annotation and enrichment analysis was carried out using g:Profiler gGOSt web-based software (http://biit.cs.ut.ee/gprofiler/, 6 June 2016, date last accessed) (Reimand et al., 2007, 2011). The g:Profiler public web server provides a comprehensive set of functional annotation tools for clustering functionally related genes according to different criteria such as Gene Ontology terms and biological pathways. Enrichment for functional terms was considered to be statistically significant, if the multiple testing corrected P-value was <0.05. A 20 Mb hemizygous deletion on the X chromosome, found in one of the POF patients, was excluded from the analysis to avoid bias.
CNV validation
Confirmation of a subset of CNVs (n = 14) was done using SYBR-Green based real-time quantitative PCR (qPCR). CNVs were selected for experimental confirmation if they met the following criteria: (i) there were no data reported in the DGV and/or CNV was present in <1% of Estonian population, and (ii) CNV harbored previously reported POF regions and genes, or contained genes that may potentially impact the age at menopause based on their biological function. Primers were custom-designed in a web-based freeware Primer3web (http://primer3.ut.ee/, 6 June 2016, date last accessed) (Untergasser et al., 2012). A full list of primers used for qPCR analysis is available upon request. The analysis was performed on a 7900 HT Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA). These data were acquired using SDS 2.2.2 software (Applied Biosystems) and further processed using qBase+ (Biogazelle, Ghent, Belgium). Analysis was performed as relative quantification using the Pfaffl method of calculation while taking into account the amplification efficiencies of each primer pair (Pfaffl, 2001). All selected CNVs have been successfully validated.
Whole-exome sequencing and data analysis
Genomic libraries were prepared according to the Illumina Nextera Rapid Capture Exome protocol (Illumina, Inc.). The captured libraries were sequenced on HiSeq2500 platform (Illumina, Inc.) with 2 × 101 bp paired-end reads, with 92.7% of bases sequenced above the quality of Q30. Demultiplexing was done with CASAVA 1.8.2. (Illumina, Inc.) allowing 1 mismatch in 8 bp index read. Number of reads varied between samples from 45 to 56 M.
Sequenced reads were aligned to the human reference genome (hg19, GRCh37) with the Burrows-Wheeler Aligner (BWA, version 0.6.1). SAMtools (version 0.1.18) was used to filter out reads marked as PCR duplicates and reads not in a proper read pair. Single-nucleotide substitutions and small indel variants were then called with GATK, after which variant sites more than 50 bp away from the nearest exome sequence capture target or with quality score <20 were filtered out. Variants were annotated using in-house developed scripts based on Ensembl API release 75, 1000 Genomes, dbSNP138, ExAC Browser, BIOBASE's Genome Trax™ database, and in-house whole-exome and whole-genome databases.
Statistical analysis
The R3.1.0 a Language and Environment (Free Software Foundation, Boston, MA, USA) was used for statistical analysis. The Welch two-sample t-test and the Wilcoxon rank sum test with continuity correction were used to assess differences in number and size for CNV calling between two SNP-arrays. Multivariate linear regression models adjusted for confounders were used to assess the associations between the number and size of CNVs and anamnestic data. A P-value of <0.05 was considered statistically significant.
Results
The association between CNV load and anamnestic data
After applying stringent filtering criteria of initial CNV calls, a total of 346 CNVs were detected on both the X chromosome and autosomal chromosomes in women with spontaneous POF. CNVs identified by both algorithms were spread over the entire genome and included 132 copy number losses (38.2%) and 214 copy number gains (61.8%).
To establish the association between the CNV load and general reproductive fitness, all identified CNVs in POF patients and women from the control group who have reached menopause (n = 1735) were subjected to a global genome-wide CNV load analysis per individual. Linear regression model adjusted by the age of menopause and genotyping array attributes did not reveal any difference in the number of CNVs detected between POF patients and controls (adjusted regression coefficient (ad r) = −0.012, P = 0.329). Similarly, there were no significant difference in the cumulative size of CNVs detected among POF patients compared with the controls (ad r = −77 658, P = 0.477). Linear regression model adjusted by genotyping array attributes revealed a negative association between the number of CNVs in the genome and the age of menopause (ad r = −0.04 CNVs per 1 year difference in menopause, P = 0.010) in a group of POF patients. When the same model was used for the control population, no association between the number of CNVs in the genome and the age of menopause was observed (ad r = −0.005 CNVs per 1 year difference in menopause, P = 0.709). Also, since the age at menopause determined the duration of fertility (r = 0.77, P < 0.00001 for POF patients and r = 0.84, P < 0.00001 for the control group), the number of CNVs was also negatively associated with the duration of fertility in POF patients (ad r = −0.04, P = 0.0008), but not in the control group (ad r = −0.016, P = 0.216), when the model was controlled by genotyping array attributes. Linear regression models were not able to detect any relationships between the number of CNVs and the length or the regularity of menstrual cycles, the number of pregnancies or live births, or parameters of body structure. The cumulative size of CNVs was positively associated with the age at first pregnancy if confounders affecting the age of pregnancy were considered in the statistical analysis (linear regression adjusted by age of menarche, number of live births and age at study, adjusted r = 80.1 kb per one additional year of the time to first pregnancy; P = 0.037). The mean age ± SD of first pregnancy was 22.5 ± 3.6 years and ranged from 15 to 36 years. The association was the same if the CNV detection genotyping array attributes were also considered (data not shown). This result suggests that overall CNV load can affect reproductive fitness in terms of the time of the first pregnancy. However, since confounding factors were not available for the control population, we were not able to perform similar association study between the cumulative sizes of the CNVs per genome with the age at first pregnancy.
Functional profiling of genes within CNV regions identifies enrichment in pathways related to the immune system functions
All identified CNVs were clustered into 95 deleted and 125 duplicated nonoverlapping copy number variation regions (CNVRs), respectively (Supplementary data, Table SI). Functional enrichment analysis on CNVRs was performed using the g:Profiler web-based software. Results of Gene Ontology (GO), KEGG pathway and Reactome (REAC) datasets with up to third hierarchical level are provided in Table II. The analysis revealed statistically significant networks related to immunological processes. In particular, a notable number of genes were involved in chemotaxis of lymphocytes (20.0% of genes, P = 2.83 × 10−2), monocytes (20.4% of genes, P = 8.49 × 10−3) and eosinophils (43.5% of genes, P = 7.68 × 10−6). Alterations in KEGG pathway of ‘chemokine signaling’ (P = 4.11 × 10−2), and REAC pathways of ‘innate immune system’ (P = 2.11 × 10−3), ‘adaptive immune system’ (P = 7.48 × 10−5) and ‘scavenging of heme from plasma’ (P = 2.41 × 10−30) were observed in our spontaneous POF cases. None of these functional pathways were affected by chromosomal structural rearrangements in women with normal age at menopause, when a similar analysis was performed for EGCUT control individuals. These results may underlie the dysregulation of immune system in patients with spontaneous POF, as numerous interactions between the immune system and ovaries are involved in the regulation of the hypothalamic–pituitary–ovarian axis and maintaining growth and regression of both follicles and corpus luteum (Haller-Kikkatalo et al., 2012).
Table II.
Significant networks identified by pathway analysis in spontaneous premature ovarian failure (POF) patients.
| Term ID | Term name | Level | Adjusted P-valuea |
|---|---|---|---|
| GO:0048247 | Lymphocyte chemotaxis | 1 | 2.83 × 10−2 |
| GO:0002548 | Monocyte chemotaxis | 1 | 8.49 × 10−3 |
| GO:0072677 | Eosinophil migration | 1 | 4.96 × 10−5 |
| GO:0048245 | Eosinophil chemotaxis | 1.1 | 7.68 × 10−6 |
| KEGG:04062 | Chemokine signaling pathway | 1 | 4.11 × 10−2 |
| REAC:168249 | Innate immune system | 1 | 2.11 × 10−3 |
| REAC:2454202 | Fc epsilon receptor (FCERI) signaling | 1.1 | 5.76 × 10−7 |
| REAC:2029480 | Fc gamma receptor (FCGR) dependent phagocytosis | 1.2 | 2.00 × 10−19 |
| REAC:166658 | Complement cascade | 1.3 | 2.41 × 10−27 |
| REAC:5653656 | Vesicle-mediated transport | 1 | 5.80 × 10−10 |
| REAC:2173782 | Binding and uptake of ligands by scavenger receptors | 1.1 | 3.68 × 10−23 |
| REAC:2168880 | Scavenging of heme from plasma | 1.1.1 | 2.41 × 10−30 |
| REAC:1280218 | Adaptive immune system | 1 | 7.48 × 10−5 |
| REAC:198933 | Immunoregulatory interactions between a lymphoid and a nonlymphoid cell | 1.1 | 2.05 × 10−15 |
Functional enrichment analysis of genes within rearranged regions was performed using the web-based g:Profiler software. Gene Ontology (GO), KEGG pathway and Reactome (REAC) datasets with adjusted P-value < 0.05 were considered to be statistically significant.
aMultiple testing corrected enrichment P-value.
Detection of potentially relevant regions for spontaneous POF
Subsequently, clustered CNVRs were screened against the DGV database to distinguish benign copy number changes from rare POF-associated CNVs. However, the exclusion of benign polymorphisms was done with caution, as genetic imbalances affecting gender development can depend on the sex of the individual, and this information is not always available for individuals included in the DGV. In addition, the Estonian population-based control group was used to exclude common variants in the Estonian population. Therefore, in subsequent case-by-case analyses, 93 out of 125 microduplications and 72 out of 95 microdeletions were classified as benign polymorphisms, and the remaining 55 CNVRs were considered to be rare copy number changes, as they were not reported in DGV nor present in the EGCUT control group (Supplementary data, Table SII). Emphasis for selecting CNVs for subsequent validation by qPCR was placed on unique aberrations that harbored previously reported POF candidate genes, or contained genes that may potentially impact the age at menopause based on their biological function. Using these criteria, 14 CNVs were selected as relevant in the susceptibility to POF, and were successfully validated by qPCR. Data from these CNVs, as well as the 20 Mb hemizygous X chromosome deletion, are summarized in Table III, while phenotypic data of CNV carriers are given in Table IV. Overlapping aberrations previously reported in POF patients were identified in five cases: two patients had duplications in the Xp22.31 region (Case14 and Case15) (Quilter et al., 2010), two patients had a 10q26.3 hemizygous deletion (Case7 and Case8) with identical breakpoints (based on microarray data), and one patient had a 15q25.2 hemizygous deletion (Case10) (McGuire et al., 2011). All of these women were unrelated according to kinship analysis. The remaining rare 11 CNVs identified in our study were considered to be novel.
Table III.
Summary of validated copy number variations (CNVs) in spontaneous premature ovarian failure (POF) cases.
| Locus | Position (hg19) | Length (kb) | CN | Probe count | Genes within CNV | Cases (n = 301) | Controls (n = 3188) | CNV carriers among cases (%) | CNV carriers among controls (%) | Case ID |
|---|---|---|---|---|---|---|---|---|---|---|
| 1q43 | chr1: 240341241–240561727 | 189.9 | 1 | 37 | FMN2 | 1 | 1 | 0.33 | 0.03 | Case1 |
| 2p13.11 | chr2: 73828493–73900329 | 71.8 | 1 | 13 | ALMS1, NAT8 | 1 | 0 | 0.33 | 0 | Case2 |
| 2q33.1 | chr2: 200250898–201845999 | 1595.1 | 3 | 451 | SATB2, FTCDNL1, C2orf69, TYW5, C2orf47, SPATS2L, KCTD18, SGOL2, AOX1, AOX2P, BZW1, CLK1, PPIL3, NIF3L1, ORC2, FAM126B | 1 | 1 | 0.33 | 0.03 | Case3 |
| 6q27 | chr6: 170713690–170890384 | 176.7 | 3 | 70 | FAM120B, PSMB1, TBP, PDCD2 | 1 | 1 | 0.33 | 0.03 | Case4 |
| 7p14.3 | chr7: 33639870–33730376 | 90.5 | 1 | 30 | BBS9 | 1 | 10 | 0.33 | 0.31 | Case5 |
| 9q22.31 | chr9: 95063947–95179836 | 115.9 | 1 | 70 | NOL8, CENPP, OGN, OMD | 1 | 1 | 0.33 | 0.03 | Case6 |
| 10q26.3c | chr10: 135256762–135379710 | 122.9 | 1 | 70 | CYP2E1, SYCE1 | 2 | 3 | 0.66 | 0.09 | Case7 Case8 |
| 12q24.31 | chr12: 125260645–125321461 | 60.8 | 3 | 13 | SCARB1 | 1 | 1 | 0.33 | 0.03 | Case9 |
| 15q25.2c | chr15: 83213963–84811815 | 1597.8 | 1 | 325 | CPEB1, AP3B2, FSD2, WHAMM, HOMER2, FAM103A1, C15orf40, BTBD1, TM6SF1, HDGFRP3, BNC1, SH3GL3, ADAMTSL3, EFTUD1P1 | 1 | 0 | 0.33 | 0 | Case10 |
| 16p13.12 | chr16: 13889247–14163635 | 274.3 | 3 | 35 | ERCC4 | 1 | 0 | 0.33 | 0 | Case11 |
| 17q12 | chr17: ×–36220373 | 1404.8 | 3 | 327 | ZNHIT3, MYO19, PIGW, GGNBP2, DHRS11, MRM1, LHX1, AATF, ACACA, C17orf78, TADA2A, DUSP14, SYNRG, DDX52, HNF1B | 1 | 8 | 0.33 | 0.25 | Case12 |
| 22q13.2 | chr22: 43122720–43500212 | 377.5 | 1 | 54 | ARFGAP3, PACSIN2, TTLL1 | 1 | 3 | 0.33 | 0.09 | Case13 |
| Xp22.31d | chrX: 6516735–8138080 | 1618.3 | 3 | 111 | HDHD1, STS, VCX, PNPLA4 | 2 | 21b | 0.66 | 0.66 | Case14 Case15 |
| Xq12 | chrX: 66905875–67475065 | 569.2 | 3 | 20 | AR, OPHN1 | 1 | 1 | 0.33 | 0.03 | Case16 |
| Xq22.1-q24 | chrX: 99931059–120328627 | 20397.6 | 1 | 2001 | SYTL4…GLUD2a | 1 | 0 | 0.33 | 0 | Case17 |
All the discrete regions have been validated by qPCR, except for 20 Mb Xq22.1-q24 deletion; del, deletion; dup, duplication; CN, copy number.
a>100 genes in total, first and last genes in the region are indicated.
bSTS gene is not covered in any of these 21 control individuals with overlapping Xp22.31 duplications.
cFirst reported by McGuire et al (2010).
dFirst reported by Quilter et al (2010); loci indicated in bold were chosen for subsequent whole-exome sequencing in corresponding CNV carriers.
Table IV.
Phenotype data of premature ovarian failure (POF) patients carrying aberrations with potentially clinical significance.
| Case ID | Locus | CN | Phenotype |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Age at study (years) | Age of menarche (years) | Age at menopause (yr) | No. of pregnancies | No. of live births | Smoking status | Concomitant diseases (age of diagnose) | |||
| Case1 | 1q43 | 1 | 71 | 18 | 40a | 4 | 2 | Never smoked | Hypothyroidism (24 yr); primary hypertension (67 yr) |
| Case2 | 2p13.11 | 1 | 53 | 11 | 39 | 3 | 2 | Never smoked | Adiposity (42 yr); hypertension with heart failure (50 yr) |
| Case3 | 2q33.1 | 3 | 41 | 11 | 33 | 5 | 2 | Never smoked | Unspecified arthrosis (35 yr) |
| Case4 | 6q27 | 3 | 39 | 12 | 24 | 2 | 1 (IVF) | Current smoker | Arthritis (24 yr); asthma (29 yr) |
| Case5 | 7p14.3 | 1 | 58 | 15 | 32 | 4 | 2 | Never smoked | Allergic contact dermatitis (45 yr); coxarthrosis and unspecified rheumatism (52 yr); post-menopausal osteoporosis (53 yr); primary hypertension (55 yr) |
| Case6 | 9q22.31 | 1 | 68 | 15 | 38 | 2 | 1 | Never smoked | – |
| Case7 | 10q26.3 | 1 | 56 | 13 | 38 | 6 | 3 | Never smoked | Allergic contact dermatitis (45 yr); hypertension with heart failure (50 yr); unspecified polyarthritis (57 yr); |
| Case8 | 10q26.3 | 1 | 69 | 18 | 30 | 0 | 0 | Never smoked | Primary bilateral gonarthrosis (49 yr); hypertension with heart failure (52 yr); familial hypercholesterolemia (62 yr); Type 2 diabetes (67 yr) |
| Case9 | 12q24.31 | 3 | 51 | 16 | 40 | 1 | 1 | Former smoker | – |
| Case10 | 15q25.2 | 1 | 55 | 13 | 38 | 3 | 2 | Never smoked | – |
| Case11 | 16p13.12 | 3 | 50 | 15 | 39 | 5 | 3 | Current smoker | Adiposity (41 yr) |
| Case12 | 17q12 | 3 | 74 | 15 | 39 | 2 | 2 | Never smoked | Hypertension with heart failure (73 yr) |
| Case13 | 22q13.2 | 1 | 47 | 13 | 37 | 0 | 0 | Current smoker | – |
| Case14 | Xp22.31 | 3 | 52 | 13 | 40 | 1 | 1 | Current smoker | Spondylosis with radiculopathy of unspecified location (33 yr) |
| Case15 | Xp22.31 | 3 | 37 | 17 | 37 | 3 | 2 | Current smoker | – |
| Case16 | Xq12 | 3 | 31 | 13 | 25 | 3 | 3 | Current smoker | – |
| Case17 | Xq22.1-q24 | 1 | 52 | 12 | 40 | 3 | 2 | Never smoked | – |
Subsequent whole-exome sequencing was performed for cases indicated in bold; yr, year.
aAge of 39 years and 6–11 months was rounded to 40 years.
Whole-exome sequencing data of six patients carrying hemizygous deletions
WES was performed on six female carriers of hemizygous deletions (Tables III and IV) that encompass genes essential for meiosis or folliculogenesis. The average read depth for sequenced exome was 82×. WES analysis did not reveal any rare POF-associated homozygous variants in genes within the CNV regions based on ExAC database and in-house whole-exome and whole-genome reference databases (Supplementary data, Table SIII). Additionally, WES identified heterozygous missense mutation in GDF9 (growth differentiation factor 9 at 5q31.1, rs61754582, c.1360C > T; p.Arg454Cys) in Case13, when analyzing whole exome data outside of CNV regions. The prevalence of GDF9 mutations in POF patients is estimated to be ∼1–4%, (Dixit et al., 2005; Laissue et al., 2006; Kovanci et al., 2007; Zhao et al., 2007); however, this particular variant is present in 0.4% of European population, thus most likely representing a nonpathogenic variant. In addition, secondary findings were identified by WES that might contribute to comorbid phenotypic features diagnosed in POF patients (Table V).
Table V.
Secondary findings identified by whole-exome sequencing in six premature ovarian failure (POF) patients.
| Case ID | Gene | Transcript | Chr | Position (hg19) | rsID | Zygosity | cDNA | Protein | ClinVar accession | Associated phenotype | Variant involved in patient's phenotypea |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case1 | SCN9A | NM_002977.3 | 2 | 167136962 | rs182650126 | Het | c.2215A>G | p.Ile739Val | SCV000191928.1 | Small fiber neuropathy | Not likely |
| Case1 | TMEM43 | NM_024334.2 | 3 | 14180731 | rs113449357 | Het | c.934C>T | p.Arg312Trp | SCV000051602.1 | Cardiomyopathy | Not likely |
| Case6 | SPINK1 | NM_003122.4 | 5 | 147207583 | rs148954387 | Het | c.194+2T>C | p.(-) | SCV000253884.1 | Hereditary pancreatitis | Likely |
| Case6 | NEBL | NM_006393.2 | 10 | 21157673 | rs137973321 | Het | c.604G>A | p.Gly202Arg | SCV000062382.3 | Cardiomyopathy | Uncertain significance |
| Case6 | MLH3 | NM_001040108.1 | 14 | 75514138 | rs28756990 | Het | c.2221G>T | p.Val741Phe | SCV000026082.1 | Endometrial carcinoma | Uncertain significance |
| Case6 | DSC2 | NM_004949.4 | 18 | 28672114 | rs144799937 | Het | c.304G>A | p.Glu102Lys | SCV000063116.3 | Cardiomyopathy | Uncertain significance |
| Case6 | GPR101 | NM_054021.1 | X | 136112910 | rs73637412 | Het | c.924G>C | p.Glu308Asp | SCV000203835.3 | Pituitary adenoma | Not likely |
| Case7 | CX3CR1 | NM_001171174.1 | 3 | 39307162 | rs3732378 | Het | c.839C>T | p.Thr280Met | SCV000028838.3 | Age-related macular degeneration | Likely |
| Case7 | JAK2 | NM_004972.3 | 9 | 5073770 | rs77375493 | Het | c.1849G>T | p.Val617Phe | NA | Thrombocythemia | Likely |
| Case7 | GPR101 | NM_054021.1 | X | 136112910 | rs73637412 | Het | c.924G>C | p.Glu308Asp | SCV000203835.3 | Pituitary adenoma | Not likely |
| Case8 | TNNC1 | NM_003280.2 | 3 | 52485426 | rs267607124 | Het | c.435C>A | p.Asp145Glu | SCV000209137.1 | Cardiomyopathy | Likely |
| Case8 | ANK2 | NM_001127493.1 | 4 | 114294462 | rs121912706 | Het | c.5434C>T | p.Arg1812Trp | SCV000223217.2 | Arrhythmia | Uncertain significance |
| Case10 | APOE | NM_000041.2 | 19 | 45412079 | rs7412 | Het | c.526C>T | p.Arg176Cys | NA | Hyperlipoproteinemia | Likely |
| Case13 | BCO1 | NM_017429.2 | 16 | 81298282 | rs119478057 | Het | c.509C>T | p.Thr170Met | SCV000025214.2 | Hypercarotenemia and vitamin A deficiency | Likely |
All data is available upon request. het, heterozygous; NA, not available.
aInvolvement of the variant on comorbid phenotypic features (not in POF syndrome) based on anamnestic data.
Discussion
The current study is the first retrospective genetic association study of spontaneous POF cases using population-based biobank samples. The main goal of this study was to investigate the role of CNVs in premature menopausal transition using phenotype and genotype data from the population-based biobank samples. We found that the overall CNV load might influence the age at which an individual transitions to menopause, probably due to an increased chance of disrupting key genes and pathways that are essential for normal ovarian function. The number of CNVs was negatively related with the age of menopause in POF cohort, meaning that in patients with premature menopause the transition occurred earlier if the genome contained more CNVs. In addition, large structural rearrangements on the X chromosome, and likely those on autosomes, may disrupt normal chromosomal pairing in meiosis, leading to germ cell death. Consequently, patients may develop POF due to the loss of germ cells (Schlessinger et al., 2002). Still, these associations were observed only in POF patients, but not in the controls, suggesting the disease-specific pattern. Therefore, more studies on healthy populations are warranted to address this question in greater detail.
In addition, it was observed that larger cumulative size of all CNVs per individual genome in POF patients was associated with later first pregnancies. The association was quite remarkable as each year of pregnancy delay was associated with 80 kb bigger size of cumulative CNV load. This analysis was controlled by confounders affecting the time of the first pregnancy. Among those, the menarche represented the potential starting time for pregnancies, the number of live births was considered to unify the choice for family size, while the age at study was considered to unify any differences in family planning between generations. As the result, these data suggest that the overall size of CNVs might be related to the fecundity of a woman, at least in the POF cohort.
Genetic profiling and functional enrichment analysis of genes within identified CNVRs of POF patients identified pathways related to immune system functions. A notable number of genes were associated with immune cells (e.g. monocytes, lymphocytes) that may influence the differentiation and maturation of germ and granulosa cells (Bukovsky et al., 1995, 2005; Koks et al., 2010). These findings are also consistent with other studies that implicate humoral and cellular autoimmune mechanisms in the etiology of POF (Hoek et al., 1997; Chernyshov et al., 2001). In addition, there was a significant alteration in the ‘scavenging of heme from plasma’ pathway (57.1% of genes, P = 2.41 × 10−30). Free heme has tissue-damaging properties, and it represents a source of redox-active iron that generates reactive oxygen species (ROS) and oxidative stress (Yamada et al., 2011). The accumulation of damage induced by ROS is believed to be involved in more rapid ovarian aging due to the reduced function of oocytes and granulosa cells (Tatone et al., 2008).
In a subsequent case-by-case analysis, we identified three previously reported aberrations and 11 novel rare microdeletions and microduplications that may contribute to spontaneous POF. As a major result, our data reveal a number of novel candidate genes that are affected by chromosomal rearrangements and may be potentially involved in the genetic pathways leading to premature menopause (Supplementary data, Table SIV). Since spontaneous POF refers to a nonspecific syndrome with many putative risk-genes/regions, the common CNVs are not expected to be prevalent among POF patients. Indeed; we saw that detected CNVs are unique to individual POF cases rather than being common to POF patient population. Therefore, identified CNVs are rare and do not succeed in statistical comparisons and P-values (all being >0.05, data not shown), and thus do not represent any statistically significant differences in the prevalence between the cases and the controls. Still, detected CNVs can potentially contribute to the onset of premature menopause based on the biological function of the genes within CNV regions. In addition; we corroborated the importance of recently discovered POF genomic regions, including hemizygous microdeletions of SYCE1 (synaptonemal complex central element protein 1 at 10q26.3, MIM*611486) and CPEB1 (cytoplasmic polyadenylation element-binding protein 1 at 15q25.2, MIM*607342) (McGuire et al., 2011). SYCE1 and CPEB1 have a crucial role in meiosis, maintaining synaptonemal complexes formed between homologous chromosomes (Bolcun-Filas et al., 2009; Zheng et al., 2010). The involvement of these genes in reproductive failure is supported by knockout mice studies, showing that knockout female mice are infertile due to meiotic arrest in meiosis I (Tay and Richter, 2001; Bolcun-Filas et al., 2009). Importantly, the deleted region on chromosome 15 also encompassed BNC1 (basonuclin 1; MIM*601930) that may also be essential for oogenesis, as oocyte morphology is affected in knockout mice, which leads to female subfertility (Ma et al., 2006).
Among the genes found within the novel microdeletions, two have a strong implication in the reproductive biology: FMN2 (formin 2 at 1q43; MIM*606373) in Case1 and ARFGAP3 (ADP-ribosylation factor GTPase activating protein 3 at 22q13.2, MIM*612439) in Case13. FMN2 plays a crucial role in cytoskeletal organization and establishment of cell polarity during oogenesis, which is essential for meiotic maturation and maintenance of oocyte asymmetry (Montaville et al., 2014). Knockout studies in mice revealed that Fmn2-deficient female mice have decreased fertility due to abnormal metaphase spindle position and abnormal first polar body formation during meiosis I (Leader et al., 2002). Notably, FMN2 was the only gene disrupted by CNV, and importance of FMN2 in human reproduction was also discussed in a case–control study of women with unexplained infertility (Ryley et al., 2005). The second gene, ARFGAP3, is an androgen-targeted gene that is thought to be involved in intracellular trafficking of proteins and in vesicular transport (Liu et al., 2001). In bovines, ARFGAP3 contributes to follicular growth, ovulation and/or luteinization (Ndiaye et al., 2005), and it might have a similar role in human. It is worth noting that whole-exome sequencing did not reveal any rare homozygous mutations relevant to POF on the remaining alleles of these deleted regions. This supports the hypothesis that haploinsufficiency of meiotic genes may facilitate germ cell loss, resulting in premature depletion of the follicular pool. In addition, secondary findings were identified by WES, and some of the conditions associated with the detected variant were also diagnosed in our POF patients (Table V). POF has an adverse long-term effect on female wellbeing, especially bone health, cardiovascular health and neurological function (Shuster et al., 2010); however, it is hard to evaluate retrospectively whether the identified variants directly contribute to the disease or the disease developed as a consequence of premature menopause.
Although the phenotypic consequences of genomic gains are usually not as straightforward as of deletions, they may impair meiosis or proper folliculogenesis by altering the gene dosage and expression level (Newman et al., 2015). Two genomic gains were detected on X chromosome, and included STS (steroid sulfatase at Xp22.31, MIM*300747) and androgen receptor (AR at Xq12, MIM*313700) genes. STS encodes steroid sulfatase that catalyses metabolic precursors for estrogens, androgens and cholesterol (Alperin and Shapiro, 1997), while androgens promote follicular growth at early stages and inhibit at final stages (Sen et al., 2014), similar to polycystic ovary syndrome (PCOS) (Azziz et al., 2009). Therefore, it is possible that duplications in these two regions may lead to imbalanced estrogen and/or androgen levels, subsequently leading to excess of follicular demise. Autosomal microduplications include meiotic gene SGOL2 (shugoshin-like 2 at 2q33.1, MIM*612425) that protects centromeric cohesion from premature separase-mediated cleavage in oocytes during meiosis I (Llano et al., 2008). Genomic gain at 6q27 encompasses TBP (TATA box binding protein, MIM*600075), and mouse studies have revealed that overexpression of TBP significantly lowers the rate of mouse oocyte progression to MII stage during meiosis, possibly due to a toxic effect on the cell (Sun et al., 2013). As a result of duplication in Case4, the overexpression of TBP can occur, causing the failure of oocyte maturation. The 6q27 region also includes PSMB1 (proteasome subunit, beta-type, 1, MIM*602017), associated with Sjögren's syndrome (Martinez-Gamboa et al., 2013), a chronic autoinflammatory disease, suggesting autoimmune reactions as the cause of this patient’s POF at the age of 24 years. SCARB1 (scavenger receptor class b, member 1 at 12q43; MIM*601040), detected in Case 9, mediates cholesterol transfer to and from high-density lipoprotein (HDL). It is noteworthy that this individual has been smoking 10 cigarettes per day for 30 years since the age of 20 years and is overweight (BMI = 30 kg/m2). SCARB1 knockout mice have abnormal HDLs, ovulation of dysfunctional oocytes and infertility (Miettinen et al., 2001). Since HDL is the only lipoprotein present in follicular fluid (Shalgi et al., 1973; Perret et al., 1985), it is possible that changes in HDL level due to SCARB1 overproduction, combined with smoking, may disturb oocyte maturation or function, and thus contribute to infertility. However, all the gene-dosage hypotheses presented here remain to be tested by future studies.
It is important to note that some CNVs may have gone undetected due to the set threshold and many high-resolution arrays do not fully cover the genome (Cooper et al., 2008). In addition to technological boundaries, our study was limited by the fact that sample selection from the EGCUT biobank was performed only based on the available anamnestic data, some of which were self-reported as opposed to data derived directly from medical records. For example, the age at menopause was self-reported on the average 20.7 years later (the duration of amenorrhea by the time of study), which may lead to reporting errors. Also, the average age at the time of the study was 57.8 ± 12.5 years; therefore, we lack detailed information on clinical, biological (hormone levels) or ultrasonographic data, retrospectively from the time of menopause. Moreover, it is not always possible to exclude every disease that could cause POF secondarily (as opposed to primary ovarian causes). The shortcomings of this study are diminished by the large cohort size for such a rare disease and the close examination of a wide range of phenotypic data in every patient.
In summary, our data provide novel insight into the association between chromosomal aberrations and premature menopause of ovarian cause. Our results highlighted the possible role of CNVs in the pathogenesis of POF. Both duplications and deletions detected in our study were associated with the POF phenotype and contained genes that are essential for reproductive function. Although functional studies are required to further delineate the contribution of identified CNVs in the genetic etiology of POF, our study confirms that DNA microarrays are a useful tool for evaluating genomic imbalances in POF patients as it offers a much higher resolution and therefore, a higher diagnostic yield compared with conventional cytogenetic methods. Finally, we conclude that using samples from population-based biobanks may be a useful and efficient way to recruit patients for large-scale CNV studies, and data collected from such studies represent a starting point for both characterization of the disease and guidance for further research.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors’ roles
A.S. and A.K.: study design; K.H.-K.: clinical characterization of samples and statistical analysis; A.S., A.K., O.T., O.Ž. and M.N.: methodology; M.N., R.M., M.K. and PA.K.: computational analysis; O.T., M.N. and O.Ž.: data interpretation; K.H.: laboratory assistance; O.T.: preparation of draft and original manuscript; K.H.-K., A.S., A.K., O.T., O.Ž. and M.N.: revision of the manuscript; A.S., A.K. and J.S.T.: Funding; A.S. and A.K.: supervision; all authors approved the original manuscript.
Funding
This study was supported by the Estonian Ministry of Education and Research (IUT20-43, IUT20-60, IUT34-16, SF0180027s10 and 9205), Enterprise Estonia (EU30020 and EU48695), Eureka's EUROSTARS Programme (NOTED, EU41564), grants from the European Union's FP7 Marie Curie Industry-Academia Partnerships and Pathways (IAPP, SARM, EU324509) and Horizon 2020 innovation programme (WIDENLIFE, 692065), Academy of Finland and the Sigrid Juselius Foundation. Funding to pay the Open Access publication charges for this article was provided by Horizon 2020 innovation programme WIDENLIFE grant #692065.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
The authors thank Kairit Mikkel, Mari-Liis Tammesoo and Steven Smit from Estonian Genome Center of the University of Tartu for providing anamnestic and genotyping data for our patients and technical help in data analyses. We are grateful to the Estonian Biocentre Genotyping Core Facility, especially Viljo Soo, for technical assistance.
References
- Aboura A, Dupas C, Tachdjian G, Portnoi MF, Bourcigaux N, Dewailly D, Frydman R, Fauser B, Ronci-Chaix N, Donadille B et al. Array comparative genomic hybridization profiling analysis reveals deoxyribonucleic acid copy number variations associated with premature ovarian failure. J Clin Endocrinol Metab 2009;94:4540–4546. [DOI] [PubMed] [Google Scholar]
- Alperin ES, Shapiro LJ. Characterization of point mutations in patients with X-linked ichthyosis. Effects on the structure and function of the steroid sulfatase protein. J Biol Chem 1997;272:20756–20763. [DOI] [PubMed] [Google Scholar]
- Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE et al. The androgen excess and PCOS society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril 2009;91:456–488. [DOI] [PubMed] [Google Scholar]
- Beke A, Piko H, Haltrich I, Csomor J, Matolcsy A, Fekete G, Rigo J, Karcagi V. Molecular cytogenetic analysis of Xq critical regions in premature ovarian failure. Mol Cytogenet 2013;6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, Benavente R, Cooke HJ. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet 2009;5:e1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukovsky A, Keenan JA, Caudle MR, Wimalasena J, Upadhyaya NB, Van Meter SE. Immunohistochemical studies of the adult human ovary: possible contribution of immune and epithelial factors to folliculogenesis. Am J Reprod Immunol 1995;33:323–340. [DOI] [PubMed] [Google Scholar]
- Bukovsky A, Caudle MR, Svetlikova M, Wimalasena J, Ayala ME, Dominguez R. Oogenesis in adult mammals, including humans: a review. Endocrine 2005;26:301–316. [DOI] [PubMed] [Google Scholar]
- Busacca M, Riparini J, Somigliana E, Oggioni G, Izzo S, Vignali M, Candiani M. Postsurgical ovarian failure after laparoscopic excision of bilateral endometriomas. Am J Obstet Gynecol 2006;195:421–425. [DOI] [PubMed] [Google Scholar]
- Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, Harrison W, Vaiman D, Ben-Neriah Z, Garcia-Tunon I et al. Mutant cohesin in premature ovarian failure. N Engl J Med 2014;370:943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyshov VP, Radysh TV, Gura IV, Tatarchuk TP, Khominskaya ZB. Immune disorders in women with premature ovarian failure in initial period. Am J Reprod Immunol 2001;46:220–225. [DOI] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an objective Bayes Hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res 2007;35:2013–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway GS, Kaltsas G, Patel A, Davies MC, Jacobs HS. Characterization of idiopathic premature ovarian failure. Fertil Steril 1996;65:337–341. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 2008;40:1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer DW, Xu H, Harlow BL. Family history as a predictor of early menopause. Fertil Steril 1995;64:740–745. [DOI] [PubMed] [Google Scholar]
- de Boer EJ, den Tonkelaar I, te Velde ER, Burger CW, van Leeuwen FE. Increased risk of early menopausal transition and natural menopause after poor response at first IVF treatment. Hum Reprod 2003;18:1544–1552. [DOI] [PubMed] [Google Scholar]
- de Bruin JP, Bovenhuis H, van Noord PA, Pearson PL, van Arendonk JA, te Velde ER, Kuurman WW, Dorland M. The role of genetic factors in age at natural menopause. Hum Reprod 2001;16:2014–2018. [DOI] [PubMed] [Google Scholar]
- Dixit H, Rao LK, Padmalatha V, Kanakavalli M, Deenadayal M, Gupta N, Chakravarty B, Singh L. Mutational screening of the coding region of growth differentiation factor 9 gene in Indian women with ovarian failure. Menopause 2005;12:749–754. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet 2006;7:85–97. [DOI] [PubMed] [Google Scholar]
- Goswami D, Conway GS. Premature ovarian failure. Hum Reprod Update 2005;11:391–410. [DOI] [PubMed] [Google Scholar]
- Group TECW. Physiopathological determinants of human infertility. Hum Reprod Update 2002;8:435–447. [DOI] [PubMed] [Google Scholar]
- Haller-Kikkatalo K, Salumets A, Uibo R. Review on autoimmune reactions in female infertility: antibodies to follicle stimulating hormone. Clin Dev Immunol 2012;2012:762541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller-Kikkatalo K, Uibo R, Kurg A, Salumets A. The prevalence and phenotypic characteristics of spontaneous premature ovarian failure: a general population registry based study. Hum Reprod 2015;30:1229–1238. [DOI] [PubMed] [Google Scholar]
- Hoek A, Schoemaker J, Drexhage HA. Premature ovarian failure and ovarian autoimmunity. Endocr Rev 1997;18:107–134. [DOI] [PubMed] [Google Scholar]
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011;13:680–685. [DOI] [PubMed] [Google Scholar]
- Knauff EA, Blauw HM, Pearson PL, Kok K, Wijmenga C, Veldink JH, van den Berg LH, Bouchard P, Fauser BC, Franke L. Copy number variants on the X chromosome in women with primary ovarian insufficiency. Fertil Steril 2011;95:1584–1588.e1. [DOI] [PubMed] [Google Scholar]
- Koks S, Velthut A, Sarapik A, Altmae S, Reinmaa E, Schalkwyk LC, Fernandes C, Lad HV, Soomets U, Jaakma U et al. The differential transcriptome and ontology profiles of floating and cumulus granulosa cells in stimulated human antral follicles. Mol Hum Reprod 2010;16:229–240. [DOI] [PubMed] [Google Scholar]
- Kovanci E, Rohozinski J, Simpson JL, Heard MJ, Bishop CE, Carson SA. Growth differentiating factor-9 mutations may be associated with premature ovarian failure. Fertil Steril 2007;87:143–146. [DOI] [PubMed] [Google Scholar]
- Lacombe A, Lee H, Zahed L, Choucair M, Muller JM, Nelson SF, Salameh W, Vilain E. Disruption of POF1B binding to nonmuscle actin filaments is associated with premature ovarian failure. Am J Hum Genet 2006;79:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, Bourcigaux N, Jacquesson L, Bouchard P, Frydman R et al. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol 2006;154:739–744. [DOI] [PubMed] [Google Scholar]
- Leader B, Lim H, Carabatsos MJ, Harrington A, Ecsedy J, Pellman D, Maas R, Leder P. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol 2002;4:921–928. [DOI] [PubMed] [Google Scholar]
- Ledig S, Ropke A, Wieacker P. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev 2010;4:225–232. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang C, Xing G, Chen Q, He F. Functional characterization of novel human ARFGAP3. FEBS Lett 2001;490:79–83. [DOI] [PubMed] [Google Scholar]
- Llano E, Gomez R, Gutierrez-Caballero C, Herran Y, Sanchez-Martin M, Vazquez-Quinones L, Hernandez T, de Alava E, Cuadrado A, Barbero JL et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev 2008;22:2400–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund KJ. Menopause and the menopausal transition. Med Clin North Am 2008;92:1253–1271, xii. [DOI] [PubMed] [Google Scholar]
- Ma J, Zeng F, Schultz RM, Tseng H. Basonuclin: a novel mammalian maternal-effect gene. Development 2006;133:2053–2062. [DOI] [PubMed] [Google Scholar]
- MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2013;42:D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gamboa L, Lesemann K, Kuckelkorn U, Scheffler S, Ghannam K, Hahne M, Gaber-Elsner T, Egerer K, Naumann L, Buttgereit F et al. Gene expression of catalytic proteasome subunits and resistance toward proteasome inhibition of B lymphocytes from patients with primary Sjögren syndrome. J Rheumatol 2013;40:663–673. [DOI] [PubMed] [Google Scholar]
- McGuire MM, Bowden W, Engel NJ, Ahn HW, Kovanci E, Rajkovic A. Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. Fertil Steril 2011;95:1595–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen HE, Rayburn H, Krieger M. Abnormal lipoprotein metabolism and reversible female infertility in HDL receptor (SR-BI)-deficient mice. J Clin Invest 2001;108:1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier-Barbarino P, Forges T, Faure GC, Bene MC. Gonadal antibodies interfering with female reproduction. Best Pract Res Clin Endocrinol Metab 2005;19:135–148. [DOI] [PubMed] [Google Scholar]
- Montaville P, Jegou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier MF. Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biol 2014;12:e1001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye K, Fayad T, Silversides DW, Sirois J, Lussier JG. Identification of downregulated messenger RNAs in bovine granulosa cells of dominant follicles following stimulation with human chorionic gonadotropin. Biol Reprod 2005;73:324–333. [DOI] [PubMed] [Google Scholar]
- Newman S, Hermetz KE, Weckselblatt B, Rudd MK. Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints. Am J Hum Genet 2015;96:208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norling A, Hirschberg AL, Rodriguez-Wallberg KA, Iwarsson E, Wedell A, Barbaro M. Identification of a duplication within the GDF9 gene and novel candidate genes for primary ovarian insufficiency (POI) by a customized high-resolution array comparative genomic hybridization platform. Hum Reprod 2014;29:1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret BP, Parinaud J, Ribbes H, Moatti JP, Pontonnier G, Chap H, Douste-Blazy L. Lipoprotein and phospholipid distribution in human follicular fluids. Fertil Steril 1985;43:405–409. [DOI] [PubMed] [Google Scholar]
- Persani L, Rossetti R, Cacciatore C. Genes involved in human premature ovarian failure. J Mol Endocrinol 2010;45:257–279. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouresmaeili F, Fazeli Z. Premature ovarian failure: a critical condition in the reproductive potential with various genetic causes. Int J Fertil Steril 2014;8:1–12. [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilter CR, Karcanias AC, Bagga MR, Duncan S, Murray A, Conway GS, Sargent CA, Affara NA. Analysis of X chromosome genomic DNA sequence copy number variation associated with premature ovarian failure (POF). Hum Reprod 2010;25:2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 2007;35:W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T, Vilo J. g:Profiler--a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res 2011;39:W307–W315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryley DA, Wu HH, Leader B, Zimon A, Reindollar RH, Gray MR. Characterization and mutation analysis of the human formin-2 (FMN2) gene in women with unexplained infertility. Fertil Steril 2005;83:1363–1371. [DOI] [PubMed] [Google Scholar]
- Schlessinger D, Herrera L, Crisponi L, Mumm S, Percesepe A, Pellegrini M, Pilia G, Forabosco A. Genes and translocations involved in POF. Am J Med Genet 2002;111:328–333. [DOI] [PubMed] [Google Scholar]
- Sen A, Prizant H, Light A, Biswas A, Hayes E, Lee HJ, Barad D, Gleicher N, Hammes SR. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc Natl Acad Sci USA 2014;111:3008–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D, Nagarajan N. Premature menopause - meeting the needs. Post Reprod Health 2014;20:62–68. [DOI] [PubMed] [Google Scholar]
- Shalgi R, Kraicer P, Rimon A, Pinto M, Soferman N. Proteins of human follicular fluid: the blood-follicle barrier. Fertil Steril 1973;24:429–434. [PubMed] [Google Scholar]
- Shelling AN. Premature ovarian failure. Reproduction 2010;140:633–641. [DOI] [PubMed] [Google Scholar]
- Shuster LT, Rhodes DJ, Gostout BS, Grossardt BR, Rocca WA. Premature menopause or early menopause: long-term health consequences. Maturitas 2010;65:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Wang XG, Ma XS, Huang XJ, Li J, Liu HL. TBP dynamics during mouse oocyte meiotic maturation and early embryo development. PLoS One 2013;8:e55425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatone C, Amicarelli F, Carbone MC, Monteleone P, Caserta D, Marci R, Artini PG, Piomboni P, Focarelli R. Cellular and molecular aspects of ovarian follicle ageing. Hum Reprod Update 2008;14:131–142. [DOI] [PubMed] [Google Scholar]
- Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell 2001;1:201–213. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3–new capabilities and interfaces. Nucleic Acids Res 2012;40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegetti W, Grazia Tibiletti M, Testa G, de Lauretis Y, Alagna F, Castoldi E, Taborelli M, Motta T, Bolis PF, Dalpra L et al. Inheritance in idiopathic premature ovarian failure: analysis of 71 cases. Hum Reprod 1998;13:1796–1800. [DOI] [PubMed] [Google Scholar]
- Vulto-van Silfhout AT, Hehir-Kwa JY, van Bon BW, Schuurs-Hoeijmakers JH, Meader S, Hellebrekers CJ, Thoonen IJ, de Brouwer AP, Brunner HG, Webber C et al. Clinical significance of de novo and inherited copy-number variation. Hum Mutat 2013;34:1679–1687. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007;17:1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Shigetomi H, Onogi A, Haruta S, Kawaguchi R, Yoshida S, Furukawa N, Nagai A, Tanase Y, Tsunemi T et al. Redox-active iron-induced oxidative stress in the pathogenesis of clear cell carcinoma of the ovary. Int J Gynecol Cancer 2011;21:1200–1207. [DOI] [PubMed] [Google Scholar]
- Zangen D, Kaufman Y, Zeligson S, Perlberg S, Fridman H, Kanaan M, Abdulhadi-Atwan M, Abu Libdeh A, Gussow A, Kisslov I et al. XX ovarian dysgenesis is caused by a PSMC3IP/HOP2 mutation that abolishes coactivation of estrogen-driven transcription. Am J Hum Genet 2011;89:572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Qin Y, Kovanci E, Simpson JL, Chen ZJ, Rajkovic A. Analyses of GDF9 mutation in 100 Chinese women with premature ovarian failure. Fertil Steril 2007;88:1474–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Griswold MD, Hassold TJ, Hunt PA, Small CL, Ye P. Predicting meiotic pathways in human fetal oogenesis. Biol Reprod 2010;82:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.