Abstract
Alishewanella sp. WH16-1 (= CCTCC M201507) is a facultative anaerobic, motile, Gram-negative, rod-shaped bacterium isolated from soil of a copper and iron mine. This strain efficiently reduces chromate (Cr6+) to the much less toxic Cr3+. In addition, it reduces sulfate (SO42−) to S2−. The S2− could react with Cd2+ to generate precipitated CdS. Thus, strain WH16-1 shows a great potential to bioremediate Cr and Cd contaimination. Here we describe the features of this organism, together with the draft genome and comparative genomic results among strain WH16-1 and other Alishewanella strains. The genome comprises 3,488,867 bp, 50.4 % G + C content, 3,132 protein-coding genes and 80 RNA genes. Both putative chromate- and sulfate-reducing genes are identified.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-016-0169-3) contains supplementary material, which is available to authorized users.
Keywords: Alishewanella, Chromate-reducing bacterium, Sulfate-reducing bacterium, Cadmium, Chromium
Introduction
The genus Alishewanella was established by Vogel et al., in 2000 with Alishewanella fetalis as the type species. It belongs to the family Alteromonadaceae of the class Gammaproteobacteria [1]. So far, Alishewanella contains six species: A. fetalis, Alishewanella aestuarii, Alishewanella jeotgali, Alishewanella agri and Alishewanella tabrizica and Alishewanella solinquinati [1–6]. The common characteristics of the genus Alishewanella are Gram-negative, rod-shaped and positive for oxidase and catalase [1–6]. Some Alishewanella strains were able to degrade pectin which is applicable in bioremediation of food industrial wastes [7–11]. Three Alishewanella strains (A. aestuarii B11T,A. jeotgaliKCTC 22429T and A. agri BL06T) have been sequenced and the pectin degradation pathway was found in their genomes [8–11]. Some strains of Alishewanella were reported to tolerate arsenic [12, 13], but the ability of Alishewanella strains to resist or transform other heavy metal(loids) have not been reported.
Alishewanella sp. WH16-1 was isolated from mining soil in 2009. This strain could resist to multiple heavy metals. During cultivation, it could efficiently reduce the toxic chromate (Cr6+) to the much less toxic and less bioavaliable Cr3+. It could also reduce sulfate (SO42−) to S2−. When Cd2+ was present, the S2− reacted with Cd2+ and precipitated as CdS. These characteristics made strain WH16-1 a great potential for bioremediate Cr and Cd contamination. In pot experiments of rice, tobacco and Chinese cabbage, with the addition of the bacterial culture, the amount of Cr and Cd in the plants decreased significantly [14]. Sequencing the genome of WH16-1 and comparing its attributes with the other Alishewanella genomes would provide a means of establishing the molecular determinants required for chromate/sulfate reduction, heavy metal resistance and pectin degradation, and for better application of these strains. Here we report the high quality draft genomic information of strain WH16-1 and compare it to the three sequenced Alishewanella genomes.
Organism information
Classification and features
Phylogenetic analysis was performed by the neighbor-joining method based on 16S rRNA gene sequences. Strain WH16-1 is closely related to A. agri BL06T (99.7 %) and A. fetalisCCUG 30811T (99.1 %) (Fig. 1). A similar result was obtained based on gyrase B gene (gyrB) sequences (Additional file 1: Figure S1). The gyrB sequences has been successfully used to establish phylogenetic relatedness in Alishewanella [1], Pseudomonas [15], Acinetobacter [16], Vibrio [17],Bacillus [18] and Shewanella [19].
Fig. 1.
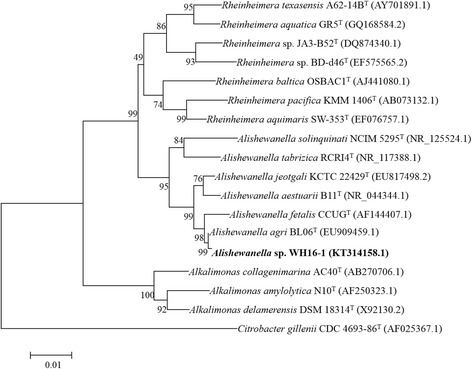
Phylogenetic tree highlighting the phylogenetic position of Alishewanella sp. WH16-1. The phylogenetic tree was constructed based on the 16S rRNA gene sequences. The analysis was inferred by MEGA 6.0 [45] with NJ algorithm and 1,000 bootstrap repetitions were computed to estimate the reliability of the tree
Strain WH16-1 is Gram-negative, facultatively anaerobic, motile and rod-shaped (0.3–0.5 × 1.2–2.0) (Fig. 2). Colonies are white, circular and raised on LB agar plate. Growth occurs at 4–40 °C, in 0–8 % (w/v) NaCl and at pH 4–11. Optimal growth occurs at 37 °C, 1 % (w/v) NaCl and at pH 6.0–8.0 (Table 1). It can grow in LB, trypticase soy broth and R2A medium. API 20NE test (bioMérieux) in combination of traditional classification methods were used to analyze the physiological and biochemical characteristics. Strain WH16-1 is positive for oxidase and catalase activities and is able to reduce nitrate to nitrite. It is positive for aesculinase, gelatinase, arginine dihydrolase and urease but is negative for indole and β-galactosidase. It can use D-sucrose and maltose as the sole carbon sources. It cannot assimilate D-glucose, L-arabinose, D-mannose, D-mannitol, N-acetylglucosamine, gluconate, capric acid, adipic acid, malic acid, trisodium citrate or phenylacetic acid. Most of these biochemical characteristics are similar to the other Alishewanella strains [1–6]. However, unlike some Alishewanella strains [8–11], strain WH16-1 cannot degrade pectin.
Fig. 2.
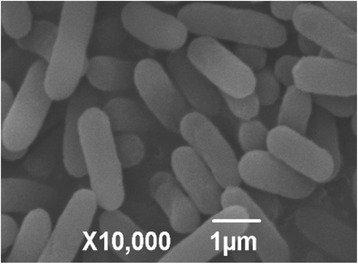
Scan electron microscope (SEM) image of Alishewanella sp. WH16-1 cells. The bar scale represents 1 μm
Table 1.
Classification and general features of Alishewanella sp. WH16-1 [47]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [48] | |
| Phylum Proteobacteria | TAS [49, 50] | ||
| Class Gammaproteobacteria | TAS [51–53] | ||
| Order Alteromonadales | TAS [52–54] | ||
| Family Alteromonadaceae | TAS [55] | ||
| Genus Alishewanella
Species Alishewanella sp. |
TAS [1] | ||
| Strain WH16-1 | |||
| Gram stain | negative | IDA | |
| Cell shape | rod | IDA | |
| Motility | motile | IDA | |
| Sporulation | non-sporulating | NAS | |
| Temperature range | 4–40 °C | IDA | |
| Optimum temperature | 37 °C | IDA | |
| pH range; Optimum | 4–11; 6–8 | IDA | |
| Carbon source | maltose, D-sucrose | IDA | |
| MIGS-6 | Habitat | soil | IDA |
| MIGS-6.3 | Salinity | 0–8 % NaCl (w/v), optimal at 1 % | IDA |
| MIGS-22 | Oxygen requirement | facultative anaerobic | IDA |
| MIGS-15 | Biotic relationship | free-living | IDA |
| MIGS-14 | Pathogenicity | non-pathogen | NAS |
| MIGS-4 | Geographic location | Huangshi city, Hubei province, China | IDA |
| MIGS-5 | Sample collection | 2009 | IDA |
| MIGS-4.1 | Latitude | N29°40′–30°15′ | IDA |
| MIGS-4.2 | Longitude | E114°31′–115°20′ | IDA |
| MIGS-4.4 | Altitude | not reported |
These evidence codes are from the Gene Ontology project [56]
IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence)
a Evidence codes
Interestingly, the strain could reduce 1 mmol/L Cr6+ (added as K2CrO4) in 36 h and remove 60 μmol/L Cd2+ (added as CdCl2) in 60 h (by the production of precipitated CdS [20] in LB liquid medium) (Fig. 3). In addition, this strain is tolerant to multi-metal(loids). The minimal inhibition concentration tests for different heavy metals were carried out on LB agar plates and incubated at 37 °C for 2 days. The MICs for K2CrO4, CdCl2, PbCl2, CuCl2 and Na3AsO3 are 45, 0.08, 10, 1 and 1 mmol/L, respectively.
Fig. 3.
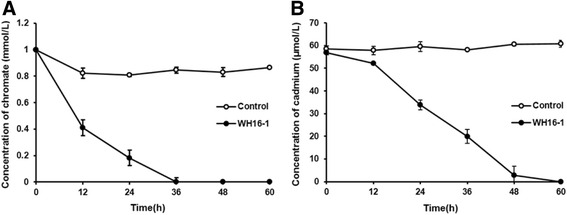
Cr6+ and Cd2+ removed by Alishewanella sp. WH16-1. Control stands for null LB medium. Strain WH16-1 was incubated until OD600 reach 1.0, and then amended with K2CrO4 (1 mmol/L) and CdCl2 (0.06 mmol/L), respectively. The cultures were removed at 12 h intervals. After centrifuging at 12,000 rpm for 2 min, the supernatant was used to determine the residual concentration of Cr6+ and Cd2+. The concentration of Cr6+ and Cd2+ were measured by the UV spectrophotometer (DU800, Beckman, CA, USA) with the colorimetric diphenylcarbazide (DPC) method [46] and the atomic absorption spectrometry AAS, respectively
Genome sequencing information
Genome project history
Strain WH16-1 was selected for genome sequencing based on its ability to reduce Cr6+ and SO42− and preliminary application for soil Cr and Cd bioremediation. Since 2009, this strain has been used in both basic and bioremediation studies and the results are very promising. It was sequenced by Majorbio Bio-pharm Technology Co., Ltd, Shanghai, China. The genome sequencing and assembly information of the project is given in Table 2. The final genome consists of 133 scaffolds with approximately 345.3 × coverage. The draft genome sequence was annotated by NCBI PGAP. The genome sequence is available in DDBJ/EMBL/GenBank under accession number LCWL00000000.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | Illumina Paired-End library (300 bp insert size) |
| MIGS-29 | Sequencing platforms | Illumina Hiseq 2000 |
| MIGS-31.2 | Fold coverage | 345.3 × |
| MIGS-30 | Assemblers | SOAPdenovo v1.05 |
| MIGS-32 | Gene calling method | GeneMarkS+ |
| Locus TAG | AAY72 | |
| Genbank ID | LCWL00000000 | |
| Genbank Date of Release | 2015.11.12 | |
| Bioproject | PRJNA283029 | |
| MIGS-13 | Source material identifier | Strain CCTCC M201507 |
| Project relevance | Bioremediation |
Growth conditions and genomic DNA preparation
A single colony of strain WH16-1 was incubated into 50 ml LB medium and grown aerobically at 37 °C for 36 h with 150 rpm shaking. The cells were collected by centrifugation. The DNA was extracted, concentrated and purified using the QiAamp kit (Qiagen, Germany). A NanoDrop Spectrophotometer 2000 was used to determine the quality and quantity of the DNA. Six micrograms of DNA was sent to Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China) for sequencing.
Genome sequencing and assembly
The genome sequencing of strain WH16-1 was performed on an Illumina Hiseq2000 [21] and assembled by Majorbio Bio-pharm Technology Co., Ltd, Shanghai, China. An Illumina standard shotgun library was constructed and sequenced, which generated 12,683,662 reads totaling 1,281,049,862 bp. All original sequence data can be found at the NCBI Sequence Read Archive [22]. The following steps were performed for removing low quality reads: (1) removed the adapter o reads; (2) cut the 5′ end bases which were not A, T, G, C; (3) filtered the reads which have a quality score lower than 20; (4) filtered the reads which contained N more than 10 %; and (5) removed the reads which have the length less than 25 bp after processed by the previous four steps. The reads were assembled into 156 contigs using SOAPdenovo v1.05 [23]. A total of 149 contigs were obtained after removing the contigs < 200 bp. The total size of the genome is 3,488,867 bp and the final assembly is based on 1,205 Mbp of Illumina data which provides a coverage of 345.3 × .
Genome annotation
The draft genome of WH16-1 was annotated through the NCBI PGAP, which combines the gene caller GeneMarkS+ [24] with the similarity-based gene detection approach. Protein function classification was performed by WebMGA [25] with E-value cutoff of 1-e10. The transmembrane helices were predicted by TMHMM v. 2.0 [26]. Signal peptides in the genome were predicted by SignalP 4.1 [27]. The translations of the predicted CDSs were also used to search against the Pfam protein family database with E-value cutoff of 1-e5 [28] and the KEGG database [29]. Internal gene clustering was performed by OrthoMCL using Match cutoff of 50 % and E-value Exponent cutoff of 1-e5 [30, 31].
Genome properties
The whole genome of strain WH16-1 is 3,488,867 bp in length, with an average G + C content of 50.4 %, and is distributed in 149 contigs (>200 bp). The genome properties and statistics are summarized in Table 3. There are 80 predicted RNA including 73 tRNA, 5 rRNAs and 2 ncRNA. In addition, a total of 3,132 protein-coding genes are identified. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3.
Nucleotide content and gene count levels of the genome
| Attribute | Genome (total) | |
|---|---|---|
| Value | % of totala | |
| Genome size (bp) | 3,488,867 | 100.00 |
| DNA coding (bp) | 3,117,033 | 89.34 |
| DNA G + C (bp) | 1,759,785 | 50.44 |
| DNA scaffolds | 133 | 100.00 |
| Contigs | 149 | 100.00 |
| Total genesb | 3,282 | |
| RNA genes | 80 | |
| Pseudo genes | 73 | |
| Protein-coding genes | 3,132 | 100.00 |
| Genes in internal clusters | 1,190 | 37.99 |
| Genes with function prediction | 2,388 | 76.25 |
| Genes assigned to COGs | 2,249 | 71.81 |
| Genes with Pfam domains | 2,710 | 86.53 |
| Genes with signal peptides | 367 | 11.72 |
| Genes with transmembrane helices | 1,101 | 35.15 |
| CRISPR repeats | 1 | |
aThe total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome
bAlso includes 73 pseudogenes, 73 tRNA genes, 5 rRNAs and 2 ncRNA
Table 4.
Number of genes associated with the 25 general COG functional categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 175 | 5.59 | Translation |
| A | 1 | 0.03 | RNA processing and modification |
| K | 153 | 4.89 | Transcription |
| L | 141 | 4.50 | Replication, recombination and repair |
| B | 2 | 0.06 | Chromatin structure and dynamics |
| D | 34 | 1.09 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.00 | Nuclear structure |
| V | 56 | 1.79 | Defense mechanisms |
| T | 216 | 6.90 | Signal transduction mechanisms |
| M | 156 | 4.98 | Cell wall/membrane biogenesis |
| N | 87 | 2.78 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 0 | 0.00 | Extracellular structures |
| U | 77 | 2.46 | Intracellular trafficking and secretion |
| O | 116 | 3.70 | Posttranslational modification, protein turnover, chaperones |
| C | 157 | 5.01 | Energy production and conversion |
| G | 90 | 2.87 | Carbohydrate transport and metabolism |
| E | 207 | 6.61 | Amino acid transport and metabolism |
| F | 62 | 1.98 | Nucleotide transport and metabolism |
| H | 132 | 4.21 | Coenzyme transport and metabolism |
| I | 85 | 2.71 | Lipid transport and metabolism |
| P | 148 | 4.73 | Inorganic ion transport and metabolism |
| Q | 41 | 1.31 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 244 | 7.79 | General function prediction only |
| S | 202 | 6.45 | Function unknown |
| - | 885 | 28.26 | Not in COGs |
aThe total is based on the total number of protein coding genes in the annotated genome
Insights from the genome sequence
Strain WH16-1 has the genes for a complete SO42− reduction pathway according to the KEGG analysis, including CysPUWA, CysN, CysD, CysC, CysH and CysIJ (Additional file 1: Figure S2; Additional file 2: Table S1). This pathway contained several steps: 1) the SO42− is uptaken by the putative CysPUWA into the cell [32]; 2) the intracellular SO42− is acetylated to adenylylsulphate (APS) by sulfate adenylyltransferases CysN and CysD [33]; 3) the APS is phosphorylated to phosphoadenylylsulphate (PAPS) by APS kinase CysC and, 4) the PAPS is reduced to sulfite (SO32−) by PAPS reductase CysH [33] and, 5) the SO32− is finally reduced to sulfide (S2−) by sulfite reductase CysIJ [33]. Strain WH16-1 was able to remove Cd2+ most probably due to the reaction between S2− and Cd2+ to form the precipitated CdS [20]. For Cr6+ reduction, a putative chromate reductase YieF was found (Additional file 2: Table S1). YieF was reported to responsible for the reduction of Cr6+ in cytoplasm [34]. An individual chromate transport gene chrA and a chromate resistance cluster including chrBAC, hp1, chrF, lppy/lpqo, hp2 and ABC transport permease gene are found in the genome (Additional file 2: Table S2) [35, 36]. Currently, we have disrupted the chrA (AAY72_02075) and the ABC transport permease genes, respectively. The chromate resistance levels were both decreased significantly in the chrA and ABC transport permease gene mutant strains (data not shown).
In addition, various heavy metal transformation and resistance determinants are identified in the genome of strain WH16-1 Several transporters (MntH, CzcA and ZntA) that might be involved in the efflux of Cd2+, Pb2+ and Zn2+ are found [37–39]. Cu2+, As3+ and Hg2+ resistance determinants are also present, such as Cu transporter ATPase [40], Cu2+ resistance system CopABCD [41], Ars [42] and Pst [43] systems for arsenic resistance and MerTPADE system for mercury resistance [44] (Additional file 2: Table S2).
Strain WH16-1 has a genome size (3.49 Mbp), similar to A. jeotgaliKCTC 22429T (3.84 Mbp), A. aestuarii B11T (3.59 Mbp) and A. agri BL06T (3.49 Mbp) [8–10] (Fig. 4). The G + C content of strain WH16-1 (50.4 %) is also consistent with the other Alishewanella strains (A. jeotgaliKCTC 22429T, 50.7 %, A. aestuarii B11T, 51 % and A. agri BL06T, 50.6 %). Strain WH16-1 shares 2,474 proteins with the other three Alishewanella genomes and has 217 strain-specific proteins (Fig. 5). The 2,474 core genes include yieF, chrA, the ten genes in the whole sulfate reduction pathway and most of the heavy metal resistance genes (Additional file 2: Table S1-S2). Strain WH16-1 possesses the higher number of chromatin resistance genes compared to the other three strains.
Fig. 4.
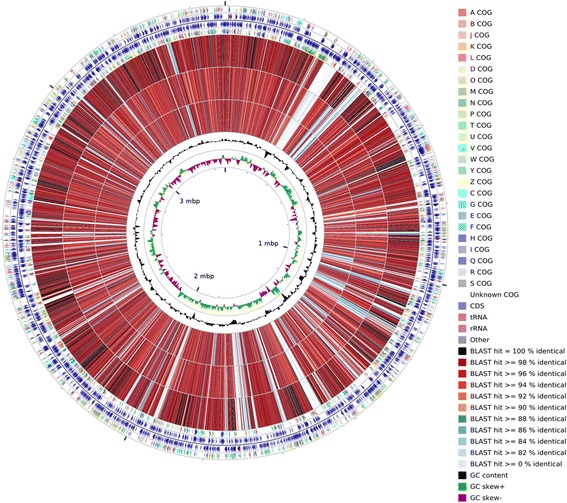
A graphical circular map of the comparison between reference strain Alishewanella sp. WH16-1 and three sequenced strains of the Alishewanella species. From outside to center, rings 1, 4 show protein-coding genes colored by COG categories on forward/reverse strand; rings 2, 3 denote genes on forward/reverse strand; rings 5, 6, 7 show the CDS vs CDS BLAST results of strain WH16-1 with those of A. agri BL06T, A. jeotgali KCTC 22429T and A. aestuarii B11T, respectively; ring 8 shows G + C % content plot and the innermost ring shows GC skew
Fig. 5.
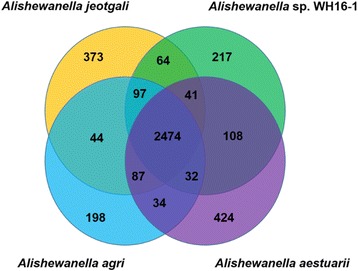
The Venn diagram depicting the core and unique genes between Alishewanella sp. WH16-1 and other three Alishewanella species (A. agri BL06T, A. jeotgali KCTC 22429T and A. aestuarii B11T)
In addition, A. agri BL06T, A. jeotgaliKCTC 22429T and A. aestuarii B11T were all reported to have the ability of degrading pectin and possess pectin degradation genes [8–11]. However, unlike strains BL06T, KCTC 22429T and B11T, strain WH16-1 was unable to degrade pectin and the pectin degradation genes are not found in its genome. Since strain WH16-1 was isolated from a heavy metal rich environment, it may be more relevant for bioremediation of heavy metal contamination. The pectin degradation genes may be lost during the evolution.
Conclusions
The genomic results of Alishewanella sp. WH16-1 reveal correlation between the gene types and some phenotypes. The strain harbors various genes responsible for sulfate transport and reduction, chromate reduction and resistance of multi-heavy metals. These observations provide insights into understand the molecular mechanisms of heavy metals. In addition, all of the analyzed Alishewanella genomes have putative sulfate and chromate reduction genes, which indicates that sulfate and chromate reduction may be the important characters of the Alishewanella strains. Thus, these strains have a great potential for application in bioremediation of heavy metal or other industrial wastes.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31470226).
Authors’ contributions
XX carried out biochemical tests, sequence analysis and preparation of the draft. JL participated in the metal resistance test and phylogenetic analysis. GZ and LL did metal reduction and metal removing tests. HW and BX conducted strain isolation. GW and SL participated in research design and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The abilities to reduce Cr6+ and immobilize Pb2+ and Cd2+ of strain WH16-1 were described in China Patent, 2015; CN 104,928,213 A [14]. Due to these abilities, strain WH16-1 has a great potential for application in bioremediation of heavy metal. All the authors of this paper and the inventors of the patent [14] declare that they have no commercial or non-commercial competing interests.
Abbreviation
- PGAP
Prokaryotic Genome Annotation Pipeline
Additional files
Figure S1. Phylogenetic relationships of Alishewanella sp. WH16-1 based on gyrB sequences. The analysis was performed by MEGA 6.0 [45] with NJ algorithm and 1,000 bootstrap repetitions were computed to estimate the reliability of the tree. The gyrB gene of strain WH16-1 is the gene sequence coding for AAY72_13600. Figure S2. The putative sulfate transport and reduction pathway in Alishewanella sp. WH16-1. APS stands for adenylylsulphate, PAPS stands for phosphoadenylylsulphate. The locus tag numbers of the predicted proteins (CysP, CysU, CysW, CysA, CysN, CysD, CysC, CysH CysJ and CysI) are AAY72_14890, AAY72_14885, AAY72_14880, AAY72_14875, AAY72_03865, AAY72_03870, AAY72_07290, AAY72_07265, AAY72_07255 and AAY72_07260, respectively. (DOCX 355 kb)
Table S1. Putative proteins involved in sulfate and chromate reduction. Table S2. Putative proteins involved in heavy metal resistance. (XLSX 14 kb)
References
- 1.Vogel BF, Venkateswaran K, Christensen H, Falsen E, Christiansen G, Gram L. Polyphasic taxonomic approach in the description of Alishewanella fetalis gen. nov., sp. nov., isolated from a human foetus. Int J Syst Evol Microbiol. 2000;50:1133–1142. doi: 10.1099/00207713-50-3-1133. [DOI] [PubMed] [Google Scholar]
- 2.Roh SW, Nam YD, Chang HW, Kim KH, Kim SM, Oh HM, Bae JW. Alishewanella aestuarii sp. nov., isolated from tidal flat sediment, and emended description of the genus Alishewanella. Int J Syst Evol Microbiol. 2009;59:421–424. doi: 10.1099/ijs.0.65643-0. [DOI] [PubMed] [Google Scholar]
- 3.Kim MS, Roh SW, Nam YD, Chang HW, Kim KH, Jung MJ, et al. Alishewanella jeotgali sp. nov., isolated from traditional fermented food, and emended description of the genus Alishewanella. Int J Syst Evol Microbiol. 2009;59:2313–2316. doi: 10.1099/ijs.0.007260-0. [DOI] [PubMed] [Google Scholar]
- 4.Kim MS, Jo SK, Roh SW, Bae JW. Alishewanella agri sp. nov., isolated from landfill soil. Int J Syst Evol Microbiol. 2010;60:2199–2203. doi: 10.1099/ijs.0.011684-0. [DOI] [PubMed] [Google Scholar]
- 5.Tarhriz V, Nematzadeh G, Vahed SZ, Hejazi MA, Hejazi MS. Alishewanella tabrizica sp. nov., isolated from Qurugöl Lake. Int J Syst Evol Microbiol. 2012;62:1986–1991. doi: 10.1099/ijs.0.031567-0. [DOI] [PubMed] [Google Scholar]
- 6.Kolekar YM, Pawar SP, Adav SS, Zheng LQ, Li WJ, et al. Alishewanella solinquinati sp. nov., isolated from soil contaminated with textile dyes. Curr Microbiol. 2013;67(4):454–459. doi: 10.1007/s00284-013-0385-7. [DOI] [PubMed] [Google Scholar]
- 7.Miran W, Nawaz M, Jang J, Lee DS. Conversion of orange peel waste biomass to bioelectricity using a mediator-less microbial fuel cell. Sci Total Environ. 2016;547:197–205. doi: 10.1016/j.scitotenv.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Jung J, Choi S, Chun J, Park W. Genome sequence of pectin-degrading Alishewanella aestuarii strain B11T, isolated from tidal flat sediment. J Bacteriol. 2012;194(19):5476. doi: 10.1128/JB.01255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Jung J, Sung JS, Chun J, Park W. Genome sequence of pectin-degrading Alishewanella agri, isolated from landfill soil. J Bacteriol. 2012;194(18):5135–5136. doi: 10.1128/JB.01129-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung J, Chun J, Park W. Genome sequence of extracellular-protease-producing Alishewanella jeotgali isolated from traditional Korean fermented seafood. J Bacteriol. 2012;194(8):2097. doi: 10.1128/JB.00153-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung J, Park W. Comparative genomic and transcriptomic analyses reveal habitat differentiation and different transcriptional responses during pectin metabolism in Alishewanella species. Appl Environ Microbiol. 2013;79:6351–6361. doi: 10.1128/AEM.02350-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah R, Jha S. Alishewanella sp. strain GIDC-5, Arsenite hyper-tolerant bacteria isolated from industrial effluent of South Gujarat, India. Chem Ecol. 2013;29(5):427–436. doi: 10.1080/02757540.2013.774379. [DOI] [Google Scholar]
- 13.Li P, Wang Y, Dai X, Zhang R, Jiang Z, Jiang D, et al. Microbial community in high arsenic shallow groundwater aquifers in Hetao Basin of Inner Mongolia, China. PLoS One. 2015;10(5):e0125844. doi: 10.1371/journal.pone.0125844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao S, Wang G, Zhou G, Xia X, Wang H. An Alishewanella strain with the ability to bioremediate heavy metal contamination. China Patent. 2015; CN 104,928,213 A.
- 15.Yamamoto S, Harayama S. PCR Amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol. 1995;61(10):3768. doi: 10.1128/aem.61.10.3768-3768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto S, Harayama S. Phylogenetic analysis of Acinetobacter strains based on the nucleotide sequences of gyrB genes and on the amino acid sequences of their products. Int J Syst Bacteriol. 1996;46(2):506–511. doi: 10.1099/00207713-46-2-506. [DOI] [PubMed] [Google Scholar]
- 17.Venkateswaran K, Dohmoto N, Harayama S. Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp. Appl Environ Microbiol. 1998;64(2):681–687. doi: 10.1128/aem.64.2.681-687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada S, Ohashi E, Agata N, Venkateswaran K. Cloning and nucleotide sequence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B. anthracis and their application to the detection of B. cereus in rice. Appl Environ Microbiol. 1999;65(4):1483–1490. doi: 10.1128/aem.65.4.1483-1490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, MacGregor BJ, et al. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int J Syst Bacteriol. 1999;49:705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- 20.Pagnanelli F, Cruz Viggi C, Toro L. Isolation and quantification of cadmium removal mechanisms in batch reactors inoculated by sulphate reducing bacteria: biosorption versus bioprecipitation. Bioresour Technol. 2010;101(9):2981–2987. doi: 10.1016/j.biortech.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 22.The NCBI Sequence Read Archive (SRA). [http://www.ncbi.nlm.nih.gov/Traces/sra/].
- 23.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 24.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–18. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogh A, Larsson BÈ, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Petersen TN, Brunak S, Heijne GV, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 28.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer S, Brunk B P, Chen F, Gao X, Harb OS, Iodice JB, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011:6–12. doi: 10.1002/0471250953.bi0612s35. PMID: 21901743 [DOI] [PMC free article] [PubMed]
- 32.Sirko A, Zatyka M, Sadowy E, Hulanicka D. Sulfate and thiosulfate transport in Escherichia coli K-12: evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J Bacteriol. 1995;177(14):4134–4136. doi: 10.1128/jb.177.14.4134-4136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol. 2000;2(2):145–177. [PubMed] [Google Scholar]
- 34.Ackerley DF, Gonzalez CF, Park CH, Blake R, Keyhan M, Matin A. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl Environ Microbiol. 2004;70(2):873–882. doi: 10.1128/AEM.70.2.873-882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branco R, Chung AP, Johnston T, Gurel V, Morais P, Zhitkovich A. The chromate-inducible chrBACF operon from the transposable element TnOtChr confers resistance to chromium (VI) and superoxide. J Bacteriol. 2008;190:6996–7003. doi: 10.1128/JB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henne KL, Nakatsu CH, Thompson DK, Konopka AE. High-level chromate resistance in Arthrobacter sp. strain FB24 requires previously uncharacterized accessory genes. BMC Microbiol. 2009;9:199. doi: 10.1186/1471-2180-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MF. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol. 2000;35(5):1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- 38.Nies DH, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci U S A. 1989;86(19):7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rensing C, Mitra B, Rosen BP. The zntA gene of Escherichia coli encodes a Zn (II)-translocating P-type ATPase. Proc Natl Acad Sci U S A. 1997;94(26):14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odermatt A, Suter H, Krapf R, Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem. 1993;268(17):12775–12779. [PubMed] [Google Scholar]
- 41.Adaikkalam V, Swarup S. Characterization of copABCD operon from a copper-sensitive Pseudomonas putida strain. Can J Microbiol. 2005;51(3):209–216. doi: 10.1139/w04-135. [DOI] [PubMed] [Google Scholar]
- 42.Kruger MC, Bertin PN, Heipieper HJ, Arsène-Ploetze F. Bacterial metabolism of environmental arsenic--mechanisms and biotechnological applications. Appl Microbiol Biotechnol. 2013;97(9):3827–3841. doi: 10.1007/s00253-013-4838-5. [DOI] [PubMed] [Google Scholar]
- 43.Rosen BP, Ajees AA, McDermott TR. Life and death with arsenic. Arsenic life: an analysis of the recent report “A bacterium that can grow by using arsenic instead of phosphorus”. Bioessays. 2011;33(5):350–357. doi: 10.1002/bies.201100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nascimento AM, Chartone-Souza E. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet Mol Res. 2003;2(1):92–101. [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monteiro MI, Fraga IC, Yallouz AV, de Oliveira NM, Ribeiro SH. Determination of total chromium traces in tannery effluents by electrothermal atomic absorption spectrometry, flame atomic absorption spectrometry and UV-visible spectrophotometric methods. Talanta. 2002;58(4):629–633. doi: 10.1016/S0039-9140(02)00317-X. [DOI] [PubMed] [Google Scholar]
- 47.Field D, Garrity GM, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woese CR, Kandler O, Weelis ML. Towards a natural system of organisms: proposal for the domains archaea, bacteria and eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrity GM, Bell JA, Phylum Lilburn T, XIV . Proteobacteria phyl nov. In: Brenner DJ, Krieg NR, Stanley JT, Garrity GM, editors. Bergey’s Manual of Sytematic Bacteriology, second edition. Vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria) New York: Springer; 2005. p. 1. [Google Scholar]
- 50.Stackebrandt E, Murray RGE, Trüper HG. Proteobacteria classis nov., a name for the phylogenetic taxon that includes the “purple bacteria and their relatives”. Int J Syst Evol Microbiol. 1988;38(3):321–325. [Google Scholar]
- 51.Garrity GM, Bell JA, Class LT, III . Gammaproteobacteria class. nov. In: Brenner DJ, Krieg NR, Stanley JT, Garrity GM, editors. Bergey’s Manual of Sytematic Bacteriology, second edition. Vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria) New York: Springer; 2005. p. 1. [Google Scholar]
- 52.Validation of publication of new names and newcombinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol. 2005;55:2235–38. [DOI] [PubMed]
- 53.Williams KP, Kelly DP. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63:2901–2906. doi: 10.1099/ijs.0.049270-0. [DOI] [PubMed] [Google Scholar]
- 54.Bowman JP, Mcmeekin TA, Order X. Alteromonadales ord. nov. In: Brenner DJ, Krieg NR, Stanley JT, Garrity GM, editors. Bergey’s Manual of Sytematic Bacteriology, second edition. Vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria) New York: Springer; 2005. p. 443. [Google Scholar]
- 55.Ivanova EP, Mikhaĭlov VV. A new family of Alteromonadaceae fam. nov., including the marine proteobacteria species Alteromonas, Pseudoalteromonas, Idiomarina and Colwellia. Microbiology. 2001;70(1):10–17. doi: 10.1023/A:1004876301036. [DOI] [PubMed] [Google Scholar]
- 56.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


