Abstract
Prebiotics are selectively fermented ingredients that result in specific changes in the composition and/or activity of the gastrointestinal microbiota, thus conferring benefit(s) upon the host health. The aim of this study was to evaluate the influence of a β (1–4) galacto-oligosaccharides formulation consisting of 90% pure GOS (GOS90), on the composition and activity of the mouse gut microbiota. Germ-free mice were colonized with microbiota from four pathogen-free wt 129 mice donors (SPF), and stools were collected during a feeding trial in which GOS90 was delivered orally for 14 days. Pyrosequencing of 16S rDNA amplicons showed that Bifidobacterium and specific Lactobacillus, Bacteroides and Clostridiales were more prevalent in GOS90-fed mice after 14 days, although the prebiotic impact on Bifidobacterium varied among individual mice. Prebiotic feeding also resulted in decreased abundance of Bacteroidales, Helicobacter and Clostridium. High-throughput quantitative PCR showed an increased abundance of Bifidobacterium adolescentis, B. pseudocatenulatum, B. lactis and B. gallicum in the prebiotic-fed mice. Control female mice showed a higher diversity (Phylogenetic Diversity PD = 15.1 ± 3.4 in stools and PD = 13.0 ± 0.6 in intestinal contents) than control males (PD = 7.8 ± 1.6 in stool samples and PD = 9.5 ± 1.0 in intestinal contents). GOS90 did not modify inflammatory biomarkers (IL-6, IL-12, IL-1β, IFN-γ and TNF-α). Decreased butyrate, acetate and lactate concentrations in stools of prebiotic fed mice suggested an increase in colonic absorption and reduced excretion. Overall, our results demonstrate that GOS90 is capable of modulating the intestinal microbiome resulting in expansion of the probiome (autochtonous commensal intestinal bacteria considered to have a beneficial influence on health).
Keywords: mouse gut microbiome, prebiotics, Bifidobacterium
Introduction
Gut microorganisms live in a symbiotic interaction with the host and co-exist in a dynamic equilibrium while contributing to host physiology. The colon microbiota comprises populations of about 1011 cells/g that represent between 500 and 1,000 different species which are involved in the host’s metabolic, physiological and immune processes (1). Recent studies have correlated diseases like inflammatory bowel diseases (IBD), obesity, colorectal cancer, diabetes and allergies with imbalances in the composition of the gut microbial community (dysbiosis) (2–5). These findings have increased the interest in modulation of the gut microbial communities through dietary strategies to maintain health. One of these strategies is the consumption of prebiotics, which are currently defined as “selectively fermented ingredients that result in specific changes in the composition and/or activity of the gastrointestinal microbiota, thus conferring benefit(s) upon the host health” (6). Dietary interventions with prebiotics have been shown to selectively stimulate the growth and/or activity of health-promoting intestinal bacteria such as species of Bifidobacterium and Lactobacillus (7–10).
Galacto-oligosaccharides (GOS) are prebiotics recognized for their health benefits like modulation of the intestinal microbiota, alleviation of constipation, enhancing mineral absorption, prevention of carcinogenesis and mitigation of allergy (11, 12). GOS can also reduce the adherence of enteropathogens to the epithelial intestinal cells by functioning as receptor decoys that mimic mucosal cell surface glycans (13–16). Moreover, GOS are currently added to infant formula to stimulate growth of intestinal bifidobacteria and lactobacilli in order to obtain the health benefits of a “breastfed-like” microbiota (9).
Studies on GOS modulation of the gut microbiome showed that the prebiotic increased bifidobacteria populations in the human gastrointestinal tract of healthy individuals at the expense of less beneficial groups of Gram-negative bacteria such as Bacteroides and Desulfovibrio spp (10, 17–19). Additionally, a study conducted by Silk et al. (20) showed that GOS specifically stimulated gut bifidobacteria and alleviated symptoms like flatulence and abdominal pain in irritable bowel syndrome (IBS) patients. Moreover, a study conducted in overweight adults fed GOS showed increased secretory IgA and decreased fecal inflammatory markers such as calprotectin and plasma C reactive protein, as well as reduced metabolic syndrome markers such as insulin, total cholesterol (TC), triglycerides and TC:HDL cholesterol ratio (19), suggesting that GOS feeding stimulates the immune response and decreases the risk of developing cardiovascular diseases.
GOS molecules are synthesized enzymatically by β-galactosidases in a reaction known as transgalactosylation (12). However, the concentration and composition of GOS can vary with the method and enzyme used for their generation, which can influence their effect on the intestinal microbiota (21). For instance, it has been reported that mono- and di-saccharides are the preferred substrate for Lactobacillus rhamnosus, while Bifidobacterium lactis grows better in tri- and tetra-saccharides since it possesses specific transport systems that will import these more efficiently (22). In a previous study, we heterologously expressed a synthetically generated version of the hexosyltransferase gene (Bht) from Sporobolomyces singularis(23). The Bht gene encodes a glycosyl-hydrolase (EC 3.2.1.21) that acts as galactosyltransferase, able to catalyze a one-step conversion of lactose to GOS. Commercial GOS formulations such as Vivinal GOS (Friesland Domo), Oligomate (Yakult) and others contain only approximately 50% GOS (w/w), and also contain residual glucose, lactose and galactose. The high purity GOS (GOS90) preparation contains 90% GOS and 10% lactose (w/w), with no residual glucose or galactose.
The aim of the present study was to evaluate the influence of GOS90 on the composition of the transient and adherent components of the microbiota and their metabolic activity in wt 129 mice.
Materials and methods
We hypothesized that the prebiotic would shift the composition and metabolic activity of the colon microbiota toward a more health-promoting community enriched with lactobacilli and bifidobacteria. To prove our hypothesis, mice of the 129 background were fed GOS90 over 14 days. Stool samples were collected at three time points and intestinal contents were collected at the end of the trial in order to characterize and compare their bacterial communities by 16S amplicon pyrosequencing. In addition, we developed and optimized a high-throughput qPCR method using the BioMark platform by Fluidigm to identify and quantify Bifidobacterium species. Targeted metabolomic and cytokine analysis were carried out to determine the impact of prebiotic feeding in the production of short-chain fatty acids (SCFAs) and inflammatory markers.
Animal housing, treatment, and sample collection
Thirteen germ-free (GF) wt 129 mice (4 males and 9 females, 8–12 weeks old) were transferred to specific pathogen-free (SPF) conditions by colonization using gavage and rectal swabs prior to the experiment without any antibiotic treatment. Numbers of animals were based on minimum that would provide statistical power to validate differences between control and treatment. This was based on experience and published literature(24). The colonization inoculum consisted of stool samples from four SPF wt 129 mice born/raised not humanized. Nine stool pellets were suspended in 5 ml of phosphate buffered saline (PBS). The particulate matter was allowed to settle for approximately 5 minutes before inoculation. Nine days after colonization, mice were randomly divided into two groups: the control standard (Purina Prolab RMH3000) diet group consisting of 4 male and 3 female mice (cages 1 and 3) and 6 GOS90-fed female mice (cages 2 and 4). We allowed 9 days for the microbiota to stabilize based on a previous study that showed that after 1 week, the gut microbiota of gavage-colonized animals were more similar to the donor community than animals that had acquired their microbiota without intervention (24). Originally, the groups were planned to be 4 males and 3 females receiving GOS90, and 6 control females, but 3 of 4 male mice would not provide stool samples on the day of the prebiotic treatment and consequently they were placed in the control group. Stool samples for those 3 mice were collected at day 2. Fifty microliters of GOS90 syrup, the equivalent of 0.26 g/kg bodyweight of GOS (not including lactose), was delivered daily orally by pipette to the prebiotic-fed group for 14 days. The prebiotic dose was selected based on a previous study(25). No carrier solution (placebo) was provided to the control mice.
All fecal samples from prebiotic-fed mice were collected at three time points during the experiment: day 1 (no dietary modulation), day 7 and day 14. Day 1 was the first day of GOS feeding and stools were collected before delivering the prebiotic. All samples were stored at −80 °C immediately after collection. Animals were euthanized by CO2 inhalation followed by cervical dislocation. Colons were removed, flushed with PBS, and cut into 3 sections – proximal, mid, and distal colon. These were each frozen in their own tube and kept at −80°C until analysis. All mouse protocols were approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill.
Generation of GOS90
Production of highly pure GOS (90% GOS, 10% lactose, w/w) was carried out by standard transgalactosylation reactions utilizing a recombinant β-hexosyltransferase (BHT) from Sporobolomyces singularis expressed in Pichia pastoris as previously described(23). Since this preparation was generated using a BHT and not a β-galactosidase, the generated product was a highly-pure β (1–4) GOS free of residual glucose and galactose. Reactions were carried out using standardized amounts of enzyme (0.5 U g/1lactose) in 5 mM sodium phosphate buffer (pH 5.0) and a similarly buffered solution containing lactose (200 g/L) at 30°C. Purity of the GOS90 product used in this study was determined as previously described (23, 27).
DNA isolation from fecal and intestinal content samples
All samples were kept frozen at −80°C until use. Prior to DNA extraction of the distal colon samples, the mucosal layer was gently scraped with a razor blade. The scraped material was placed in a microtube. At that time, DNA from fecal samples and intestinal samples (scraped material) was extracted using the QIAmp DNA Stool kit (Qiagen, Hilden, Germany). Modifications in the disruption step were made to the supplied protocol to ensure lysis of Gram-positive bacteria. In short, 200 mg of sample were transferred to a microtube containing 0.2 g of autoclaved glass beads (11 micron in diameter, Sigma) and 1.4 mL of ASL solution (Qiagen). Next, samples were homogenized in a TissueLyser II instrument (Qiagen, Valencia, Ca) at 25 Hz for 2 min. Following steps were performed according to the manufacturer’s protocol.
Barcoded 16S rRNA PCR and pyrosequencing
Amplicon pyrosequencing of the 16S ribosomal gene was performed as previously described (28, 29). Amplification of the V1–V2 region of the bacterial 16S rDNA gene was performed on total DNA from fecal and intestinal samples. Master mixes for these reactions included the Qiagen HotStar HiFidelity Polymerase Kit (Qiagen, Valencia CA) with a forward primer composed of the Roche Titanium Fusion Primer A (5’-CCATCTCATCCCTGCGTGTCTCCGACTCAG -3’), a 10 bp Multiplex Identifier (MID) sequence (Roche, Indianapolis, IN) unique to each sample and the universal bacteria primer 8F (5'-AGAGTTTGATCCTGGCTCAG-3') (30). The reverse primer was composed of the Roche Titanium Primer B (5’-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG -3’), the identical 10 bp MID sequence as the forward primer and the reverse primer 338R (5’-GCTGCCTCCCGTAGGAGT-3’) (31). We also performed amplicon sequencing of fragments generated by PCR amplification using the Bifidobacterium-specific primer B-8F (5’-AGGGTTCGATTCTGGCTCAG-3’) (32) paired with the 338R as the reverse primer for specific detection of this genus. For intestinal content samples we used a combination (4:1) of the primers 8F and Bifido-8F with 338R as the reverse primer as described (32). PCR products were gel-purified individually using the E-Gel Electrophoresis System (Life Technologies, Invitrogen), and standardized prior to pooling. The 16S rDNA amplicons from the pooled sample were sequenced on a Roche 454 Genome Sequencer FLX Titanium instrument in the Microbiome Core Facility, Chapel Hill, NC using the GS FLX Titanium XLR70 sequencing reagents and protocols.
Amplicon sequencing data analysis
16S rRNA amplicon sequencing data was analyzed using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline (33). Sequences were de-multiplexed, filtered for quality control and grouped into OTUs (Operational Taxonomic Units) at a 97% level to approximate species-level phylotypes using UCLUST with standard parameters (34). Next, chimeras and singletons were removed using ChimeraSlayer within QIIME (35). To ensure an even sampling depth, a random selection of 2,700 sequences (matching the sample with lowest number of reads) from each sample was used for rarefaction analysis to determine alpha diversity metrics (observed species and Phylogenetic Diversity indexes) on rarefied OTU tables. Beta diversity and principal coordinates analysis (PCoA) were also calculated within QIIME using Weighted and Unweighted Unifrac distances (36) between samples at a depth of 2,700 sequences per sample to evaluate dissimilarities between samples. Sequencing data obtained in this work has been registered as a bio project in NCBI (BioProject ID PRJNA291486)
PICRUSt analysis of 16S amplicon sequencing data
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (http://picrust.github.com/picrust/) is a tool that predicts metagenome functional content from 16S rRNA amplicon sequencing input data. OTUs were picked to be used in PICRUSt using a Closed Reference OTU picking against the reference genome Greengenes database Version 13_8 (http://greengenes.secondgenome.com/). OTUs were assigned at 97% identity within QIIME using the UCLUST method (34).
Bifidobacterium species detection by 24.192 Dynamic array
The Access Array AA 24.192 (Fluidigm Corporation, San Francisco, CA, USA) in the Advanced Analytics Core at UNC (NC, USA) was used to detect specific Bifidobacterium species. The bacterial strains listed in Table 1 were used as controls in the study. All strains were grown in MRS + 0.5% cysteine at 37°C under anaerobic conditions (80% N2,10% CO2, and 10%, H2) in an anaerobic chamber (Coy Laboratory Products, MI, USA). Primers defined in previous studies (37–40) were used to amplify the 16S ribosomal RNA and the chaperonin GroEL genes (Table 2).
Table 1.
Bifidobacterium strains used in the present study as positive and negative controls.
| Bifidobacterium strain | Sample original ID | Origin |
|---|---|---|
| B. longum longum | NCK 1575 | Kindly provided by T. Klaenhammer (NC State University) |
| B. longum infantis | ATCC 15697 | ATCC |
| B. pseudocatenulatum | Human stool sample | Kindly provided by T. Keku (UNC-Chapel Hill) |
| B. bifidum | ATCC 29521 | ATCC |
| B. breve | ATCC 15700 | ATCC |
| B. lactis | NCK 1573 | Kindly provided by T. Klaenhammer (NC State University) |
| B. adolescentis | Human stool sample | Kindly provided by T. Keku (UNC-Chapel Hill) |
| B. bifidum | NCK 2083 | Kindly provided by T. Klaenhammer (NC State University) |
Table 2.
Specific primers used in the study to target different bacterial groups and species of Bifidobacterium based on the 16S and groEL gene.
| Target species | Primer | Target gene |
Sequence (5′ to 3′) | Amplicon size |
Reference |
|---|---|---|---|---|---|
| All bacteria | Uni_V4_F | 16S | CAGCAGCCGCGGTAATAC | 389 | (39) |
| Uni_V4_R | CCGTCAATTTCTTTGAGTTT | ||||
| All bacteria | Uni_F | 16S | GTGSTGCAYGGYYGTCGTCA | 194 | (41) |
| Uni_R | ACGTCRTCCMCNCCTTCCTC | ||||
| All bacteria | 8F | 16S | AGAGTTTGATCCTGGCTCAG | 330 | (30), (31), (32) |
| 8F_B | AGGGTTCGATTCTGGCTCAG | ||||
| Uni_338_R | GCTGCCTCCCGTAGGAGT | ||||
| Actinobacteria | Actino_235_F | 16S | GCGKCCTATCAGCTTGTT | 283 | (42) |
| Actino_567_R | CCGCCTACGAGCYCTTTACGC | ||||
| Bifidobacteriaceae | Bifidobac_F | 16S | CTCCTGGAAACGGGTGG | 442 | (39) |
| Bifidobac_R | CTTCACACCRGACGCG | ||||
| Bifidobacterium | Bifido_5 | 16S | GATTCTGGCTCAGGATGAACGC | 236 | (43) |
| Bifido_3 | CTGATAGGACGCGACCCCAT | ||||
| B. adolescentis | Biado_1a | 16S | CTCCAGTTGGATGCATGTC | 279 | (44) |
| Biado_1b | TCCAGTTGACCGCATGGT | ||||
| Biado_2 | CGAAGGCTTGCTCCCAGT | ||||
| B. longum | Longum_5 | 16S | TTCCAGTTGATCGCATGGTCTTCT | 277 | (43) |
| Longum_3 | GGCTACCCGTCGAAGCCACG | ||||
| B. catenulatum group | BiCATg_1 | 16S | CGGATGCTCCGACTCCT | 285 | (44) |
| BiCATg_2 | CGAAGGCTTGCTCCCGAT | ||||
| B. bifidum | Bifidum_F | 16S | TGA CCG ACC TGC CCC ATG CT | 110 | (45) |
| Bifidum_R | CCC ATC CCA CGC CGA TAG AAT | ||||
| B. breve | BiBRE_F | 16S | CCGGATGCTCCATCACAC | 288 | (44) |
| BiBRE_R | ACAAAGTGCCTTGCTCCCT | ||||
| B. animalis | ISR_lactis_F | ISR | ATCCGAACTGAGACCGGTT | 382 | (40) |
| ISR_lactis_R | GCATGTTGCCAGCGGGTGA | ||||
| B. angulatum | BiAng_1 | 16S | CAGTCCATCGCATGGTGGT | 280 | (44) |
| BiAng_2 | GAAGGCTTGCTCCCCAAC | ||||
| B. dentium | BiDEN_1 | 16S | ATCCCGGGGGTTCGCCT | 387 | (44) |
| BiDEN_2 | GAAGGGCTTGCTCCCGA | ||||
| B. gallicum | BiGAL_1 | 16S | TAATACCGGATGTTCCGCTC | 303 | (44) |
| BiGAL_2 | ACATCCCCGAAAGGACGC | ||||
| B. adolescentis | B_ado_F | groEL | CTCCGCCGCTGATCCGGAAGTCG | 268 | (46) |
| B_ado_R | AACCAACTCGGCGATGTGGACGACA | ||||
| B.longum | B_lon_F | groEL | CGGCGTYGTGACCGTTGAAGAC | 259 | (46) |
| B_lon_R | TGYTTCGCCRTCGACGTCCTCA | ||||
| Catenulatum group | B_cat_F | groEL | GGCTATCGTCAAGGAGCTCA | 188 | (46) |
| B_cat_R | AGTCCAGATCCAAACCGAAAC | ||||
| B. bifidum | B_bif_F | groEL | CTCCGCAGCCGACCCCGAGGTT | 233 | (46) |
| B_bif_R | TGGAAACCTTGCCGGAGGTCAGG | ||||
| B. breve | B_bre_F | groEL | GCTCGTCGTTGCCGCCAAGGACGTT | 272 | (46) |
| B_bre_R | ACAGAATGTACGGATCCTCGAGCACG | ||||
| B. animalis | B_ani_F | groEL | CACCAATGCGGAAGACCAG | 184 | (46) |
| B_ani_R | GTTGTTGAGAATCAGCGTGG | ||||
| B. angulatum | B_ang_F | groEL | CTGTCCTCCCAGCAGGACGTGGTC | 97 | (46) |
| B_ang_R | GCGCTTCGCCGTCAACGTCTTCGG | ||||
| B. dentium | B_den_F | groEL | GGCCCAGTCTTTGGTGCATGAAGGCC | 364 | (46) |
| B_den_R | GTCTTCGAGCACCGCGGTCTGGTCC | ||||
| B. gallicum | B_gal_F | groEL | AGCTCGTCAAGTCCGCCAAGC | 188 | (46) |
| B_gal_R | CATACCTTCGGTGAACTCGAGG |
Primers targeted the following: domain Bacteria, phylum Actinobacteria, genus Bifidobacterium and the Bifidobacterium species listed in Table 2. All primers were tested by standard PCR and conventional quantitative PCR using pure cultures of the strains listed in Table 1 and a pool of the strains at the same concentration. Standard PCR was performed using Qiagen HotStar HiFidelity PCR reagents and 50 ng of genomic DNA per reaction. PCR reactions were performed in duplicate under the following conditions: initial denaturation at 95°C for 1 minute, followed by 35 cycles of 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, followed by a final 5 minute extension at 72°C. Conventional qPCR was performed using an ABI Prism 7000 (Applied Biosystems). The reaction mixture (20 µl) contained 2X power SYBRGreen PCR Master Mix, 200 nM of each primer and 2 µl of the DNA template. qPCR reactions were performed in triplicate under the following conditions: initial holding stage at 50°C for 2 minutes and 95°C for 10 minutes, followed by 35 cycles of 95°C for 15 s and 60°C for 1 min.
DNA from stool samples at a concentration of 25 ng/µl was used for high-throughput qPCR. To increase the template target yield, a pre-amplification (specific target amplification, STA) was performed before the Fluidigm Real-Time PCR according to recommendations from the manufacturer. This step consisted of a multiplex PCR reaction with a pool of the 13 primer sets. The reaction was performed using the following conditions: initial annealing at 95°C for 2 min, followed by 12 cycles of 95°C for 15 s and 60°C 4 min. After the STA reaction, products were diluted 5 times with TE buffer. Microfluidic qPCR was performed using a BioMark HD reader with a Dynamic Array 24.192 chip (Fluidigm). The 24.192 chip was processed following the manufacturer instructions. Briefly, 3 µl of the sample premix (2X SsoFast Evagreen supermix, 20X DNA binding Dye and 1.35 µl 5-fold diluted STA products) and 3 µl of the assay premix (2X assay loading buffer, 1X DNA suspension buffer, and 10µM each primer) were loaded into the chip. Finally, Real-Time PCR was performed following the next conditions: 95°C 60 s, followed by 35 cycles of 96°C for 5 s and 60°C for 45 s.
In order to validate the method and obtain the Pearson’s correlation between the two diferent primer sets (16S and GroEL) used in this study, pure bacterial cultures of Bifidobacterium species (Table 1) at three different concentrations (25ng/µl, 2.5 ng/µl and 0.25ng/µ) were analyzed with their respective primers sets.
High-throughput qPCR data analysis
Raw data were normalized using the Livak method (47). Cq values for each sample were normalized against their respective Cq value obtained from Universal primers using the equation: Ratio (reference/target) = 2−Ct (ref)-Ct (target).
Targeted metabolome analysis
Target metabolites included 5 short-chain fatty acids (SCFAs: acetate, propanoate, isobutyrate, butyrate, valerate), and other relevant metabolites: succinate, glycerol, lactate, glucose, aminobutyrate and galactose. Briefly, approximately 40 mg of fecal samples were weighed and extracted with 1ml of water. 4-chlorophenylalanine and 13C-labeled hexanoic acid were used as internal standards. After centrifugation, a volume of 500 µl was used for SCFA analysis with propyl chloroformate (PCF) derivatization, and a volume of 150 µl was used for analysis of other metabolites with methoxyamine and BSTFA derivatization. The derivates were analyzed with gas chromatograph-mass spectrometer (GC-MS) analysis at UNCG Metabolomics Facility (Kannapolis, NC).
Cytokine Analysis
Colon tissues (proximal and medium regions) were collected in 700 µl of sterile phosphate-buffered saline (PBS) supplemented with complete EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA) and 0.1% IGEPAL. Colon tissues weighing between 300mg and 600mg were homogenized in a TissueLyser (Qiagen, Valencia CA) for 5 min at 30 Hz at room temperature. Colon homogenates were centrifuged twice at 15,000 × g for 20 min at 4°C to remove cell debris, and the supernatants were aliquoted and stored at −80°C. Supernatants were used for cytokine analysis using an ELISA kit (Qiagen, Valencia CA) following the procedures recommended by the manufacturer. The cytokines analyzed were IFN-γ, IL-1β, IL-12, IL-6, IL-10 and TNF-α. Values were expressed as pg/mg protein. The mucosal protein content was determined using the Bradford assay (BioRad; Richmond, CA).
Statistical analyses
To identify bacterial taxa and biological pathways significantly impacted by prebiotic feeding, we used the non-parametric Mann-Whitney and Steel Dwass All Pairs test, which accounts for multiple comparisons, in JMP Genomics (SAS JMP Genomics 5.0). Differences in the abundance of bacterial taxa and pathways were analyzed between control and GOS90 at day 7 and at day 14. To test differences in microbial communities we performed Analyses of Similarity (ANOSIM) to calculate R and P values using the phylogeny-based unweighted UniFrac distance metric. For metabolomics and cytokine analysis, unpaired t tests were used to compare metabolites concentration between feeding groups (SAS JMP Genomics 5.0). P-values of less than 0.05 were considered significant.
Results
Microbiome composition of stools and intestinal contents of wt 129 mice
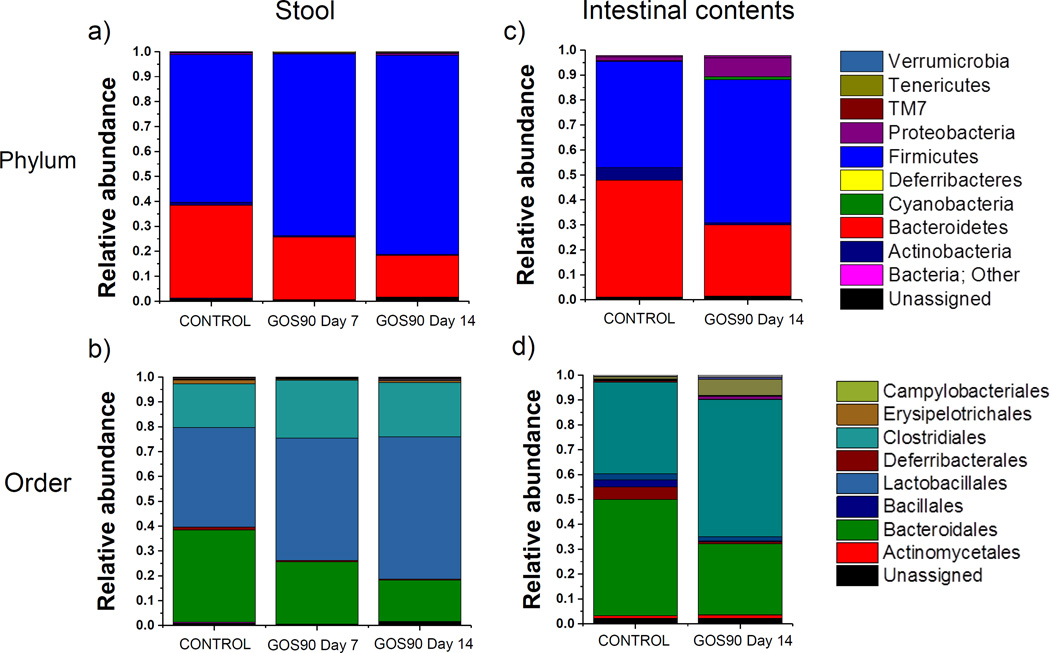
To assess the influence of GOS90 on the gut microbiome, we performed a community structure analysis of fecal samples of wt 129 SPF mice whose diet was supplemented or not (control) with 1% (0.26 g/kg/day) of the prebiotic for 14 days (Figure 1). Pyrosequencing of the regions V1–V2 of the 16S rRNA gene from 39 fecal samples using the universal bacterial primers 8F and 338R resulted in an average of 4,316±1,095 sequences per sample. A total of 172,636 sequences were assigned to 3,366 Operational Taxonomic Units (OTUs) at ≥ 97% similarity, clustering into 78 genera, 18 classes and 9 phyla. The overall composition of the fecal microbiome without dietary supplementation (n= 27) was dominated by the phyla Firmicutes (59.2% ± 22.4%) and Bacteroidetes (37% ± 23.6%) (Figure 2a), in accordance with previous reports of the gut microbiome composition of wt 129 mice (48). Other phyla detected included Deferribacteres (1.9% ± 2.0%), Proteobacteria (0.8% ± 0.6%) and Actinobacteria (0.6% ± 0.6%) (Figure 2a). Representatives of the Tenericutes and TM7 phyla were also detected at low levels (<0.5%). At the order level, the predominant groups were Lactobacillales (40% ± 25.8%), Bacteroidales (37% ± 23.6%) and Clostridiales (17.6% ± 15.4%) (Figure 2b). Composition of the gut microbiome of individual mice showed that Bacteroidetes was the phylum predominant in males while Firmicutes was more abundant in female mice at day 1, with the exception of mice C2M3 and C4M3 (Supplementary Figure 1).
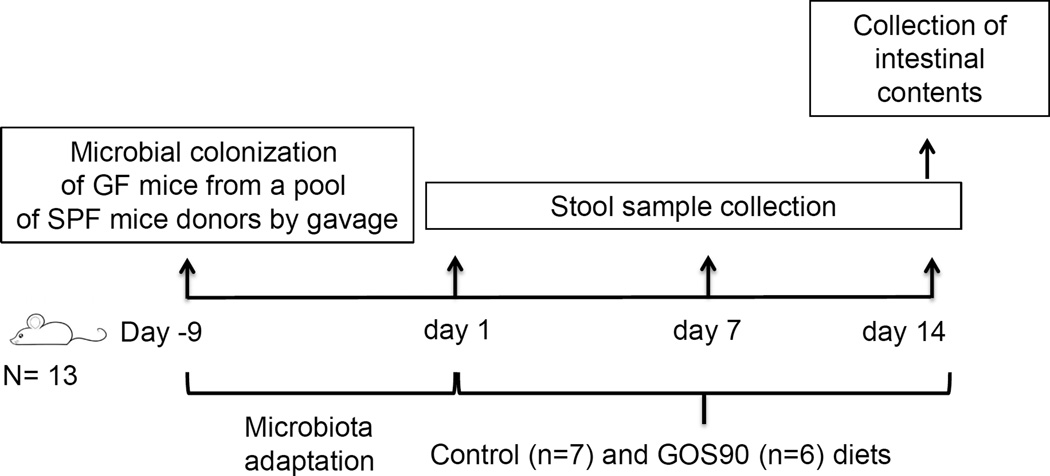
Figure 1. Study experimental design.
Thirteen germ-free mice (GF) were colonized by gavage from a pool of specific pathogen-free (SPF) donors. After 9 days, GOS90 was delivered orally to six mice for 14 days. Fecal samples were collected at three time points (1, 7 and 14 days) and intestinal contents were collected at day 14. Day in negative numbers indicate days before starting the feeding trial.
Figure 2. Microbiome composition of stools and intestinal contents.
Phylum and order level distributions of fecal (a,b) and intestinal contents (c,d) microbiota by feeding group (control and GOS90) at day 7 and 14 of the study. The control group included all stool samples from mice not fed the prebiotic. Universal 8F-338R primers were used for stool samples and a combination of the universal and Bifidobacterium-modified primer set were used for the intestinal contents.
Sequencing of 16S rRNA amplicons from intestinal samples using a mixture (4:1) of forward primers 8F and B8F for the specific detection of Bifidobacterium as described by Martinez et al. (32) yielded a total of 50,716 sequences that were assigned to 362 OTUs at ≥ 97% similarity. As in stools, Firmicutes and Bacteroidetes were the dominant taxa but proportions of both phyla were similar (46.7% ± 15.2% and 42.5% ± 18.3%, respectively; Figure 2c). Abundance of Deferribacteres (5.1% ± 8.9%) and Proteobacteria (1.6%±1.3%) was higher in intestinal contents than fecal samples (1.9% ± 2.0% and 0.7% ± 0.6% respectively) although differences did not reach statistical significance. Interestingly, abundance of the order Lactobacillales in intestinal contents was low (2.5% ± 2.1%) compared to stool samples (40% ± 17.5%), while abundance of Clostridiales was higher in intestinal contents (36.0% ± 16.3%) than in fecal samples (17.0% ± 12.3%).
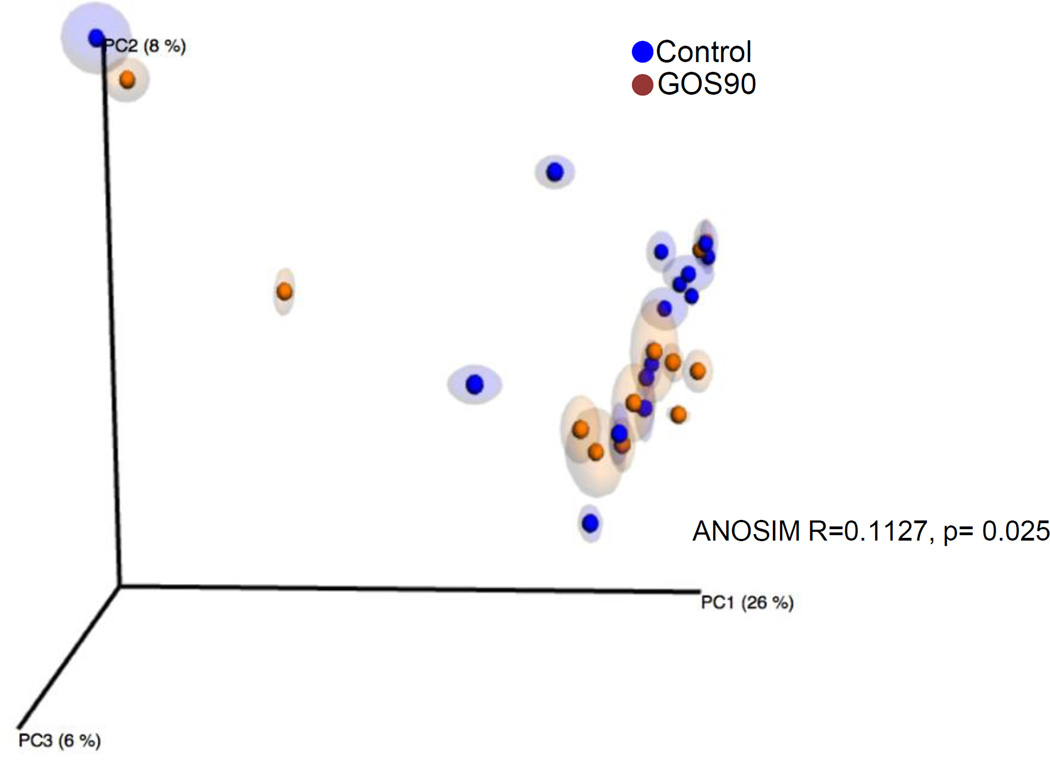
Alpha diversity analysis performed in 2,700 randomly selected sequences per sample showed that the fecal microbiome diversity and richness increased over time in prebiotic-fed mice (Table 3). A small, but statistically significant effect on variables was observed for treatment suggesting that the fecal microbiome composition was modified by the prebiotic (ANOSIM R = 0.1127, p= 0.025) (Figure 3).
Table 3.
Phylogenetic diversity and species richness of control and GOS90 samples at days 7 and 14. Universal 8F-338R primers were used for 16S amplicon sequencing of stool samples and a combination of the universal and Bifidobacterium-modified primer set (4:1) were used for intestinal contents.
| Stool samples | Intestinal contents | |||
|---|---|---|---|---|
| Phylogenetic diversity |
Species richness |
Phylogenetic diversity |
Species richness |
|
| Control | 15.0 ± 6.1 | 138.2 ± 72.9 | 10.6 ± 2.0 | 117.0 ± 56.8 |
| GOS90 Day 7 | 16.5 ± 4.7 | 157.8 ± 60.8 | - | - |
| GOS90 Day 14 | 17.5 ± 4.3 | 170.5 ± 65.0 | 10.9 ± 2.9 | 124.4 ± 59.9 |
Figure 3. Unweighted Principle Coordinate Analysis (PCoA) plot of fecal microbiome from control (n=15) and prebiotic-fed female mice (n=12).
Each dot represents one fecal sample. All time points were included in the analysis. ANOSIM R and P values are indicated in the Figure.
Sex had an apparent effect on the fecal microbiota of wt 129 mice
In our study, unweighted PCoA analysis of samples showed a clear clustering of samples by sex (Supplemental Figure 2a and 2b). However, since we applied the treatment per pen, and we had only one pen per interaction of factors, data was insufficient to draw significant conclusions. Control males had significantly lower bacterial diversity (PD = 7.8 ± 1.6 in stools and PD = 9.5 ± 1.0 in intestinal contents) compared to control females (PD = 15.1 ± 3.4 in stools and PD = 13.0 ± 0.6 in intestinal contents). We identified significant differences in the abundance of fifteen genera belonging to the phyla Actinobacteria, Proteobacteria, Bacteroidetes, Firmicutes and Tenericutes between genders (Supplemental Figure 2). Specifically, the relative abundance of nine genera (Anaeroplasma, Helicobacter, Butiricicoccus, Roseburia, Coprococcus, Lactobacillus, Alistipes, Odoribacter and Asaccharobacter) was significantly higher (between 1- and 80-fold) in females than males. In contrast, abundance of eight genera (Parasuterella, Coprobacillus, Anaerostipes, Anaerovorax, Hidrogenoanaerobacterium, Eubacterium, Enterococcus and Bacteroides was higher (between 1.3- and 10-fold) in the microbiota of male mice. Since, no male mice were included in the prebiotic-fed group and to identify the GOS90-specific impact on the gut microbiota, only samples from female mice (mice without dietary modification [n=15], GOS90-fed mice at days 7 [n=6] and 14 [n=6]) were considered for further analysis.
Dietary GOS90 influenced the composition of the gut microbiome
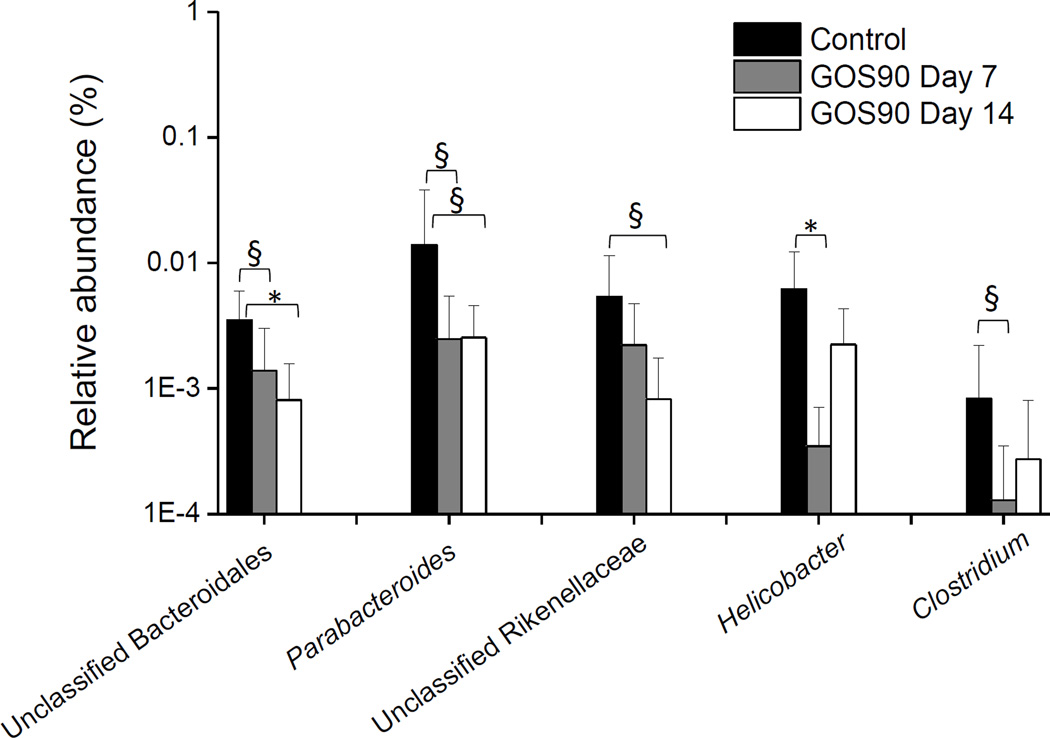
GOS90 feeding resulted in statistically insignificant increases in relative abundance of Firmicutes (Control = 70% ± 20.8%; GOS90 = 80% ± 17.5%), especially Lactobacillales (Control = 48% ± 29.11%; GOS90 = 57%± 29.8%), and decreases of Bacteroidetes (Control = 25% ± 20.9%, GOS90 = 16.7% ± 15.7%), especially the order Bacteroidales (Figure 2). We observed a high inter-individual variation, but despite this variation, GOS90 feeding was associated with an underrepresentation of three Bacteroidales taxa: unclassified Bacteroidales (Steel Dwass All Pairs p=0.02), Parabacteroides (p=0.1) and unclassified Rikenellaceae (p=0.07) after 14 days (Figure 4). In addition, Helicobacter (p=0.005), Ruminococcus (p=0.02) and Clostridium (p=0.1) were underrepresented at days 7 and 14 compared to the control.
Figure 4. Mean relative abundance of underrepresented genera at days 7 (n=6) and 14 (n=6) of GOS90 feeding compared to control samples (n=15).
Error bars indicate standard deviations (Steel-Dwass All Pairs, * p ≤ 0.05, § p ≤ 0.1).
Analysis at OTU level showed that 14 out of 15 Lactobacillus OTUs impacted by GOS showed an increased abundance (Supplementary Figure 3). Likewise, although total proportional amounts of Bacteroidetes (Bacteroidales) declined after 14 days, GOS90 increased abundance of specific Bacteroides OTUs at days 7 and 14. Additionally, within Clostridiales we observed an increased abundance of six Lachnospiraceae OTUs, three Ruminococcaceae OTUs, and one Clostridium OTU at the end of the feeding trial.
The analysis of intestinal contents with a combination of Universal and Bifidobacterium-specific primers resulted in similar changes at the phylum level as stools, with increased abundance of Firmicutes and decreased Bacteroidetes in prebiotic-fed mice. Due to the small sample size (n=5) owing to depletion of 4 samples (original n=9 female mice) we did not perform statistical analyses on abundance of bacterial taxa of intestinal samples from female mice. However, we observed that, unlike stools that showed an increase in the order Lactobacillales, in intestinal contents there was an increased abundance of the order Clostridiales (Figure 2d) and, similar to stool samples, we observed a marked reduction in the abundance of unclassified Bacteroidales (control = 0.02 ± 0.007; GOS90 = 0.005 ± 0.002).
Abundance of Bifidobacterium species in the fecal microbiome of prebiotic-fed mice showed high inter-individual variation
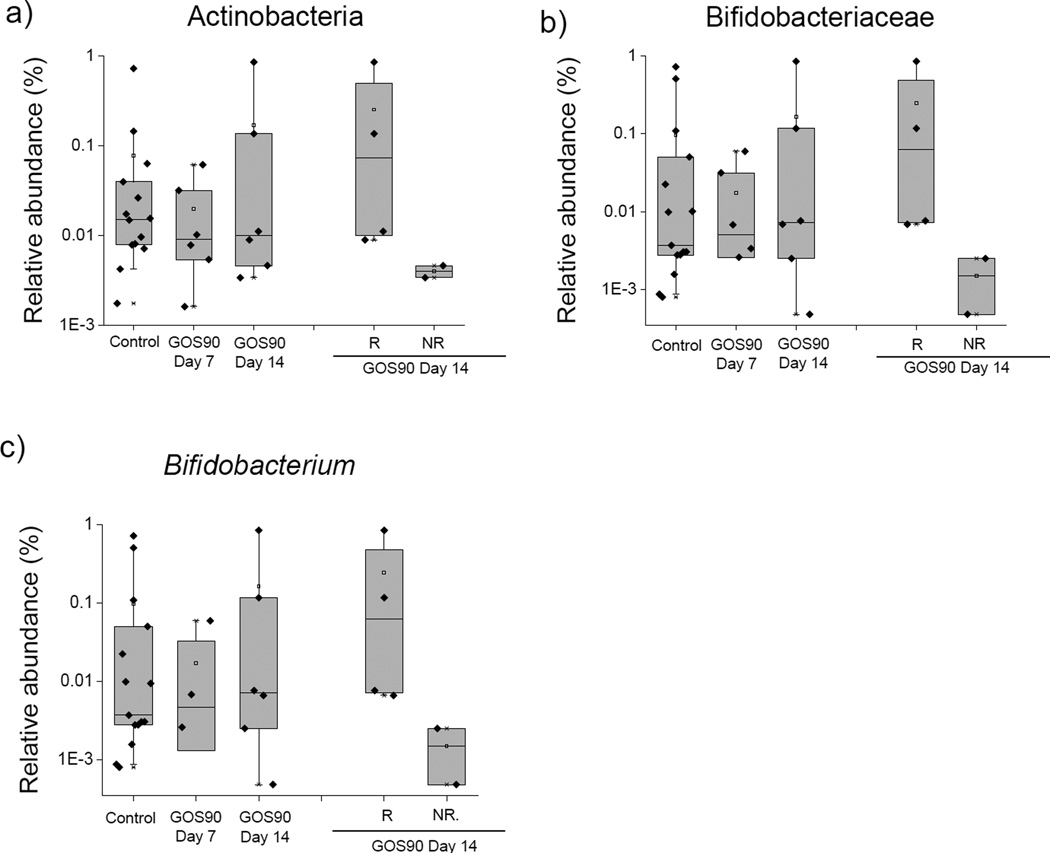
Since the universal primers used in our first 16S amplicon sequencing experiment have been reported to under-detect Bifidobacterium (49), extensively reported to be increased by GOS feeding (10, 18, 19), we performed additional amplicon pyrosequencing of fecal samples using Bifidobacterium-specific primers (32). Sequencing resulted in an average of 3,328 ± 1,319 sequences per sample (149,962 sequences in total), which were assigned to 769 OTUs. Thirty-six OTUs were specifically assigned to the phylum Actinobacteria and nine to the genus Bifidobacterium. We observed an increasing trend in the abundance of Actinobacteria, Bifidobacteriaceae and Bifidobacterium associated with GOS90 feeding (Figure 5). A closer examination of samples at day 14 revealed that the effect of GOS90 varied among individual mice. From day 7 to 14, abundance of Bifidobacterium increased 2.5- to 15-fold in four out of six GOS90-fed mice while abundance of this group decreased in the other two animals. Following the designation by Davis et al. (10), we denominated the animals that showed increased abundance of Bifidobacterium “responders” and the ones that showed no increase “non-responders” (Figure 5).
Figure 5. Mean relative abundances of a) phylum Actinobacteria, b) family Bifidobacteria and c) genus Bifidobacterium by feeding group at days 7 (n= 6) and 14 (n=6).
Data was stratified in responders (n=4) showing increased abundance of Bifidobacterium in response to prebiotic feeding, and non-responders (n=2), showing no change in response to prebiotic feeding. Error bars indicate the 10th and 90th percentile. Steel-Dwass All Pairs test *p ≤ 0.05, § ≤ 0.1.
Bifidobacterium adolescentis, B. catenulatum, B. lactis, and B. gallicum were increased in 4 out of 6 prebiotic-fed mice
We next aimed to identify specific Bifidobacterium species affected by GOS90. Primers specific for nine species of Bifidobacterium, genus Bifidobacterium, family Bifidobacteriaceae and phylum Actinobacteria were used to quantify bacterial groups by high-throughput qPCR using the Access Array 192.24 (Fluidigm®). We used two primer sets for each taxon, targeting the 16S ribosomal gene and the chaperonin groEL. Positive correlations (Pearson’s r > 0.5) were observed between the two methods for 16S and groEL primers sets (Supplementary Figure 4)
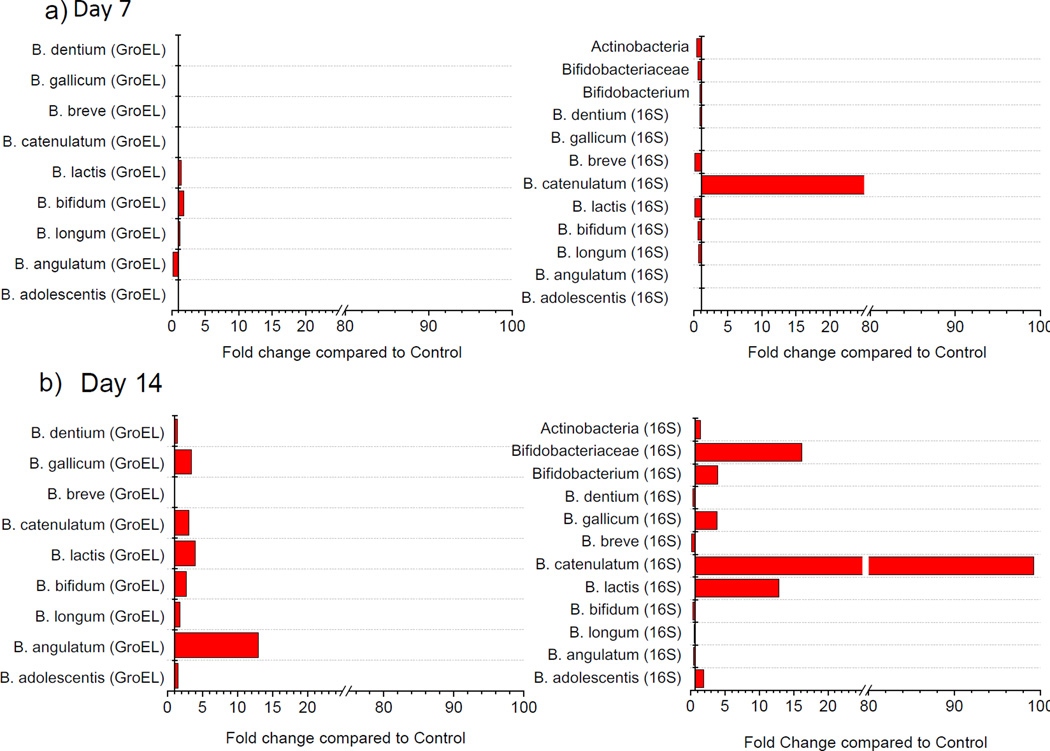
Data showed an increased abundance of Bifidobacteriaceae and Bifidobacterium (13- and 4-fold, respectively) after 14 days (Figure 6) as well as a marked increase in the abundance of B. adolescentis, B. catenulatum, B. lactis and B. gallicum. The B. catenulatum group (B. catenulatum and B. pseudocatenulatum) exhibited the largest increase in the GOS-fed group.
Figure 6. Absolute quantification of Bifidobacterium species by high-throughput qPCR.
Changes in abundance between GOS90 (n=12) and controls (n=15) are shown at a) day 7 (n=6) and b) day 14 (n=6). Left panels show data generated using GroEL-specific primers, right panels show data generated using 16S rRNA gene-specific primers.
A linear fit analysis performed to assess the correlation between 16S rRNA and groEL data for detection of Bifidobacterium species showed Pearson’s r correlation values > 0.70 for B. longum, B. adolescentis, B. gallicum and B. lactis, while lower correlation indices (Pearson’s r < 0.70) were observed for B. catenulatum, B. angulatum, B. dentium, B. bifidum and B. breve (Supplementary Figure 5). Data suggest that all primers targeting the groEL gene were adequate for all Bifidobacterium species with the exception of B. breve, while primers targeting the 16S rRNA gene were less optimal for B. angulatum, B. longum and B. bifidum.
Predictive analysis of metabolic functions associated with GOS90 feeding
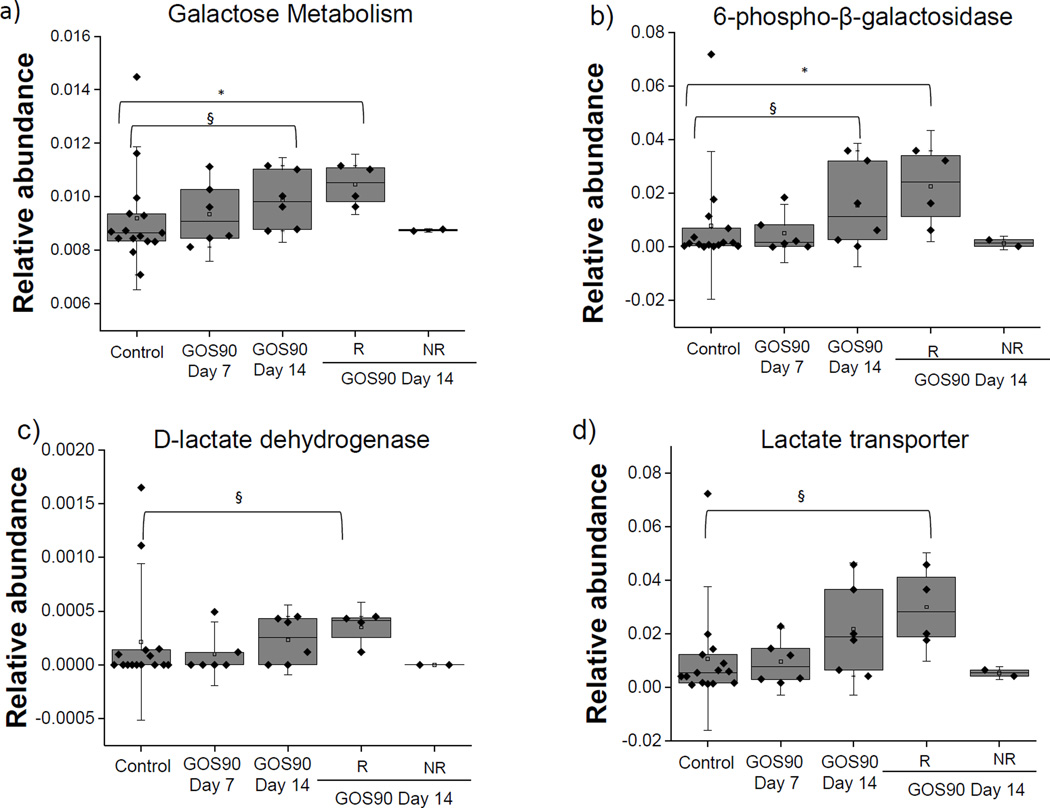
We next used PICRUSt to predict metabolic pathways and enzymes influenced by GOS. We identified a total of 328 pathways in all samples (control and prebiotics). We observed a non significant (Steel Dwass All Pairs p=0.14) over-representation of the galactose metabolism pathway in the GOS90 group at the end of the trial (Figure 7a). When data were stratified into responders and non-responders, a clear over-representation of this pathway was observed in responders (p=0.01), while representation of this pathway in non-responders was similar to the control group (Figure 7a). Similar patterns were observed for the starch and sucrose metabolism pathways (data not shown).
Figure 7. Relative abundances of specific KEGG pathways and enzymes impacted by GOS90 feeding.
a) Galactose metabolism pathway, b) 6-phospho-beta-galactosidase, c) D-lactate dehydrogenase and d) lactate transporter. Control n=15, GOS90 at day 7 n=6, GOS90 at day 14 n=6, Responders to GOS90 at day 14 (n=4) and Non-responders to GOS90 at day 14 (n=2). Boxes represent data between 25th and 75th percentile. Error bars indicate the 10th and 90th percentile. Steel-Dwass All Pairs tests § p ≤ 0.1, *p ≤ 0.05.
GOS90 feeding also resulted in the predicted over-representation of a D-lactate dehydrogenase and a lactate transporter from the LctP family (Figure 7b), and one gene involved in the metabolism of galactose, 6-phospho-β-galactosidase [EC 3.2.1.85] (Figure 7c). When data were further divided into responders and non-responders, differences became significant or close to significant for responders (Figures 7b and 7c).
Stool of prebiotic fed mice showed reduced concentrations of acetate, lactate and butyrate
Intermediate and end metabolites from GOS utilization (SCFAs, succinate, glycerol, lactate, glucose, aminobutyrate and galactose) were measured in stool samples by chromatography to determine if the prebiotic influenced SCFA generation. Butyrate, lactate, acetate, propanoate, glucose and glycerol were detected at their highest concentration in both groups at day 1 (no dietary modulation). Levels of isobutyrate, valerate, 2-aminobutyrate, succinate and galactose were on average below 1 µmol/g fecal sample (Table 4). We observed a significantly lower abundance of acetate (unpaired t-test p=0.03), and borderline significant decreases of lactate (p=0.09), and butyrate (p=0.1) in prebiotic-fed mice compared to control mice. The same results were observed when data from responders and non-responders were analyzed separately (not shown).
Table 4.
Concentration of fecal metabolites from control and GOS90-fed mice at the end of the feeding trial. Significant differences (unpaired t-tests p<0.1) between controls and prebiotic-fed mice and between controls and prebiotic responders are bold.
| Metabolite | Control (n =15) |
GOS90 (n = 6) |
GOS90 (Responders) (n = 4) |
p value (control vs GOS90) |
P value (control vs GOS90 responders) |
|---|---|---|---|---|---|
| Acetate | 6.93 ± 4.02 | 4.17 ± 1.65 | 3.52 ± 1.52 | 0.03 | 0.02 |
| Propanoate | 6.12 ± 5.39 | 3.19 ± 2.38 | 3.26 ± 2.90 | 0.17 | 0.16 |
| Isobutyrate | 0.24 ± 0.28 | 0.15 ± 0.19 | 0.08 ± 0.09 | 0.28 | 0.10 |
| Butyrate | 16.13 ± 8.99 | 10.41 ± 5.64 | 9.30 ± 5.41 | 0.10 | 0.06 |
| Valerate | 0.14 ± 0.16 | 0.11 ± 0.09 | 0.04 ± 0.02 | 0.58 | 0.05 |
| Lactate | 7.36 ± 6.90 | 2.22 ± 3.83 | 2.73 ± 4.54 | 0.09 | 0.11 |
| 2-aminobutyrate | 0.14 ± 0.06 | 0.13 ± 0.10 | 0.13 ± 0.13 | 0.90 | 0.99 |
| 4-aminobutyrate | 2.15 ± 0.93 | 1.61 ± 0.85 | 1.33 ± 0.86 | 0.40 | 0.08 |
| Glycerol | 3.62 ± 3.07 | 2.55 ± 2.69 | 3.26 ± 2.97 | 0.71 | 0.81 |
| Succinate | 0.12 ± 0.10 | 0.15 ± 0.08 | 0.16 ± 0.09 | 0.36 | 0.39 |
| Galactose | 0.90 ± 0.71 | 0.64 ± 0.57 | 0.79 ± 0.63 | 0.62 | 0.74 |
| Glucose | 4.64 ± 3.22 | 3.34 ± 3.12 | 4.00 ± 3.57 | 0.57 | 0.71 |
GOS90 did not modify inflammatory biomarkers
Pro-inflammatory cytokines IL-6, IL-12, IL-1β, IFN-γ, TNF-α and the anti-inflammatory cytokine IL-10 were quantified by ELISA in the proximal and distal colon of the control and GOS90 groups. No significant differences were detected between groups; however, we observed different cytokine profiles in the proximal and distal colon in both groups. Specifically, in the distal colon the pro-inflammatory cytokines IL-6 and IL-12 were detected at lower levels while levels of TNF-α were higher. Levels of IL-1β and IFN-γ in both colon sections, and IL-10 in the distal colon, were below the method’s detection limits (Table 5).
Table 5.
Cytokine levels of the proximal and distal colon of control (n=3) and GOS90-fed female mice at day 14 (n=6). Quantification was performed by ELISA. Values are expressed as pg/mg protein.
| Proximal colon | Distal colon | |||
|---|---|---|---|---|
| Control | GOS90 | Control | GOS90 | |
| IL-12 | 19.37 ± 14.57 | 16.7 ± 10.94 | 6.50 ± 3.38 | 6.00 ± 14.95 |
| IL-6 | 14.71 ± 10.98 | 34.38 ± 20.91 | 9.40 ± 6.17 | 14.09 ± 14.95 |
| TNF-α | 20.80 ± 8.46 | 22.34 ± 6.01 | 47.92 ± 6.39 | 47.24 ± 9.63 |
| IL-10 | 10.74 ± 14.71 | 4.83 ± 7.44 | <LOD | <LOD |
| IFN-γ | <LOD | <LOD | <LOD | <LOD |
| IL-1β | <LOD | <LOD | <LOD | <LOD |
LOD: Limit of detection
Discussion
The aim of this study was to evaluate the impact of a highly pure GOS formulation (GOS90)(23), composed of GOS (90%) and lactose (10%) on the gut microbiome of wt 129 mice. No significant differences were observed in starting values at day 1. In addition, since we intended to focus our analysis on the longitudinal effect of GOS90 rather than cross-sectional differences, we performed most comparisons between the GOS90 group at 7 and 14 days and the overall control. Our data showed that GOS90 feeding for 14 days resulted in an increase of fecal relative abundance of the bifidobacteria Bifidobacterium adolescentis, B. catenulatum, B. lactis, B. gallicum and specific OTUs of Lactobacillus, Bacteroidetes and Clostridiales. Duration of treatment was chosen based on previously published research in humans and mice, which reported range treatments from 7 days (50, 51) to 12 weeks (10). Our study showed an specific increase of B. catenulatum and a decrease of Clostridium at day 7 of the feeding trial in agreement with the study by Nakayama and Oish (50),which reported that, in mice, 7 days of GOS treatment were enough to identify changes in the microbiota that resulted in decreases of Clostridium and E. coli and increases in Bifidobacterium abundances.
Our approach used a dose of 0.26 g GOS90/kg weight administered daily over 14 days. Previous human studies reported a prebiotic impact on gut microbiome composition at doses ranging from 4 to 20 g/day with inulin, fructo-oligosaccharides (FOS) and GOS (52). With FOS or inulin doses higher than 15 g/day (equivalent to 0.21 g/kg weight/day) inducing flatulence or abdominal bloating (53, 54). Most of the studies with GOS in humans used a recommended dosage of 8 to 15 g/day (0.11 to 0.21 g/kg weight/day) (11), although the bifidogenic effect has been observed at doses as low as 5 g/day (10, 17, 55). In animal studies, increased abundance of Bifidobacterium and Lactobacillus has been reported in mice fed 1 g of GOS/kg weight(56) and with diets supplemented with Oligomate at a 1% concentration (57). The impact of GOS on the gut microbiome of laboratory rats has not been studied but animals administered between 2.5 and 5g/kg weight/day of GOS showed no significant adverse toxicological effects attributable to the prebiotic (55).
A fiber-rich diet has been associated with increased microbial diversity (58), while a low gut microbial diversity has been correlated with recurrent Clostridium difficile infection, Crohn’s disease, and obesity (59, 60). In this study, we observed a marginal, statistically insignificant increase in diversity and species richness in GOS90-fed mice. These results suggest that longer periods of prebiotic feeding could result in a higher bacterial diversity.
Interestingly, we observed gender-specific differences in the fecal microbiome of control mice, with lower bacterial diversity in males and an over-representation of the butyrate producers Roseburia and Butyricicoccus in female mice. A study by Bernbom et al. showed that the fecal microbiota of SPF rats and human microbiota-associated rats clustered according to the gender of the host animal (61). Our results are consistent also with the observations by Markle et al. who suggested that sex hormones could influence microbial populations (62). Moreover, a type 1 diabetes study in non-obese diabetic (NOD) mice showed a hormone-supported increase of selected microbial taxa, which may work as a positive-feedback mechanism contributing to the sexual dimorphism of autoimmune diseases (63). More recently, Bolnick et al.(64) analyzed associations between gut microbiota and sex*diet, and showed that microbial responses to diet depended on sex in laboratory mice fed either a control diet or a high fat diet. These and our data warrant more studies to elucidate the mechanisms driving differences in the gut microbiota associated with sex*diet.
Impact of GOS90 on the composition of the fecal microbiome varied between individual mice. Moreover, since in our study control mice did not receive any carrier solution, it is possible that stress had an additional impact on GOS90 fed mice. Specifically, four out of six treated mice (responders) showed an increase of up to 15-fold in Bifidobacterium abundance while two animals (non-responders) showed no response. The selective response to prebiotics has been previously reported in humans (10, 17). Also, the enrichment of bifidobacteria at the expense of other groups such as Bacteroides has been reported in humans fed GOS (10, 19). In addition, in our study we detected a reduced representation of other Bacteroidetes taxa including unclassified Bacteroidales and Parabacteroides. Species of these taxa are often associated with opportunistic infections, mainly intra-abdominal and systemic, as well as harboring some antimicrobial resistance traits that could be transferred to other strains (65).
Since 16S amplicon sequencing methods do not have enough sensitivity to identify bacterial taxa at the species level, we developed and optimized a high-throughput real-time qPCR assay using the Biomark instrument by Fluidigm to quantify bifidobacterial species. Although we observed an overall positive correlation between primers targeting the 16S rRNA and groEL genes for detection of Bifidobacterium species, our analysis suggests that primers targeting the groEL gene efficiently detected and quantified the species selected in our study with the exception of B. lactis. Conversely, primers targeting the 16S rRNA gene were less optimal for B. angulatum, B. longum and B. bifidum. The groEL gene has been reported to be more discriminative for the detection and quantification of Bifidobacterium species (46), in part because it is present in one copy per genome. We observed striking differences in the levels of detection of the B. catenulatum group with the two different primer sets, which are explained by the different specificity of each set. The groEL primer is specific for B. catenulatum, while the oligonucleotide targeting the 16S ribosomal gene can detect both B. catenulatum and B. pseudocatenulatum. This result indicates a more pronounced expansion of B. pseudocatenulatum in response to the prebiotic.
Data obtained with this new platform confirmed pyrosequencing results and showed an increased abundance of B. pseudocatenulatum, B. gallicum, B. lactis and B. angulatum in prebiotic-fed mice at day 14. The most dramatic effect was observed in the B. catenulatum group (B. catenulatum and B. pseudocatenulatum) with a 100-fold expansion. Significant GOS-induced increases of this group have been also reported in humans (10). In fact, a study that tested the growth of Bifidobacterium species with different prebiotics showed that, of those tested, GOS was the only prebiotic to support growth of B. pseudocatenulatum (66). An increase of 13-fold was also observed in B. angulatum; β-galactosidases extracted from B. angulatum have been used for the utilization and generation of GOS with high concentrations of lactose (67). Finally, we also observed proliferation of B. lactis, a species widely used as a probiotic capable of using GOS (68, 69). In addition, synbiotics—where this species and GOS act synergistically—have been used in different clinical studies proving their beneficial effect on health (70, 71).
In this study, we observed a decreased abundance of Helicobacter and Clostridium in GOS90-fed mice. Previous studies have reported a reduction of colitis severity in Smad3-deficient mice treated with the pathogen Helicobacter as well as reduction of Clostridium by GOS in vitro and in mice (72–74). In addition to their impact on gut bacterial taxa, prebiotics have been shown to enhance host health by directly inhibiting adherence of pathogens to the host epithelial cell surface (75) and modulating the colon microenvironment, which results in a reduction of intestinal infections (13, 76). The role of Helicobacter in gastric cancer is well-documented (77, 78) and recent reports suggest a potential role of this bacterium in IBD (79–81). Likewise, Clostridium has been correlated with increased incidence and growth of colonic tumors in animal studies (82, 83). Further studies are needed to elucidate the role and potential mechanisms involved in reduction of pathogenic species by the prebiotic used in this study.
Short-chain fatty acids (SCFAs), especially butyrate, have an important role in host health since they function as energy source for host tissues, have antimicrobial activity and have anti-inflammatory and anti-proliferative properties (84, 85). In the present study, we observed a significant decrease in the fecal concentration of acetate, lactate, and butyrate in prebiotic-fed mice. SCFAs’ fecal concentration reflects the amount remaining after colonic absorption. Although an inhibited microbial production of these SCFAs cannot be excluded, the reduction of these metabolites could reflect an increase in colonic absorption and reduced excretion. In fact, Vogt and Wolever reported a negative correlation between acetate fecal concentration and absorption suggesting that fecal SCFA concentrations may better reflect colonic SCFA absorption than production (86). Undoubtedly, these results warrant further experiments to elucidate generation and fate of SCFA in the colon of GOS90-fed animals.
We used PICRUSt to infer metagenome functional content from 16S rRNA amplicon sequencing input data. Since GOS are composed of galactose and glucose molecules joined in a β-configuration, we specifically looked at pathways and enzymatic functions involved in galactose metabolism. We observed an enrichment of the galactose metabolism pathways in the prebiotic group that became more evident when only responders were included in the analysis. We also observed an over-representation of pathways involved in the metabolism of starch and sucrose, which suggest that GOS-metabolizing bacteria could also have the ability to use other prebiotics such as resistant starch. In fact, studies have reported increases in the relative abundance of Bifidobacterium and Lactobacillus when starch was used as prebiotic (89, 90). A lactate dehydrogenase, an enzyme that catalyzes the conversion from pyruvate to lactate, and a lactate transporter from the LctP family were also over-represented in the prebiotic group. We also observed a significant over-representation of the 6-phospho-beta-galactosidase [EC 3.2.1.85], an enzyme involved in the utilization of GOS that hydrolyzes the 6-phospho-β-D-galactosides previously phosphorylated and translocated by PTS transporters. Otherwise, we did not observe significant changes in β-galactosidases that are more commonly used by some lactic acid bacteria or by Bifidobacterium (91). This could be explained by the GOS90 fermentation by other species whose abundance was higher than Bifidobacterium and that use mainly the PEP-PTS system to ferment galactose, species such as Streptococcus, Lactococcus and some species of Lactobacillus, like L. casei and L. acidophilus (92, 93).
Studies have shown that prebiotics can attenuate the pro-inflammatory response in intestinal epithelial cells in animals model of disease (94–96). Modulation of inflammatory biomarkers by GOS90 was not observed in prebiotic fed mice due most probably to the fact that this study was conducted in healthy wt mice with no inflammatory conditions. Overall, in this study we demonstrated that GOS90 has the potential to modulate the gut microbiota and its metabolites, specifically enriching for Bifidobacterium and Lactobacillus and promoting a reduction in Bacteroidales and potential pathogens such as Clostridium and Helicobacter in mice. Future studies will evaluate this prebiotic alone or in conjunction with probiotics in animal models of inflammation and colon cancer to evaluate its potential role in prevention and treatment.
Supplementary Material
Acknowledgments
A. Monteagudo-Mera was supported by a fellowship from the Foundation Alfonso Martin Escudero (Spain). We are grateful to Scott Magness and Carlton Anderson at the UNC Advanced Analytics Core for their support. The Microbiome Core Facility and the Advanced Analytics Core are supported in part by the NIH/National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK34987.
References
- 1.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, Finlay BB. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dibaise J, Frank DN, Mathur R FRCPC. Impact of the gut microbiotaon the development of obesity: Current concepts. Am J Gastreoenterol Sppl. 2012;1:22–27. [Google Scholar]
- 5.Rowland IR. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des. 2009;15:1524–1527. doi: 10.2174/138161209788168191. [DOI] [PubMed] [Google Scholar]
- 6.Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev. 2004;17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 7.Sangwan V, Tomar SK, Singh RR, Singh AK, Ali B. Galactooligosaccharides: novel components of designer foods. J Food Sci. 2011;76:R103–R111. doi: 10.1111/j.1750-3841.2011.02131.x. [DOI] [PubMed] [Google Scholar]
- 8.Watson D, O'Connell Motherway M, Schoterman MH, van Neerven RJ, Nauta A, van Sinderen D. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol. 2013;114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 9.Ben XM, Li J, Feng ZT, Shi SY, Lu YD, Chen R, et al. Low level of galactooligosaccharide in infant formula stimulates growth of intestinal Bifidobacteria and Lactobacilli. World J Gastroenterol. 2008;14:6564–6568. doi: 10.3748/wjg.14.6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis LM, Martinez I, Walter J, Goin C, Hutkins RW. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One. 2011;6:e25200. doi: 10.1371/journal.pone.0025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macfarlane GT, Steed H, Macfarlane S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol. 2008;104:305–344. doi: 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- 12.Tzortzis G, Vulevic J. Galacto-oligosaccharide. In: Charalampopoulos D, RA R, editors. Prebiotics and Probiotics: Science and Technology. Guildford: Springer; 2009. pp. 207–243. [Google Scholar]
- 13.Shoaf K, Mulvey GL, Armstrong GD, Hutkins RW. Prebiotic galactooligosaccharides reduce adherence of enteropathogenic Escherichia coli to tissue culture cells. Infect Immun. 2006;74:6920–6928. doi: 10.1128/IAI.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzortzis G, Goulas AK, Gee JM, Gibson GR. A novel galactooligosaccharide mixture increases the bifidobacterial population numbers in a continuous in vitro fermentation system and in the proximal colonic contents of pigs in vivo. J Nutr. 2005;135:1726–1731. doi: 10.1093/jn/135.7.1726. [DOI] [PubMed] [Google Scholar]
- 15.Searle LE, Best A, Nunez A, Salguero FJ, Johnson L, Weyer U, Dugdale AH, Cooley WA, Carter B, Jones G, Tzortzis G, Woodward MJ, La Ragione RM. A mixture containing galactooligosaccharide, produced by the enzymic activity of Bifidobacterium bifidum, reduces Salmonella enterica serovar Typhimurium infection in mice. J Med Microbiol. 2009;58:37–48. doi: 10.1099/jmm.0.004390-0. [DOI] [PubMed] [Google Scholar]
- 16.Quintero M, Maldonado M, Perez-Munoz M, Jimenez R, Fangman T, Rupnow J, Wittke A, Russell M, Hutkins R. Adherence inhibition of Cronobacter sakazakii to intestinal epithelial cells by prebiotic oligosaccharides. Curr Microbiol. 2011;62:1448–1454. doi: 10.1007/s00284-011-9882-8. [DOI] [PubMed] [Google Scholar]
- 17.Davis LM, Martinez I, Walter J, Hutkins R. A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. Int J Food Microbiol. 2010;144:285–292. doi: 10.1016/j.ijfoodmicro.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Walton GE, van den Heuvel EG, Kosters MH, Rastall RA, Tuohy KM, Gibson GR. A randomised crossover study investigating the effects of galacto-oligosaccharides on the faecal microbiota in men and women over 50 years of age. Br J Nutr. 2012;107:1466–1475. doi: 10.1017/S0007114511004697. [DOI] [PubMed] [Google Scholar]
- 19.Vulevic J, Juric A, Tzortzis G, Gibson GR. A Mixture of trans-Galactooligosaccharides Reduces Markers of Metabolic Syndrome and Modulates the Fecal Microbiota and Immune Function of Overweight Adults. J Nutr. 2013;143:324–331. doi: 10.3945/jn.112.166132. [DOI] [PubMed] [Google Scholar]
- 20.Silk DB, Davis A, Vulevic J, Tzortzis G, Gibson GR. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther. 2009;29:508–518. doi: 10.1111/j.1365-2036.2008.03911.x. [DOI] [PubMed] [Google Scholar]
- 21.Rastall RA, Maitin V. Prebiotics and synbiotics: towards the next generation. Curr Opin Biotechnol. 2002;13:490–496. doi: 10.1016/s0958-1669(02)00365-8. [DOI] [PubMed] [Google Scholar]
- 22.Sangwan V, Tomar SK, Singh RR, Singh AK, Ali B. Galactooligosaccharides: novel components of designer foods. J Food Sci. 2011;76:R103–R111. doi: 10.1111/j.1750-3841.2011.02131.x. [DOI] [PubMed] [Google Scholar]
- 23.Dagher SF, Azcarate-Peril MA, Bruno-Barcena JM. Heterologous expression of a bioactive beta-hexosyltransferase, an enzyme producer of prebiotics, from Sporobolomyces singularis. Applied and Environmental Microbiology. 2013;79:1241–1249. doi: 10.1128/AEM.03491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCafferty J, Muhlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–2125. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, Possemiers S, Van Holle A, Francois P, de Vos WM, Delzenne NM, Schrenzel J, Cani PD. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dagher SF, Azcarate-Peril MA, Bruno-Barcena JM. Heterologous expression of a bioactive beta-hexosyltransferase, an enzyme producer of prebiotics, from Sporobolomyces singularis. Appl Environ Microbiol. 2013;79:1241–1249. doi: 10.1128/AEM.03491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruno-Barcena JM, Azcarate-Peril MA. Galacto-oligosaccharides and Colorectal Cancer: Feeding our Intestinal Probiome. J Funct Foods. 2015;12:92–108. doi: 10.1016/j.jff.2014.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devine AA, Gonzalez A, Speck KE, Knight R, Helmrath M, Lund PK, Azcarate-Peril MA. Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PLoS One. 2013;8:e73140. doi: 10.1371/journal.pone.0073140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Frontiers in cellular and infection microbiology. 2015;5:3. doi: 10.3389/fcimb.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One. 2010;5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome research. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Junick J, Blaut M. Quantification of human fecal bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl Environ Microbiol. 78:2613–2622. doi: 10.1128/AEM.07749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70:167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermann-Bank ML, Skovgaard K, Stockmarr A, Larsen N, Molbak L. The Gut Microbiotassay: a high-throughput qPCR approach combinable with next generation sequencing to study gut microbial diversity. BMC Genomics. 2013;14:788. doi: 10.1186/1471-2164-14-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon H-S, Yang E-H, Lee S-H, Yeon S-W, Kang B-H, Kim T-Y. Rapid identification of potentially probiotic Bifidobacterium species by multiplex PCR using species-specific primers based on the region extending from 16S rRNA through 23 S rRNA. FEMS Microbiology Letters. 2006;250:55–62. doi: 10.1016/j.femsle.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 41.Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, Arai H, Tanimoto I, Nishimura F, Takashiba S. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 2003;39:81–86. doi: 10.1016/S0928-8244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- 42.Hermann-Bank ML, Skovgaard K, Stockmarr A, Larsen N, Molbak L. The Gut Microbiotassay: a high-throughput qPCR approach combinable with next generation sequencing to study gut microbial diversity. BMC Genomics. 14:788. doi: 10.1186/1471-2164-14-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gueimonde M, Tolkko S, Korpimaki T, Salminen S. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl Environ Microbiol. 2004;70:4165–4169. doi: 10.1128/AEM.70.7.4165-4169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinne MM, Gueimonde M, Kalliomaki M, Hoppu U, Salminen SJ, Isolauri E. Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota. FEMS Immunol Med Microbiol. 2005;43:59–65. doi: 10.1016/j.femsim.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Junick J, Blaut M. Quantification of human fecal bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl Environ Microbiol. 2012;78:2613–2622. doi: 10.1128/AEM.07749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 48.Ohland CL, Kish L, Bell H, Thiesen A, Hotte N, Pankiv E, Madsen KL. Effects of Lactobacillus helveticus on murine behavior are dependent on diet and genotype and correlate with alterations in the gut microbiome. Psychoneuroendocrinology. 2013;38:1738–1747. doi: 10.1016/j.psyneuen.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Martinez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, Moriyama EN, Walter J. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakayama T, Oishi K. Influence of coffee (Coffea arabica) and galacto-oligosaccharide consumption on intestinal microbiota and the host responses. FEMS Microbiol Lett. 2013;343:161–168. doi: 10.1111/1574-6968.12142. [DOI] [PubMed] [Google Scholar]
- 51.Ito M, Deguchi Y, Miyamori A, Matsumoto K, Kikuchi H, Matsumoto K, Kobayashi Y, Yajima T, Kan T. Effects of administration of galactooligosaccharides on the human faecal microflora, stool weight and abdominal sensation. Microbial Ecology in Health and Disease. 1990;3:285–292. [Google Scholar]
- 52.Tuohy KM, Rouzaud GC, Bruck WM, Gibson GR. Modulation of the human gut microflora towards improved health using prebiotics--assessment of efficacy. Curr Pharm Des. 2005;11:75–90. doi: 10.2174/1381612053382331. [DOI] [PubMed] [Google Scholar]
- 53.Pedersen A, Sandstrom B, Van Amelsvoort JM. The effect of ingestion of inulin on blood lipids and gastrointestinal symptoms in healthy females. Br J Nutr. 1997;78:215–222. doi: 10.1079/bjn19970141. [DOI] [PubMed] [Google Scholar]
- 54.Stone-Dorshow T, Levitt MD. Gaseous response to ingestion of a poorly absorbed fructo-oligosaccharide sweetener. Am J Clin Nutr. 1987;46:61–65. doi: 10.1093/ajcn/46.1.61. [DOI] [PubMed] [Google Scholar]
- 55.Anthony JC, Merriman TN, Heimbach JT. 90-day oral (gavage) study in rats with galactooligosaccharides syrup. Food Chem Toxicol. 2006;44:819–826. doi: 10.1016/j.fct.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 56.Pan XD, Chen FQ, Wu TX, Tang HG, Zhao ZY. Prebiotic oligosaccharides change the concentrations of short-chain fatty acids and the microbial population of mouse bowel. Journal of Zhejiang University. Science. B. 2009;10:258–263. doi: 10.1631/jzus.B0820261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos A, San Mauro M, Diaz DM. Prebiotics and their long-term influence on the microbial populations of the mouse bowel. Food Microbiol. 2006;23:498–503. doi: 10.1016/j.fm.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 58.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 60.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernbom N, Norrung B, Saadbye P, Molbak L, Vogensen FK, Licht TR. Comparison of methods and animal models commonly used for investigation of fecal microbiota: effects of time, host and gender. J Microbiol Methods. 2006;66:87–95. doi: 10.1016/j.mimet.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 62.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 63.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, Lusis AJ, Knight R, Caporaso JG, Svanback R. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014;5:4500. doi: 10.1038/ncomms5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boente RF, Ferreira LQ, Falcao LS, Miranda KR, Guimaraes PL, Santos-Filho J, Vieira JM, Barroso DE, Emond JP, Ferreira EO, Paula GR, Domingues RM. Detection of resistance genes and susceptibility patterns in Bacteroides and Parabacteroides strains. Anaerobe. 2010;16:190–194. doi: 10.1016/j.anaerobe.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 66.Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol. 2014;87:30–40. doi: 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 67.Rabiu BA, Jay AJ, Gibson GR, Rastall RA. Synthesis and fermentation properties of novel galacto-oligosaccharides by beta-galactosidases from Bifidobacterium species. Appl Environ Microbiol. 2001;67:2526–2530. doi: 10.1128/AEM.67.6.2526-2530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gopal PK, Ramsay R, Sullivan PA, Smart JB. Growth promoting effects of galactooligosaccharides on B. lactis HN010 (DR10) and L. rhamnosus HN001 (DR20) American Journal of Clinical Nutrition. 2001;73:496S–496S. [Google Scholar]
- 69.Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol. 2013;79:4304–4315. doi: 10.1128/AEM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prasad J, Sazawal S, Dhingra U, Gopal PK. Detection of viable Bifidobacterium lactis HN019 (DR10TM) in stools of children during a synbiotic dietary intervention trial. International Dairy Journal. 2013;30:64–67. [Google Scholar]
- 71.Martinez RC, Aynaou AE, Albrecht S, Schols HA, De Martinis EC, Zoetendal EG, Venema K, Saad SM, Smidt H. In vitro evaluation of gastrointestinal survival of Lactobacillus amylovorus DSM 16698 alone and combined with galactooligosaccharides, milk and/or Bifidobacterium animalis subsp. lactis Bb-12. Int J Food Microbiol. 149:152–158. doi: 10.1016/j.ijfoodmicro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 72.Morishita Y, Oowada T, Ozaki A, Mizutani T. Galactooligosaccharide in combination with Bifidobacterium and bacteroides affects the population of Clostridium perfringens in the intestine of gnotobiotic mice. Nutrition Research. 2002:1333–1341. [Google Scholar]
- 73.Hopkins MJ, Macfarlane GT. Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl Environ Microbiol. 2003;69:1920–1927. doi: 10.1128/AEM.69.4.1920-1927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gopalakrishnan A, Clinthorne JF, Rondini EA, McCaskey SJ, Gurzell EA, Langohr IM, Gardner EM, Fenton JI. Supplementation with galacto-oligosaccharides increases the percentage of NK cells and reduces colitis severity in Smad3-deficient mice. J Nutr. 2012;142:1336–1342. doi: 10.3945/jn.111.154732. [DOI] [PubMed] [Google Scholar]
- 75.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annual Review of Nutrition. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 76.Arslanoglu S, Moro GE, Schmitt J, Tandoi L, Rizzardi S, Boehm G. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J Nutr. 2008;138:1091–1095. doi: 10.1093/jn/138.6.1091. [DOI] [PubMed] [Google Scholar]
- 77.Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to pylori. Int J Cancer. 2014;136:487–490. doi: 10.1002/ijc.28999. [DOI] [PubMed] [Google Scholar]
- 78.Polk DB, Peek RM., Jr Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laharie D, Asencio C, Asselineau J, Bulois P, Bourreille A, Moreau J, Bonjean P, Lamarque D, Pariente A, Soule JC, Charachon A, Coffin B, Perez P, Megraud F, Zerbib F. Association between entero-hepatic Helicobacter species and Crohn's disease: a prospective cross-sectional study. Aliment Pharmacol Ther. 2009;30:283–293. doi: 10.1111/j.1365-2036.2009.04034.x. [DOI] [PubMed] [Google Scholar]
- 80.Hansen R, Hold GL, Berry SH, Thomson JM, El-Sakka NE, Bisset WM, Mahdi G, Murray GI, Helms PJ, El-Omar EM. The prevalence of non-pylori Helicobacter organisms in paediatric inflammatory bowel disease. Gut. 2009;58:A289. [Google Scholar]
- 81.Whary MT, Danon SJ, Feng Y, Ge Z, Sundina N, Ng V, Taylor NS, Rogers AB, Fox JG. Rapid onset of ulcerative typhlocolitis in B6.129P2-IL10tm1Cgn (IL-10−/−) mice infected with Helicobacter trogontum is associated with decreased colonization by altered Schaedler's flora. Infect Immun. 2006;74:6615–6623. doi: 10.1128/IAI.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Onoue M, Kado S, Sakaitani Y, Uchida K, Morotomi M. Specific species of intestinal bacteria influence the induction of aberrant crypt foci by 1,2-dimethylhydrazine in rats. Cancer Lett. 1997;113:179–186. doi: 10.1016/s0304-3835(97)04698-3. [DOI] [PubMed] [Google Scholar]
- 83.Horie H, Kanazawa K, Okada M, Narushima S, Itoh K, Terada A. Effects of intestinal bacteria on the development of colonic neoplasm: an experimental study. Eur J Cancer Prev. 1999;8:237–245. doi: 10.1097/00008469-199906000-00012. [DOI] [PubMed] [Google Scholar]
- 84.Vinolo MA, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly YM, Stephens L, Hawkins PT, Curi R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One. 2011;6:e21205. doi: 10.1371/journal.pone.0021205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 86.Vogt JA, Wolever TM. Fecal acetate is inversely related to acetate absorption from the human rectum and distal colon. J Nutr. 2003;133:3145–3148. doi: 10.1093/jn/133.10.3145. [DOI] [PubMed] [Google Scholar]
- 87.Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- 89.Senevirathne RN, Janes M, Keenan MJ, Martin RJ, Raggio AM, McCutcheon KL, Tulley RT, Zhou J. Dietary resistant starch increases Bifidobacterium spp./ Lactobacillus spp and Clostridia spp. in the gut of mice fed low and moderate fat diets. FASEB J. 2009;28 [Google Scholar]
- 90.Silvi S, Rumney CJ, Cresci A, Rowland IR. Resistant starch modifies gut microflora and microbial metabolism in human flora-associated rats inoculated with faeces from Italian and UK donors. J Appl Microbiol. 1999;86:521–530. doi: 10.1046/j.1365-2672.1999.00696.x. [DOI] [PubMed] [Google Scholar]
- 91.Schwab C, Ganzle M. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol Lett. 2011;315:141–148. doi: 10.1111/j.1574-6968.2010.02185.x. [DOI] [PubMed] [Google Scholar]
- 92.Kanatani K, Tahara T, Yoshida K, Miura H, Sakamoto M, Oshimura M. Plasmid-linked Galactose Utilization by Lactobacillus acidophilus TK8912. Biosci. Biotech. Biochem. 1992;56:826–827. doi: 10.1271/bbb.56.826. [DOI] [PubMed] [Google Scholar]
- 93.Chassy BM, Thompson J. Regulation and characterization of the galactose-phosphoenolpyruvate-dependent phosphotransferase system in Lactobacillus casei. J Bacteriol. 1983;154:1204–1214. doi: 10.1128/jb.154.3.1204-1214.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tanabe S, Hochi S. Oral administration of a galactooligosaccharide preparation inhibits development of atopic dermatitis-like skin lesions in NC/Nga mice. Int J Mol Med. 2010;25:331–336. doi: 10.3892/ijmm_00000349. [DOI] [PubMed] [Google Scholar]
- 95.Komiyama Y, Andoh A, Fujiwara D, Ohmae H, Araki Y, Fujiyama Y, Mitsuyama K, Kanauchi O. New prebiotics from rice bran ameliorate inflammation in murine colitis models through the modulation of intestinal homeostasis and the mucosal immune system. Scand J Gastroenterol. 2011;46:40–52. doi: 10.3109/00365521.2010.513062. [DOI] [PubMed] [Google Scholar]
- 96.Gourbeyre P, Desbuards N, Gremy G, Tranquet O, Champ M, Denery-Papini S, Bodinier M. Perinatal and postweaning exposure to galactooligosaccharides/inulin prebiotics induced biomarkers linked to tolerance mechanism in a mouse model of strong allergic sensitization. J Agric Food Chem. 2013;61:6311–6320. doi: 10.1021/jf305315g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.