SUMMARY
Early B cell development is characterized by large scale Igh locus contraction prior to V(D)J recombination to facilitate a highly diverse Ig repertoire. However, an understanding of the molecular architecture that mediates locus contraction remains unclear. We have combined high resolution chromosome conformation capture (3C) techniques with 3D DNA FISH to identify three conserved topological sub-domains. Each of these topological folds encompasses a major VH gene family that become juxtaposed in pro-B cells via Mb-scale chromatin looping. The transcription factor Pax5 organizes the sub-domain that spans the VHJ558 gene family. In its absence the J558 VH genes fail to associate with the proximal VH genes, thereby providing a plausible explanation for reduced VHJ558 gene rearrangements in Pax5-deficient pro-B cells. We propose that Igh locus contraction is the cumulative effect of several independently controlled chromatin sub-domains that provide the structural infrastructure to coordinate optimal antigen receptor assembly.
INTRODUCTION
The mechanisms that govern V gene usage in VDJ rearrangements are central to understanding the formation of the BCR and TCR repertoires. Chromatin conformation and coordinated chromosomal movements govern the clustering of genes in transcription machines and the matrix of interactions specifying regulatory element associations. The Igh locus undergoes several different chromosomal movements that ensure developmental-stage and lineage specific DNA recombination and transcription including relocation from the nuclear periphery to the center and re-organization of the Igh locus chromatin topology during B cell ontogeny (Fuxa et al., 2004; Kosak et al., 2002; Sayegh et al., 2005). In the mouse, there are ~100 functional VH gene segments that are scattered over 2.5 mega-bases (Mb) of the Igh locus that must recombine with a rearranged DJH element assembled from 1 of 8–12 DH and 1 of 4 JH gene segments. In primary pro-B cells of the bone marrow (BM), RAG recombinase, mediates V(D)J or VJ joining for both Ig H and L chain genes. However, the molecular mechanism by which the distal VH genes gain spatial proximity to the rearranged DHJH gene segments remains obscure.
Chromatin compaction has been studied extensively by cytological methods. Three dimensional (3D) DNA fluorescent in situ hybridization (FISH) studies in pro-B cells indicate that the Igh locus contracts and this process is inferred to juxtapose distal VH genes near to proximal DH segments to promote V(D)J joining (Fuxa et al., 2004; Jhunjhunwala et al., 2008; Kosak et al., 2002). Locus contraction requires the transcriptional regulators, Pax5, YY1 and Ikaros (Fuxa et al., 2004; Liu et al., 2007; Reynaud et al., 2008). Loss of Igh locus compaction is correlated with the biased usage of the proximal VH gene segments (Hesslein et al., 2003). The degrees of locus compaction are inferred from relationships of interprobe nuclear distances versus genomic distances. However, FISH based measurements have limited resolution (100–1000 nm) and it has been difficult to ascertain the identity of specific DNA sequences that mediate locus contraction. The advent of chromosome conformation capture (3C) and related methods allows examination of pairwise chromatin interactions at the molecular level (~1–100 nm) in cell populations (Gibcus and Dekker, 2013). 3C based methods can delineate long range chromatin looping interactions and have been successfully used to reveal large scale chromatin organizations that are congruent with FISH studies (Bickmore and van Steensel, 2013). However, looping interactions specifying locus contraction remain poorly defined and one recent study has suggested that distal VH gene contacts with DHJH elements are stochastic (Medvedovic et al., 2013).
Chromosomes are organized into higher order spatial architectures of multiple length scales (Gibcus and Dekker, 2013). Independent compartments of euchromatin and heterochromatin form at intermediate length scales of 1–10 Mb within chromosomal territories (Lieberman-Aiden et al., 2009). Chromatin is further organized into Mb sized topologically associating domains (TADs) that represent spatial zones of high frequency self-interacting chromatin contacts (Dixon et al., 2012; Nora et al., 2012). Many TADs show a high degree of alignment with discrete transcriptionally repressive nuclear lamina-associated domains (LADs) that occur at variable stages of development (Nora et al., 2012). Although TADs are conserved between mouse and human and are invariant during development, focal facultative chromatin folding regulating gene expression can occur on the sub-Mb scale without changing TAD organization (Dixon et al., 2012; Nora et al., 2012). We reasoned that mapping Igh locus chromatin topologies might allow identification of functional long-range interactions and their underlying loop anchors that mediate locus contraction.
Here we examine the relationship between higher order chromatin structure and gene function at several levels using the 3C based methodology, chromosome conformation capture carbon-copy (5C) in combination with 3D DNA FISH. We find that the Igh locus spans a multi-Mb sized topological fold that in turn corresponds remarkably well to the previously described Igh LAD (Zullo et al., 2012). We find a nested hierarchy of constitutive chromatin interactions that serve to structure conserved sub-topologies within the Igh topological fold that range in size from ~0.44 to 1.1 Mb. At the pro-B cell stage of development these sub-domains achieve spatial proximity by means of chromatin looping over multi-Mb-scale distances to collectively generate locus contraction. We find that the transcription factor, Pax5 is required for specific pro-B cell looping interactions and these contacts are independent of the cis-regulatory element, Eμ. We provide a molecular definition of locus contraction by identifying loop anchor sites that are key mediators of this process and consider the functional implications of hierarchical layers of chromatin folding and compaction on VH gene usage and IgH diversity.
RESULTS
The Igh locus is organized as a compartment with several sub-topological domains
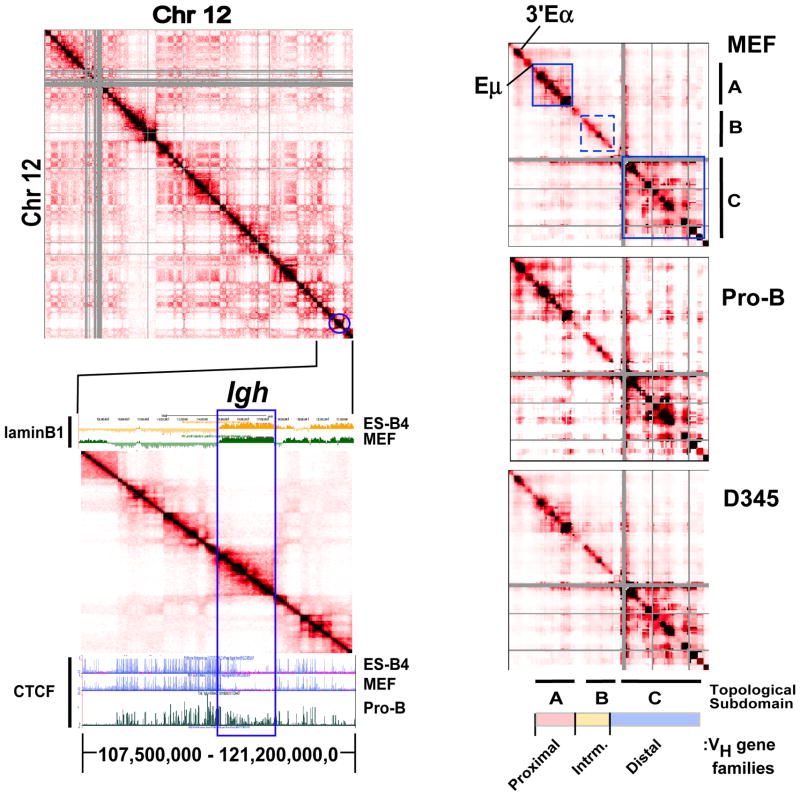
Comprehensive genome wide mapping of chromatin architecture using HiC, is based on proximity ligation chromosome conformation capture (3C) assays, and permits visualization of higher order chromosomal landscapes (Lieberman-Aiden et al., 2009). To gain insight into the chromatin organization of the Igh locus, HiC data for chromosome 12 from an AMuLV pro-B cell line (Zhang et al., 2012) and ES cells (Dixon et al., 2012) generated in a different studies were analyzed. The 2D interaction matrices constructed using 100 kilobase binning analyses shows all pairwise interaction frequencies captured by HiC along chromosome 12 for the pro-B cell line (Fig. 1A, upper panel). The diagonal reflects interaction frequencies along the chromosome between fragments that are neighbors in the genome whereas color off the diagonal represents long range looping interactions. The frequency of interactions is inversely related to the genomic distance separating the pair of restriction fragments. The Igh locus, indicated by the blue circle, is located close to the telomeric end of chromosome 12 (Fig. 1A, upper panel). TADs are discrete blocks, spanning on average 0.8 Mb in mammalian cells and range up to several Mb, within which the pairwise contacts are relatively frequent and are relatively conserved between cell types (Dixon et al., 2012; Nora et al., 2012). TADs are visible along the diagonal of chromosome 12 and are readily evident in an expanded view of the HiC heatmap (Fig. 1A, lower panel) (Fig. 1A). Compartments are large chromosomal domains spanning several Mb that preferentially interact with other compartments (Lieberman-Aiden et al., 2009) and are visible as a plaid pattern off the diagonal (Fig. 1A, upper panel). Compartments tend to be cell type- or differentiation stage specific. The Igh locus is encompassed within a single 2.9 Mb self-associating domain that is pro-B cell stage specific as it is absent in ES cells (Fig. 1A, lower panel; also see Fig. S1). However, the Igh domain is also a relatively autonomous unit since it interacts infrequently with other compartments (Fig. 1A, upper panel). Thus, Igh topological fold is largely insulated from other chromosome 12 contacts. We refer to the spatial arrangement of the Igh locus as an insulated topological fold (i-TOF).
Figure 1. The Igh locus partitions into a 2.9 Mb TAD and conserved topological sub-domains.
A) (Upper panel) Normalized HiC interaction frequencies from an AMuLV pro-B cell line (adapted from Zhang et al 2012 (Zhang et al., 2012)) representing chromosome 12 is displayed as a 2D heatmap binned at 100 kb resolution. HiC sequencing reads are indicated by the color (red) intensity from interacting pairs of Hind III fragments. Gray pixels indicate areas of low uniquely mapped reads or regions with many large restriction fragments (>100 kb). (Lower panel) HiC data from chromosome 12 (107,500,000–121,200,000,0; mm9) is shown aligned to a UCSC Genome Browser snap-shot showing ChIP-seq for CTCF (Lin et al., 2012) and Dam-ID lamin B1 data (Peric-Hupkes et al., 2010) in the indicated cell types. B) 5C analyses of the 2.9 Mb Igh locus in MEF, Rag deficient pro-B cells and the AMuLV pro-B cell line, D345. Normalized 5C data underwent binning analysis (bin size, 150,000 kb; step size,15 kb). Topological sub-domains A, and C are indicated by the solid blue outline and B by dashed blue lines). The heat maps are scaled as follows: MEF 37–1840, Rag2−/− pro B cells: 19–1254, D345: 89–6355.
The Igh locus is associated with the nuclear lamina (NL) in non-lymphoid cells and multipotent progenitors but relocates away from the NL to the transcriptionally permissive nuclear center in lineage committed pro-B cells (Zullo et al., 2012). We investigated the relationship between the Igh i-TOF and LAD by leveraging previously constructed maps of genome-NL interactions established using laminB1 (LMNB1) DNA adenine methyltransferase identification (DamID) protocols (Peric-Hupkes et al., 2010; Zullo et al., 2012). We aligned LMNB1-DamID maps along with HiC data from a segment of chromosome 12 that spans the Igh locus and its flanking regions (Fig. 1A, lower panel). In ESC and MEF the Igh LAD is essentially coincident with the Igh locus as indicated by the blue rectangle (Fig. 1A, lower panel; also see Fig. S2, Table S1). Furthermore, in MEF and ESC, the activating histone modifications H3K4me1 and H3K27Ac are underrepresented throughout the transcriptionally silent Igh LAD whereas these marks are present in flanking regions (Fig. S2). In marked contrast, H3K4me1 is enriched across the transcriptionally active Igh locus in pro-B cells (Fig. S2) and the LAD is absent (Zullo et al., 2012). Our finding that the Igh LAD and i-TOF are coincident is intriguing since it implies that developmentally regulated DNA elements differentially contribute to chromatin organizational features supporting LAD and i-TOF scaffolds.
A counterpoint to the loss of Igh LAD NL-association in pro-B cells is the acquisition of CTCF and cohesin (Rad21) binding. CTCF is an architectural protein that contributes to the establishment of a 3D spatial organization of chromatin fibers and to Igh locus compaction (Degner et al., 2011; Guo et al., 2011a). Cohesin is a highly conserved multiprotein complex that includes Rad21, functions during DNA replication to mediate the association of sister chromatids and is an interaction partner with CTCF (Seitan and Merkenschlager, 2011). Using publically available data sets we observe that CTCF decorates the Igh locus in pro-B cells and its binding largely coincides with the cohesin subunit, Rad21, in accord with earlier studies (Fig. 1a, lower panel; also see Fig. S2) (Degner et al., 2009). CTCF occupancy is greatly diminished in MEF, as previously noted (Degner et al., 2009) as well as in ESC, and thymus (Fig. 1, lower panel, and Fig. S1). The absence of CTCF binding was not due to a technical failure of the assay since CTCF abundance was high in region flanking the Igh locus for ESC, MEF and thymus (Fig. 1A, lower panel and Fig. S1). Genomewide CTCF binding occurs both at invariant sites that are conserved between species and tissues and at sites that show cell type specific distribution (Cuddapah et al., 2009). It is striking therefore, that occupancy of the Igh locus with CTCF, Rad21 and activating histone marks occurs across the entire i-TOF and is contemporaneous with disengagement from the NL. These observations strongly imply that the Igh LAD provides a regulated framework in which alterations in nuclear positioning and chromatin structure occur in a domain wide fashion during B cell development.
Unique topological substructures in the Igh locus
To examine cell type specific patterns of higher order chromatin organization in the Igh locus at high resolution, we performed 5C in a massively parallel manner at distinct stages of B cell development. 5C, a high throughput derivative of 3C, involves selective amplification of chromatin interactions captured by proximity ligation and permits definition of chromatin structure at the level of single restriction fragments (Fig. S3A). An alternating forward and reverse 5C primer design interrogated in cis over 12,000 potential Igh long range interactions and 525 interactions at the chromosome 5 gene desert, using two biological replicates of MEFs and Rag deficient pro-B cells, as described (Kumar et al., 2013) and an Abelson transformed Rag2-deficient pro-B cell line, D345 (Fig. S3B). We assessed our 5C results by examining the consistency between biological replicates. Heatmaps of our raw 5C data were similar for biological replicates at both the Igh locus and the chromosome 5 gene desert (Fig. S3C). 5C signals were well correlated between biological replicates (pro-B 1 versus pro-B 2, Spearman correlation coefficient 0.723; MEF1 versus MEF2, Spearman correlation coefficient 0.888) when considering counts for all fragment combinations and indicate that the 5C libraries are high quality and consistent between replicates.
We visualized the normalized 5C heatmaps for MEF, Rag2-deficient pro-B cells and the D345 cell line (Fig. 1B; also see Table S2). The 5C data was submitted to binning analyses over 150 kilobases (kb) revealing that high density chromatin contacts occurred preferentially within two sub-domains A and C, ~0.44 and 1.2 Mb, respectively, indicated by the intense red color and demarcated by the blue boxes (Fig. 1B). A weaker set of interactions occurred within sub-domain B (indicated by the dashed blue box). The overall locations and sizes of the sub-domains are conserved in MEFs, pro-B cells and D345 cells indicating constancy of genomic folding patterns in different cell types (Fig. 1B). However, in primary pro-B cells sub-domain A also appears to encompass the 3′ end of the Igh locus including the B cell specific 3′Eα enhancer and highlights potential lineage specific looping interactions (Fig. 1B). The disposition of the 3′Eα enhancer is examined in greater detail in the sections below where direct sample to sample comparison of 5C interaction frequencies are analyzed.
We next examined the distribution of VH gene families with respect to Igh topological folds. The C57Bl/6 mouse strain contains 195 VH genes that are arrayed across ~2.4 Mb of the Igh locus (Johnston et al., 2006). The J558 gene cluster, located at the distal 5′-end of the locus is the largest grouping of VH exons (Fig. 1B). The 7183 VH gene family comprises the next largest group (~0.4 Mb) and is situated at the proximal end of the locus closely abutting the DH segment cluster. The intermediate VH genes are comprised by ten smaller families of VH genes (~0.5 Mb) located between the 7183 and J558 families (Fig. 1B). Topological sub-domain A spans from around the intronic Eμ, through the JH and DH clusters and includes proximal 7183 VH genes (Fig. 1B). Notably, in pro-B cells, sub-domain A extends from 3′Eα enhancer and extends through the proximal 7183 VH genes (Fig. 1B). The 3′ end of topological sub-domain C is close to the boundary for the J558 genes (Johnston et al., 2006). The two highly structured topological blocks, A and C, are separated by the less structured B interval spanning most of the intermediate VH genes (Fig. 1B). Thus, VH gene families are discretely segregated within specific topological folds. Conservation of the topological fold structure of the Igh locus in MEF and pro-B cells suggests that many chromatin interactions are constitutive and conserved through mouse development. Looping interactions unique to pro-B cells could be obscured by a plethora of constitutive interactions common to MEF and primary pro-B cells.
Igh locus spatial organization defined by constitutive and cell type specific chromatin contacts
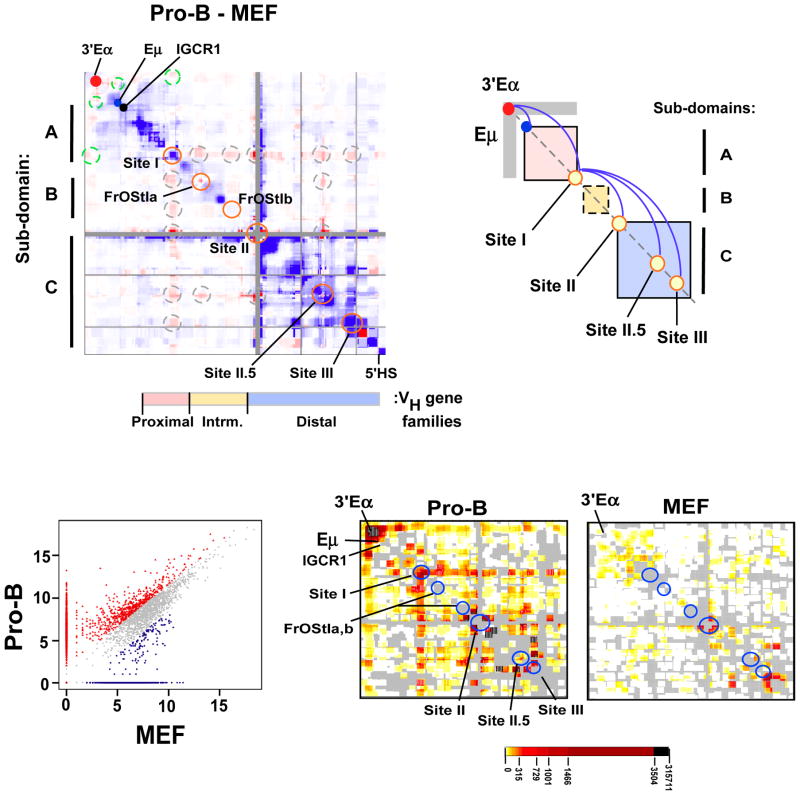
We explored whether focal facultative chromatin contacts between subdomains A (DH/JH) and C (distal VH) contribute to unique chromatin folding in pro-B cells. First, we constructed a difference heatmap generated by subtracting MEF from primary pro-B 5C normalized datasets to identify pro-B cell- (red intensities) and MEF- (blue intensities) specific interactions (Fig. 2A). The difference heatmap allows visualization of the relative frequencies of looping interactions within pro-B and MEF chromatin samples. The heatmap is aligned with the VH gene families and the Igh topological sub-domains A–C, oriented along the X and Y axis, respectively. We found two groups of very long range pro-B cell- specific interactions that are indicated by the dashed circles off the diagonal (Fig. 2A). The first group (green dashed circles) is anchored by 3′Eα and associates with several sites including Eμ and Site I located at the 3′- and 5′ ends of topological sub-domain A, respectively (Fig. 2A). The second group of contacts, anchored at Site I (gray dashed circles), are termed meta-long range loops, (or meta-loops) as they span exceptionally long distances between topological sub-domains (Fig. 2A). Looping contacts anchored at Sites I, II, II.5, III, Friend of Site I a (FrOStI a) and FrOStI b are enriched in the primary proB cells (orange circles along the diagonal), are not evident in MEF and are distinct from interactions previously noted (Guo et al., 2011a; Jhunjhunwala et al., 2008) (Fig. 2A). Site I anchors chromatin looping with sites in sub-domain B (FrOStI a and FrOStI b), and sub-domain C (Sites II, II.5 and III) containing the intermediate and distal VH genes, respectively (Fig. 2A,B). The simultaneous engagement of these loop anchors that span the entire Igh locus may be responsible for locus contraction. Sites I, II, III and FrOStIa display CTCF, Rad21 and Mediator1 binding. Detailed analyses of these sites are underway and will be reported separately.
Figure 2. Developmentally programmed reorganization of the Igh locus chromatin architecture in pro-B cells.
A) A difference plot (pro-B cells minus MEF) was calculated using normalized 5C signals (150 kb bins, 15 kb step). Elevated 5C reads in pro-B (red intensities), or MEF (blue intensities) or constitutive (white). Dots are: 3′Eα (red), Eμ (blue) and IGCR1 (black). Looping interactions associated with 3′Eα (dashed green circles). Sites I (115,100,000–115,239,000; 139kb), II (115,949,000–116,069,000; 120kb), II.5 (116,665,000–116,770,000; 105kb), III (116,784,000–116,874,000; 90kb) and FrOStIa (115,411,000–115,451,000; 40kb), FrOStIb (115,819,000–115,859,000; 40kb) are indicated along the diagonal (orange circles) and looping interactions off the diagonal are shown (dashed gray circles). Genomic coordinates are chr12, mm9. VH gene families and topological sub-domains A–C are indicated. B) A diagram of topological sub-domains A (pink), B (yellow), C (blue) boxes with Eμ and 3′Eα, as indicated. Sites I–III are shown (yellow circles). Gray rectangular arms extending from 3′Eα indicate increased interactions in pro-B cells. The pro-B cell specific loops (blue arches). C) Scatterplot of 5C interactions (colored dots) (constitutive, gray; pro-B cell, red; MEF, blue). Cell type specific interactions were >3-fold above the constitutive frequency (see Suppl. Methods). D) The pro-B and MEF specific 5C interactions defined in (B) are plotted as 2D heatmaps (100 kb bins, 10kb step). The frequency of 5C reads is indicated by the color key and gray indicates the absence of interactions.
To further examine the involvement of Sites I–III in defining pro-B cell chromatin topologies we normalized the 5C data sets using the chromosome 5 gene desert interactions as a denominator (SI; Table S3, S,4 S5; Fig. S4). Chromatin interactions within the gene desert do not change in different cell types. Our method produces an interaction frequency that is directly comparable between experiments and permits detection of significant fragment-to-fragment looping contacts above the expected background interactions. Igh interactions are identified which are common to both MEF and pro-B cells (gray dots), or preferentially expressed in pro-B cells (red dots) or MEF cells (blue dots) (Fig. 2C). Constitutive 5C interactions (66%; gray dots) differed less than 3-fold between MEF and pro-B cells whereas, interactions that displayed a greater than a 3-fold difference were defined as cell type specific. Interactions that were exclusively detected in pro-B or MEF cells are located along the X- or Y axis, respectively (Fig. 2C). We examined the pro-B and MEF specific interactions in 2D heatmaps to discover cell type specific looping interactions. We found that interactions involving 3′Eα were elevated in pro-B cells and were nearly undetectable in MEF (Fig. 2D). Frequent chromatin contacts are detected in pro-B cells between 3′Eα with Eμ, IGCR1 and Site I, and Site I with FrOStIa, FrOStIb, Site II.5 and Site III (Fig. 2D). We and others have previously noted pro-B cell specific chromatin looping anchored at 3′Eα and associated with Eμ and IGCR1 (Degner et al., 2009; Guo et al., 2011a; Guo et al., 2011b; Kumar et al., 2013). Our findings indicate that locus contraction results from pro-B cell specific focal facultative chromatin associations through Site I and not from random compaction of the locus (Fig. 2B).
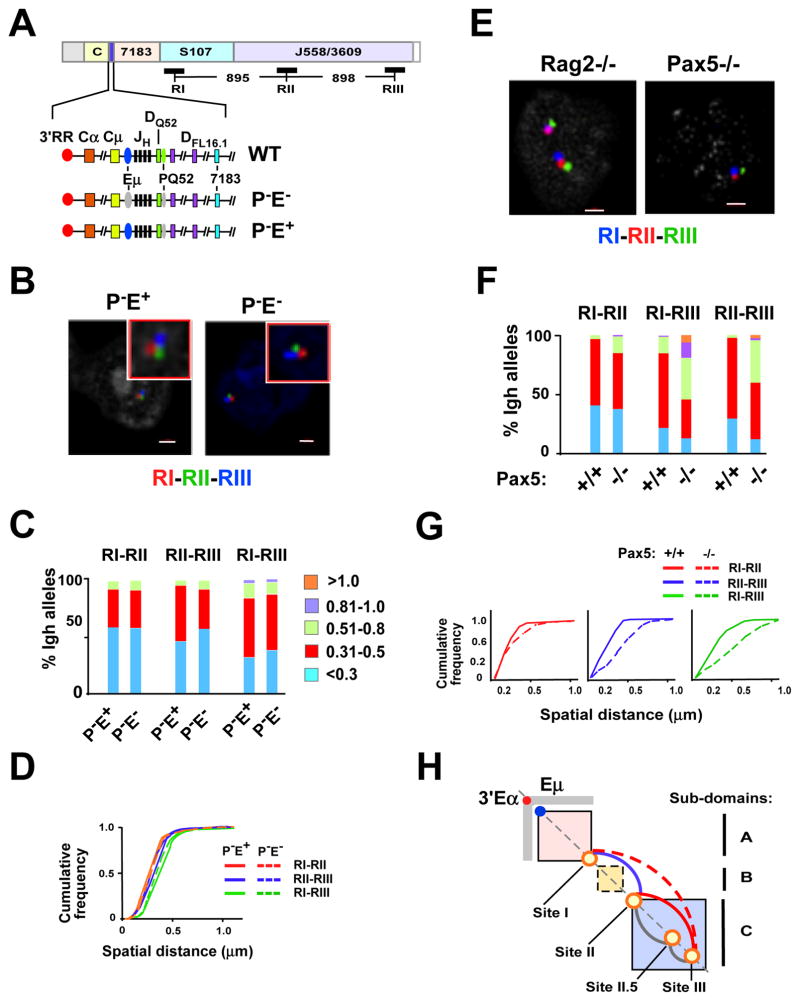
Spatial proximity of loop-attachment Sites I–III is pro-B cell specific
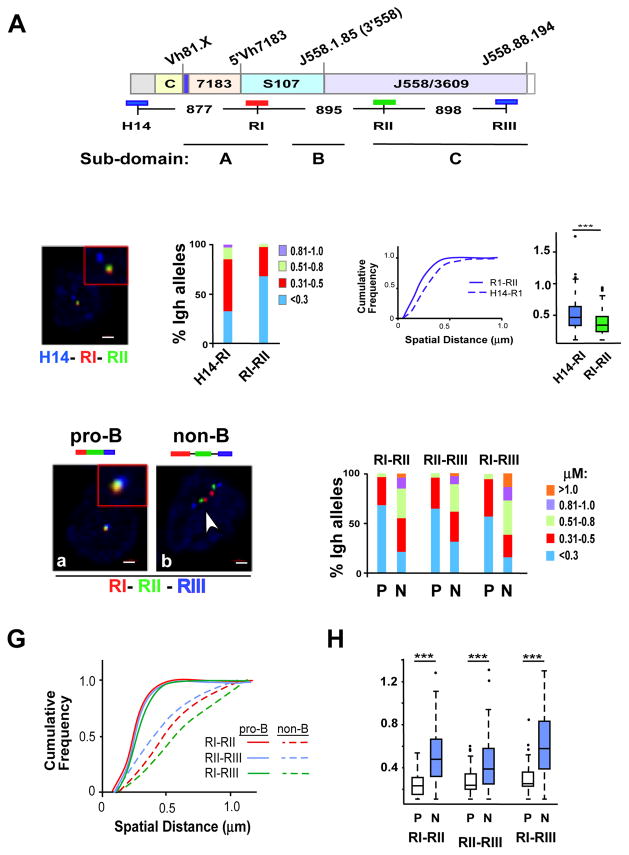
Assays based on 3C detect the average chromatin interaction frequency in a cell population. We used 3D DNA FISH to independently assess the chromatin interaction frequency between specific anchor sites in single cells. We first tested the prediction that two loci within a TAD are spatially closer to each other than two loci that are separated by the same genomic distance but situated in adjacent domains. For this we used bacterial artificial chromosome (BAC) probes H14, RI and RII that are separated by 877kb (H14-RI) and 895 kb (RI–RII), respectively (Fig. 3A) in quantitative 3D-FISH analyses. H14 maps 100 kb outside the 2.9 Mb Igh TAD indicated by Hi-C, and RI and RII correspond closely to Sites I and II identified by 5C (Fig. 3A). Quantitative three color 3D-FISH analyses in primary pro-B cells from RAG2-deficient mice showed that spatial distances between RI–RII probes were, on average, significantly smaller than H14-RI inter-probe distances (Fig. 3B–D). These observations confirmed that Sites I and II lay within the Igh i-TOF, whereas H14 did not. The frequent superposition of probes RI and RII in pro-B cells (approximately 70% of nuclei examined) strongly supported the identification of long-distance interactions between Sites I and II by 5C.
Figure 3. Sites I, II and III are loop attachment sites in pro-B cells.
A) Diagram of the C57Bl/6 Igh locus is shown with genomic distances (Johnston et al., 2006). The BAC probes used in FISH studies are indicated by the blue, red and green lines below the Igh locus. Topological sub-domains A (114,665,117–115,239,000), B (115,239,000–115,949,000), and C (116,069,000–116,874,000) and all genomic coordinates are chr12, mm9. The Igh locus contains approximately 100 VH gene segments that span 2.4 Mb. The interspersed distal VH segments at the 5′-end of the locus are comprised of the J558 and 3609 families. The 7183 family is located at the proximal end of the locus. The intermediate VH segments are comprised of the S107 family along with 9 smaller VH families. Representative VH gene segments are positioned above the locus, Vh81.X (Vh7183.2.3; 114816717–114817010), 5’7183 (Vh7183.20.37; 115097360–115097653), 3’558 (VhJ558.1.85; 115707286–115707582), (VhJ558.88.194; 117238239–117238532). The vertical blue bar on the locus located between the constant region genes (CH) and 7183 VH segments contains the joining (JH) and diversity (DH) gene segments. The CH genes encode Igh isotypes and span an additional 220kb. Not shown: Eμ is located between the JH and CH gene segments and 3′Eα an enhancer at the 3′-end of the locus. B) Three color FISH using purified bone marrow Rag2 deficient pro-B cells. BAC probes are indicated in (A), and color-coded as indicated. A representative nucleus is shown. Probes labeled with Alexa Fluor 594 (red), 488 (green) and Alexa Fluor 697 (blue) were hybridized to fixed pro-B cells. Probe signals were from epifluorescence microscopy and the distances between probes were computed as described (Jhunjhunwala et al., 2008). Probes combinations were red and green (RI–RII), red and blue (H14-RI). C) Quantitation of FISH data. The distance between the red, green and blue FISH signals shown in (B) for 100–300 alleles were divided into 4 categories: <0.3, 0.31–0.5, 0.51–0.8, 0.81–1.0 μm. The percentage of Igh alleles in each category was determined (y axis) for each probe combination and is represented in different colors. Probe combinations are indicated on the x-axis below the histograms. Purified pro-B cells from at least three mice were used for each experiment. D) (Left panel) Cumulative frequency analyses are for each probe combination. (Right panel) Boxplots indicate the first and third quartiles and the band inside the box is the median distance between the probes. The whiskers indicate the 9th and 91st percentiles for probe distances and outliers are indicated by the dots. P values were calculated using the Mann-Whitney U test. *** indicates p < 0.0001. E) Three color FISH using purified BM pro-B and non-B cells on the Rag2 deficient background. Cells are from at least three mice. BAC probes are shown in (A). Probes combinations were red and green (RI–RII), red and blue (RI–RIII) and green and blue (RII–RIII). F) Quantitation of FISH data. 3D FISH signals shown in (E) for ~100–300 alleles were quantitated as described in (C). Probe combinations are shown above the histograms. G,H) P values are calculated in Kolmogorov-Smirnov and the Wilcoxon tests (see Table S6). Cumulative frequency analyses (G) and boxplots (H) are shown for each probe combination in each cell type.
5C analysis indicated that long distance interactions between Sites I, II and III were enriched in pro-B cells and under-represented in MEFs (Fig. 2). To independently assess cell lineage specificity of these interactions, we performed 3D-FISH using Rag2-deficient pro-B and non-B lineage cells. BAC probe RIII corresponded closely to Site III identified by 5C and the three probes were separated by equidistant genomic intervals (Fig. 3A). All three pair-wise FISH signals between probes RI–RII, RII–RIII and RI–RIII were essentially super-imposed in 60–80% of pro-B cell alleles (Fig. 3E–H; also see Table S6). This was most remarkable for RI–RIII interactions where the probes are located almost 2Mb apart. In contrast, probes RI–RIII were spatially clustered in only approximately 18% of non-B lineage cells (Fig. 3E,F), suggesting a major reduction in co-location. However, there is also a degree of intrinsic association that was independent of cell lineage. The conserved topological sub-domains that are supported by constitutive chromatin interactions in MEFs and pro-B cells may be the basis for under-representation, but not absence, of these interactions.
To further examine the proposition that Sites I-II-III represented preferred pro-B-specific interaction sites, we used a probe, R.I.5, that lies between RI and RII in FISH assays (Fig. S5A). If looping is random, then genomic proximity of probes RI.5-RII (separated by 451kb) would favor their pairwise association compared to RI–RII (separated by 895kb) or RII-RII.5 (separated by 685kb). Site II.5, co-locates with FISH probe RII.5, is situated in subdomain C and is predicted to interact with Site II based on our 5C analyses (Fig. 3). We found that RI–RII and RII-RII.5 probe interactions were similar in pro-B cell alleles. Notably, the spatial separation between R1.5-RII was significantly greater than either of the other two interactions (Fig. S5B,C), further strengthening the notion that Sites I, II and III represented preferred, rather than random interaction sites.
Spatial organization of the Igh locus is configured by three-way looping of Sites I-II-III
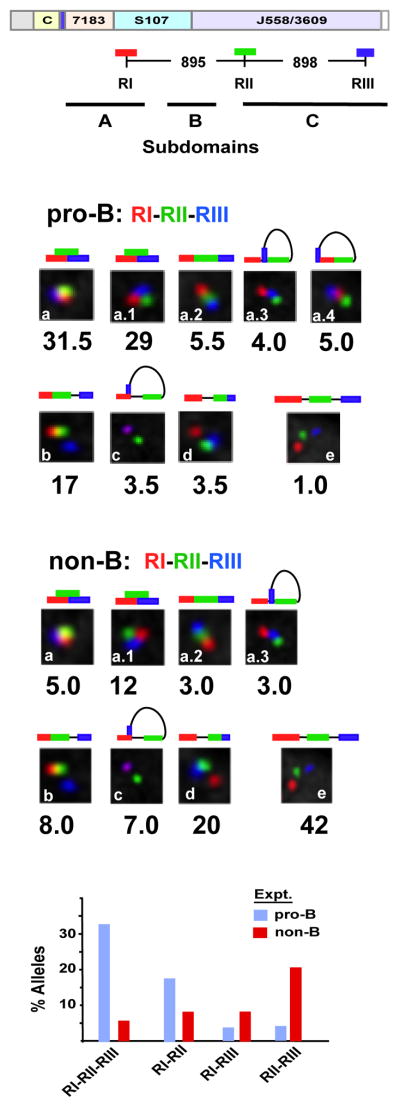
Because 5C assays assess chromatin interactions in populations of cells our 5C studies did not distinguish between pair-wise or more complex interactions among multiple interacting sites. However, the high frequency of pair-wise interactions between Sites I–II, II–III and I–III, strongly suggested that these sites interact with each other simultaneously (Fig. 3). To directly assess the frequency of three-way long-distance interactions we performed three color 3D-FISH with RI (red), RII (green) and RIII (blue) in primary pro-B cells (Fig. 4A). Guidelines used to score allele configurations in three color FISH assays are described (Fig. S6). Our FISH studies indicated that loop anchor Sites I, II and III were superimposed in three-way interactions in a high proportion (31.5%) of pro-B cell alleles and were spatially close on an additional 29% of alleles (Figure 4B, insets a and a.1). Superimposed probes are indicative of molecular contacts (Belmont, 2014). Conversely, probes RI-RII-RIII were spatially separated on only 1% of alleles in pro-B cells (Fig. 4B, inset e). We infer that Sites I–III engage in three-way interactions in a high proportion of pro-B cells.
Figure 4. Sites I-II-III are selectively superimposed in pro-B cells.
Three color FISH data for ~200 nuclei for each probe set. Allele configurations and probe distances are defined in Figure S6. Purified pro-B cells from at least three mice for each of two experiments. P values are derived in Kolmogorov-Smirnov and the Wilcoxon tests (see Table S6). A) A schematic of the WT Igh locus with genomic distances between BAC probes. The probes combinations are RI (red), RII (green) and RIII (blue). The topological sub-domains A, B and C that are derived from the 5C studies shown in Figure 2 are arrayed at bottom. B,C) BAC probes RI, RII and RIII, labeled as indicated, were hybridized simultaneously to Rag2-deficient pro-B cells (B) or non-B cells (C) from bone marrow of Rag2-deficient mice. The percentage of Igh alleles in various spatial configurations as schematized above each inset is indicated. D) Histograms summarizing the three color FISH results shown in (b) and (c) using the probe combinations RI, RII and RIII for pro-B cells (blue bars) and non-B lineage cells (red bars) (x-axis). The percentage of alleles in which two- or three-probes are superimposed or in very close contact are indicated (y axis).
The pattern of probe interactions found in pro-B cells was reversed in primary non-B lineage cells from the bone marrow. Here probes RI-RII-RIII overlapped on only 5.0% of alleles, relative to 31% in pro-B cells, and were spatially close on an additional 12% alleles, as compared to 29% in pro-B cells (Fig. 4C, insets a and a.1). Moreover, in non-B lineage cells the most prevalent configuration of alleles positioned probes RI-RII-RIII at a distance (42% compared to 1% in pro-B cells) (Fig. 4C, inset e), presumably reflecting linear un-contracted loci in these cells. Notably, probes RII–RIII are pair-wise associated in 20% of alleles of non-B lineage cells whereas this configuration occurs in only 3.5% of pro-B cell alleles (Fig. 4B, inset d and Fig. 4C, inset d). The presence of frequent RII–RIII interactions in non-B lineage cells implies that the J558 VH gene family is compacted in accord with the 5C findings (Fig. 2). We conclude that spatial aggregation of Sites I-II-III is pro-B-selective and that these interactions may comprise important determinants of VH locus compaction (Fig. 4D).
Meta-loop formation through Sites I, II and III is Eμ independent
The tissue specific Eμ enhancer is required for V(D)J joining (Afshar et al., 2006) and for mediating a subset of long range chromatin interactions that contribute to locus compaction (Guo et al., 2011a). Earlier studies showed that Eμ interacted with two sites in the VH region (Guo et al., 2011a). One of these sites, 5’7183, lies close to RI identified in this study; the other, referred to as 3’558, lies between RI and RII (Fig. 3A, 5A). We had proposed that Eμ-5’7183 and Eμ-3’558 associations bring the VH region close to the DJH part of the locus to facilitate VH recombination (Guo et al., 2011a). Our current observations suggested two possibilities: 1) that RI-II-III interactions were a part of Eμ-dependent locus contraction or 2) that RI-II-III interactions folded the VH domain independent of Eμ. To distinguish between these possibilities we performed three-color 3D FISH to measure RI-II-III associations on Igh alleles with defined deletions.
Figure 5. Locus contraction mediated by Sites I-II-III is Eμ independent but requires Pax5.
A) A diagram of the Igh locus indicates the location and distances between the three FISH probes RI, RII and RIII. The lower three lines show an expanded section of the Igh locus from 3′RR through the DH cluster and ending with the proximal VH gene segment, 7183. Eμ (blue oval) is a transcription enhancer located in the JH-Cμ intron and PQ52 (green oval) is a promoter associated with the 3′ most DH gene segment, DQ52. Deletions of PQ52 (P−) and/or Eμ (E−) are indicated (gray ovals). B) Three color FISH using purified Rag deficient pro-B cells that are deleted for PQ52 (P-E+) and for both PQ52 and Eμ (PE−). BAC probes are RI (red), RII (green) and RIII (blue). A representative nucleus is shown for each genotype. Probes contacts were red and green (RI–RII), red and blue (RI–RIII) and green and blue (RII–RIII). C) Quantitation of FISH data. The distance between the red, green and blue FISH signals shown in (B) for ~200 nuclei were divided into 5 categories: <0.3, 0.31–0.5, 0.51–0.8, 0.81–1.0, >1.0 μm. The percentage of Igh alleles in each category (y axis) for each Igh genotype (x-axis) is represented in different colors. Probe combinations are shown above the histograms. Purified pro-B cells from at least three mice were used for each experiment. Data are from at least two independent experiments. D) Cumulative frequency analyses are shown for each probe combination and the genotypes are indicated. P values are defined by Kolmogorov-Smirnov and Wilcoxon tests (see Table S6). E) Three color FISH using Rag2−/− or Pax5−/− pro-B cells and BAC probes are as in (A) and were labeled as follows: RI (blue), RII (red) and RIII (green). Representative nuclei from each genotype are shown. F) Distances between the probe signals were analyzed in ~100 nuclei. G) Cumulative frequency analyses are shown. P values are summarized in Table S6. H) A diagram of Igh topological subdomains A (pink box), B (yellow box) and C (blue box). Eμ (blue dot), 3′Eα (red dot) and Sites I, II, II.5 and III (orange circles) are located along the diagonal. The gray rectangular arms extending from 3′Eα indicate increased 5C interactions in pro-B cells. Loops that anchored at Sites I, II, II.5 and III and that are Pax5-dependent (red, dashed red), Pax5-independent (blue), and not tested (gray).
We used P−E+ alleles that lack a promoter associated with the 3′ most DH gene segment DQ52 and P−E− alleles that lack both the DQ52 promoter and Eμ (Afshar et al., 2006). P−E+ alleles behave indistinguishably from WT Igh alleles in most epigenetic and locus conformation assays, whereas P−E− alleles are substantially different (Guo et al., 2011a). We therefore attribute differences between these two alleles to the presence, or absence, of Eμ. Both Igh genotypes were examined in a RAG2-deficient context to preserve the locus in an unchanged configuration. Using purified primary pro-B cells we found that FISH signals from RI (red), RII (green), and RIII (blue) probes were virtually superimposable for both P-E+ or P-E− Igh alleles (Fig. 5B and quantified in C and D; also see Table S6). We conclude that interactions between Sites I–III represent Eμ-independent looping within the VH locus.
Site I–III mediated looping is Pax5-dependent
Contraction of the Igh locus in pro-B cells is altered by the transcription factor, Pax5 (Fuxa et al., 2004). This effect is thought to be mediated by Pax5 binding to a series of 14 Pax5-associated intergenic repeat (PAIR) sequences that are spread across 750 kb spanning the distal J558 VH genes. PAIR motifs are comprised of Pax5, E2A and CTCF binding sites (Ebert et al., 2011). Sites II.5 and III overlap with PAIR 6–8 and 10–11, respectively. PAX5 occupancy has been detected at these sites in vivo (Ebert et al., 2011). However, the means by which Pax5 binding to PAIR elements contracts the locus remains unclear. Because specific chromatin interaction sites that mediate Igh locus contraction have not been identified, one working model is that locus contraction occurs by mechanisms that are distinct from classical chromatin looping (Medvedovic et al., 2013). Therefore, it was of significant interest to determine the role of Pax5 in chromatin looping at Sites I, II, III. For this we carried out FISH with probes RI (blue), RII (red), and RIII (green) in pro-B cells from RAG2-deficient and Pax5-deficient mice (Fig. 5A,E). We found that Pax5-deficiency had little impact on RI–RII pair-wise association, but significantly disrupted interactions between Sites II–III as well as Sites I–III (Fig. 5F,G; also see Table S6). We conclude that Pax5 ensures interactions between Sites II and III (Fig. 5H, red line). When this interaction is disrupted in the absence of Pax5 the anchor Sites I–II are released from Site III, thereby altering the spatial distance between Sites I and III. That is, Pax5-independent interactions between Site I–II and Pax5-dependent interactions between Site II–III brings all three sites together in pro-B cells. Alternatively, Site I may also directly interact with Site III in a Pax5-dependent manner (Fig. 5H, dashed red line). Our three-color FISH studies indicate that interactions between Sites I–III and II–III are infrequent whereas the three-way association of Sites I-II–III is the most prevalent looping configuration in pro-B cell nuclei (Fig. 4D). Looping between Sites I and II is the next most frequent interaction and also very prevalent (Fig. 4D). Thus, we favor a model in which Pax5 facilitates locus contraction by promoting looping of Site III with Sites I and II (Fig. 5H).
DISCUSSION
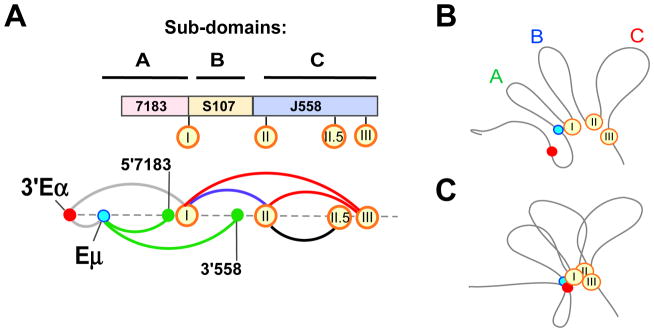
Our studies define several levels of topological fine structure within the IgH TAD/LAD that provide unique insights into the 3D architecture of the locus. First, HiC studies indicate that the Igh locus is encompassed within an i-TOF at the pro-B cell stage of development. Second, 5C studies reveal three distinct topological subdomains in pro-B cells and MEFs (Fig. 6). Subdomain A, that covers approximately 440 kb, extends from Eμ to the newly identified Site I. Subdomain C covers ~1.1 Mb and extends between newly identified Sites II and III. Extensive intra-domain contacts are evident within these two domains. The region between Sites I and II, that we refer to as subdomain B, differs from the other two domains in having many fewer intra-domain contacts.
Figure 6. Site I coordinates VH locus contraction.
A) (Top) Topological sub-domains A–C and VH gene families are drawn approximately to scale. Sites I, II, II.5 and III (orange circles) are aligned with their positions in topological subdomains and with respect to VH gene families. (Bottom) Meta-looping interactions anchored at Sites I, II, II.5 and III (orange circles) are Pax5-dependent (red arc), independent (blue arc) and not tested (black arc). Eμ-dependent loops (green arcs) (Guo et al., 2011a). 3′Eα loops (gray arc) with Eμ (Kumar et al., 2013) and Site I. B,C) Diagrams of sub-domain A, B and C. Sites I, II, and III (orange circles). Dots indicate 3′Eα (red), Eμ (teal). B) Non-B cells: topological sub-domains A, B and C are in an extended configuration. C) In pro-B cells, Site I loops with Sites II and III and with Eμ and 3′Eα to contract the locus.
Subdomain A, that encompasses proximal VH genes, in MEFs becomes subsumed within a larger subdomain that extends from Site I to near the 3′Eα enhancer at the 3′ end of the Igh locus in pro-B cells. These differences reflect chromatin interactions with the B lineage-specific Eμ and 3′Eα enhancers in pro-B cells. Eμ and 3′Eα were previously shown to interact together as well as with the 5′ end of the VH7183 gene family (Guo et al., 2011a; Kumar et al., 2013). Several earlier observations have shown that proximal VH genes are regulated differently from the rest of the VH genes. Proximal VH genes recombine normally whereas distal VH gene recombination is reduced in Pax5-, YY1-, and Ezh2-deficient pro-B cells (Fuxa et al., 2004; Hesslein et al., 2003; Liu et al., 2007; Reynaud et al., 2008). Thus, encapsulation of proximal VH genes together with the DH/JH part of the locus within subdomain A may, in part, distinguish them from distal VH genes that lie within subdomain C.
Intra-domain contacts within subdomain C occur in pro-B cells and MEFs. Earlier evidence suggests that CTCF, cohesin and Mediator-1 anchor constitutive architectural chromatin contacts between and within constitutive topological subdomains (Phillips-Cremins et al., 2013). However, the Igh locus, including the expanse of subdomain C, is devoid of these factors prior to the pro-B cell stage of development. We infer that constitutive topologies may be mediated by other, as yet undefined, means.
Third, our 5C and FISH studies identify long-range, B lineage-specific interactions between subdomains (Fig. 6A, blue and red arcs). These interactions, that we term meta-loops, serve to bring the three subdomains into spatial proximity in pro-B cells (Fig. 6B,C). The importance of interactions between Sites I, II and III is revealed in the juxtaposition of all three sites in approximately 60% of pro-B cells compared to <20% non-B cells. As a consequence, the entire VH region is brought into proximity of the DH/JH region of the locus to enable comparable access of all VH gene segments for recombination. It is intriguing that a small proportion Sites I, II and II are also in close proximity in non-B cells. One possibility is that this is stochastic. An interesting alternate possibility is that these regions are favored sites of interaction in non-B cells as well, and it is only the frequency of interaction that changes in proB cells. In other words, pro-B cells utilize constitutively-favored interactions to maximize locus compaction. The binding of architectural proteins, such as CTCF and Pax5, at Sites I, II and III in pro-B cells may facilitate such interactions.
Fourth, we demonstrate that Pax5 is essential to bring Site III into contact with Sites I and II (Fig. 6A, red arcs). Since Site I–II interactions are unaffected in Pax5-deficient pro-B cells, we conclude that Pax5 primarily drives interactions with Site III. It is noteworthy that the 14 PAIR elements that were proposed to mediate locus compaction via Pax5 are all located in the genomic region spanning subdomain C and PAIR motifs 10 and 11 overlap with Site III (Ebert et al., 2011). In the absence of Site III interactions VHJ558 gene segments (sub-domain C) fail to gain proximity to Site I and sub-domain A providing a plausible explanation for reduced VHJ558 rearrangements in Pax5-deficient pro-B cells. We also note that VH locus compaction, revealed as three-way interaction between Sites I, II and III does not require the intronic enhancer Eμ.
Since the pioneering studies of Kosak et al. it is clear that the 5′ and 3′ ends of the Igh locus are closer in pro-B cells compared to other non-B lineage cells (Kosak et al., 2002). The term locus compaction/contraction is frequently used to refer to this phenomenon. A recent HiC study was interpreted to suggest that in pro-B cells the 3′ end regulatory region (3′RR) located at the Igh locus boundary may function as a super-anchor that creates proximity between proximal and distal gene segments based on numerous CTCF directed looping interactions along the length of the locus {Benner, 2015 #832}. However, HiC analyses are relatively low resolution due to sequencing depth limitations in conjunction with very large binning windows (≥100kb) and therefore lack the sensitivity to identify interactions between specific motifs or determine whether those contacts are direct or indirect. An important conceptual point that emerges from our high resolution 5C studies is that locus compaction is the result of several overlapping phenomena that are mediated by discrete and independent chromatin loops. We propose that meta-loops serve to compact the conserved topological folds of sub-domains A–C. The topological folds may gain their structure in pro-B cells via CTCF-dependent short-range locus compaction. Thus, partial locus compaction and its consequences are evident in special circumstances. Pax5-induced locus compaction (Fuxa et al., 2004) is focused on Site III and only affects distal VH genes, whereas Eμ-dependent locus compaction (Guo et al., 2011a) affects interaction between the 3′ Igh region (sub-domain A) and the VH region (Fig. 6A, green arcs). Accordingly, only distal VH gene recombination is affected in Pax5-deficient cells (Hesslein et al., 2003). Because Eμ deletion is pleiotropic its effect on VH recombination per se is difficult to deconvolute. In summary, we propose that the Igh locus is contracted by means of meta-loops that are anchored at Sites I–III and are mediated by distinct genetic factors.
EXPERIMENTAL PROCEDURES
Mice and primary cell culture and cell lines and FISH
P-E+ and P-E− mice were previously described (Afshar et al., 2006). Rag deficient mice on the C57BL/6 background were from Jackson Laboratories or maintained in colonies at the University of Illinois College of Medicine, or the NIA, NIH. All mouse procedures were approved by the Institutional Animal Care Committee of the University of Illinois College of Medicine, or the NIA, NIH animal facilities, in accordance with protocols approved by the UIC or NIA Institutional Animal Care and Use Committees. For all other pro-B cell analyses, Rag2−/− pro-B cells were cultured as previously described (Sayegh et al., 2005) with minor modifications. CD19+ cells were isolated using anti-CD19 coupled magnetic beads (Miltenyi) and cultured in the presence of IL7 (1% vol/vol supernatant of a J558L cell line stably expressing IL7) for 4 days. Pax5-deficient pro-B cells (kindly provided by Dr. J. Pongubala). The Abelson-MuLV transformed pro-B cell line was D345 (Ji et al., 2010). FISH was performed as described (Guo et al., 2011a). Additional experimental details are available online in Supplementary Experimental Methods.
Hi-C and 5C libraries and analyses
HiC libraries derived from A-MuLV transformed cell lines were previously described (Bredemeyer et al., 2006; Zhang et al., 2012). 5C library construction was performed as described (Dostie and Dekker, 2007; Kumar et al., 2013) (Supplementary Experimental Methods).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (RO1AI052400, R21AI117687 to A.K., HG003143 and HG00459 to J.D, GM079804 to J.L.), National Science Foundation (DBI 1062328 and MCB-1415589 to J.L.); the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (to A.K. and J.L.), the W.M Keck Foundation (to J.D), Intramural Research Program of the National Institute on Aging (Baltimore, MD) (to R.S.). We thank G. Gursoy for helpful discussions and Dr. J. Pongubala for the Pax5-deficient pro-B cell line.
Footnotes
The authors declare that they have no competing financial interests.
AUTHORS CONTRIBUTIONS
Conceptualization, A.L.K.; Methodology, A.L.K., R.W., R.S. and J.D.; Investigation, R.W., L.M., T.G.; Writing- Original Draft, A.L.K.; Writing – Review & Editing A.L.K. and R.S.; Funding Acquisition, A.L.K. and R.S., Formal Analysis, D.R., J.L., J.D. and B.L.; Resources, C.G, S.D., W.W., K.G.B.; Supervision, A.L.K. and R.S.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afshar R, Pierce S, Bolland DJ, Corcoran A, Oltz EM. Regulation of IgH gene assembly: role of the intronic enhancer and 5′DQ52 region in targeting DHJH recombination. J Immunol. 2006;176:2439–2447. doi: 10.4049/jimmunol.176.4.2439. [DOI] [PubMed] [Google Scholar]
- Belmont AS. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr Opin Cell Biol. 2014;26:69–78. doi: 10.1016/j.ceb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011a;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011b;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kumar S, Wuerffel R, Achour I, Lajoie B, Sen R, Dekker J, Feeney AJ, Kenter AL. Flexible ordering of antibody class switch and V(D)J joining during B-cell ontogeny. Genes Dev. 2013;27:2439–2444. doi: 10.1101/gad.227165.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, Shi Y. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, Sun Q, Huis In ’t Veld PJ, Guo C, Yoon HS, et al. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity. 2013;39:229–244. doi: 10.1016/j.immuni.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh CE, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Merkenschlager M. Cohesin and chromatin organisation. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.