Abstract
Standardization of biorepository best practices will enhance the quality of translational biomedical research utilizing patient-derived biobank specimens. Harmonization of pathology quality assurance procedures for biobank accessions has lagged behind other avenues of biospecimen research and biobank development. Comprehension of the cellular content of biorepository specimens is important for discovery of tissue-specific clinically relevant biomarkers for diagnosis and treatment. While rapidly emerging technologies in molecular analyses and data mining create focus on appropriate measures for minimizing pre-analytic artifact-inducing variables, less attention gets paid to annotating the constituent make up of biospecimens for more effective specimen selection by biobank clients. Both pre-analytic tissue processing and a specimen's composition influence acquisition of relevant macromolecules for downstream assays. Pathologist review of biorepository submissions, particularly tissues as part of quality assurance procedures, helps to ensure that the intended target cells are present and in sufficient quantity in accessioned specimens. This manual procedure can be tedious and subjective. Incorporating digital pathology into biobank quality assurance procedures, using automated pattern recognition morphometric image analysis to quantify tissue feature areas in digital whole slide images of tissue sections, can minimize variability and subjectivity associated with routine pathologic evaluations in biorepositories. Whole-slide images and pathologist-reviewed morphometric analyses can be provided to researchers to guide specimen selection. Harmonization of pathology quality assurance methods that minimize subjectivity and improve reproducibility among collections would facilitate research-relevant specimen selection by investigators and could facilitate information sharing in an integrated network approach to biobanking.
Keywords: biological specimens bank, standards; biomarkers, isolation and purification; morphometry; quantitative pathology; pattern recognition image analysis; computer-assisted diagnosis
Introduction
Biobanks are repositories of biospecimens from multiple patients with defined diseases. These collections encompass a range of disease manifestations and provide materials for discovery of molecular factors associated with disease initiation and progression. Conventionally, proper biobank operation includes planning about what to collect (diseased specimens vs. healthy control), harmonization of sample collection/processing protocols, and proper storage of specimens (1-5). Types of specimens stored in biobanks frequently include blood/serum/plasma, snap frozen tissues, tissue frozen for cryosectioning, and formalin-fixed paraffin-embedded tissues (6). Timely sample processing is the first step to reduce degradation of constituent molecules, ensuring a high quality specimen while minimizing pre-analytic artifacts (7, 8). Newly available fixatives such as those permitting multiple downstream applications from a single fixative (e.g. PaxGene, Qiagen), may improve efficiencies at surgical/tissue collection sites. Considerable attention has been devoted to developing procedures for appropriate collection and optimum preservation methods including the time to preservation, sample sizes, fixation/ storage conditions and most recently, markers useful for assessing sample quality (6, 9-18). Such attention is reflected in the release of standard operating procedures (SOPs) by the National Cancer Institute Cancer Human Biobank (caHUB), which conducts tissue collection for The NIH Genotype-Tissue Expression (GTEx) Project (19). However, considerable effort is still required to translate these concepts into consistent practices that minimize circumstances contributing to pre-analytical artifacts, as many contravening logistical demands of patient care exist. Several of these factors and practices are the subject of other reviews (6, 20-22).
Emphasis on sample collection and processing protocols must be accompanied by other equally important biobank quality assurance (QA) steps to aid successful utilization of biobanked specimens. Such QA procedures should be designed to promote the appropriate selection of relevant and representative biospecimens that contain detectable and clinically pertinent molecular constituents, thereby fostering meaningful translational biomedical research. Specimens for biobanking are usually derived from tissues excised/collected for diagnostic or treatment purposes. A portion of these samples will be submitted for diagnostic evaluation and the remaining tissue may then be subdivided to generate an adequate number of biobank specimens that can be preserved in a variety of ways. Some tissue subdivision may result in insufficiency or absence of the designated diseased cells. A survey of tumor biospecimens revealed inadequate presence of cancer target tissue more frequently than desired; only 59% of specimens, on average, contained >65% tumor cellularity, while 17% had no evidence of tumors (23). Since validating tissue signatures and targetable biomarkers is a focus of translational research utilizing biobanking materials, these results exemplify the importance of sample screening and annotation in biobanks, to provide perspective on specimen constituents. Extraction of materials (DNA, RNA or protein) for molecular analyses typically employs entire portions of lesion tissue obtained from a biorepository in an attempt to discover mutations or DNA/RNA/protein signatures of the disease of interest via techniques such as sequencing, microarray, or mass spectrometry. In order to derive disease-relevant macromolecules for discovery, sufficient quantity/percentage of target tissues must be harbored by the tissue mass and not be obscured due to dilutional effects of other more plentiful tissue constituents.
The initial phase of genomic study of The Cancer Genome Atlas (TCGA) mandated 80% tumor components in analyzed tissue or 60% when second generation sequencing platforms were applied (24). However, the origin (cancer, stroma, or blood) of the various differentially expressed proteins (between cancer and normal controls) derived in follow-on proteomic studies on these same samples were questioned (First Annual Scientific Symposium of the Clinical Proteomic Tumor Analysis Consortium (November 13, 2013, Bethesda, MD (25))). The findings corroborated the profound influence that heterogeneous cell phenotypes harbored in medical specimens may have on downstream molecular data obtained. Thus, information on the tissue components present in specimens will be beneficial to investigators at the time tissues are selected for research. Knowing what tissue constituents make up a patient specimen and to what degree these features are present in the specimen, aids appropriate sample selection for successful downstream applications. Therefore, confirmation of the presence of designated tissue types in biorepository specimens through a program of QA that includes pathology assessments of acquisitions, helps to guide selection of research-relevant samples and should be an important goal in biobanking QA procedures.
Elements of biobank pathology QA
The application of pathology for biobank specimen QA is not fully standardized, and information about today's biobanking practices is often not easy to ascertain (26). The pathology QA approach is at the discretion of the biorepository or contributor (27). Typically specimens are sectioned for histopathology and evaluated by a pathologist practicing in an institution associated with the biobank. In general, pathology-based biorepository QA programs employ single tissue sections from biobank specimens as a surrogate measure of the various tissue components in the rest of the tissue biobank tissue sample (27). Histopathology assessment provides valuable intelligence about specimen composition and includes diagnostic corroboration (20). However, such practice is subjective and can be associated with intra- and inter-observer variation.
A set of QA procedures was instituted for a cancer biobank that derives specimens from multiple treatment centers (28), which can serve as a model for integrating pathology QA best practices to promote optimal biobank quality and utilization. Analogous to the caHUB biorepository network, the CCOGC biobank collection includes frozen and fixed tumor specimens, blood, serum, urine, genomic DNA and RNA and tumor-adjacent normal tissues from several types of neoplasms (29). Tumor tissue and corresponding normal tissue are processed within five minutes of the samples being removed from the body, consistent with optimal preservation approaches (26). Each tumor is subdivided into “pea-sized segments” that are then fixed and stored in several different ways, including formalin, cryopreservation media, and flash freezing in liquid nitrogen (CCOGC protocols for tissue collection, processing, and preservation are available (30)).
For pathology-based QA, caHUB recommends pathologist examination on all incoming samples and second pathologist reviews are conducted on 10% of cases. The CCOGC protocol provides a model for a two-tiered approach, an extension from caHUB protocols. Specimens considered representative for each tumor type are sampled upon receipt in the first tier QA to provide a prospective overview survey of banked specimens. The second tier QA is triggered post hoc, when an investigator requests samples from those specimens not included in the survey analysis during their original accession. Procedures set up in first tier QA are applied to confirm the original diagnoses and evaluate the tissue composition in each requested sample prior to specimen distribution. For the initial overview survey steps, the first tier QA protocol stipulates prospective analyses from up to 50 patients for each tumor type as they are accessioned. Formalin-fixed tumor specimens selected by the biobank were processed as paraffin-embedded hematoxylin and eosin stained tissue sections and analyzed by histopathology and quantitative morphometric image analysis to provide (A) histopathological corroboration of submitted diagnoses, and (B) estimates of the tissue area occupied by tumor and stroma assessed from specimen tissue sections in order to depict information on the composition of the tissue banked. The morphometric data were obtained following published methods (27), and are discussed below. While this approach differs in minor ways from other pathology biobank QA practices (6, 18), it can be less resource intensive at the outset to implement, and has the added advantage of enhancing pathology QA reproducibility and throughput, while providing detailed biospecimen content annotations once established. Additional steps in first tier QA included evaluation of nucleic acid integrity of snap frozen tissues, whole blood, and DNA/RNA samples obtained from material matched with some of the specimens examined in the pathology QA process (27, 29).
The tissue QA diagnosis in the CCOGC survey evaluation did not completely agree with the original diagnosis submitted to the biobank for some specimens. In each of those cases, additional tissue sections from the biobank accessions were stained and reviewed histopathologically to corroborate submission diagnoses further. Unresolved discrepant histopathology diagnoses in the QA procedures triggered a review by a panel of 8 or more pathologists in order to obtain a final consensus diagnosis, which in some cases differed from the submitted diagnosis. Such differences may arise from tissue and tumor heterogeneity, rather than connoting a total absence of neoplastic disease in the patient or an incorrect diagnosis. The particular tissues reviewed at biobanks may be derived from a portion of excised tissues that were subdivided at the clinical site for banking purposes, and thus differ phenotypically from the tissues originally used for clinical diagnostic purposes (31).
As applied, the CCOGC biobank first tier QA procedures established diagnostic corroboration for 295 of 331 (89%) cancer specimens across 7 cancer types. Tumor tissue was not evident in 16 of 331 specimens (4.8%). Significant discrepancies to the submitted diagnoses were documented in 34 of 331 (10.2%) tumor specimens surveyed, including the 16 in which no tumor tissue was evident in tissue sections. Through these QA procedures, we found lack of diagnostic agreement and /or target cancer tissue in specimens of five of seven cancer types. It is worthwhile noting that, while this analysis provides substantial information on biobank specimens, tissue sections are not necessarily representative of the entire portions of the tumor submitted to the biorepository. It is possible for tumor to be present in other portion(s) of the banked specimens even if not apparent in the QA specimen. A quality assessment of the nucleic acid yield and integrity from 188 snap frozen biobank specimens across various tumor types revealed that over 90% of tumor sample RNA and DNA and 99% of blood DNA samples were of sufficient quality for molecular applications (29). The outcome of this survey highlights the importance of QA in biobanks and the attention to be paid by researchers when selecting biobank samples.
QA steps that include pathologist evaluation of every specimen result in significantly tedious workloads. Estimates of the proportions of various tissue features in specimens made solely by histopathologic visual inspection are subjective. Intra- and inter-observer variability is unavoidable when different (or the same) pathologists carry out biobank specimen assessments over time. This operator-related variability is not entirely unmanageable, however. Digital pathology, utilizing automated slide scanning and computer-assisted morphometric image analysis, can be a valuable solution to improving reproducibility in specimen composition annotations. Furthermore, the image analysis output provides a means for biobanks to annotate quantitative estimates of specimen content. With standardized image analysis protocols facilitating improved throughput, such pathology functions can be accomplished more objectively, but not devoid of pathologist oversight (32).
Digital pathology can be incorporated into biobanking QA procedures
Digital microscopy enables an entire histological slide to be optically scanned creating a digitized image data file of a whole slide image (WSI) that can be stored in a database. Pathologists and researchers can then retrieve the images through a networked database and navigate the slide images on a computer screen for visual inspection in a manner similar to microscopic examination (33-35). For routine use, tissue mounted on glass slides are commonly scanned at 20× magnification and studies have shown such resolution (~0.43 micrometer per pixel) proves sufficient for many diagnostic applications (32, 36). Similar diagnostic accuracy has been achieved when pathologists compared diagnoses obtained using conventional glass slides with digital WSI; this outcome indicates that the high resolution WSI captured by scanners are adequate for many evaluations and diagnostic uses, as evidenced by recent adoption of the technology for clinical diagnosis in Canada; however such applications are not currently approved for primary clinical diagnosis in the U.S. (37). In addition to image viewing/sharing, WSI can be utilized in image analyses by incorporating algorithms constructed for pattern recognition with color identification/quantification functions (31, 32, 38). Digital pathology is transforming pathology practice and is increasingly being integrated into teaching/education/training, diagnosis (consultation) and biobank quality assurance procedures (32, 39-43). Annotating digital WSIs is now recommended as part of routine pathology examination, in addition to traditional glass slide viewing (18, 27). Several manufacturers have digital pathology products on the market supporting slide scanning and image analyses (reviewed in (44)).
Annotating biobank specimen tissue components using image analysis to guide selection
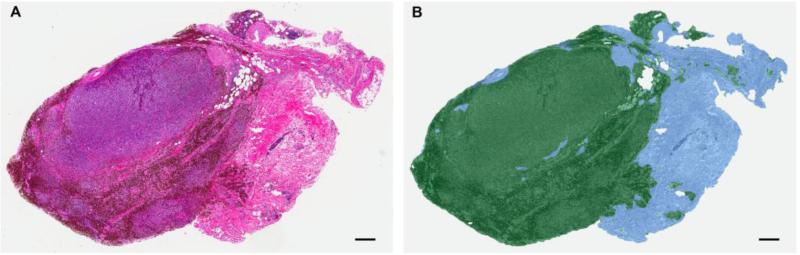
We previously developed and applied unique cancer type-specific computer-assisted pattern recognition image analysis (PRIA) algorithms to cancer biorepository specimens (27). This readily adoptable process can be included as a part of foundational pathology QA. In this QA procedure, algorithms are created to discriminate various tissue types in individual WSIs. Due to the variation in tissue components present among different types of cancers, it is impossible to create one algorithm to analyze an entire biobank collection for various lesions, hence cancer type-specific algorithms are necessary to conduct optimum analyses. Areas containing representative features of each type of tissue present in a particular cancer are selected from WSI for training an algorithm; the approach follows parameters similar to those featured in caHUB collection guidelines (27, 38, 45, 46). Annotating biobank specimens with evidence of their component cellular makeup involved designing algorithms with mean training accuracies of > 98% and with > 95 % sensitivity and specificity. Post analytic processing algorithms are evaluated for fitness-for-purpose by assessing image mark-ups for appropriate tissue feature segmentation and quantitation. Optimized image analysis algorithms appropriate for each cancer type were applied to WSIs of biobank specimens to analyze and measure the areas of various tissue components (Fig. 1). PRIA provides data for annotations delineating relevant proportions of neoplastic cells and stromal elements present in biorepository specimens. Results of tumor/stroma computational areas and percentages (e.g., Table 1) can be made available to prospective clients in the biobank database. The approach demonstrated the feasibility of annotating complex mixtures of cells/tissues comprising individual tissue specimens in a more objective manner than traditional pathologist visual assessments, minimizing the inter-/intra observer variability (27). We believe that such annotations can help investigators choose the particular specimens that have sufficient characteristics to meet their research objectives.
Figure 1.
Automated pattern recognition image analysis approach for quantifying tissue features in biobank tissue specimens as part of pathology quality assurance. Unique algorithms were developed for each tumor type. A. Sub-gross photomicrograph of an optically scanned melanoma tissue section image used for morphometric analysis is displayed as a representative example. Hematoxylin and eosin stain. B. Tissue section shown in (A) after image analysis, displayed with pseudo-color mark-up revealing tissue areas segmented as cancer (green, 68.6%) and stroma (blue, 31.4%). Image reproduced with permission from the Journal of Biomolecular Techniques (JBT), ©Association of Biomolecular Resource Facilities, www.abrf.org (27).
Table 1.
| Patient Numbera | Diagnosisb | Tissue Area (mm2) | Tumor Area (mm2) | Stroma Area (mm2) | Percent Tumor | Percent Stroma |
|---|---|---|---|---|---|---|
| 101A | Melanoma | 13.08 | 8.97 | 4.11 | 68.60 | 31.40 |
Patient identifier assigned by repository.
Diagnosis – Cancer diagnosis corroborated by a pathologist as part of quality assurance.
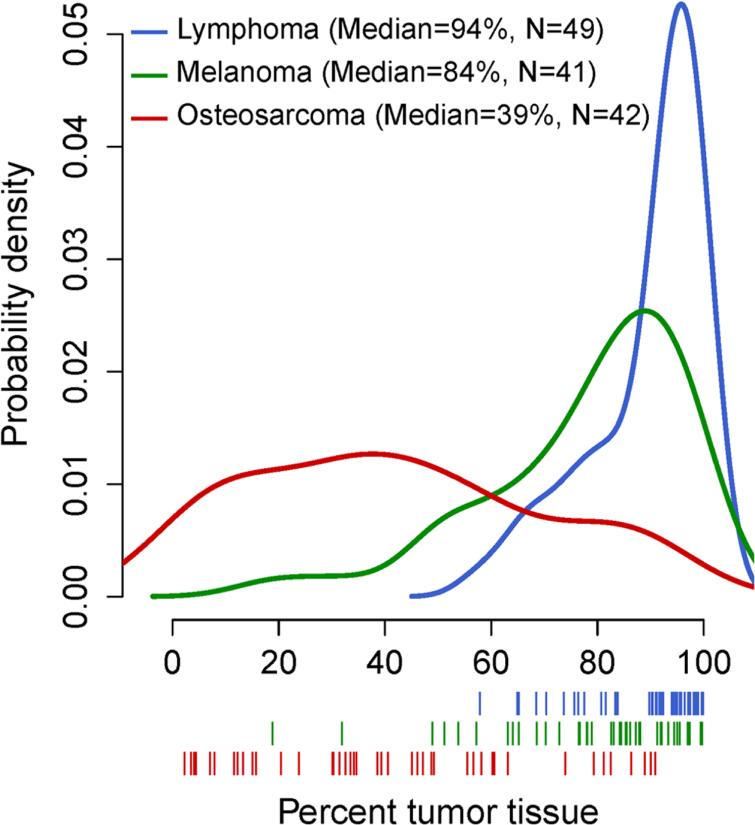
The morphometric data from tissue sections used for pathology QA not only provides information about characteristics of individual biorepository acquisitions (Table 1), but also permits summative frequency distribution analyses of the collective findings for a particular tumor type (Fig. 2). Such analyses provide perspective insight on the relative contributions of tumor and stroma that an investigator may encounter for each cancer type across the biorepository collection, including the untested portions of banked specimens. For example, a number of osteosarcoma specimens in the CCOGC biobank would likely not meet the caHUB guidelines of a minimum of 70% tumor since this analysis documented that the median portion of biorepository tissue area occupied by tumor in the osteosarcoma patients was 39%, with specimens harboring a broad range of tumor tissue content (interquartile range = 44%) (Fig. 2). The vast majority of lymphoma specimens, on the other hand, are likely to contain >70% tumor. Ideally, the morphometric data on component make up of specimens could be linked for correlation with the image files and other biobank specimen data, augmenting recommendations outlined by the International Society for Biological and Environmental Repositories (18).
Figure 2.
Frequency distribution of the percentages of tumors in cancer biorepository tissue sections by cancer type using kernel density estimation, for lymphoma (blue lines), melanoma (green lines), and osteosarcoma (red lines). Single tissue sections chosen randomly from all patients with analyzable specimens were used in these analyses (N, shown on figure). Below the x-axis, estimates of tumor tissue percentage from each patient are shown as vertical lines. Image reproduced with permission from the Journal of Biomolecular Techniques (JBT), ©Association of Biomolecular Resource Facilities, www.abrf.org. (27).
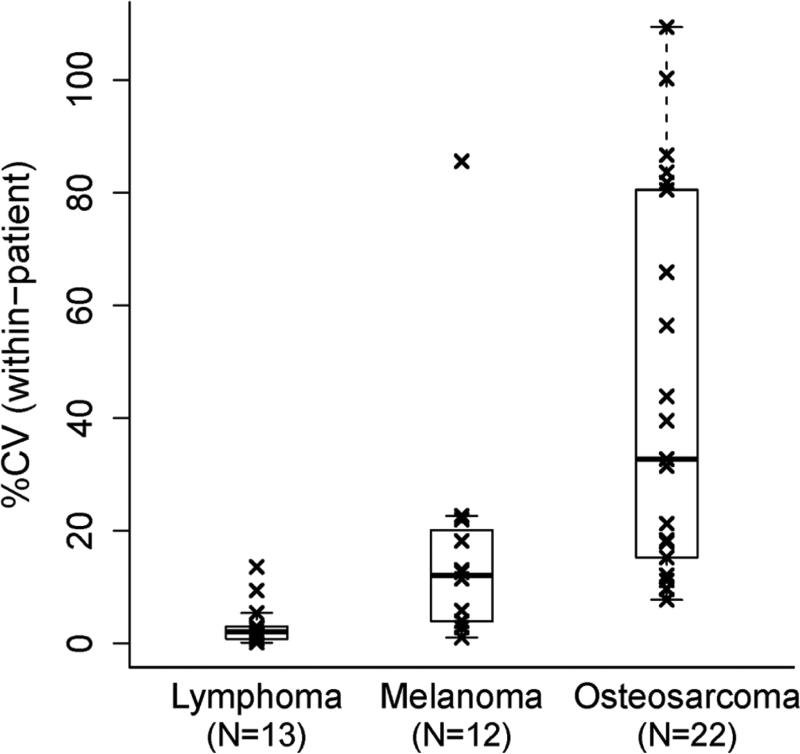
Single tissue sections from complex 3-dimensional lesions, such as tumors, provide a means for a definitive clinical diagnosis in many patients requiring biopsy. Biorepository pathology QA programs similarly rely on evaluations of single tissue sections from a biobank specimen for diagnostic corroboration and for assessment of the distribution of tissue components present in biobank samples. The degree to which tissue section surface area assessments predict the various components of a volumetric tissue specimen in biorepositories is not exact. A survey evaluating inter-sample variation by comparing subdivided tumors from the same patients having either lymphoma, melanoma or osteosarcoma, demonstrated that the within-patient inter-sample variability can be cancer type dependent (Fig. 3). The median coefficient of variations (CV) among replicate portions of individual patient tumors of 2%, 12%, and 33% were identified for lymphomas, melanomas, and osteosarcomas, respectively. Cancer is heterogeneous with varying degrees of tumor cells and admixed supporting tissues throughout a mass (20, 47). However, our results indicate that morphometric image analysis estimates of tumor burden area, as demonstrated previously, reasonably predict tissue components for some cancer types (27).
Figure 3.
Dispersion analyses of the proportion of cancer biorepository tissue sections occupied by tumor. Box-whisker plots show distributions of the within-patient coefficient of variation (%CV) of the tumor tissue percentage estimates for three cancer types. Specimen comparisons represent multiple portions from the same cancer masses (subdivided for biobanking purposes) excised for each patient's initial diagnosis. The line in the middle of the box represents the median, with the lower and upper edges showing the first and third quartiles, respectively. The whiskers extend to the extreme values and the outliers are the coefficients of variation exceeding 1.5*IQR (interquartile range). Image reproduced with permission from the Journal of Biomolecular Techniques (JBT), ©Association of Biomolecular Resource Facilities, www.abrf.org. (27).
Utility of biobank tissue feature area annotations
Mapping biobank tissue components and measuring their areas (percentages) in tissue sections for guiding both the appropriate selection of biobank specimens and the processes needed to obtain tissue target of interest has been suggested as prudent practice (27). This concept has been adopted by the caHUB in its Annotating Digitally Scanned Microscope Slides standard operating procedure recommendations for 2012 (48); the protocol provides for measuring tissue and tumor areas using digital tools. A special annotation is applied when viable tumor is < 70% of tissue specimen. Automating this process by using PRIA can serve to improve precision and throughput.
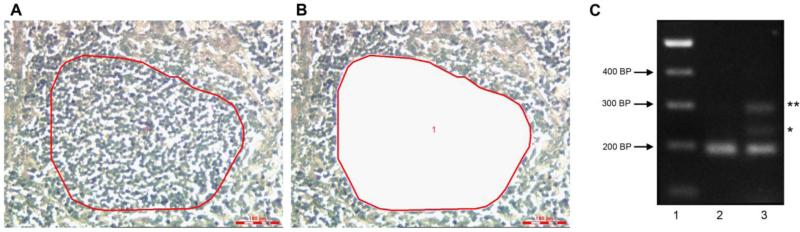
We carried out a pilot analysis to illustrate how the ratios of tissue components mapped using automated PRIA during the QA process can foreshadow target discovery. The selected specimen obtained for this purpose from the biorepository represented a sarcoma (mast cell tumor) characterized as having 62% of tissue area occupied by tumor. This predicted, generally less than optimal, density of cancer cells was obtained based upon our pathology QA process using a PRIA algorithm designed to recognize and quantify tumor and non-tumor (tumor associated stroma and adjacent non-neoplastic tissues) areas. Approximately 10 - 25% of these tumors harbor an internal tandem duplication (ITD) mutation in exon 11 of c-kit, which is associated with higher-grade tumors (49, 50). A conventional PCR was carried out in an attempt to evaluate the potential for this mutation in c-kit. The initial analysis included DNA extracted from a whole portion of this tumor mass. While PCR on DNA extracted from this portion of tumor mass failed to uncover evidence of a mutation, a 45 bp-insertion ITD mutation in the neoplasm was revealed using 1/10th of the DNA prepared from 1mm2 of microdissected tumor tissue, in which a population of cancer cells was enriched and separated from surrounding tumor microenvironment and stroma (Fig 4). This result is consistent with biorepository guidelines recommending a minimum for 65-70% tumor cell content for downstream analysis (51, 52).
Figure 4.
Application of biorepository tissue morphometric annotation for guidance of sample selection and processing for downstream molecular analysis. A. Photomicrograph of a subject sarcoma tissue section (having 62% of tissue area occupied by tumor), depicting an area of tumor intended for laser microdissection (red circle) for separating and enriching a sample population of neoplastic cells from surrounding non-tumor connective tissue. B. Same field of view as in A, following tissue microdissection, indicating the absence of cancer cell population having been dissected away from surrounding non-tumor tissue remaining on the section (clear space). C. Gel electrophoresis of PCR amplified DNA extracted from a sarcoma harboring an internal tandem duplication (ITD) mutation of c-kit. Lane 1, DNA ladder in basepairs (bp). Lane 2, c-kit amplicon indicating the normal wild-type gene amplified in DNA prepared from a portion of whole specimen. Lane 3, detection of ITD (*) in DNA extracted from laser microdissected tumor cells as shown in B) (** heterodimer of wild type and mutated PCR products). PCR protocol adapted from Webster et.al. (49). This finding was confirmed by sequence analysis (not shown).
Conclusion and future directions
PRIA and other pathology QA methods can be translated directly to human tissue biobank quality assurance protocols to provide improved consistency, while minimizing variability associated with routine biorepository pathologic evaluations (27, 53). Hundreds of biobanks are currently being developed across North America by individuals, clinics, hospitals, foundations, companies, universities and other institutions, providing the potential to form cooperative networks (6). In recognition of the disparate nature of these endeavors and the historical approach to individual biospecimen collections that has contributed to relatively low usage of recently developed biobanks (54), the National Cancer Institute has launched a National Cancer Informatics Program initiative. Using tools stemming from this initiative, biobanks can develop approaches to scale biorepository specimen annotation for integration with “clinical, imaging and genomic data to support multidisciplinary investigations” (52). The conceptual approach reviewed in the present report for providing specimen feature quantitation using automated throughput for morphometric tissue image analysis creates a more precise means for annotating biobank accessions and fits nicely into this initiative. Comprehension of biospecimen composition is necessary for successful molecular profiling (8, 55). The use of digital pathology and image analysis to annotate biorepository specimens would appear to strengthen the potential for biomarker discovery by helping to guide the selection of study-appropriate specimens by investigators.
How well suited is today's automated digital pathology image analysis for application to biobanking QA? Some further technical development would be beneficial to its use in biobanking. Application of computer-assisted diagnosis technology to histopathology tissue sections was initially adapted from satellite multispectral pattern recognition image analysis genetic-based algorithms used for geospatial analyses (56). Acceptable reproducibility of histopathology PRIA was demonstrated previously through objective comparisons to well established manual image segmentation, using repeated measures analyses (32). Suitable diagnostic agreement can be achieved with PRIA, particularly by employing multiple algorithms and by focusing on limited phenotypic ranges and few numbers of tissue features. Such PRIA shortcomings, however, would benefit from further optimization of technology development (32). Additionally, image analysis is influenced by histopathology slide staining tincture, which varies from laboratory to laboratory. Such staining inconsistencies among laboratories can influence the need for retraining some algorithms on locally available biospecimens in order to optimize algorithms for proper image analyses. Unified image viewing platforms would help make access to digital pathology imaging files more widespread. An NCI initiative in fostering Digital Imaging and Communications in Medicine (DICOM) compatibility may further this need.
The advent of digital pathology and image analysis contributes to the informatics infrastructure necessary to facilitate the appropriate utilization of biobank specimens by the research community. Pathologists will continue to need to evaluate image analyses data output while performing other aspects of biorepository quality assurance to advance this. Pathologists and laboratory scientists/directors supporting the development of tissue biorepositories can strengthen resources and create protocols furthering the harmonization of QA measures.
Highlights.
importance of pathology quality assurance in biobank management
the impact of tissue components on downstream research findings
how digital pathology and automated image analysis can improve biobanking QA
improvements include throughput, reproducibility and specimen features quantitation
improvements guide more optimal biospecimen selection
Acknowledgements
This research was supported by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, Bethesda, Maryland. The authors appreciate the scholarly contributions of Jennifer Dwyer, Shelley Hoover and Josh Webster.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hullsiek KH, George M, Brown SK. Designing and managing a flexible and dynamic biorepository system: a 15-year perspective from the CPCRA, ESPRIT, and INSIGHT clinical trial networks. Current opinion in HIV and AIDS. 2010 Nov;5(6):538–44. doi: 10.1097/COH.0b013e32833f2058. PubMed PMID: 20978398. Pubmed Central PMCID: PMC3039520. Epub 2010/10/28. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grizzle WE, Bell WC, Sexton KC. Issues in collecting, processing and storing human tissues and associated information to support biomedical research. Cancer biomarkers : section A of Disease markers. 2010;9(1-6):531–49. doi: 10.3233/CBM-2011-0183. PubMed PMID: 22112494 Pubmed Central PMCID: PMC3445033. Epub 2011/11/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaffney EF, Madden D, Thomas GA. The human side of cancer biobanking. Methods in molecular biology (Clifton, NJ) 2012;823:59–77. doi: 10.1007/978-1-60327-216-2_5. PubMed PMID: 22081339. Epub 2011/11/15. eng. [DOI] [PubMed] [Google Scholar]
- 4.Vaught J, Lockhart NC. The evolution of biobanking best practices. Clinica chimica acta; international journal of clinical chemistry. 2012 Oct 9;413(19-20):1569–75. doi: 10.1016/j.cca.2012.04.030. PubMed PMID: 22579478. Pubmed Central PMCID: PMC3409343. Epub 2012/05/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hveem K. Creation of a new prospective research biobank: the example of HUNT3. Methods in molecular biology (Clifton, NJ) 2011;675:231–9. doi: 10.1007/978-1-59745-423-0_12. PubMed PMID: 20949393 Epub 2010/10/16. eng. [DOI] [PubMed] [Google Scholar]
- 6.Hewitt RE. Biobanking: the foundation of personalized medicine. Current opinion in oncology. 2011 Jan;23(1):112–9. doi: 10.1097/CCO.0b013e32834161b8. PubMed PMID: 21076300. Epub 2010/11/16. eng. [DOI] [PubMed] [Google Scholar]
- 7.Spruessel A, Steimann G, Jung M, Lee SA, Carr T, Fentz AK, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques. 2004 Jun;36(6):1030–7. doi: 10.2144/04366RR04. PubMed PMID: 15211754 eng. [DOI] [PubMed] [Google Scholar]
- 8.Botling J, Micke P. Biobanking of fresh frozen tissue from clinical surgical specimens: transport logistics, sample selection, and histologic characterization. Methods in molecular biology (Clifton, NJ) 2011;675:299–306. doi: 10.1007/978-1-59745-423-0_16. PubMed PMID: 20949397. [DOI] [PubMed] [Google Scholar]
- 9.Kang HJ, Jeon SY, Park JS, Yun JY, Kil HN, Hong WK, et al. Identification of Clinical Biomarkers for Pre-Analytical Quality Control of Blood Samples. Biopreserv Biobank. 2013 Apr;11(2):94–100. doi: 10.1089/bio.2012.0051. PubMed PMID: 23634248. Pubmed Central PMCID: PMC3629782. Epub 2013/05/02. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin P, Peter A, Franken H, Zhao X, Neukamm SS, Rosenbaum L, et al. Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clinical chemistry. 2013 May;59(5):833–45. doi: 10.1373/clinchem.2012.199257. PubMed PMID: 23386698. Epub 2013/02/07. eng. [DOI] [PubMed] [Google Scholar]
- 11.Betsou F, Barnes R, Burke T, Coppola D, Desouza Y, Eliason J, et al. Human biospecimen research: experimental protocol and quality control tools. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009 Apr;18(4):1017–25. doi: 10.1158/1055-9965.EPI-08-1231. PubMed PMID: 19336543. Epub 2009/04/02. eng. [DOI] [PubMed] [Google Scholar]
- 12.Palmirotta R, De Marchis ML, Ludovici G, Leone B, Savonarola A, Ialongo C, et al. Impact of preanalytical handling and timing for peripheral blood mononuclear cells isolation and RNA studies: the experience of the Interinstitutional Multidisciplinary BioBank (BioBIM). The International journal of biological markers. 2012 Apr-Jun;27(2):e90–8. doi: 10.5301/JBM.2012.9235. PubMed PMID: 22562396. Epub 2012/05/09. eng. [DOI] [PubMed] [Google Scholar]
- 13.Mee BC, Carroll P, Donatello S, Connolly E, Griffin M, Dunne B, et al. Maintaining Breast Cancer Specimen Integrity and Individual or Simultaneous Extraction of Quality DNA, RNA, and Proteins from Allprotect-Stabilized and Nonstabilized Tissue Samples. Biopreserv Biobank. 2011 Dec;9(4):389–98. doi: 10.1089/bio.2011.0034. PubMed PMID: 23386926. Pubmed Central PMCID: PMC3558729. Epub 2011/12/01. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore HM, Kelly A, Jewell SD, McShane LM, Clark DP, Greenspan R, et al. Biospecimen Reporting for Improved Study Quality. Biopreserv Biobank. 2011 Apr;9(1):57–70. doi: 10.1089/bio.2010.0036. PubMed PMID: 21826252. Pubmed Central PMCID: PMC3142856. ENG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanni U, Spila A, Riondino S, Valente MG, Somma P, Iacoboni M, et al. RFID as a new ICT tool to monitor specimen life cycle and quality control in a biobank. The International journal of biological markers. 2011 Apr-Jun;26(2):129–35. doi: 10.5301/JBM.2011.8323. PubMed PMID: 21574153 Epub 2011/05/17. eng. [DOI] [PubMed] [Google Scholar]
- 16.Hallmans G, Vaught JB. Best practices for establishing a biobank. Methods in molecular biology (Clifton, NJ) 2011;675:241–60. doi: 10.1007/978-1-59745-423-0_13. PubMed PMID: 20949394. Epub 2010/10/16. eng. [DOI] [PubMed] [Google Scholar]
- 17.Peakman T, Elliott P. Current standards for the storage of human samples in biobanks. Genome medicine. 2010;2(10):72. doi: 10.1186/gm193. PubMed PMID: 20923579. Pubmed Central PMCID: PMC2988449. Epub 2010/10/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.2012 Best practices for repositories, Collection, storage, retrieval, and distribution of biological materials for research, International society for biological and environmental repositories. Biopreservation and Biobanking. 2012;10(2):79–161. doi: 10.1089/bio.2012.1022. [DOI] [PubMed] [Google Scholar]
- 19. [2013 Nov. 26]; http://biospecimens.cancer.gov/resources/sops/library.asp. GTEx SOP Library.
- 20.Shevde LA, Riker AI. Current concepts in biobanking: development and implementation of a tissue repository. Frontiers in bioscience. 2009;1:188–93. doi: 10.2741/s18. PubMed PMID: 19482694. [DOI] [PubMed] [Google Scholar]
- 21.Schrohl AS, Wurtz S, Kohn E, Banks RE, Nielsen HJ, Sweep FC, et al. Banking of biological fluids for studies of disease-associated protein biomarkers. Molecular & cellular proteomics : MCP. 2008 Oct;7(10):2061–6. doi: 10.1074/mcp.R800010-MCP200. PubMed PMID: 18676364. Pubmed Central PMCID: PMC2559931. Epub 2008/08/05. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malm J, Fehniger TE, Danmyr P, Vegvari A, Welinder C, Lindberg H, et al. Developments in biobanking workflow standardization providing sample integrity and stability. Journal of proteomics. 2013 Jul 13; doi: 10.1016/j.jprot.2013.06.035. PubMed PMID: 23856607. Epub 2013/07/17. Eng. [DOI] [PubMed] [Google Scholar]
- 23.Sandusky G, Dumaual C, Cheng L. Review paper: Human tissues for discovery biomarker pharmaceutical research: the experience of the Indiana University Simon Cancer Center-Lilly Research Labs Tissue/Fluid BioBank. Vet Pathol. 2009 Jan;46(1):2–9. doi: 10.1354/vp.46-1-2. PubMed PMID: 19112109. eng. [DOI] [PubMed] [Google Scholar]
- 24. [2013 Nov. 26];TCGA Tissue Sample Requirements: High Quality Requirements Yield High Quality Data. http://cancergenome.nih.gov/cancersselected/biospeccriteria.
- 25. [2013 Nov. 26];Clinical Proteomic Tumor Analysis Consortium. http://proteomics.cancer.gov/programs/cptacnetwork.
- 26.McDonald SA. Principles of Research Tissue Banking and Specimen Evaluation from the Pathologist's Perspective. Biopreserv Biobank. 2010 Dec;8(4):197–201. doi: 10.1089/bio.2010.0018. PubMed PMID: 23386923. Pubmed Central PMCID: PMC3562469. ENG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster JD, Simpson ER, Michalowski AM, Hoover SB, Simpson RM. Quantifying histological features of cancer biospecimens for biobanking quality assurance using automated morphometric pattern recognition image analysis algorithms. J Biomol Tech. 2011 Sep;22(3):108–18. PubMed PMID: 21966258. Pubmed Central PMCID: PMC3165860. eng. [PMC free article] [PubMed] [Google Scholar]
- 28. [2013 Oct. 1]; (internet) cn. Pfizer-CCOGC Biospecimen Repository:Available from: http://www.ccogc.net.
- 29.Thomas R SM, Mochizuki H, Williams C, Poorman K, Kennedy K, Mazcko C, Modiano JF, Breen M. Quality control and quality assurance of canine biological specimens available through the Pfizer-CCOGC, Inc. national biorepository for comparative oncology studies. (abstract).. 105th Am Assoc Cancer Res Annu Mtg; San Diego, CA. April 5, 2014; 2014;unpublished data. [Google Scholar]
- 30.ccogc.net/collection.html [2013 Oct. 1]; Pfizer-CCOGC Biospecimen Repository:Available from: http://www.ccogc.net/collection.html.
- 31.Simpson RM, Bastian BC, Michael HT, Webster JD, Prasad ML, Conway CM, et al. Sporadic naturally occurring melanoma in dogs as a preclinical model for human melanoma. Pigment Cell Melanoma Res. 2013 Oct 15; doi: 10.1111/pcmr.12185. PubMed PMID: 24128326, DOI: 101111/pcmr.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webster JD, Michalowski AM, Dwyer JE, Corps KN, Wei BR, Juopperi T, et al. Investigation into diagnostic agreement using automated computer-assisted histopathology pattern recognition image analysis. J Pathol Inform. 2012;3:18. doi: 10.4103/2153-3539.95130. PubMed PMID: 22616030. Pubmed Central PMCID: PMC3352619. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jara-Lazaro AR, Thamboo TP, Teh M, Tan PH. Digital pathology: exploring its applications in diagnostic surgical pathology practice. Pathology. 2010;42(6):512–8. doi: 10.3109/00313025.2010.508787. PubMed PMID: 20854068. [DOI] [PubMed] [Google Scholar]
- 34.Al-Janabi S, Huisman A, Van Diest PJ. Digital pathology: current status and future perspectives. Histopathology. 2012 Jul;61(1):1–9. doi: 10.1111/j.1365-2559.2011.03814.x. PubMed PMID: 21477260. [DOI] [PubMed] [Google Scholar]
- 35.Mulrane L, Rexhepaj E, Penney S, Callanan JJ, Gallagher WM. Automated image analysis in histopathology: a valuable tool in medical diagnostics. Expert review of molecular diagnostics. 2008 Nov;8(6):707–25. doi: 10.1586/14737159.8.6.707. PubMed PMID: 18999923. [DOI] [PubMed] [Google Scholar]
- 36.Stathonikos N, Veta M, Huisman A, van Diest PJ. Going fully digital: Perspective of a Dutch academic pathology lab. J Pathol Inform. 2013;4:15. doi: 10.4103/2153-3539.114206. PubMed PMID: 23858390 Pubmed Central PMCID: PMC3709427. Epub 2013/07/17. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al Habeeb A, Evans A, Ghazarian D. Virtual microscopy using whole-slide imaging as an enabler for teledermatopathology: A paired consultant validation study. J Pathol Inform. 2012;3:2. doi: 10.4103/2153-3539.93399. PubMed PMID: 22439122. Pubmed Central PMCID: PMC3307226. Epub 2012/03/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan DJ, Brandstedt J, Rexhepaj E, Foley M, Ponten F, Uhlen M, et al. Tumour-specific HMG-CoAR is an independent predictor of recurrence free survival in epithelial ovarian cancer. BMC cancer. 2010;10:125. doi: 10.1186/1471-2407-10-125. PubMed PMID: 20359358. Pubmed Central PMCID: 3087316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brachtel E, Yagi Y. Digital imaging in pathology--current applications and challenges. Journal of biophotonics. 2012 Apr;5(4):327–35. doi: 10.1002/jbio.201100103. PubMed PMID: 22213680. Epub 2012/01/04. eng. [DOI] [PubMed] [Google Scholar]
- 40.Cornish TC, Swapp RE, Kaplan KJ. Whole-slide imaging: routine pathologic diagnosis. Advances in anatomic pathology. 2012 May;19(3):152–9. doi: 10.1097/PAP.0b013e318253459e. PubMed PMID: 22498580 Epub 2012/04/14. eng. [DOI] [PubMed] [Google Scholar]
- 41.Krenács IZ T, Micsik T, et al. Digital microscopy the upcoming revolution in histopathology teaching, diagnostics, research and quality assurance. In: Méndez-Vilas A, Álvarez JD, editors. Microscopy: Science, Technology, Applications and Education 2. Formatex Research Center; Badajoz: 2010. pp. 965–77. [Google Scholar]
- 42.Pantanowitz L, Szymas J, Yagi Y, Wilbur D. Whole slide imaging for educational purposes. J Pathol Inform. 2012;3:46. doi: 10.4103/2153-3539.104908. PubMed PMID: 23372987. Pubmed Central PMCID: PMC3551531. Epub 2013/02/02. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teodorovic I, Isabelle M, Carbone A, Passioukov A, Lejeune S, Jamine D, et al. TuBaFrost 6: virtual microscopy in virtual tumour banking. European journal of cancer (Oxford, England : 1990) 2006 Dec;42(18):3110–6. doi: 10.1016/j.ejca.2006.04.033. PubMed PMID: 17027253. Epub 2006/10/10. eng. [DOI] [PubMed] [Google Scholar]
- 44.Isse K, Lesniak A, Grama K, Roysam B, Minervini MI, Demetris AJ. Digital transplantation pathology: combining whole slide imaging, multiplex staining and automated image analysis. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012 Jan;12(1):27–37. doi: 10.1111/j.1600-6143.2011.03797.x. PubMed PMID: 22053785. Pubmed Central PMCID: PMC3627485. Epub 2011/11/08. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.http://cahub.cancer.gov/.CaHub [2013 Nov. 30];The Cancer Human Biobank. available from: http://cahub.cancer.gov/
- 46.Angeletti C, Harvey NR, Khomitch V, Fischer AH, Levenson RM, Rimm DL. Detection of malignancy in cytology specimens using spectral-spatial analysis. Laboratory investigation; a journal of technical methods and pathology. 2005 Dec;85(12):1555–64. doi: 10.1038/labinvest.3700357. PubMed PMID: 16200074. [DOI] [PubMed] [Google Scholar]
- 47.Espina V, Heiby M, Pierobon M, Liotta LA. Laser capture microdissection technology. Expert review of molecular diagnostics. 2007 Sep;7(5):647–57. doi: 10.1586/14737159.7.5.647. PubMed PMID: 17892370. [DOI] [PubMed] [Google Scholar]
- 48. [2013 Nov. 26];Annotating Digitally Scanned Microscope Slides (PR-0002-W1 v1.0.0) http://biospecimens.cancer.gov/resources/sops/GTEx_SOPs/V/A/Annotating%20 Digitally%20Scanned%20Microscope%20Slides%20(PR-0002-W1%20v1.0.0).pdf.
- 49.Webster JD, Yuzbasiyan-Gurkan V, Miller RA, Kaneene JB, Kiupel M. Cellular proliferation in canine cutaneous mast cell tumors: associations with c-KIT and its role in prognostication. Vet Pathol. 2007 May;44(3):298–308. doi: 10.1354/vp.44-3-298. PubMed PMID: 17491070. eng. [DOI] [PubMed] [Google Scholar]
- 50.Letard S, Yang Y, Hanssens K, Palmerini F, Leventhal PS, Guery S, et al. Gain-of-function mutations in the extracellular domain of KIT are common in canine mast cell tumors. Molecular cancer research : MCR. 2008 Jul;6(7):1137–45. doi: 10.1158/1541-7786.MCR-08-0067. PubMed PMID: 18644978. [DOI] [PubMed] [Google Scholar]
- 51.Sandusky GE, Teheny KH, Esterman M, Hanson J, Williams SD. Quality control of human tissues--experience from the Indiana University Cancer Center-Lilly Research Labs human tissue bank. Cell and tissue banking. 2007;8(4):287–95. doi: 10.1007/s10561-007-9037-0. PubMed PMID: 17387635 Epub 2007/03/28. eng. [DOI] [PubMed] [Google Scholar]
- 52. [2013 Oct. 1]; (internet) cnngnb-i-ra. NCI Center for Biomedical Informatics and Information Technology:Available from: http://cbiit.nci.nih.gov/ncip/biomedical-informatics-resources/applications.
- 53.Potts SJ, Young GD, Voelker FA. The role and impact of quantitative discovery pathology. Drug discovery today. 2010 Nov;15(21-22):943–50. doi: 10.1016/j.drudis.2010.09.001. PubMed PMID: 20946967. [DOI] [PubMed] [Google Scholar]
- 54.Henderson GE, Cadigan RJ, Edwards TP, Conlon I, Nelson AG, Evans JP, et al. Characterizing biobank organizations in the U.S.: results from a national survey. Genome medicine. 2013 Jan 25;5(1):3. doi: 10.1186/gm407. PubMed PMID: 23351549. Pubmed Central PMCID: PMC3706795. Epub 2013/01/29. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riegman PH, Dinjens WN, Oosterhuis JW. Biobanking for interdisciplinary clinical research. Pathobiology. 2007;74(4):239–44. doi: 10.1159/000104451. PubMed PMID: 17709966. eng. [DOI] [PubMed] [Google Scholar]
- 56.genie.lanl.gov [2013 Oct. 1];GENetic Imagery Exploitation, Operated by Los Alamos National Security, LLC for the U.S. Department of Energy's NNSA. Available from: http://genie.lanl.gov/