Abstract
Dengue is a major threat for public health in tropical and subtropical countries around the world. In the absence of a licensed vaccine and effective antiviral therapies, control measures have been based on education activities and vector elimination. Current efforts for developing a vaccine are both promising and troubling. At the advent of the introduction of a tetravalent dengue vaccine, molecular surveillance of the circulating genotypes in different geographical regions has gained considerable importance. A growing body of in vitro, preclinical, and clinical phase studies suggest that vaccine conferred protection in a geographical area could depends on the coincidence of the dengue virus genotypes included in the vaccine and those circulating. In this review we present the state-of-the-art in this field, highlighting the need of deeper knowledge on neutralizing immune response for making decisions about future vaccine approval and the potential need for different vaccine composition for regional administration.
Keywords: dengue virus, vaccines, neutralizing antibodies, genotypes, surveillance
Abbreviations
- DENV
dengue viruses
- DC-SIGN
Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin
- Hsp70
heat shock protein 70
- Hsp90
heat shock protein 90
- ssRNA+
single-stranded positive-sense RNA viruses
- MRCA
more recent common ancestor
- UTR
untranslated region
- E
envelope protein
- M
membrane protein
- NS1
non-structural protein 1
- ADE
antibody-dependent enhancement
Introduction
Dengue officially causes 50–100 million infections and 22 000 deaths per year around the world.1 Recent cartographic approaches have estimated to be around 390 million infections per year becoming the most important arthropod-borne viral disease in more than 100 tropical and subtropical countries where 2.5 billion people live at risk of infection.2
Invasive species of mosquitoes, Aedes aegypti and Ae. albopictus serve as vectors leading to rapid worldwide spread of the disease. The disease is caused by dengue viruses (DENV), which are members of the Flavivirus genus (Flaviviridae family) causing asymptomatic, mild (dengue with or without warning signs), or severe disease (severe dengue), sometimes leading to death.1 The infection in humans starts when DENV reaches cells of the mononuclear phagocyte lineage by interaction with some of the proposed receptors (DC-SIGN, heparan sulfate, Hsp70, Hsp90, etc.) and the viral particle is internalized by receptor-mediated endocytosis. Subsequently, low pH-dependent membrane fusion and uncoating lead to viral RNA release, polyprotein translation and processing, viral RNA synthesis by the replicase complex, virus assembly and in the endoplasmic reticulum and Golgi, maturation in the Golgi, and release of progeny viruses.3
There are no specific treatments available for dengue and the development of a vaccine has been limited by there factors: first the huge antigenic and genetic diversity of the virus, the lack of cross-protection immunity among DENV serotypes and eventually genotypes, and the host immune interactions that have been associated with disease severity.4 Several tetravalent vaccine candidates are currently under development. Though the vaccine studies have shown some promising outcome, the overall studies are not encouraging and needs a lot of research.
Because of the existence of enormous intra-serotype genetic diversity, the possibility of cross-protection after vaccination is questionable. Hence the vaccine composition should be based on circulating strains of DENV. A more intense DENV genotype surveillance must be conducted in those countries where vaccine candidates are planed. The results from such surveillance should be available in real-time for helping decision makers about potentially different vaccine composition for administration in the different regions, a novel challenge for vaccine developers attempting a worldwide coverage.
DENV Genetic Diversity
DENVs are enveloped single-stranded positive-sense RNA viruses (ssRNA+) whose genomes encode for a viral RNA-dependent RNA polymerase lacking proofreading activity5 that leads to very high substitution rates, rapid divergence, and the existence of at least four serotypes (DENV-1 to -4) with high intra-serotype genetic diversity.6 Differences in phylogeny-based estimations of DENV substitution rates depending of prior assumptions (strict or relaxed molecular clock, changes in the distribution of variable and invariable sites across the phylogeny, full-length genome or gene-based analysis) have been reported.7 The rates for DENV-1 range from 4.55 × 10−4 to 9.08 × 10−4 substitutions per site per year (subs/site/year); for DENV-2 the rates range from 6.07 × 10−4 to 9.84 × 10−4 subs/site/year; for DENV-3 the nucleotide substitution rates range from 9.01 × 10−4 to 10.40 × 10−4 subs/site/year, and estimations for DENV-4 range from 6.02 × 10−4 to 10.63 × 10−4 subs/site/year.6,8 Based on these rates and the coalescent theory, the more recent common ancestor (MRCA) of the 4 DENV serotypes could have existed more than 1000 y ago.9 DENV-4 was probably the first diverging serotype, followed by DENV-2 (around 350 y ago) and finally DENV-1 (125 y ago) and -3 (100 y ago).9
The 4 DENV serotypes were first defined by their antigenic properties as members of the DENV serocomplex.10 However, the accumulation of genetic diversity during the last centuries has led to inter-serotype genetic distances even higher than those observed among different species within the Flavivirus genus.9 The spread of the DENV serotypes around the world has also allowed the accumulation of intra-serotype genetic variation and the emergence of different monophyletic groups (genotypes) in the different geographic regions of the world (Fig. 1).11 Although genetic diversity has accumulated along the whole viral genome in which the structural and non-structural genes and untranslated regions (UTRs) are critical in one or more steps of the virus life cycle,5 the envelope gene has raised more attention because of the role of the envelope (E) protein in virus attachment and entry into the cell, as well as membrane fusion and interaction with the immune system.5
Figure 1.
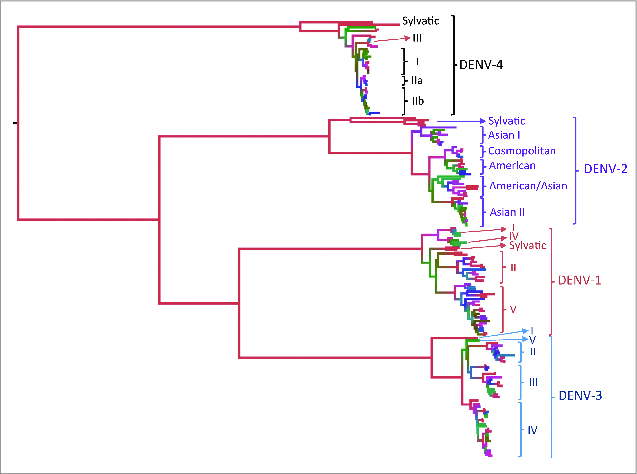
Phylogenetic tree of the 4 DENV serotypes based on the complete (1479–1485 nt) envelope gene of 289 viral isolates worldwide. The intra-serotype genetic diversity has allowed the designation of genotypes within each DENV serotype.37-40
The Envelope Protein
The E protein of DENV is a membrane-anchored glycoprotein of approximately 53 kDa that forms homodimers which are organized into rafts, each containing 3 parallel dimers in the mature virus.12 The arrangement of these rafts with the viral membrane (M) proteins and host-derived lipid membrane leads to formation of the viral coat with icosahedral-like symmetry. The E protein plays important roles in the life cycle of the DENV and in the stimulation of host protective immunity. The E protein has a major region known as the ectodomain (soluble fragment containing residues 1–394) and a minor membrane-anchored insoluble region (residues 395–495).13 The soluble fragment contains 3 structural domains (I, II, and III), which have been extensively characterized.13 Domains I and II function as a molecular hinge for E protein reorganization at low pH.14 Additionally, domain II contains the highly conserved fusion peptide responsible for virus-mediated cell-membrane fusion.15 Domain III forms an immunoglobulin-like fold containing putative receptor-binding motifs involved in receptor recognition, attachment and virus entry into the host cell. Several studies suggest that immune sera of DENV-infected patients contain several antibody populations which target the different antigenic epitopes exposed on both, the virion surface (e.g., M and E), and nonstructural proteins secreted during viral infection (e.g., viral protein NS1).16
Virus Neutralization and Vaccine Development
DENV E is the main protein involved in the immunological response and the induction of neutralizing antibodies.5 The antigenic epitopes of the E protein recognized by neutralizing antibodies during infection in humans have been mapped in all 3 domains. Antibodies targeting domain I/II have been correlated with cross-reactive immunity to the 4 DENV serotypes and weak neutralization potency,17 while those targeting domain III (an epitope localized on the lateral ridge and other located on the center of the A strand) have been correlated with a strong neutralizing activity.18 DENV-specific antibodies in human immune sera are mainly cross-reactive and weakly neutralizing with a very low proportion having strong neutralizing activity against only one serotype.19
Several in vitro and in vivo studies suggest important differences in the efficacy of antibody-mediated neutralization, due to the inability of certain antibodies to interact with the epitopes exposed by a virus belonging to a different genotype within the same serotype.20-22 Although several hypotheses are plausible, it is possible that the low efficacy of a tetravalent vaccine to protect against DENV-2, during a phase 2b clinical trial in Thailand could be due to the fact that a different DENV-2 genotype circulated during that period of time.23
DENV envelope gene has genetic variation large enough even to allow performing genotype discrimination and phylogeographic studies. It is therefore expected that the naturally accumulated variation allow DENV to escape a previously acquired immunity to a certain DENV genotype especially when a DENV strain with different genotype was used for the first challenge. In vitro, it is possible to obtain neutralization escape mutants of DENV through selection during serial passages of a wild type virus in the presence of low doses of monoclonal neutralizing antibodies.24 Because accumulation of mutations could lead to antigenic drift, the potential escape of a particular DENV genotype from vaccine-elicited antibodies may contribute to disease severity, which occurs by the broadly accepted mechanism of antibody-dependent enhancement (ADE).4
Antibody-Dependent Enhancement
ADE has been postulated as the best explanation for severe outcomes of dengue. This hypothesis is based on evidence in vitro and in vivo suggesting that DENV infection is enhanced by the administration of DENV-immune sera or monoclonal antibodies in cell cultures and monkeys.25-27 Additional evidence arises from epidemiological observations that infection in humans with one DENV serotype confers long-term protection against that serotype, but not against any other serotype. Cross-reactive antibodies can recognize and attach the virus belonging to a heterologous serotype in a non-neutralizing way. This cross-reaction could enhance the virus uptake by Fc receptor-bearing cells where they can quickly replicate,28 invade the lymph nodes and cause higher viremia that have been correlated with disease severity.29
In case the administered vaccine does not confer protection, another major concern emerges from the fact that the ADE could lead to a severe outcome of the disease.14 We are therefore encouraged to define the extent of protection against the different serotypes and genotypes and the geographic distribution and continuous surveillance before vaccine approval and subsequent vaccine composition.
Global Distribution of the Different DENV Genotypes
The intra-serotype genetic variation of DENV in the form of genotypes determined by sequencing was first reported in the early 1990s for DENV-1 and -2.30 Sequence availability during the last 2 decades allowed the high-resolution genotyping which were named according to their distribution.11 In spite of intense microevolution of DENV, the worldwide distribution of the DENV genotypes has been stable through the time (Fig. 2) with only few important changes. Although viruses belonging to a specific genotype may be reported in places, which are distant from each other, these are frequently considered imported cases and several factors limit these from becoming established.31
Figure 2.
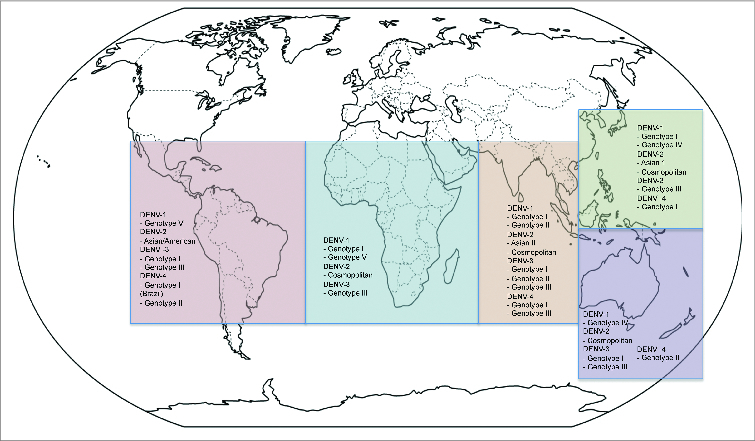
Global distribution of the different DENV genotypes. Strains of the different DENV genotypes circulating during the period 2001–2014 were included. The DENV strains from imported cases where disease establishment has not been demonstrated were excluded from the analysis.
The Native American genotype of DENV-2 co-circulated in several Latin-American countries, and was finally replaced by the Asian/American genotype during 1990s.32 Also, despite the circulation of DENV-3 genotype III in the Americas for a long period, the co/circulation with DENV-3 genotype I was recently reported for a short period of time in Brazil, Colombia, and Ecuador.31,33,34
Although genotype co-circulation and replacement are not very common phenomena in the Americas, these are frequently observed in several countries in Southeast Asia and South Pacific.35,36 Because of globalization, commercial relationships, and tourism, no country is exempt of importing and establishing novel DENV strains of any genotype. It is therefore important to maintain active genotype surveillance in all the endemic countries where the DENV is transmitted and vaccination is planed in the near future.
Conclusion
Major efforts in DENV research are currently focused on designing/producing and licensing a tetravalent vaccine. It is therefore important to understand the role of DENV genetic variability in vaccine efficacy. If the results of vaccine candidates in clinical trials continue showing low efficacy, future vaccine approaches should consider the design of vaccines for regional administration whose antigenic and genetic composition are based on genotype surveillance.
References
- 1. Dengue WHO. guidelines for diagnosis, treatment, prevention and control - new edition. Geneva: World Health Organization; 2009. 147p p. [PubMed] [Google Scholar]
- 2. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. . The global distribution and burden of dengue. Nature 2013; 496:504-7; PMID:23563266; http://dx.doi.org/ 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clyde K, Kyle JL, Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J Virol 2006; 80:11418-31; PMID:16928749; http://dx.doi.org/ 10.1128/JVI.01257-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Halstead SB. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev Infect Dis 1989; 11(Suppl 4):S830-9; PMID:2665015; http://dx.doi.org/ 10.1093/clinids/11.Supplement_4.S830 [DOI] [PubMed] [Google Scholar]
- 5. Lindenbach DB, Thiel H-J, Rice CM. Flaviviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed Philadelphia: Wolkers Kluwer/Lippincott Williams and Wilkins; 2007. p 1101-1152. [Google Scholar]
- 6. Twiddy SS, Holmes EC, Rambaut A. Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol 2003; 20:122-9; PMID:12519914; http://dx.doi.org/ 10.1093/molbev/msg010 [DOI] [PubMed] [Google Scholar]
- 7. Dunham EJ, Holmes EC. Inferring the timescale of dengue virus evolution under realistic models of DNA substitution. J Mol Evol 2007; 64:656-61; PMID:17541679; http://dx.doi.org/ 10.1007/s00239-006-0278-5 [DOI] [PubMed] [Google Scholar]
- 8. Allicock OM, Lemey P, Tatem AJ, Pybus OG, Bennett SN, Mueller BA, Suchard MA, Foster JE, Rambaut A, Carrington CV. Phylogeography and population dynamics of dengue viruses in the Americas. Mol Biol Evol 2012; 29:1533-43; PMID:22319149; http://dx.doi.org/ 10.1093/molbev/msr320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol 2003; 3:19-28; PMID:12797969; http://dx.doi.org/ 10.1016/S1567-1348(03)00004-2 [DOI] [PubMed] [Google Scholar]
- 10. Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol 1989; 70:37-43; PMID:2543738; http://dx.doi.org/ 10.1099/0022-1317-70-1-37 [DOI] [PubMed] [Google Scholar]
- 11. Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res 2003; 59:315-41; PMID:14696333; http://dx.doi.org/ 10.1016/S0065-3527(03)59009-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure 2004; 12:1607-18; PMID:15341726; http://dx.doi.org/ 10.1016/j.str.2004.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A 2003; 100:6986-91; PMID:12759475; http://dx.doi.org/ 10.1073/pnas.0832193100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Messer WB, de Alwis R, Yount BL, Royal SR, Huynh JP, Smith SA, Crowe JE, Jr., Doranz BJ, Kahle KM, Pfaff JM, et al. . Dengue virus envelope protein domain I/II hinge determines long-lived serotype-specific dengue immunity. Proc Natl Acad Sci U S A 2014; 111:1939-44; PMID:24385585; http://dx.doi.org/ 10.1073/pnas.1317350111 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Huang CY, Butrapet S, Moss KJ, Childers T, Erb SM, Calvert AE, Silengo SJ, Kinney RM, Blair CD, Roehrig JT. The dengue virus type 2 envelope protein fusion peptide is essential for membrane fusion. Virology 2010; 396:305-15; PMID:19913272; http://dx.doi.org/ 10.1016/j.virol.2009.10.027 [DOI] [PubMed] [Google Scholar]
- 16. Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res 2003; 59:141-75; PMID:14696329; http://dx.doi.org/ 10.1016/S0065-3527(03)59005-4 [DOI] [PubMed] [Google Scholar]
- 17. Crill WD, Chang GJ. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J Virol 2004; 78:13975-86; PMID:15564505; http://dx.doi.org/ 10.1128/JVI.78.24.13975-13986.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, et al. . Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol 2007; 81:12816-26; PMID:17881453; http://dx.doi.org/ 10.1128/JVI.00432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. . The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 2010; 8:271-83; PMID:20833378; http://dx.doi.org/ 10.1016/j.chom.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, Diamond MS. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol 2010; 84:10630-43; PMID:20702644; http://dx.doi.org/ 10.1128/JVI.01190-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, Baric RS. Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl Trop Dis 2012; 6:e1486; PMID:22389731; http://dx.doi.org/ 10.1371/journal.pntd.0001486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zulueta A, Martín J, Hermida L, Alvarez M, Valdés I, Prado I, Chinea G, Rosario D, Guillén G, Guzmán MG. Amino acid changes in the recombinant Dengue 3 Envelope domain III determine its antigenicity and immunogenicity in mice. Virus Res 2006; 121:65-73; PMID:16781791; http://dx.doi.org/ 10.1016/j.virusres.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 23. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, et al. . Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 2012; 380:1559-67; PMID:22975340; http://dx.doi.org/ 10.1016/S0140-6736(12)61428-7 [DOI] [PubMed] [Google Scholar]
- 24. Lim HY, Ng ML. A different mode of entry by dengue-2 neutralisation escape mutant virus. Arch Virol 1999; 144:989-95; PMID:10416380; http://dx.doi.org/ 10.1007/s007050050561 [DOI] [PubMed] [Google Scholar]
- 25. Halstead SB. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis 1979; 140:527-33; PMID:117061; http://dx.doi.org/ 10.1093/infdis/140.4.527 [DOI] [PubMed] [Google Scholar]
- 26. Littaua R, Kurane I, Ennis FA. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J Immunol 1990; 144:3183-6; PMID:2139079 [PubMed] [Google Scholar]
- 27. Goncalvez AP, Engle RE, St Claire M, Purcell RH, Lai CJ. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc Natl Acad Sci U S A 2007; 104:9422-7; PMID:17517625; http://dx.doi.org/ 10.1073/pnas.0703498104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kliks SC, Nimmanitya S, Nisalak A, Burke DS. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am J Trop Med Hyg 1988; 38:411-9; PMID:3354774 [DOI] [PubMed] [Google Scholar]
- 29. Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, et al. . Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis 2000; 181:2-9; PMID:10608744; http://dx.doi.org/ 10.1086/315215 [DOI] [PubMed] [Google Scholar]
- 30. Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 1990; 174:479-93; PMID:2129562; http://dx.doi.org/ 10.1016/0042-6822(90)90102-W [DOI] [PubMed] [Google Scholar]
- 31. Domingo C, Niedrig M, Gascón J, Palacios G, Reyes N, Malo MJ, Wichmann O, Ruiz J, Schultze D, Schunk M, et al. . Molecular surveillance of circulating dengue genotypes through European travelers. J Travel Med 2011; 18:183-90; PMID:21539658; http://dx.doi.org/ 10.1111/j.1708-8305.2011.00501.x [DOI] [PubMed] [Google Scholar]
- 32. Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RM, da Rosa AT. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 1997; 230:244-51; PMID:9143280; http://dx.doi.org/ 10.1006/viro.1997.8504 [DOI] [PubMed] [Google Scholar]
- 33. Barcelos Figueiredo L, Batista Cecílio A, Portela Ferreira G, Paiva Drumond B, Germano de Oliveira J, Bonjardim CA, Peregrino Ferreira PC, Kroon EG. Dengue virus 3 genotype 1 associated with dengue fever and dengue hemorrhagic fever, Brazil. Emerg Infect Dis 2008; 14:314-6; PMID:18258129; http://dx.doi.org/ 10.3201/eid1402.070278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Usme-Ciro JA, Mendez JA, Tenorio A, Rey GJ, Domingo C, Gallego-Gomez JC. Simultaneous circulation of genotypes I and III of dengue virus 3 in Colombia. Virol J 2008; 5:101; PMID:18764951; http://dx.doi.org/ 10.1186/1743-422X-5-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rabaa MA, Ty Hang VT, Wills B, Farrar J, Simmons CP, Holmes EC. Phylogeography of recently emerged DENV-2 in southern Viet Nam. PLoS Negl Trop Dis 2010; 4:e766; PMID:20668540; http://dx.doi.org/ 10.1371/journal.pntd.0000766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. A-Nuegoonpipat A, Berlioz-Arthaud A, Chow V, Endy T, Lowry K, Mai Q, Ninh TU, Pyke A, Reid M, Reynes JM, et al. . Sustained transmission of dengue virus type 1 in the Pacific due to repeated introductions of different Asian strains. Virology 2004; 329:505-12; PMID:15518827; http://dx.doi.org/ 10.1016/j.virol.2004.08.029 [DOI] [PubMed] [Google Scholar]
- 37. Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, Liprandi F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology 2002; 303:110-9; PMID:12482662; http://dx.doi.org/ 10.1006/viro.2002.1686 [DOI] [PubMed] [Google Scholar]
- 38. Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology 2002; 298:63-72; PMID:12093174; http://dx.doi.org/ 10.1006/viro.2002.1447 [DOI] [PubMed] [Google Scholar]
- 39. Uzcategui NY, Comach G, Camacho D, Salcedo M, Cabello de Quintana M, Jimenez M, Sierra G, Cuello de Uzcategui R, James WS, Turner S, et al. . Molecular epidemiology of dengue virus type 3 in Venezuela. J Gen Virol 2003; 84:1569-75; PMID:12771427; http://dx.doi.org/ 10.1099/vir.0.18807-0 [DOI] [PubMed] [Google Scholar]
- 40. Klungthong C, Zhang C, Mammen MP, Jr., Ubol S, Holmes EC. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 2004; 329:168-79; PMID:15476884; http://dx.doi.org/ 10.1016/j.virol.2004.08.003 [DOI] [PubMed] [Google Scholar]


