1 Rationale for Live Cell Imaging of mRNA
Recent advances in mRNA visualization technology have placed biomedical imaging at an interface between molecular biology and cellular biology. It is now increasingly possible to study in vivo gene expression at the transcript level. Analyses of mRNA expression, movement, interactions, and localization will enhance understanding of cellular responses to various conditions and will complement studies of protein expression.
The ability of mRNAs to move in the cytoplasm of eukaryotic cells is essential for sorting sites of synthesis for specific proteins. Studies analyzing the mechanisms of mRNA localization have demonstrated the existence of complex mRNA transport systems. The cytoskeleton is usually required for mRNA localization, and, in a few instances, molecular motors have been shown to be associated with localized mRNA and are required for their movements (Bertrand et al. 1998; Schnorrer et al. 2000; Cha et al. 2001; Wilkie and Davis 2001). In yeast, ASH1 mRNPs are directly associated with a specific myosin motor, and their movements require both this motor and actin filaments (Bertrand et al. 1998). In the Drosophila blastocyst, apically localized mRNAs are thought to be associated with dynein, since their movements require both dynein and microtubules (Schnorrer et al. 2000; Cha et al. 2001). These and other studies have provided strong support for the idea that localized mRNAs are actively transported on cytoskeletal filaments.
In contrast to localized mRNAs, nonlocalized ones are thought to be distributed homogeneously throughout the cytoplasm. Nevertheless, they could still be capable of movement. For instance, following nuclear export, they must leave the perinuclear area to be translated throughout the cytoplasm. Studies with inert tracers, such as fluorescent dextrans or ficolls, suggested that particles with sizes equivalent to some mRNPs have limited diffusion in the cytoplasm (Luby-Phelps et al. 1987). Therefore, even nonlocalized mRNA molecules may require an active transport process. Transport of localized and nonlocalized mRNA should differ, to eventually generate their particular distributions. Thus, study of mRNA transport processes will elucidate behavioral differences among mRNA subpopulations.
Furthermore, live cell mRNA imaging technology may add new information about where and when transcription and translation occur, and can provide a single cell expression profile that cannot be achieved with microchip or biochemical analyses. Simultaneous analysis of multiple transcription sites can provide a single cell profile of gene expression that can be linked precisely to cellular morphology (Levsky et al. 2002) and observation of these transcription sites in living cells will provide novel information about the time course of gene expression in normal and pathological samples.
There are potential medical applications for mRNA imaging, extending from studies of disease mechanism to diagnostic and treatment applications. One of the most significant contributions of mRNA studies to the identification of a disease mechanism was the analysis of trinucleotide repeat transcript foci in nuclei of myotonic dystrophy cells and tissues (Taneja et al. 1995; Davis et al. 1997). This study revealed that myotonic dystrophy (DM) protein kinase transcripts containing increased numbers of CTG repeats accumulate in the nucleus in stable, long-lived clusters. Nuclear accumulation of mutant transcripts is consistent with a model of DM loss of function due to lack of nuclear export, along with possible dominant-negative effects on the export of other mRNA export. Another disease mechanism that can be studied using mRNA visualization is the process of metastasis. Localization patterns of certain mRNAs such as b-actin appear to be related to metastatic capability; nonmetastatic tumor cells differ in their mRNA localization and motility characteristics compared to metastatic tumor cells (Shestakova et al. 1999). Analyses of mRNAs are also being applied to the study of Fragile X Mental Retardation, in which the RNA binding protein, fragile X mental retardation protein (FMRP), is disrupted, providing a disconnect between mRNA regulation and normal neuronal development (Ashley et al. 1993; Siomi et al. 1993). Translational dysregulation of mRNAs normally associated with FMRP may be the proximal cause of fragile X syndrome, and one proposed model for FMRP function is that it shuttles mRNAs from the nucleus to postsynaptic sites where transcripts are held in a translationally inactive form until further stimulation alters FMRP function (Brown et al. 2001). Several other mRNA binding proteins have been shown to play a role in mRNA trafficking in developing neurons. These neuronal mRNA binding proteins include zipcode binding proteins one and two (ZBP-1, ZBP-2; Ross et al. 1997; Gu et al. 2002), which play a role in the dendritic trafficking of b actin mRNA. Live cell mRNA studies can also provide important insights in virology. Subcellular localization and tracking of viral sequences, combined with labeling of relevant proteins and other structures, will elucidate viral host interaction mechanisms. These studies will also help clarify the cellular lifecycle of viral RNA as well as host cell defense mechanism and may correlate cell phenotype with viral RNA load as infection progresses.
Messenger RNA detection in individual, living cells has several advantages over other techniques for the study of gene expression. For example, in comparison to population analyses, such as Northern blotting, single cell studies can describe the amount of virus in individual infected cells, or the percentage of cells that are infected. By imaging mRNA in living cells instead of fixed cells, it is possible to determine the timing of mRNA expression and dynamics, as well as different patterns of mRNA movement and interactions throughout the lifetime of a transcript. Because mRNA movement appears to be a rapid, rare event, the ability to examine a series of time points becomes important for the observation of mRNA transit. In addition, live cell imaging avoids the possible introduction of artifact during cell fixation.
For years, the major limitation of imaging mRNA in living cells has been a dearth of available technology. Recent years have yielded significant advances in four major categories of mRNA visualization technology, including microinjection of fluorescent RNA (Ainger et al. 1993; Ferrandon et al. 1994), hybridization of fluorescent oligonucleotide probes (Politz et al. 1998, 1999), the use of cell-permeant dyes which bind RNA, and sequence-specific mRNA recognition by a GFP fusion protein (Bertrand et al. 1998). Each of these technologies (Fig. 1, by permission) will be presented, but the focus of this article will be the sequence-specific recognition of mRNA by GFP fused to the mRNA binding protein MS2.
Fig. 1.
Existing technology for the visualization of mRNA in living cells, including images obtained using those technologies (Knowles et al. 1996, 1997; Bertrand et al. 1998; Theurkauf et al. 1998; Politz et al. 1999; Lorenz et al. 2000; Dirks et al. 2001; Perlette et al. 2001; Privat et al. 2001)
2 Existing Technologies for the Visualization of RNA in Living Cells
2.1 Microinjection of Fluorescent RNA
Fluorescent RNA can be synthesized by in vitro transcription with phage polymerases in the presence of fluorescent nucleotide analogs for microinjection into living cells (Wilkie and Davis 2001). Likewise, transcription with a mixture of unmodified and amino-allyl nucleotides generates RNA that can be chemically coupled to activated fluorophores (Wang et al. 1991). Because amino-allyl nucleotides are incorporated at a higher frequency than fluorescent analogs, very bright RNA can be obtained. The main disadvantage of microinjecting fluorescently labeled mRNA is the introduction of nonendogenous material into the cell. Nevertheless, this approach has proven successful in a number of cases to reveal routes of trafficking of localized RNAs (Ainger et al. 1993; Theurkauf and Hazelrigg 1998; Wilkie and Davis 2001).
2.2 Fluorescent Oligonucleotide Probes (FIVH-Fluorescent In Vivo Hybridization)
Fluorescent oligonucleotide probes have been developed to follow unmodified, endogenous cellular RNA. An oligonucleotide complementary to the desired sequence is covalently linked to fluorochromes and can contain chemical modifications that increase cell penetration and hybrid stability. Oligonucleotides are delivered across the plasma membrane and hybridize with the target sequence to identify RNA directly in living cells (Politz et al. 1995, 1999). Six categories of fluorescent oligonucleotide probes exist, including conventional DNA probes (Politz et al. 1995; Lorenz et al. 2000), caged DNA probes (Politz et al. 1999; Pederson 2001), linear 2′ O methyl (2′OMe) RNA probes (CarmoFonseca et al. 1991; Dirks et al. 2001; Molenaar et al. 2001), peptide nucleic acid (PNA) probes (Dirks et al. 2001), molecular beacons (Tyagi et al. 1996, 1998; Sokol et al. 1998; Molenaar et al. 2001), and probes with a linked fluorophore whose fluorescence properties change upon association with the target mRNA strand (Thuong et al. 1987; Privat et al. 2001). Advantages of oligodeoxynucleotide (ODN) probes are their ability to detect endogenous mRNA, their relative stability (as long as 18 h, Politz et al. 1995), and the ease with which they are introduced into the cell. ODNs can be used to perform fluorescence resonance energy transfer (FRET) by simultaneously introducing two oligonucleotides labeled with different fluorophores and recording the loss of donor fluorescence and/or gain of acceptor fluorescence. A caveat of antisense oligodeoxyribonucleotides is that they can prevent translation by blocking ribosome movement along the mRNA or by inducing RNase H cleavage of the RNA/oligo hybrid, altering expression of target mRNA. In addition, fluorescent oligonucleotides have been found to localize exclusively to the nucleus in some cases (Dirks et al. 2001).
2.3 Cell Permeant Dyes
SYTO-14 is a membrane-permeable nucleic acid stain that has been used to study RNA in living cells (Knowles et al. 1996, 1997). SYTO-14 is advantageous because it will detect the endogenous RNA population, it is not limited to the nucleus, and its use requires no subsequent intervention after introduction into cells. A major disadvantage of cell permeant RNA binding dyes is their lack of specificity due to identical recognition of all RNAs.
2.4 RNA Tagged with Sequence-Specific GFP Fusion Proteins
It is possible to visualize a specific transcript by fusion of GFP with a sequence-specific RNA binding protein, such as the coat protein of phage MS2, and insertion of the RNA sequence recognized by this protein in the RNA of interest. When both the MS2-GFP fusion protein and the reporter RNA are co-expressed, MS2-GFP binds to the reporter in living cells and GFP fluorescence constitutes a reliable indicator of RNA localization (Bertrand et al. 1998). To remove the noise of unbound MS2-GFP, it is possible either to express a low level of MS2-GFP such that most of the MS2-GFP molecules are bound to the RNA, or to include a localization signal in the MS2-GFP protein which will exclude unbound molecules from the area of interest (Bertrand et al. 1998). This technique has several advantages: (1) it is possible to visualize RNA that is synthesized and processed by the cell; (2) it is possible to use the technique in organisms that cannot be microinjected, or that do not allow penetration of macromolecules; (3) it is possible to increase the sensitivity of RNA detection by multimerizing the binding site in the RNA reporter. The detection limit of this technique has reached the level of single mRNA molecules through the use of MS2 repeats containing 6 to 24 binding sites (i.e., 12–48 GFP molecules; Fusco et al. 2003). An analogous detection system has been designed to visualize genes in living nuclei, using a lac i-GFP fusion protein and an array of lac o binding sites inserted in the genome. This system first demonstrated the limits of detection, and requires as little as 60 GFP molecules (Robinett et al. 1996). The combination of these two systems, for the simultaneous visualization of gene activation and mRNA trafficking in living cells, is in progress.
However, the MS2-GFP system does involve the visualization of a nonendogenous mRNA, introduces a nonendogenous protein into the RNA movement process, and does not allow cellular control of expression levels. While the first two disadvantages are inherent to the MS2 labeling system, the third disadvantage can be addressed by design of inducible cell lines with specific control of MS2 reporter levels. Alternatively, an endogenous GFP-labeled RNA binding protein can be used. Theurkauf and Hazelrigg tagged bicoid mRNA with a GFP-exuperantia fusion (Theurkauf et al. 1998). While this system avoids introduction of an exogenous protein other than GFP, it is difficult to determine whether a protein that is known to bind an mRNA is, in fact, persistently bound to that mRNA, thus presenting the question of whether all GFP fusion protein movements are indicative of mRNA movements. The MS2-GFP-labeling system provides a stable link between GFP and mRNA (Kd = 39nM, in vitro; Lago et al. 1998), allowing reliable identification of mRNA particles throughout trafficking behavior. Importantly, when the fusion protein contains a nuclear localization signal (NLS), it will be targeted to the nucleus and only be introduced into the cytoplasm by its association with an exported RNA. Finally, the multimerization of the MS2-GFP allows accumulation of a fluorescent signal sufficient to characterize single molecules.
3 Analysis of Single RNA Dynamics in Living Cells
Following successful visualization of the RNA of interest, one must choose a method for analysis of RNA dynamics. Two general techniques have been used: photobleaching and fluorescent correlation spectroscopy. Photobleaching techniques involve irradiation with intense light leading to the irreversible inactivation of fluorophores (Axelrod et al. 1976; Phair and Misteli 2000). Fluorescence correlation spectroscopy (FCS) is a fluctuation analysis method used to measure molecular transport and chemical kinetics by counting the molecules entering and leaving a small interrogation volume over successive time intervals (Wang et al. 1991). When single RNA molecules are detectable, direct tracking of individual RNA by time-lapse microscopy is applicable. These results yield a population profile of RNA movements.
4 Time-Lapse Microscopy/Particle Tracking
It therefore becomes possible to use time-lapse microscopy to follow RNA movements. This requires either single molecule sensitivity, or the concentration of many RNA molecules in a multi-molecule aggregate, as previously observed in some cases (Ainger et al. 1993; Ferrandon et al. 1994; Bertrand et al. 1998; Wilkie et al. 2001). The single molecule dynamics measured in this method are complementary to the population dynamics obtained through fluorescence recovery after photobleaching (FRAP) or fluorescence correlation spectroscopy (FCS). Diffusion coefficients can be obtained from time-lapse data although analysis of a large number of molecules is required, but in addition, the temporal sequence of movements can provide novel information. For example, if a molecule is alternatively immobile and mobile on a relatively short time-scale, photobleaching techniques may average these behaviors while both will be apparent by time-lapse microscopy. Likewise, FCS would only see a small window of the behavior profile of single molecules. The ability to follow the fate of a molecule can reveal an ordered sequence of events. In addition, time-lapse techniques can reveal rare, but functionally important behaviors that could not be detected otherwise. For instance, ASH1 mRNA visualized in the cytoplasm of living yeast (Bertrand et al. 1998) is localized at the bud tip, after transport by a specific myosin motor on actin cables. However, because the transport is very rapid and lasts for less than a minute of the entire cellular lifetime, the proportion of actively transported mRNA molecules is on average very low and does not exceed a few percent of the total molecules. Such a minor subset of behavior in a population would very likely not be detected by photobleaching and FCS techniques.
When single molecule resolution is achieved, a detailed analysis of individual particle movements can yield information related to the mechanism of mRNA transport. Analysis includes collection of position, velocity, and acceleration values for the determination of movement type, such as active transport, Brownian motion, or corralled diffusion. In the case of active transport, movement analysis can help identify candidate motors involved in transport. For example, if a particle is observed to display actin dependent movement at a constant speed of approximately 200–400 nm/sec, a myosin motor is likely to be involved (Cheney et al. 1993). Other interesting characteristics of mRNA particles that can be obtained from time-lapse images in living cells include particle size, location, diffusion coefficient, mean squared displacement, and relation to other cellular components, such as the cytoskeleton, all of which can be useful in determining the mechanism of mRNA movement and localization. When images are of a high quality, but particles can still not be tracked, it is possible to perform two-dimensional deconvolutions on movies to further improve signal to noise ratio and facilitate particle tracking (Huygens, Bitplane, The Netherlands).
We summarize below the results obtained using the described imaging techniques to study cytoplasmic mRNAs. In the cytoplasm, an increasing number of specific transcripts are being studied in living cells. These studies are significant because they provide unique information about the dynamics of mRNA movement and help elucidate the mechanisms of mRNA particle assembly, transport, and localization as well as crucial mRNA interactions with proteins and the cytoskeleton. This information can be obtained in single cells for endogenous, engineered and viral transcripts, greatly widening our current understanding of mRNA behavior.
5 In Vivo mRNA Analyses in the Cytoplasm
In yeast, ASH1 mRNA was visualized in live Saccharomyces cerevisiae, in the first use of the MS2-GFP labeling system. The GFPASH1 particle required specific ASH1 sequences for formation and localization to the bud tip (Bertrand et al. 1998). Mutations in the ASH1 -associated She proteins were found to disrupt particle localization, and She2 and She3 mutants also inhibited particle formation. Video microscopy showed that She1p/Myo4p moved mRNA particles to the bud tip at velocities of 200–440 nm/sec. Further studies of the ASH1 mRNA transport process (Beach et al. 1999) showed that in cells lacking Bud6p/Aip3p or She5p/Bni1p, ASH1 mRNA particles traveled to the bud, but failed to remain at the bud tip, revealing a distinction between mRNA transport and localization similar to that described in oligodendrocytes (Ainger et al. 1997). These findings not only characterized the movement of the ASH1 mRNA particle, but also contributed significantly to the identification of proteins, including a motor, that are required for ASH1 particle transport.
One method for the visualization of mRNA in living cells using GFP is the use of GFP-tagged RNA binding proteins. The dynamic behavior of bicoid mRNA in living Drosophila oocytes was visualized with such a mechanism using a GFP-exuperantia fusion (Theurkauf et al. 1998). The results of this study elaborate the relationship between mRNA transport and the cytoskeleton, demonstrating that particle movement can alternate through phases of cytoskeleton dependence and independence.
Additional information regarding the bicoid localization mechanism came from microinjection of fluorescent bicoid transcripts into Drosophila oocytes (Cha et al. 2001). Interestingly, fluorescent bicoid mRNA injected into the oocyte displays nonpolar microtubule-dependent transport to the closest cortical surface, but bicoid mRNA injected into the nurse cell cytoplasm, withdrawn, and injected into a second oocyte shows microtubule-dependent transport to the anterior cortex. This study identified the requirement of a nurse cell cytoplasmic component for proper trafficking of bicoid mRNA. It is interesting to note that a component that is present in the nurse cell cytoplasm allows reconnection of bicoid mRNA to the microtubule network after a microtubule independent transit through the ring canal, indicating that transcripts bear some mark, possibly a bound protein, indicating their prior location even as they continue transit into other compartments. Thus, transcripts in a given compartment may be able to be distinguished by their past behavior based on their present set of interaction partners. This concept of transcript historesis has also been proposed with respect to the behavior of CaMKII granules (Rook et al. 2000). In this case, it was suggested that granule location at a specific point in time reflected the activation history of a synapse, allowing identification of stimuli that were in the past.
Injection of fluorescently labeled wingless and pair-rule transcripts into living Drosophila syncytial blastoderm embryos (Wilkie et al. 2001) showed that fluorescent apical RNAs specifically assemble into particles that approach the minus end of microtubules using the motor protein cytoplasmic dynein and its associated dynactin complex.
In neurons, the movement of endogenous RNA was directly visualized in neuronal processes of living cells using the intercalating dye SYTO 14 (Knowles et al. 1996, 1997). One of the first studies in which fluorescently labeled mRNA was microinjected into the cytoplasm of living cells was performed by Ainger et al. (1993), who microinjected myelin basic protein (MBP) mRNA into neurites and observed the formation of granules which were transported along oligodendrocytes and localized to the myelin compartment. More recent analyses of A2RE RNA granule movements, performed with rapid confocal line scanning through a single granule on a microtubule, demonstrated a rapid pattern of back and forth vibrations along the microtubule axis. Through mean squared displacement analysis of these vibrations, it was observed that A2RE granules undergo “corralled diffusion”, or movement of a defined distance before reversal of direction (Carson et al. 2001). The movements of GFPlabeled CAMKII mRNA particles were observed in rat hippocampal neurons, in the first use of the MS2-GFP system in somatic cells. Interestingly, this study showed that anterograde particle movements increased in response to neuronal depolarization, in comparison to oscillatory and retrograde movements (Rook et al. 2000). Examination of GFP-labeled b actin zipcode binding protein 1 (ZBP-1) provided insights regarding the translocation of b actin mRNA in neurons (Zhang et al. 2001). Live cell imaging of EGFP-ZBP1 revealed that GFPlabeled particles move in a rapid, bidirectional fashion that is reduced tenfold by antisense oligonucleotides to the b actin zipcode.
In cell lines, the behavior of transiently expressed c-fos mRNA was observed in living Cos-7 cells through microinjection of fluorescently labeled oligonucleotides (Tsuji et al. 2001). Microinjection of molecular beacons targeted to the vav protooncogene in K562 human leukemia cells followed by confocal microscopy within 15 min of microinjection demonstrated that molecular beacons could detect transcripts in the cytoplasm of living cells, with a detection limit of ~10 molecules of mRNA (Sokol et al. 1998). A later study showed that most beacon hybridization occurred within 10 min of microinjection and maximum intensity was obtained by 15 min and maintained until 40 min after microinjection. (Perlette and Tan 2001).
To better understand the mechanisms controlling mRNA movements, we developed a method for visualizing single RNA molecules in living mammalian cells in real time. This method uses the MS2-GFP fusion protein and a reporter mRNA containing tandemly repeated MS2 binding sites, inserted between LacZ and SV40 sequences (Bertrand et al. 1998), followed by quantitative in situ hybridization. When the fusion protein was coexpressed with the reporter mRNA, the MS2-GFP bound to the mRNA was exported to the cytoplasm. When the MS2-GFP fusion protein was expressed alone, a nuclear localization signal confined the protein to the nucleus. We used Cos cells in this study because of their optical properties.
Discrete particles of GFP-labeled mRNA were resolvable only when 24 MS2 sites were in the reporter mRNA, not when either 6 or 12 MS2 sites were present. To quantify the number of mRNA molecules in each GFP-labeled particle, we imaged concentrations of purified GFP to determine the amount of light emitted by a given number of GFP molecules, and we calculated the number of GFP molecules in each GFP-labeled mRNA particle (Fusco et al. 2003). Quantification of the fluorescence from more than 600 GFP-labeled mRNA particles in eight cells showed that most GFP-labeled mRNA particles contained 20–50 molecules of GFP, with an average of 33. Since the MS2 protein binds as a dimer (Valegard et al. 1994), this measurement coincided with the expected number of GFP molecules labeling a single molecule of reporter RNA, assuming that most of the 24 MS2 sites were bound. This quantification procedure was independently validated by using a laco/GFPlaci system for calibration of the GFP signal (Robinett et al. 1996). To exclude the possibility that clusters of mRNA molecules sparsely labeled with GFP were misidentified as single mRNA molecules, we performed in situ hybridization to a single target on the mRNA reporter (Femino et al. 1998). Cells were simultaneously hybridized with a Cy5-labeled probe to a region between MS2 binding sites and a single Cy3-labeled probe to the LacZ portion of the mRNA. GFP, Cy5, and Cy3 were detected concurrently in single cells, allowing quantification of the number of probes colocalized with individual GFP particles (Fusco et al. 2003). Analysis of 125 multicolor particles in five cells showed that the majority of multicolored particles were single molecules, containing one Cy3 probe and eight MS2 probes. This validates the independent conclusion, from the analysis described above that the majority of the GFP-labeled mRNA particles detected in the cytoplasm contained a single RNA molecule. There were a small number of GFP particles containing more than a single RNA molecule present in every cell. The single-molecule GFP particles were distinct from the RNA “granules” observed in a variety of cell types, which are much brighter and larger and, although not quantitated, apparently contain multiple RNA molecules (Davis and Ish-Horowicz 1991; Ainger et al. 1993; Bertrand et al. 1998; Theurkauf and Hazelrigg 1998; Rook et al. 2000; Wilkie and Davis 2001; Kloc et al. 2002; Farina et al. 2003).
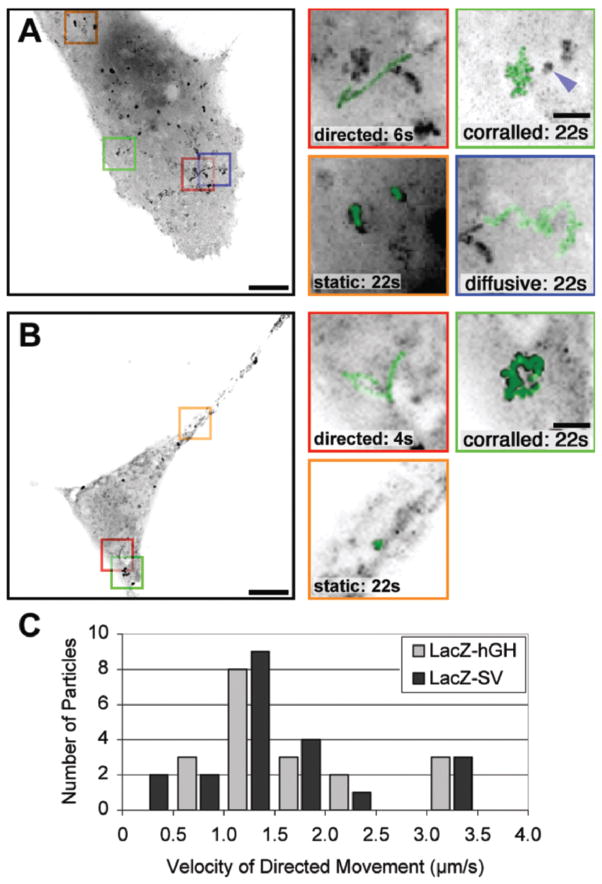
Figure 2 shows the movements of the single mRNA particles recorded in living cells at a rate of nine images per second. This high frequency was required because some mRNA molecules exceeded 1 mm/s (see below). The mRNAs contained the coding sequence of LacZ, the MS2 sites, and the 3′ UTR of either the human growth hormone gene or SV40 (reporters LacZ-24-hGH and LacZ-24-SV, respectively).
Fig. 2.
Dynamics of mRNA molecules in the cytoplasm of mammalian cells (reproduced from Fusco et al. 2003). All images were obtained at a rate of nine images per second, for periods of 22 s, and were deconvolved. A Movements of LacZ-24-hGH mRNAs. Cos cells transiently expressing LacZ-24-hGH mRNAs and the MS2-GFP protein were imaged live. Left a maximum intensity image projection of 200 time frames on one image (“maximal projection”). The scale bar represents 10 mm. Right panel magnifications: the scale bar represents 2 mm mRNA track superimposed (green) from each of the indicated boxed regions. The blue arrow points to a “static” particle in the vicinity of a “corralled” mRNA. B Movements of LacZ-24-SV mRNAs. Cos cells were transiently cotransfected with pRSV-Z-24-SV and pMS2-GFP and were imaged as in A. The scale bar represents 10 mm. Right panel magnifications: track of mRNA movement superimposed (green) on an enlargement from each of the indicated boxed regions. The scale bar represents 2 mm. C Velocities of directed motion. For each directed movement of either the LacZ-24-hGH or the LacZ-24-SV mRNA particles, the mean velocity was calculated. The corresponding histograms show a peak at 1.0–1.5 mm/s (Knowles et al. 1996, 1997; Bertrand et al. 1998; Theurkauf et al. 1998; Politz et al. 1999; Lorenz et al. 2000; Dirks et al. 2001; Perlette et al. 2001; Privat et al. 2001)
Dynamic behavior of b actin 3′ UTR mRNA was compared to that of human growth hormone (hGH) and SV40 transcripts in Cos7 cells. These three transcripts were chosen because b actin mRNA, hGH mRNA, and SV40 mRNA represent transcripts of three distinct categories: endogenous sequences that have been observed to localize, endogenous sequences that have not been observed to localize, and nonendogenous sequences that have not been observed to localize, respectively. Regardless of any specific cytoplasmic distribution, individual mRNA molecules exhibit rapid and directional movements on microtubules. In addition to directional movements, all three transcript types displayed static, diffusional, and corralled diffusional behavior. Disruption of the cytoskeleton with drugs showed that microtubules and microfilaments are involved in the types of mRNA movements we have observed, which included complete immobility and corralled and nonrestricted diffusion. Individual mRNA molecules switched frequently among these movements, suggesting that mRNAs undergo continuous cycles of anchoring, diffusion, and active transport. Interestingly, transcripts containing the b actin 3′ UTR were observed to undergo directed movement more often than SV40 and hGH transcripts, indicating that the mechanism for mRNA localization may involve a sequence-dependent increase in motor-like movements. While Cos7 cells do not themselves localize b actin mRNA, they do contain the b actin 3′ UTR binding protein, ZBP-1, which may be sufficient to enable the primary steps of localization and the observed increase in sequence-dependent directed movements.
To better determine the behavioral characteristics that underlie mRNA localization, it is necessary to study this process directly in primary cells, such as fibroblasts or neurons, where localization is known to occur. These studies are difficult because of the low transfection efficiency of primary cells and the thickness of their cytoplasm, which complicates particle tracking. Nevertheless, such studies are in progress (Fusco, unpubl. data) and it is hoped that they will yield significant insights into the mechanism of mRNA localization in somatic cells, along with the role of this process in wound healing, metastasis, and neuronal development.
6 Conclusion
The field of live cell mRNA analysis is in a period of reorientation, slowly shifting focus from the development of new visualization technology to the application of that technology to important biological and medical questions. Significant advances have been made already, including the detailed characterization of mRNA transport and localization particles in yeast (Bertrand et al. 1998) and in Drosophila oocytes (Theurkauf and Hazelrigg 1998; Cha et al. 2001), signal transduction mechanisms related to mRNA transport in neurons (Rook et al. 2000) and in fibroblasts (Latham et al. 2001), and sequence dependent movement differences in mammalian cell lines (Fusco et al. 2003). Despite these advances, major questions remain. For example, where are the multi-molecule mRNA “granules” formed, in the nucleus or the cytoplasm? In the cytoplasm, what is the mechanism of mRNA localization, and does it include a sequence-specific process of directed movement, of packaging, of anchoring, or some combination of these processes? Is the mechanism the same for all localized transcripts and what are the signal transduction pathways involved in stimulating localization? Finally, what are the medical consequences of disrupted mRNA trafficking and the medical applications of altered mRNA trafficking? As the technology described above becomes better practised and applied, we will doubtless find answers to many of these questions and advance our understanding of molecular and cellular biology in the process. Furthermore, the future will yield new developments in imaging cells in live animals (Farina et al. 1998) and will eventually unveil the details of the RNA lifecycle.
Acknowledgments
The authors thank Shailesh Shenoy for his help with the figures. Supported by NIH GM 54887.
References
- Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262(5133):563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Cernan S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond AI. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 1991;10:1863–1873. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JH, Cui H, Barbarese E. The balance of power in RNA trafficking. Curr Opin Neurobiol. 2001;11:558–563. doi: 10.1016/s0959-4388(00)00249-x. [DOI] [PubMed] [Google Scholar]
- Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106:35–46. doi: 10.1016/s0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- Cheney RE, O’Shea MK, Heuser JE, Coelho MV, Wolenski JS, Espreafico EM, Forscher P, Larson RE, Mooseker MS. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- Davis I, Ish-Horowicz D. Apical localization of pairrule transcripts requires 3′ sequences and limits protein diffusion in the Drosophilia blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RW, Molenaar C, Tanke HJ. Methods for visualizing RNA processing and transport pathways in living cells. Histochem Cell Biol. 2001;115:3–11. doi: 10.1007/s004180000214. [DOI] [PubMed] [Google Scholar]
- Farina KL, Wyckoff JB, Rivera J, Lee H, Segall JE, Condeelis JS, Jones JG. Cell motility of tumor cells visualized in living intact primary tumors using green fluorescent protein. Cancer Res. 1998;58:2528–2532. [PubMed] [Google Scholar]
- Farina KL, Huettelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate b-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D. Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard J, Singer RH, Bertrand E. Sequence dependent movement of single mRNA molecules in the cytoplasm of living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Zearfoss NR, Etkin LD. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck RJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RB, Kosik KS. Neurotrophin-3 signals redistribute RNA in neurons. Proc Natl Acad Sci USA. 1997;94:14804–14808. doi: 10.1073/pnas.94.26.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago H, Fonseca SA, Murray JB, Stonehouse NJ, Stockley PG. Dissecting the key recognition features of the MS2 bacteriophage translational repression complex. Nuc Acid Res. 1998;26(5):1337–1344. doi: 10.1093/nar/26.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham VM, Yu EH, Tullio AN, Adelstein RS, Singer RH. Rho-dependent signaling pathway operating through myosin localizes beta-actin mRNA in fibroblasts. Curr Biol. 2001;11:1010–1016. doi: 10.1016/s0960-9822(01)00291-3. [DOI] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo R, Singer RH. Single-cell gene expression profiling. Science. 2002;297(5582):836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- Lorenz P, Misteli T, Baker BF, Bennett CF, Spector DL. Nucleocytoplasmic shuttling: a novel in vivo property of antisense phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 2000;28:582–592. doi: 10.1093/nar/28.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K, Castle PE, Taylor DL, Lanni F. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc Natl Acad Sci USA. 1987;84:4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar C, Marras SA, Slats JCM, Truffert J-C, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:E89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Fluorescent RNA cytochemistry: tracking gene transcripts in living cells. Nucleic Acids Res. 2001;29:1013–1016. doi: 10.1093/nar/29.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlette J, Tan W. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Politz JC, Taneja KL, Singer RH. Characterization of hybridization between synthetic oligodeoxynucleotides and RNA in living cells. Nucleic Acids Res. 1995;23:4946–4953. doi: 10.1093/nar/23.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Browne ES, Wolf DE, Pederson T. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc Natl Acad Sci USA. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly (A) RNA throughout the interchromatin space in living cells. Curr Biol. 1999;9:285–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- Privat E, Melvin T, Asseline U, Vigny P. Oligonucleotide-conjugated thiazole orange probes as “light-up” probes for messenger ribonucleic acid molecules in living cells. Photochem Photobiol. 2001;74:532–541. doi: 10.1562/0031-8655(2001)074<0532:octopa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Marray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a betaactin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F, Bohmann K, Nusslein-Volhard C. The molecular motor dynein is involved in targeting swallow and biocoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol. 2000;2:185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- Shestakova EA, Wyckoff J, Jones J, Singer RH, Condeelis J. Correlation of beta-actin messenger RNA localization with metastatic potential in rat adenocarcinoma cell lines. Cancer Res. 1999;59:1202–1205. [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA RNA hybridization in living cells. Proc Natl Acad Sci USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Hazelrigg TI. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multi-step anterior localization pathway. Development. 1998;125:3655–3666. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- Thuong NT, Asseline U, Roig V, Takasugi M, Helene C. Oligo (alpha-deoxynucleotide) s covalently linked to intercalating agents: differential binding to riboand deoxyribopolynucleotides and stability towards nuclease digestion. Proc Natl Acad Sci USA. 1987;84:5129–5133. doi: 10.1073/pnas.84.15.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A, Sato Y, Hirano M, Suga T, Koshimoto H, Taguchi T, Ohsuka S. Development of a time-resolved fluorometric method for observing hybridization in living cells using fluorescence resonance energy transfer. Biophys J. 2001;81:501–515. doi: 10.1016/S0006-3495(01)75717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Lijas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- Wang J, Cao LG, Wang YL, Pederson T. Localization of pre-messenger RNA at discrete nuclear sites. Proc Natl Acad Sci USA. 1991;88:7391–7395. doi: 10.1073/pnas.88.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]