Abstract
Follicular thyroid carcinoma (FTC) and benign follicular adenoma (FA) are indistinguishable by preoperative diagnosis due to their similar histological features. Here we report the first RNA sequencing study of these tumors, with data for 30 minimally invasive FTCs (miFTCs) and 25 FAs. We also compared 77 classical papillary thyroid carcinomas (cPTCs) and 48 follicular variant of PTCs (FVPTCs) to observe the differences in their molecular properties. Mutations in H/K/NRAS, DICER1, EIF1AX, IDH1, PTEN, SOS1, and SPOP were identified in miFTC or FA. We identified a low frequency of fusion genes in miFTC (only one, PAX8–PPARG), but a high frequency of that in PTC (17.60%). The frequencies of BRAFV600E and H/K/NRAS mutations were substantially different in miFTC and cPTC, and those of FVPTC were intermediate between miFTC and cPTC. Gene expression analysis demonstrated three molecular subtypes regardless of their histological features, including Non–BRAF–Non–RAS (NBNR), as well as BRAF–like and RAS–like. The novel molecular subtype, NBNR, was associated with DICER1, EIF1AX, IDH1, PTEN, SOS1, SPOP, and PAX8–PPARG. The transcriptome of miFTC or encapsulated FVPTC was indistinguishable from that of FA, providing a molecular explanation for the similarly indolent behavior of these tumors. We identified upregulation of genes that are related to mitochondrial biogenesis including ESRRA and PPARGC1A in oncocytic follicular thyroid neoplasm. Arm-level copy number variations were correlated to histological and molecular characteristics. These results expanded the current molecular understanding of thyroid cancer and may lead to new diagnostic and therapeutic approaches to the disease.
Author Summary
Recently, The Cancer Genome Atlas proposed an improved classification of the subtypes of papillary thyroid carcinoma (PTC) based on gene expression profiles, which better represents cell signaling and differentiation. However, a molecular characterization of follicular thyroid carcinoma (FTC), which has a greater tendency for hematogenous spread to lung and bone is not yet fully elucidated. In this study, we describe the first RNA sequencing data of minimally invasive FTC (miFTC) and benign follicular adenoma (FA), which cause diagnostic difficulties due to their similar histological features. Additionally, classical PTC and follicular variant of PTC (FVPTC) were sequenced to compare their transcriptional and mutational landscape. BRAF, H/K/NRAS, fusion genes, and copy number variations were associated with tumor histology. Based on gene expression profiles, thyroid tumors were classified as three molecular subtypes regardless of histological subtypes, BRAF–like, RAS–like, and Non–BRAF–Non–RAS. In particular, we found identical gene expression profiles between miFTC, FA, and encapsulated FVPTC. Oncocytic follicular thyroid tumors have gene expression signatures related to mitochondrial biogenesis including ESRRA and PPARGC1A. These results expanded the current molecular understanding of thyroid cancer to its follicular types.
Introduction
Most thyroid cancers are classified as either classical papillary thyroid carcinoma (cPTC), follicular variant of PTC (FVPTC), or follicular thyroid carcinoma (FTC) based on histological architecture [1]. However, the distinction between follicular-patterned thyroid tumors, such as FVPTC, FTC, and benign follicular adenoma (FA), still remains as a challenging problem [2]. Moreover, FTC and FA are indistinguishable by preoperative diagnosis as in practice they are often jointly referred to as follicular thyroid neoplasm (FTN) [3].
FTC accounts for approximately 10% of all thyroid cancers [4] and is known to harbor H/K/NRAS mutations, which are one of the molecular markers used for diagnosis [5]. However, H/K/NRAS mutations are also found in FVPTC and FA [6,7]. Therefore, these mutations are not sufficient as predictors of pure follicular histology or malignant potential in thyroid cancer.
The recent publication of The Cancer Genome Atlas (TCGA) studied molecular characteristics of PTC including the subtypes of classical type, tall cell variant, and follicular variant [8]. It was the first comprehensive pan-genomic study of thyroid cancer. They concluded that classification with two molecular subtypes, BRAFV600E–like and RAS–like, represents the underlying signaling and differentiation properties better than pathological classifications. However, the analysis of TCGA was confined to subtypes of PTC and molecular characterization of FTC has not been performed.
In particular, the TCGA study demonstrated that the mitogen-activated protein kinase (MAPK) signaling pathway in PTC, as well as differentiation of thyroid cells, was differently regulated depending on molecular subtypes. There are some other reports about differential activation of the MAPK signaling pathway through several different genetic events such as RET/PTC fusions, BRAF, and H/K/NRAS point mutations [9,10]. The initiation of those genetic alterations likely depends on some triggering factor such as radiation or chemical elements [11–15]. However, the association between clinical risk factors and genetic alterations has not been fully understood yet.
We have performed a comprehensive RNA sequencing (RNA-seq) analysis to reveal the molecular characteristics of thyroid cancer including minimally invasive FTC (miFTC) and FA, and investigated their association to clinical data. Since there is no preceding large-scale RNA-seq study on miFTC and FA, we expect that our result will facilitate the discovery of new diagnostic and therapeutic approaches to thyroid cancer.
Results
Driver mutations of thyroid tumors
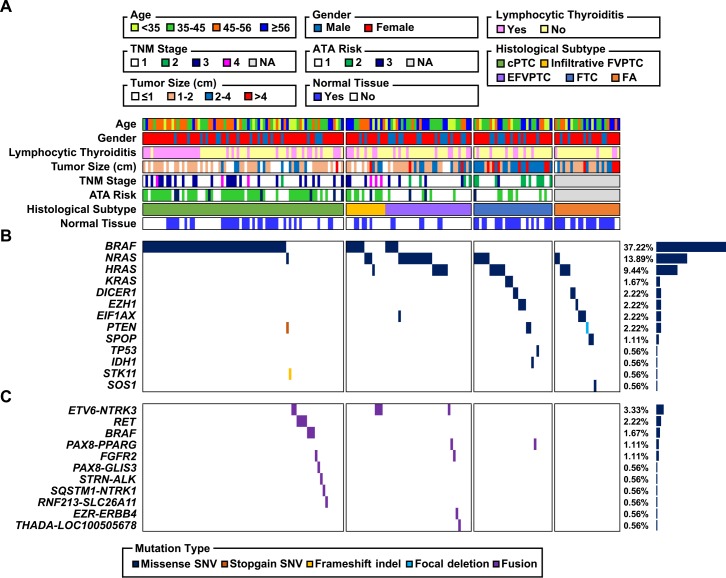
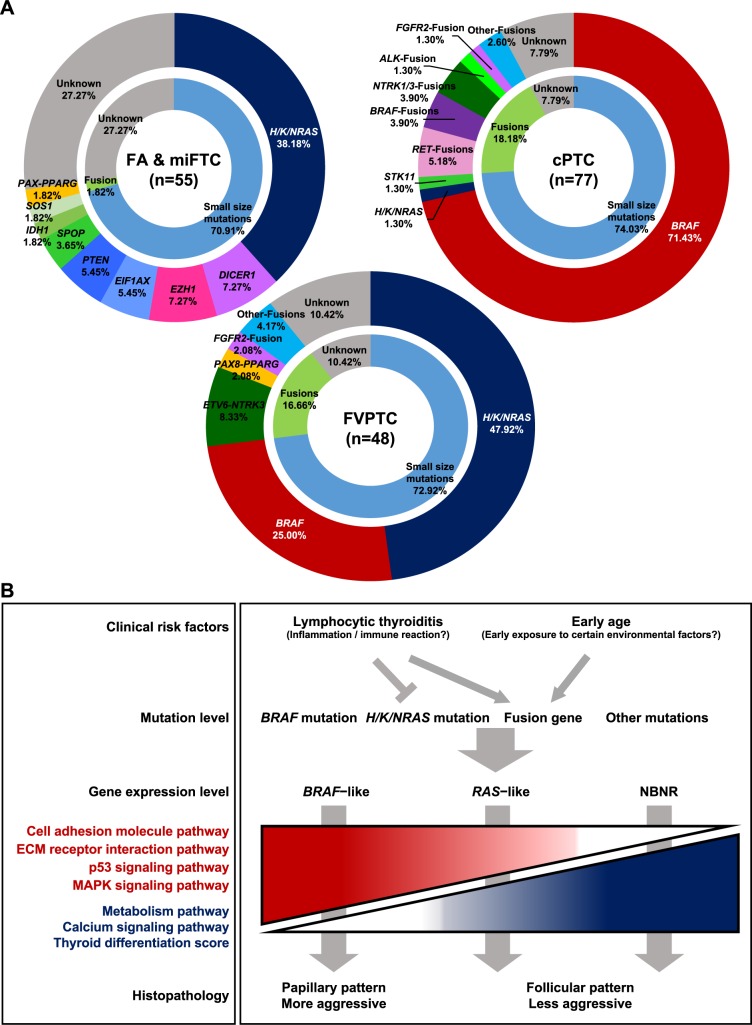
The mutational landscape of 180 thyroid tumors including 25 FAs, 30 miFTCs, 48 FVPTCs, and 77 cPTCs is illustrated in Fig 1. Mutations in well-known cancer driver genes (BRAF and H/K/NRAS) and fusion gene rearrangements were identified in 37.22%, 25.00%, and 12.78% of total tumors, respectively. The patterns of genetic alteration differed between PTC and FTN; most fusion genes were observed in PTC (17.60% and 1.82% in PTC and FTN, respectively; p = 0.002), while most mutations except BRAFV600E and H/K/NRAS were found in FTN (32.73% and 0.80% in FTN and PTC, respectively; p < 0.0001). BRAFV600E was only identified in PTC and its frequency differed between cPTC and FVPTC (71.43% and 25.00%, respectively; p < 0.0001). Many H/K/NRAS mutations were identified in FVPTC, miFTC, and FA (47.92%, 50.00%, and 24.00%, respectively). Only 1.30% of cPTC harbored NRAS mutations.
Fig 1. The mutational landscape of thyroid tumors.
Each column represents an individual sample. (A) Age, gender, the presence of lymphocytic thyroiditis, tumor size, TNM Classification of Malignant Tumors stage, American Thyroid Association risk stratification, availability of matched normal tissue, and histological subtype. (B) Frequency of small size mutation by gene (right) and distribution of mutation across the 180 tumors (middle). (C) Frequency of fusion mutation by gene (right) and distribution of mutation across the 180 tumors (middle).
Four tumors (6.67% of miFTC and 8.00% of FA) harbored somatic DICER1 mutations (E1705Q, D1810H, E1813G, and E1813Q; S1A Fig). These mutations were mutually exclusive with BRAFV600E and H/K/NRAS mutations in FA as well as miFTC. The expression level of DICER1 was increased with these somatic mutations (S1B Fig). Among these, two mutations were previously reported in TCGA study (D1810H and E1813G in TCGA-EL-A3GO and TCGA-EL-A3D5, respectively). They also tended to be mutually exclusive with BRAF and H/K/NRAS mutations (S1C Fig). Several mutations in the Ribonuclease III domain of DICER1 were previously reported in PTC and other types of cancer [16–19], but DICER1E1705Q mutation was first to be identified in thyroid tumor. We found three missense mutations in EIF1AX (G9V, R13C, and R13L) which was recently proposed as a driver gene in PTC (S1C Fig) [8]. These mutations were occurred more often in FA than in PTC (12.00% and 0.80%, respectively; p = 0.015) and they were mutually exclusive with BRAFV600E and H/K/NRAS mutations. In addition, there were IDH1R132C and two PTEN missense mutations (V343E and V175A) in miFTC. Also, one FA sample appeared to have a somatic focal deletion of PTEN based on its lacked expression in tumor. Furthermore, we suggest some novel driver candidates: SOS1N233Y, SPOPP94R, EZH1Q571R, EZH1Y642F, and STK11R86fs. SOS1N233Y was identified as a recurrent hotspot in several cancers including uterine endometrial carcinoma, lung adenocarcinoma, and cancer cell lines [20]. SPOPP94R was localized to the MATH domain and most somatic mutations in SPOP occurred in this domain [21,22]. EZH1 is a member of the Polycomb group protein complex which are important components for prevention of cancer stem cell development [23]. In the TCGA dataset, SPOPP94R and EZH1Y642F tended to be mutually exclusive with BRAF and H/K/NRAS mutations (S1C Fig). Mutations in STK11 were also reported in other types of cancer including poorly differentiated and anaplastic thyroid carcinoma [24,25]. Those mutations, SPOPP94R, EZH1Q571R, DICER1E1813G, DICER1E1813Q, and EIF1AXR13C, were confirmed as somatic mutations by polymerase chain reaction (PCR) and Sanger sequencing in tumor and matched normal tissues (S2A Fig).
We described all predicted fusion genes, breakpoint regions, and expression levels in S1 Table. All fusion genes including novel candidates were mutually exclusive with other mutations. Previously reported fusion genes in thyroid cancer, ETV6–NTRK3 (4.80% in PTC), CCDC6–RET (2.40% in PTC), NCOA4–RET (0.80% in PTC), SQSTM1–NTRK1 (0.80% in PTC), STRN–ALK (0.80% in PTC), and PAX8–PPARG (0.80% and 1.82% in PTC and FTN, respectively), were also identified [8]. ALK, RET, and NTRK1 represented aberrant overexpression after fusion gene breakpoint (S3A Fig). Moreover, ETV6–NTRK3 and STRN–ALK were validated by fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) staining, respectively (S3B and S3C Fig). In case of BRAF, we identified two novel candidate fusion genes, PICALM–BRAF (0.80% in PTC) and NFYA–BRAF (0.80% in PTC), in addition to formerly reported SND1–BRAF (0.80% in PTC) [8]. PICALM–BRAF was validated by reverse transcriptase PCR (RT-PCR) and Sanger sequencing (S2B Fig). Additionally, we suggest other fusion gene candidates such as EZR–ERBB4, FGFR2–KIAA1598, FGFR2–WARS, PAX8–GLIS3, THADA–LOC100505678, and RNF213–SLC26A11. From the above fusion gene candidates, EZR–ERBB4, FGFR2–KIAA1598, and RNF213–SLC26A11 were identified in other types of cancer [26–28]. ERBB4 also had aberrant overexpression after fusion gene breakpoint (S3A Fig). THADA rearrangement was previously reported in FA and PTC [8,29]. PAX8 and GLIS3 are both related to thyroid metabolism and function [8].
Association between clinical risk factors and genetic alterations
Each subject showed different combinations of clinical risk factors such as age, smoking, alcohol drinking, menopausal status, and the presence of lymphocytic thyroiditis (LT). To investigate the association of these risk factors with genetic alterations, we categorized the patients into three groups: 1) small size mutation, 2) fusion gene, and 3) driver-unknown (Table 1). The average age of the fusion gene group (39.2 ± 13.1) was younger than driver-unknown (52.3 ± 14.9) and small size mutation (47.4 ± 12.1) groups (p = 0.002). Moreover, tumors with fusion gene were found more frequently in young adults (20.00% of subjects age < 45 yrs. and 6.26% of subjects age ≥ 45 yrs.; p = 0.006). The percentage of pre-menopausal women in the fusion gene group (75.00%) was higher than driver-unknown (23.53%) and small size mutation (55.00%) groups (p = 0.01). Patients harboring fusion gene were less likely to smoke than others, but it was not statistically significant. Also, the percentage of patients who drink alcohol was not different among the groups. Patients harboring H/K/RAS mutations had a lower frequency of LT (11.11%), which was defined by histologic findings in normal thyroid parenchyma, than BRAFV600E (37.31%; p = 0.002) and fusion gene (47.83%; p = 0.0004) groups. The fusion gene group showed higher frequency of LT compared with driver-unknown (26.92%) and other mutation (21.05%) groups, although this was not statistically significant.
Table 1. Comparison of clinical risk factors among the groups with different types of mutation.
| Variable | Driver-unknown | Small size mutation | Fusion gene | p-value a | p-value b | |||
|---|---|---|---|---|---|---|---|---|
| Total | BRAF | H/K/NRAS | Others | |||||
| N | 26 | 131 | 67 | 45 | 19 | 23 | ||
| Age | 52.3 ± 14.9c | 47.3 ± 12.1c | 47.2 ± 12.0 | 48.1 ± 12.5 | 45.8 ± 12.1 | 39.2 ± 13.1 | 0.002 | 0.010 |
| Sex (female) | 21 (80.77) | 93 (70.99) | 50 (74.62) | 28 (62.22) | 15 (78.95) | 17 (73.91) | 0.587 | 0.425 |
| Pre-menopause (regular) | 4/17 (23.53)c | 44/80 (55.00) | 20/38 (52.63) | 14/27 (51.85) | 10/15 (66.67) | 12/16 (75.00) | 0.010 | 0.038 |
| Smoking (current + ex) | 3 (11.54) | 20/130 (15.38) | 9 (13.43) | 7/44 (15.91) | 4 (21.05) | 1 (4.35) | 0.431 | 0.578 |
| Male | 1/5 (20.00) | 16/38 (42.11) | 8/17 (47.05) | 6/17 (35.29) | 2/4 (50.00) | 1/6 (16.66) | 0.446 | 0.659 |
| Female | 2/21 (9.52) | 4/92 (4.35) | 1/50 (2.00) | 1/27 (3.70) | 2/15 (13.33) | 0/17 (0.00) | 0.400 | 0.172 |
| Drinking (current) | 7 (26.92) | 40/130 (30.77) | 16 (23.88) | 16/44 (36.36) | 8 (42.11) | 7 (30.43) | 0.926 | 0.482 |
| Lymphocytic thyroiditis | 7 (26.92) | 34 (25.95) | 25 (37.31) | 5 (11.11)c,d | 4 (21.05) | 11 (47.83) | 0.100 | 0.008 |
a p-value for comparison among Driver-unknown, Total, and Fusion gene mutation groups.
b p-value for comparison among Driver-unknown, BRAF, H/K/NRAS, Others, and Fusion gene mutation groups.
c Significantly different from “Fusion gene mutation” group. (p < 0.05 for post-hoc Bonferroni test)
d Significantly different from “BRAF” group. (p = 0.002 for post-hoc Bonferroni test)
Gene expression analysis on thyroid tumors
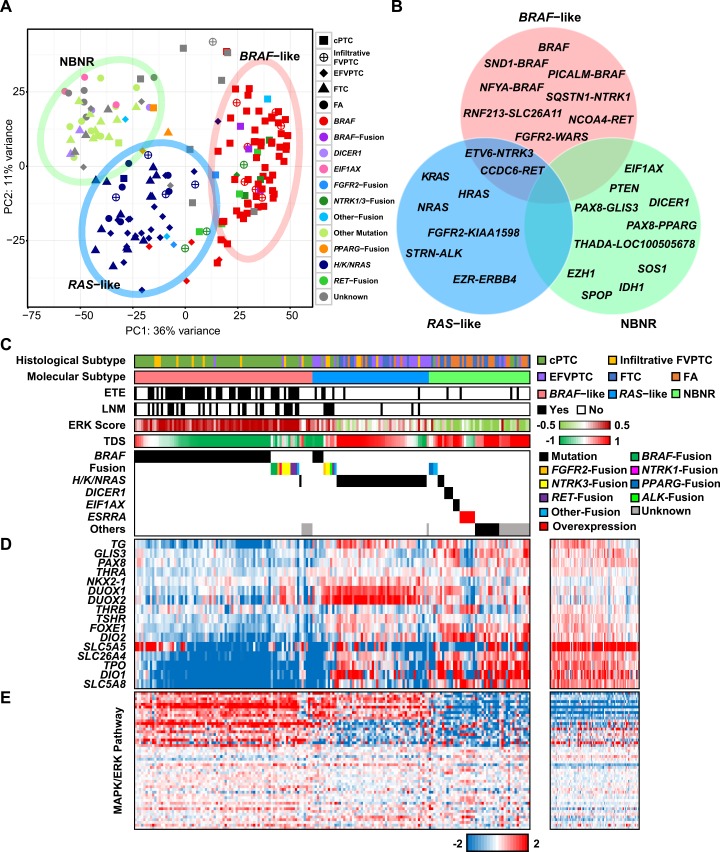
The result of K-means clustering via principal component analysis (PCA) on all study subjects is shown in S4A Fig. Tumor and normal tissues were distinctively separated in the PC2 axis even though some of them were grouped together in one of the K-means cluster. This cluster was associated with LT which was observed in 28.89% of study subjects (S4B Fig). Samples with BRAFV600E mutation and LT were also separated from samples with BRAFV600E mutation and without LT when the same analysis was conducted with only tumors (S4C and S4D Fig). In case of TCGA dataset, we were not able to distinguish an LT derived cluster although 22.89% of specimens harbored LT (S4E Fig). The inconsistent result between TCGA and the current study could be raised from different gene set usage for each analysis; we used the Ensembl gene set instead of the UCSC gene set which was used in TCGA study. Within the most variable 500 genes in the Ensembl gene set applied to PCA, 91 genes were associated with immunoglobulin and only four genes were overlapped with the UCSC gene set. In order to decrease the gene expression variation affected by LT and increase that derived from oncogenic signal transduction, we used genes covered by the UCSC gene set for molecular classification. With this approach, we obtained three molecular subtypes in relation to oncogenic signal transduction: BRAF–like, RAS–like, which were proposed by TCGA, and a third which we refer to as Non–BRAF–Non–RAS (NBNR). The three molecular subtypes that we identified showed a clear separation of samples by driver genes (Fig 2A). We could get exceedingly similar result when the same analysis was performed on TCGA dataset (S4F Fig). As the effect of gene expression derived by BRAF and H/K/NRAS was overwhelming in PCA due to their huge sample size, this analysis was performed on a partial TCGA dataset.
Fig 2. Gene expression analysis on thyroid tumors.
(A) The result of K-means clustering via PCA. Three distinct molecular subtypes were found: BRAF–like, RAS–like, and NBNR. Each cluster was represented by a 95.00% confidence ellipse. (B) Driver gene of three molecular subtypes. (C) Histological subtype, molecular subtype, the presence of extrathyroidal extension, the presence of lymph node metastasis, ERK score, TDS, and driver gene in individual samples. 180 tumors were sorted by molecular subtype, driver gene, and high to low TDS. (D) The heat maps represent expression level of 16 thyroid metabolism and function genes and (E) the MAPK signaling pathway genes in tumor (middle) and normal (right) specimens. Genes were clustered by K-means clustering algorithm (K = 3).
BRAF–like consisted of BRAFV600E and fusion genes (PICALM–BRAF, NFYA–BRAF, SND1–BRAF, FGFR2–WARS, ETV6–NTRK3, SQSTM1–NTRK1, CCDC6–RET, NCOA4–RET, and RNF213–SLC26A11). None of FTN was clustered into BRAF–like because of their skewed proportion of BRAFV600E and fusion genes. RAS–like consisted of H/K/NRAS and fusion genes (STRN–ALK, EZR–ERRB4, FGFR2–KIAA1598, ETV6–NTRK3, and CCDC6–RET). Lastly, NBNR was associated with DICER1, EIF1AX, IDH1, PTEN, PAX8–PPARG, and other driver gene candidates (Fig 2B).
The aggressive pathologic characteristics, lymph node metastasis (LNM) and extrathyroidal extension (ETE) were correlated with the 3 molecular subtypes (Fig 2C); higher frequency of LNM (37.04%) or ETE (61.73%) was found in the BRAF–like group, while less or no LNM or ETE was observed in the RAS–like group (15.09% of LNM, 11.32% of ETE) or NBNR group (0.00% of LNM, 8.70% of ETE) (For both categories; p < 0.0001).
To measure differentiation of thyroid cells and activation of the MAPK signaling pathway in three molecular subtypes, we implemented two scoring methods that were introduced by TCGA study: thyroid differentiation score (TDS) and ERK score (Fig 2C) [8]. Most BRAF–like tumors had low TDS, while RAS–like and NBNR tumors had high TDS. There was a strong negative correlation between TDS and molecular subtype classification (Pearson correlation coefficient = -0.66). The low level of TDS was derived from decreased expression level of 16 thyroid metabolism and function genes [8]. Many of these 16 genes were downregulated in BRAF–like, while RAS–like and NBNR maintained stable gene expression levels. In BRAF–like, significantly downregulated genes were DIO1, DIO2, TPO, SLC26A4, and SLC5A8. DUOX1 and DUOX2 were increased in RAS–like. On the other hand, NBNR had no differentially regulated gene except ESRRA overexpressed tumors (See “The characteristic gene expression of oncocytic FTN” section), which represented downregulation of several genes: DIO1, FOXE1, GLIS3, PAX8, and SLC5A5 (Fig 2D).
The involvement of constitutive activation of the MAPK signaling pathway in the pathogenesis of PTC is well established [9]. ERK score strongly represented activation level of MAPK signaling pathway and there was very strong positive correlation between ERK score and molecular subtype classification (Pearson correlation coefficient = 0.80). As discussed in TCGA study, ERK score was highly elevated in most BRAF–like, but not in RAS–like samples. Although RAS–like represented lower ERK score than BRAF–like, it had some activated genes in the MAPK signaling pathway. However, NBNR did not have activated genes as represented by the ERK score (Fig 2E).
Gene expression analysis on follicular-patterned thyroid tumors
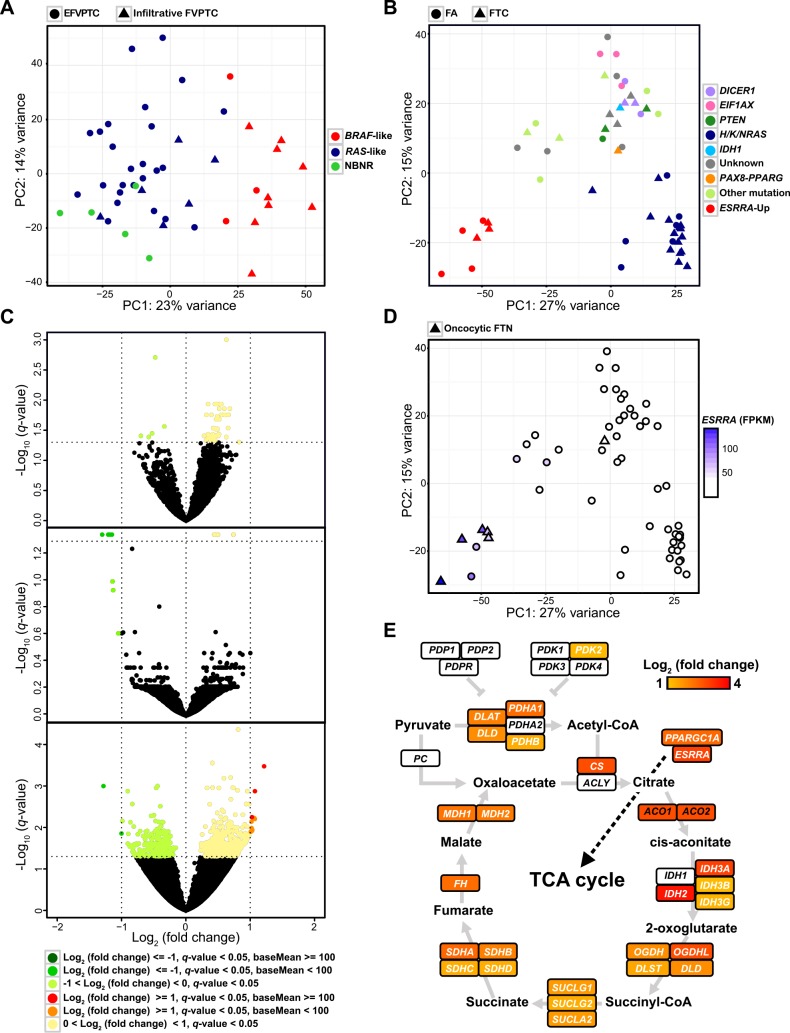
The mutational profile of miFTC and FA were very similar to each other. Moreover, that of EFVPTC was also similar to FTNs, while that of infiltrative FVPTC was similar to cPTC (Fig 1). All these tumors are follicular-patterned, which are occasionally hard to distinguish from one another. To identify the transcriptional difference among these follicular-patterned thyroid tumors, we performed PCA and differentially expressed gene (DEG) analysis.
In PCA performed on EFVPTC and infiltrative FVPTC, PC1 axis clearly divided tumors which were classified as BRAF–like and RAS–like/NBNR (Fig 3A). EFVPTC was mainly associated with RAS–like/NBNR rather than infiltrative FVPTC (p = 0.0004). When we performed PCA on miFTC and FA which are hard to distinguish by pathological examination, we could not find any cluster nor PC axis that separates miFTC and FA. Several clusters and PC axes divided those tumors, but all groups consisted of miFTC and FA. (Fig 3B). The lower right corner and the upper central group were associated with H/K/NRAS and other driver genes (e.g., DICER1, EIF1AX, IDH1, PTEN, and PAX8–PPARG), respectively. DEG analysis also confirmed that miFTC and FA did not have significant transcriptional difference. Moreover, the transcriptome of EFVPTC which shows indolent behavior was also indistinguishable from miFTC and FA (Fig 3C).
Fig 3. Gene expression analysis on follicular-patterned thyroid tumors.
(A) The result of PCA on FVPTC. (B) The result of PCA on miFTC and FA. (C) The volcano plots represent identical gene expression among three subtypes: miFTC and FA (upper), FA and EFVPTC (middle), and miFTC and EFVPTC (lower). All analysis was restricted to RAS–like tumors. (D) ESRRA expression level of miFTC and FA. (E) The characteristic gene expression of oncocytic FTN. ESRRA, PPARGC1A, genes in TCA cycle were represented by Log2 (fold change). The illustration was generated based on a figure from Davis et al and KEGG pathway [34,54].
The characteristic gene expression of oncocytic FTN
We identified the increased ESRRA expression level of tumors in the lower left corner group in PCA performed on miFTC and FA (Fig 3B and 3D). Pathway enrichment analysis [30] on chemical and genetic perturbations database showed that DEGs of the aforementioned cluster harbored genes that were upregulated by ESRRA and were related to mitochondria (S2 Table). Remarkably, most of those tumors were oncocytic FTN (p < 0.0001); 83.33% of oncocytic FTN (five out of six) was clustered into ESRRA overexpression group (Fig 3D).
Oncocytic FTN is characterized by remarkable accumulation of mitochondria [31]. In those tumors, expression level of ESRRA showed very strong positive correlation with expression level of PPARGC1A (Pearson correlation coefficient = 0.83 using FPKM). Both ESRRA and PPARGC1A are key regulators of mitochondrial biogenesis [32,33]. DEG analysis demonstrated that the majority of genes in citric acid cycle (TCA cycle) were dramatically upregulated in oncocytic FTN (Fig 3D). All of the oncocytic FTNs were classified as NBNR (Fig 2C).
Pathway analysis on molecular subtypes
To investigate detailed gene expression signatures in the three molecular subtypes, we performed pathway enrichment analysis on DEGs of each molecular subtype using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database [34]. The top 20 most significantly enriched KEGG pathways of upregulated and downregulated genes of each molecular subtype are illustrated in S5 Fig and S6 Fig, respectively. In BRAF–like, pathways including cell adhesion molecules (CAMs), the extracellular matrix (ECM) receptor interaction, and focal adhesion were remarkably upregulated. The involvement of these pathways in the carcinogenesis of thyroid nodules and cancer invasiveness had been reported previously [35–37]. Moreover, the risk assessments based on TNM Classification of Malignant Tumors (TNM) stage and American Thyroid Association (ATA) risk stratification supported that BRAF–like is more aggressive than other molecular subtypes (p = 0.030 and p = 0.001, respectively). The p53 and MAPK signaling pathways were upregulated in both BRAF–like and RAS–like but not in most of NBNR. Numerous metabolism and calcium signaling pathways were downregulated in BRAF–like, and these pathways were barely downregulated in other molecular subtypes.
Novel molecular subtype NBNR is composed of diverse kinds of driver genes and they had different gene expression profiles depending on the types of mutated or overexpressed gene. As we mentioned earlier, upregulated genes of ESRRA overexpressed tumors were significantly enriched for pathways related to TCA cycle, oxidative phosphorylation (OXPHOS), PPAR signaling, and several metabolisms. Moreover, tumors with PAX8–PPARG rearrangement also showed increased metabolism and PPAR signaling pathways. The Wnt and mTOR signaling pathways were enriched in DICER1 and EIF1AX mutated tumors, respectively.
Copy number variation in thyroid tumors
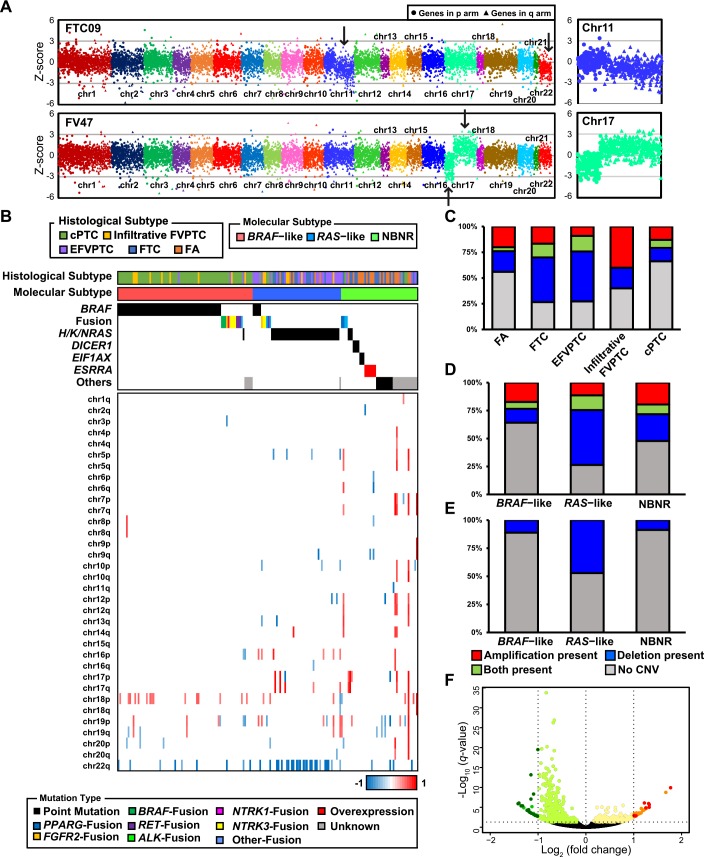
To identify arm-level copy number variations (CNVs) of thyroid tumors, we defined arm-level jointly regulated blocks (JRBs) which demonstrate colocalization of overexpressed and underexpressed chromosome arms. Our previous study demonstrated high correlation between JRB and CNV status in cancer genomes [38]. In this study, we modified our former method to define arm-level JRBs. We successfully predicted aberration of chromosome arms which represent arm-level amplification and deletion (Fig 4A). We illustrated the CNV landscape of all thyroid tumors in Fig 4B. cPTC had the lowest percentage of arm-level CNV, while miFTC, EFVPTC, and infiltrative FVPTC had high percentage of that. FA showed a lower percentage of arm-level CNV than miFTC (Fig 4C). It was reported that chromosome 12 is more frequently amplified in FA and indolent tumors than aggressive tumors [39]. We also identified amplification of chromosome 12 in FTN but not in PTCs (p = 0.008). The percentage of arm-level deletion was higher in RAS–like than BRAF–like and NBNR (Fig 4D). Similar to previous reports [8,39], deletion of chromosome 22q was the most frequently identified arm-level CNV in RAS–like (Fig 4E; p < 0.0001). DEG test on RAS–like tumors with or without chromosome 22q deletion confirmed the reliability of detecting CNV by our approach. The moderately downregulated genes (-1 < Log2 (fold change) < 0, q-value < 0.05) were enriched to chromosome 22q when positional gene set enrichment was performed (Fig 4F and S3 Table). However, there was no difference in other clinicopathological features whether chromosome 22q deletion occurred in RAS–like or not; only multifocality was more significantly frequent (S4 Table; p = 0.037). In BRAF–like, amplification of chromosome 18p was more frequent than other molecular subtypes, but it was not statistically significant (p = 0.051). The ratio of LT was elevated when chromosome 18p was amplified (S5 Table; p = 0.032,).
Fig 4. The CNV landscape of thyroid tumors.
(A) The examples of arm-level duplication and deletion were pointed by arrow. Enlarged images of chromosome 11q deletion (upper right) and chromosome 17 with deletion and duplication in p and q arm (lower right) were provided. (B) The distribution of CNVs across 180 tumors. The chromosome arms without CNV across all tumors were excluded. Specimens were sorted in the same manner as Fig 2. (C) The percentage of CNV in each histological subtype. (D) The percentage of CNV in each molecular subtype. (E) The percentage of chromosome 22q deletion in each molecular subtype. (F) The volcano plot shows massive number of moderately downregulated genes in RAS–like tumors with chromosome 22q deletion. Positional gene set enrichment result of these genes is provided in S4 Table.
Discussion
Recently, the genomic landscape of PTC has been well investigated [8]. This study reduced the rate of unknown oncogenic drivers in subtypes of PTC from 25% to 3.5% through discoveries of somatic alterations including EIFIAX, PPM1D, CHEK2, and diverse fusion genes. However, the transcriptional and mutational landscape of miFTC, which has a greater tendency for hematogenous spread to lung and bone, is yet to be widely explored. In the present study, we performed RNA-seq on miFTC and FA, in addition to cPTC and FVPTC.
We identified driver genes in 72.73%, 89.58%, and 92.21% of FTN, FVPTC, and cPTC samples, respectively (Fig 5A). The patterns of genetic alteration differed between histological subtypes. cPTC and miFTC showed considerably different patterns of genetic alteration to each other. However, FVPTC represented an intermediate mutational status between cPTC and miFTC; EFVPTC and infiltrative FVPTC were similar to miFTC and cPTC, respectively. Furthermore, miFTC and FVPTC have higher percentages of arm-level CNVs than cPTC (Fig 4C). This is consistent with previous studies that described a higher fraction of somatic copy number alterations in FVPTC and FTN than cPTC [8,40]. Taken together, our result suggests that different genetic alterations could lead to different tumor histology. In addition, we found that FA has a lower percentage of arm-level CNVs than miFTC. This result supports the hypothesis that FA is a preneoplastic condition of miFTC despite the similar patterns of genetic alteration between them.
Fig 5. The overview of the present study.
(A) The driver mutations in thyroid tumors. Each pie chart represents the distribution of driver mutations identified in FTN, FVPTC, and cPTC, respectively. (B) A schematic model of thyroid cancer progression integrating clinical risk factors, mutational, transcriptional profile, and clinical presentation. Tumor aggressiveness was determined by the presence of invasive pathologic characteristics of ETE or LNM.
It has been suggested that several clinical risk factors including smoking [41], alcohol drinking [42], LT [43,44], menopausal status [45], genetic predisposition [46], and early exposure to radiation [11,13,14] affect the development of thyroid cancer. However, there are few studies considering genetic alterations and clinical risk factors at the same time [12]. Therefore, we analyzed the possible association between types of genetic alteration and clinical risk factors to investigate the etiology of thyroid cancer (Table 1). The recent reports from Chernobyl cohort well demonstrated a relationship between fusion gene and thyroid cancer. [47,48]. Although there was no history of radiation exposure in our subjects, the younger age of the fusion gene group than other groups may reflects the involvement of environmental or genetic factors to the development of chromosomal rearrangement inducing thyroid cancer. Hashimoto’s thyroiditis is the main etiology of LT which is related to inflammation and immune reactions observed in thyroid. In this study, an elevated tendency of LT in the fusion gene group was shown and negative association between LT and H/K/NRAS mutation was also identified. These results suggest an etiologic role of LT in thyroid cancer development. Based on our findings, we suggest that some risk factors influence the types of genetic alteration. We believe that further study would allow better understanding of thyroid cancer development.
Based on transcriptional landscape, 180 tumors were classified as BRAF–like, RAS–like, and NBNR (Fig 2A). Our result in PTCs has similar context to the TCGA study, which classified subtypes of PTC as BRAFV600E–like and RAS–like. It was reported that FVFTCs in TCGA, which are classified as RAS–like, were often misdiagnosed as FTC by pathologists [49]. Moreover, as we mentioned earlier, arm-level copy number alterations were frequently observed in FVPTC similarly to FTC as well as H/K/NRAS mutations [40]. The aforementioned issues raised a question regarding the distinction between FTC and FVPTC in the point of biological and clinical relevance. In our analysis, EFVPTC and infiltrative FVPTC showed different mutational and transcriptional characteristics to each other (Fig 1 and 3A). EFVPTC, which was recently re-classified as “noninvasive follicular thyroid neoplasm with papillary-like nuclear features” according to its indolent features [50] had highly similar gene expression profiles to miFTC or FA, (Fig 3C). This result emphasizes that re-classification of thyroid cancers based on their mutational and transcriptional characteristics may be beneficial for stratified medicine.
One of the goals of this study was to discover molecular markers to distinguish miFTC and FA. Differential diagnosis for FTC and FA is important for decisions to undergo surgery in clinic, but it is almost impossible due to their highly similar cytological features at present [2]. Several researchers have suggested markers based on gene expression levels [51–53], but they are not widely adopted. In our analysis, we could not find any significant transcriptional difference between miFTC and FA (Fig 3B and 3C). These results again suggest that miFTC is indolent and it could be treated minimally. However, the transcriptional difference between widely invasive FTC (wiFTC) and miFTC/FA is yet to be investigated as there was no wiFTC in the current study.
Most tumors harboring EIF1AX mutations and PAX8–PPARG rearrangement were classified as RAS–like in TCGA study. However, they were distinguished from RAS–like and were classified as NBNR according to current and TCGA datasets (Fig 2A and S4F Fig). Traditionally, thyroid cancer is well known to be associated with activation of the MAPK signaling pathway [9,10]. Our results suggested that NBNR involves totally different mechanism and pathways (Fig 2E). Furthermore, NBNR exhibited distinct gene expression profiles within the class (S5 Fig). We believe that accumulating data would lead to more effective molecular classification and to discovery of therapeutic targets.
In BRAF–like, higher activation of ECM receptor interaction, CAMs, p53, and MAPK signaling pathways than other molecular subtypes was identified (S5 Fig). Furthermore, low level of TDS and downregulation of several metabolism pathways supported poor clinical presentation in BRAF–like (Fig 2C and S6 Fig). We could not establish the clinical impact of molecular subtypes on locoregional recurrence (n = 1), distant metastasis (n = 4), and cancer-specific mortality (n = 0) due to the short median follow-up of 37 months (1–100 months) and low percentage of advanced thyroid cancer. However, the other aggressive pathologic characteristics LNM and ETE were observed much more frequently in the BRAF–like group (Fig 2C), demonstrating its association with clinical presentation or aggressiveness.
Collectively, we propose a schematic model of thyroid cancer progression integrating clinical risk factors, mutational and transcriptional landscape, and clinical presentation (Fig 5B).
The underlying mechanism of mitochondria accumulation in oncocytic FTN has not been elucidated clearly. We deduced that oncocytic FTN had distinct transcriptome among thyroid tumors containing extremely upregulated mitochondria-related metabolic pathways (Fig 3D and 3E, and S5 Fig). This feature was in agreement with a recent study on eosinophilic chromophobe renal cell carcinoma which is also characterized by densely packed mitochondria [54]. The stimulation of mitochondrial biogenesis and OXPHOS by ESRRA and PPARGC1A is well established [32,33] and upregulation of two genes supported mitochondria accumulation in oncocytic FTN. The stimulation of high expression level of ESRRA and PPARGC1A is not fully studied here. However, we believe that our findings could provide important clues to understand the role of mitochondrial biogenesis in oncocytoma. Recently, there was a study that suggested mechanism of oncocytic thyroid tumor development [55]. They demonstrated that many oncocytic thyroid tumors harbored copy number gained mitochondrial biogenesis genes including ESRRA.
In summary, this study demonstrates the transcriptional and mutational landscape of miFTC and FA together with cPTC and FVPTC. We revealed that thyroid cancers developed by different types of genetic alteration could be classified as three molecular subtypes (BRAF–like, RAS–like, and NBNR) based on gene expression profiles. The three molecular subtypes showed difference in chromosomal aberration, cell proliferation, differentiation, intracellular signaling, and metabolism. We propose that reclassification of thyroid tumors, especially follicular-patterned ones, on the basis of molecular characteristics would provide novel diagnostic implications.
Materials and Methods
Ethics statement
This study was approved by the institutional review board of Seoul National University Hospital, in accordance with the Declaration of Helsinki (approved ID: H-1108-041-372). Written informed consent was obtained from each subject.
Patients
Specimens from 180 patients (49 men and 131 women; 47 ± 13 years of age) whose fresh frozen thyroid tissue after thyroid surgery were collected from March 2007 to January 2014. We could collect 180 tumor tissue samples (25 FAs, 30 FTCs, 48 FVPTCs, and 77 cPTCs) and 81 paired-normal tissue samples that matched with their tumor tissues. The diagnosis of each sample was determined based on pathological findings from thyroid specimens obtained after thyroidectomy. The clinical information of study subjects is shown in S6 Table. There were no patients who were exposed radiation previously.
Pathological diagnosis
Pathological slides were reviewed by a specialized pathologist. cPTC was defined if the tumor has well-formed papillae with fibrovascular cores and characteristic nuclear features of papillary carcinoma. FVPTC was defined as a PTC with predominantly a follicular growth pattern more than 50%, no well-formed papillae. There are two subtypes of FVPTC: infiltrative FVPTC and EFVPTC regarding the tumor border—infiltrative border or a pushing border with smooth outlines and a capsule, respectively. Capsular invasion was identified in only two cases in EFVPTC and there was not capsular invasion in the other FVPTCs. Therefore, we did not categorize encapsulated FVPTC into two subgroups regarding capsular invasion. miFTC was diagnosed if the tumor is encapsulated by capsular invasion and/or small-caliber sized angioinvasion. FA was diagnosed with no capsular invasion and angioinvasion [56].
RNA sequencing
Extraction of RNA from frozen tissues was performed using the QIAcube and RNeasy Mini Kit (Qiagen, Hilden, Germany) or the Easy Spin RNA extraction kit (Intron, Daejeon, Korea) when tissue volume was small but high product yield was needed. RNA was assessed for quality and concentration measurement using an RNA 6000 Nano LabChip on a 2100 Bioanalyzer (Agilent Inc., Palo Alto, CA). The sequencing libraries were sequenced on a HiSeq 2000 platform (Illumina, San Diego, CA).
The sequenced paired-end reads were aligned to GRCh37.p13 human reference genome using STAR 2-pass method [57,58] and PCR duplicates were removed by Picard MarkDuplicate (http://picard.sourceforge.net). Filtered reads were further processed for variant calling using best-practice of GATK (https://software.broadinstitute.org/gatk/best-practices/), which includes insertion/deletion (indel) realignment and base quality score recalibration [59]. S8 Table shows a summary of sequencing throughput and alignment yield in our study subjects.
SNV and indel calling
We called somatic single-nucleotide variants (SNVs) from 81 matched samples using MuTect [60]. For non-matched samples, we applied SNV calling using the single sample mode of MuTect and GATK’s HaplotypeCaller. Moreover, GATK’s HaplotypeCaller was also used for indel detection. All variants called in these manners were annotated with information from several databases using ANNOVAR [61]. Furthermore, we used GATK’s DepthOfCoverage for counting alternative allele of mutation hotspots in common oncogenes.
To discover driver mutations in thyroid cancers, we applied additional filtration criteria to variant calls, as follows: 1) not or rarely shown in public databases of normal individuals, such as Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/), 1000 Genomes projects [62] and Exome Sequencing Project 6500 (http://evs.gs.washington.edu/EVS/) (MAF ≤ 0.0001 for ExAC and ≤ 0.01 for other databases); 2) nonsilent SNVs (nonsynonymous and splice-site) and frameshift indels; 3) genes that were annotated in COSMIC70 or PTC dataset of TCGA project. Driver candidates in TCGA were examined using cBioPortal for Cancer Genomics [63].
Fusion gene mutation analysis
To discover fusion genes in thyroid cancers, we used MOJO (https://github.com/cband/MOJO) with TCGA GAF 3.0 reference. For filtering false positive calls, we applied further filtration to predicted calls: 1) fusion genes only shown in tumor samples; 2) discordant read pairs between gene A and B ≥ 2; and 3) genomic distance between predicted coordinates ≥ 100 kb or two genes located on different chromosomes.
Validation of novel driver candidates
To validate the mutation of novel driver candidates, gDNA was extracted using QIAamp DNA Kits and QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) from fresh-frozen tissues, or formalin-fixed, paraffin-embedded (FFPE) tissue specimens. DNA was quantified using a Nanodrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, USA) and used as template for PCR amplification. Amplification primers were designed with Primer3 [64]. PCR was performed on GeneAmp PCR System 9700 (Applied Biosystems; Life Technologies, Carlsbad, USA) using Hotstar Taq polymerase (Qiagen) as follows; 15 minutes at 95°C for initial denaturation, then 40 cycles at 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 60 seconds, then 5 minutes at 72°C for final extension. 20 ng of gDNA was used to amplify. Amplified products were purified with DNA purification kit (HiYield DNA fragment extraction kit, Real Genomics, New Taipei City, Taiwan), and then analyzed on an Applied Biosystems 3730XL DNA Sequencing Facility (Applied Biosystems). PCR primer and DNA sequencing services were provided by Cosmo Genetech (Cosmo Genetech, Seoul, Korea). The sequencing results were analyzed using ABI Sequencing analysis software 5.2 (Applied Biosystems). The primer sequences used in this study provided in S8 Table.
Validation of novel fusion gene candidates
RT-PCR and Sanger sequencing were performed across the fusion break points to identify the exact fusion junction of PICALM–BRAF. The tissue blocks were cut into 4μm slides, and total RNA from FFPE samples was isolated using a tissue kit (Maxwell 16 LEV RNA FFPE purification kit, Promega, Madison, USA) and an automatic extractor (Maxwell MDx 16, Promega). RT-PCR was performed on GeneAmp 9700 using Hotstar Taq polymerase (Qiagen) as follows;15 minutes at 95°C for initial denaturation, then 45 cycles at 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 60 seconds, then 5 minutes at 72°C for final extension. 30 ng of cDNA synthesized from total RNA was used to analyze. All PCR products were sequenced on both strands using the same primers and BigDye Terminator v3.1 Cycle Sequencing kits and a 3730 DNA analyzer (Applied Biosystems). The specific fusion primers for PICALM–BRAF provided in S8 Table.
Fluorescence in situ hybridization
To evaluate ETV6 rearrangement, we performed FISH analysis on FFPE tumor tissues using the Vysis LSI ETV6 spectrum Orange/Green probe (Abbot Molecular, Illinois, USA). These commercially-available probes are designed as a dual-color probe where the two regions across the break-point. For microscopic evaluation, at least 100 intact and nonoverlapping cell nuclei were scored for the presence of a split signal using a Zeiss Axio Imager with appropriate filters. Pictures were captured using a digital microscope camera ProgRes MF (Jenoptik, Germany) and analyzed with the Isis software (MetaSystems, Germany). The signal pattern interpretation was as follows: interphase nucleus with two colocalized green/orange fusion signals identified normal chromosomes, while a separated orange and green signals and green/orange fusion signals indicated rearranged gene. The positive threshold was defined as more than 10% of signals split and/or isolated orange signal in 100 tumor cells.
Immunohistochemistry
To verity STRN–ALK, IHC staining was performed on FFPE tissue sections that were 4 m thick using an automated immunostainer (Leica Microsystems, Milton Keynes, UK). Briefly, the slides were heated for 20 min at 100°C in Epitope retrieval solution, pH 9.0 (Leica Microsystems). The slides were then incubated with a monoclonal mouse anti-human ALK antibody (Novocastra, Newcastle Upon Tyne, UK) at a dilution of 1:25. This antibody was raised against a C-terminal portion of the tyrosine kinase domain of ALK and was intended for the qualitative identification of ALK molecules in paraffin sections by light microscopy. Staining intensity was scored as 0 (no staining), 1+ (weak cytoplasmic staining without any background staining), and 2+ (strong cytoplasmic staining). Tumors with 1+ or 2+ expression in more than 10% of the tumor cells were deemed positive for ALK protein expression. For ALK IHC-positive cases, we subsequently performed IHC using an antibody against phosphorylated ALK (phosphor Y1507, Abcam, Cambridge, MA, USA) at a dilution of 1:100.
Gene expression profiling and differentially expressed gene analysis
According to Ensembl gene set, we counted the number of reads aligned to each gene using HTSeq-count and normalized them via regularized log (rlog) transformation method of DESeq2 [65,66]. In this study, DEGs were determined by DESeq2 to have q-value < 0.05, |Log2 (fold change)| ≥ 1, and baseMean ≥ 100. The calculated p-values were adjusted to q-values for multiple testing using the Benjamini–Hochberg correction. The normalized gene expression values were applied to PCA using the most variable 500 genes. For heatmap display, the centered rlog values were applied to the K-means clustering algorithm using cluster 3.0 [67]. To identify pathways that were significantly enriched in DEGs, we applied them to the Molecular Signatures Database 5.0 [30].
Thyroid differentiation score and ERK signature
As described by TCGA study, we calculated TDS using 16 thyroid metabolism and function genes. The rlog values from DESeq2 were first median-centered across 180 tumor samples, and then average values across the 16 genes in each tumor were determined as TDS.
To numerically represent activation level of MAPK signaling pathway, we implemented and modified ERK score calculation from TCGA. We applied identical method that was described in TDS calculation using 52 MAPK signaling pathway genes [68].
Jointly regulated block analysis
For JRB analysis, we selected genes in autosomes that have average FPKM ≥ 1.5 and were classified as protein-coding gene in Ensembl database. After that, we sorted genes by chromosomal coordinate and applied three normalization steps as follows: 1) Log (FPKM) of gene (gene A) in ith tumor sample was Z-score transformed:
where μ and σ represent average and standard deviation of Log (FPKM) of 81 normal tissues.
2) Z-score of gene (gene A) in ith tumor sample was Z-score transformed:
where μi and σi represent average and standard deviation of Z-score of ith tumor sample.
3) median Z-score of each chromosome arm of ith tumor sample was median-centered by subtracting the median Z-score of all chromosome arms. After normalization steps, we defined arm with median-centered Z-score ≥ 0.5 and ≤ -0.5 as overexpressed and underexpressed JRB, respectively.
Statistical analyses of clinical data
All statistical analyses were performed using SPSS version 20.0 (IBM Co, Armonk, NY, USA). Data are presented either as frequencies (%) or as mean ± standard deviation. Comparisons of categorical variables were performed using either the Pearson’s χ2 or Fisher’s exact test (if the number was < 5), and the independent t-test was used for continuous variables. Adjusted p-values for age and sex were obtained by the binomial or multinomial logistic regression analyses for categorical variables and by either the linear regression or analysis of covariance (ANCOVA) for continuous variables. A post-hoc Bonferroni test were used to determine which groups have statistically different proportion of clinical risk factors. Statistical significance was defined as two-sided p-values < 0.05.
Data availability
We submitted all the sequenced paired-end reads to EBI European Nucleotide Archive database with accession number PRJEB11591 (Direct access: http://www.ebi.ac.uk/ena/data/view/PRJEB11591).
Supporting Information
(A) The amino acid coordinate of Ribonuclease III domain and distribution of mutations in DICER1. (B) The expression level of DICER1 in normal, tumors with other mutations, and tumors with DICER1 mutations. (C) The distribution of mutation across TCGA cases. Each column represents individual specimen. Right matrix was omitted due to there is no overlapped mutation. (D) The amino acid coordinate and distribution of mutations of EIF1AX.
(PDF)
(A) PCR and Sanger sequencing results of mutations in driver candidates. Upper and lower panel represent results of matched normal and tumor tissues, respectively. Left and right panel represent results of forward and reverse sequences, respectively. (B) RT-PCR and Sanger sequencing result of novel PICALM–BRAF fusion gene.
(PDF)
(A) Aberrant overexpression of fusion genes. (B) FISH result of ETV6–NTRK3. The separated green and orange signals and green/orange fusion signals indicate rearranged gene. (C) Photomicrograph image of ALK IHC result. IHC showed strong cytoplasmic staining in periphery of tumor.
(PDF)
The result of K-means clustering via PCA on (A) All normal and tumor tissues using Ensembl gene set (Marked as histological subtype and driver gene). (B) All normal and tumor tissues using Ensembl gene set (Marked as LT and driver gene). (C) All tumors using Ensembl gene set (Marked as histological subtype and driver gene). (D) All tumors using Ensembl gene set (Marked as LT and driver gene). (E) Whole TCGA dataset using UCSC gene set. (F) Partial TCGA dataset (Two DICER1, four EIF1AX, four PAX8–PPARG, 15 H/K/NRAS, and 20 BRAF mutated samples were included) using UCSC gene set. Each cluster was represented by a 95.00% confidence ellipse.
(PDF)
The significantly enriched KEGG pathways are marked by–log (q-value).
(PDF)
The significantly enriched KEGG pathways are marked by–log (q-value).
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files. Sequenced paired-end reads are available in EBI European Nucleotide Archive database with accession number PRJEB11591 (Direct access: http://www.ebi.ac.uk/ena/data/view/PRJEB11591).
Funding Statement
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI13C1927). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schlumberger MJ (1998) Papillary and follicular thyroid carcinoma. N Engl J Med 338: 297–306. [DOI] [PubMed] [Google Scholar]
- 2.Schmid KW, Farid NR (2006) How to define follicular thyroid carcinoma? Virchows Arch 448: 385–393. [DOI] [PubMed] [Google Scholar]
- 3.Sherman SI (2003) Thyroid carcinoma. Lancet 361: 501–511. [DOI] [PubMed] [Google Scholar]
- 4.Correa P, Chen VW (1995) Endocrine gland cancer. Cancer 75: 338–352. [DOI] [PubMed] [Google Scholar]
- 5.Vasko V, Ferrand M, Di Cristofaro J, Carayon P, Henry JF, et al. (2003) Specific pattern of RAS oncogene mutations in follicular thyroid tumors. J Clin Endocrinol Metab 88: 2745–2752. [DOI] [PubMed] [Google Scholar]
- 6.Fukahori M, Yoshida A, Hayashi H, Yoshihara M, Matsukuma S, et al. (2012) The associations between RAS mutations and clinical characteristics in follicular thyroid tumors: new insights from a single center and a large patient cohort. Thyroid 22: 683–689. 10.1089/thy.2011.0261 [DOI] [PubMed] [Google Scholar]
- 7.Zhu Z, Gandhi M, Nikiforova MN, Fischer AH, Nikiforov YE (2003) Molecular profile and clinical-pathologic features of the follicular variant of papillary thyroid carcinoma. An unusually high prevalence of ras mutations. Am J Clin Pathol 120: 71–77. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network (2014) Integrated genomic characterization of papillary thyroid carcinoma. Cell 159: 676–690. 10.1016/j.cell.2014.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montagut C, Settleman J (2009) Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett 283: 125–134. 10.1016/j.canlet.2009.01.022 [DOI] [PubMed] [Google Scholar]
- 10.Melillo RM, Castellone MD, Guarino V, De Falco V, Cirafici AM, et al. (2005) The RET/PTC-RAS-BRAF linear signaling cascade mediates the motile and mitogenic phenotype of thyroid cancer cells. J Clin Invest 115: 1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Leeman-Neill RJ, Kelly LM, Liu P, Brenner AV, Little MP, et al. (2014) ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer 120: 799–807. 10.1002/cncr.28484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan H, Ji M, Bao R, Yu H, Wang Y, et al. (2009) Association of high iodine intake with the T1799A BRAF mutation in papillary thyroid cancer. J Clin Endocrinol Metab 94: 1612–1617. 10.1210/jc.2008-2390 [DOI] [PubMed] [Google Scholar]
- 13.Klugbauer S, Lengfelder E, Demidchik EP, Rabes HM (1995) High prevalence of RET rearrangement in thyroid tumors of children from Belarus after the Chernobyl reactor accident. Oncogene 11: 2459–2467. [PubMed] [Google Scholar]
- 14.Fugazzola L, Pilotti S, Pinchera A, Vorontsova TV, Mondellini P, et al. (1995) Oncogenic rearrangements of the RET proto-oncogene in papillary thyroid carcinomas from children exposed to the Chernobyl nuclear accident. Cancer Res 55: 5617–5620. [PubMed] [Google Scholar]
- 15.Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R (2013) Worldwide Increasing Incidence of Thyroid Cancer: Update on Epidemiology and Risk Factors. Journal of Cancer Epidemiology 2013: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Kock L, Sabbaghian N, Soglio DB, Guillerman RP, Park BK, et al. (2014) Exploring the association Between DICER1 mutations and differentiated thyroid carcinoma. J Clin Endocrinol Metab 99: E1072–1077. 10.1210/jc.2013-4206 [DOI] [PubMed] [Google Scholar]
- 17.Costa V, Esposito R, Ziviello C, Sepe R, Bim LV, et al. (2015) New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma. Oncotarget 6: 11242–11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, et al. (2012) Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 366: 234–242. 10.1056/NEJMoa1102903 [DOI] [PubMed] [Google Scholar]
- 19.Witkowski L, Mattina J, Schonberger S, Murray MJ, Choong CS, et al. (2013) DICER1 hotspot mutations in non-epithelial gonadal tumours. Br J Cancer 109: 2744–2750. 10.1038/bjc.2013.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, et al. (2016) Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 34: 155–163. 10.1038/nbt.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, et al. (2012) Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 44: 1310–1315. 10.1038/ng.2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani RS (2014) The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discov Today 19: 1498–1502. 10.1016/j.drudis.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauvageau M, Sauvageau G (2010) Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313. 10.1016/j.stem.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su GH, Hruban RH, Bansal RK, Bova GS, Tang DJ, et al. (1999) Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol 154: 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guldberg P, thor Straten P, Ahrenkiel V, Seremet T, Kirkin AF, et al. (1999) Somatic mutation of the Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma. Oncogene 18: 1777–1780. [DOI] [PubMed] [Google Scholar]
- 26.Nakaoku T, Tsuta K, Ichikawa H, Shiraishi K, Sakamoto H, et al. (2014) Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 20: 3087–3093. 10.1158/1078-0432.CCR-14-0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, et al. (2014) New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist 19: 235–242. 10.1634/theoncologist.2013-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou JB, Zhang T, Wang BF, Gao HZ, Xu X (2013) Identification of a novel gene fusion RNF213SLC26A11 in chronic myeloid leukemia by RNA-Seq. Mol Med Rep 7: 591–597. 10.3892/mmr.2012.1183 [DOI] [PubMed] [Google Scholar]
- 29.Rippe V, Drieschner N, Meiboom M, Murua Escobar H, Bonk U, et al. (2003) Identification of a gene rearranged by 2p21 aberrations in thyroid adenomas. Oncogene 22: 6111–6114. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maximo V, Sobrinho-Simoes M (2000) Hurthle cell tumours of the thyroid. A review with emphasis on mitochondrial abnormalities with clinical relevance. Virchows Arch 437: 107–115. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, et al. (2004) The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A 101: 6472–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, et al. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124. [DOI] [PubMed] [Google Scholar]
- 34.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, et al. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chudova D, Wilde JI, Wang ET, Wang H, Rabbee N, et al. (2010) Molecular classification of thyroid nodules using high-dimensionality genomic data. J Clin Endocrinol Metab 95: 5296–5304. 10.1210/jc.2010-1087 [DOI] [PubMed] [Google Scholar]
- 36.Shan Z, Li G, Zhan Q, Li D (2012) Gadd45a inhibits cell migration and invasion by altering the global RNA expression. Cancer Biol Ther 13: 1112–1122. 10.4161/cbt.21186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nucera C, Lawler J, Parangi S (2011) BRAF(V600E) and microenvironment in thyroid cancer: a functional link to drive cancer progression. Cancer Res 71: 2417–2422. 10.1158/0008-5472.CAN-10-3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, et al. (2012) The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 22: 2109–2119. 10.1101/gr.145144.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H-s, Wilsbach K, Marti A, Najafian A, Meeker AK, et al. (2015) DNA copy number variation and driver mutation patterns of follicular thyroid tumors [abstract]. In: Proceedings of the 106th Annual Meeting of the American Association for Cancer Research; 2015 April 18–22; Philadelphia (PA): AACR; 2015 Abstract nr 4268.
- 40.Wreesmann VB, Ghossein RA, Hezel M, Banerjee D, Shaha AR, et al. (2004) Follicular variant of papillary thyroid carcinoma: genome-wide appraisal of a controversial entity. Genes Chromosomes Cancer 40: 355–364. [DOI] [PubMed] [Google Scholar]
- 41.Mack WJ, Preston-Martin S, Dal Maso L, Galanti R, Xiang M, et al. (2003) A pooled analysis of case-control studies of thyroid cancer: cigarette smoking and consumption of alcohol, coffee, and tea. Cancer Causes Control 14: 773–785. [DOI] [PubMed] [Google Scholar]
- 42.Rossing MA, Cushing KL, Voigt LF, Wicklund KG, Daling JR (2000) Risk of papillary thyroid cancer in women in relation to smoking and alcohol consumption. Epidemiology 11: 49–54. [DOI] [PubMed] [Google Scholar]
- 43.Repplinger D, Bargren A, Zhang YW, Adler JT, Haymart M, et al. (2008) Is Hashimoto's thyroiditis a risk factor for papillary thyroid cancer? J Surg Res 150: 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirtschafter A, Schmidt R, Rosen D, Kundu N, Santoro M, et al. (1997) Expression of the RET/PTC fusion gene as a marker for papillary carcinoma in Hashimoto's thyroiditis. Laryngoscope 107: 95–100. [DOI] [PubMed] [Google Scholar]
- 45.Cao Y, Wang Z, Gu J, Hu F, Qi Y, et al. (2015) Reproductive Factors but Not Hormonal Factors Associated with Thyroid Cancer Risk: A Systematic Review and Meta-Analysis. BioMed Research International 2015: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohler A, Chen B, Gemignani F, Elisei R, Romei C, et al. (2013) Genome-wide association study on differentiated thyroid cancer. J Clin Endocrinol Metab 98: E1674–1681. 10.1210/jc.2013-1941 [DOI] [PubMed] [Google Scholar]
- 47.Rabes HM, Demidchik EP, Sidorow JD, Lengfelder E, Beimfohr C, et al. (2000) Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-chernobyl papillary thyroid carcinomas: biological, phenotypic, and clinical implications. Clin Cancer Res 6: 1093–1103. [PubMed] [Google Scholar]
- 48.Ricarte-Filho JC, Li S, Garcia-Rendueles ME, Montero-Conde C, Voza F, et al. (2013) Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J Clin Invest 123: 4935–4944. 10.1172/JCI69766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asa SL, Giordano TJ, LiVolsi VA (2015) Implications of the TCGA genomic characterization of papillary thyroid carcinoma for thyroid pathology: does follicular variant papillary thyroid carcinoma exist? Thyroid 25: 1–2. 10.1089/thy.2014.0540 [DOI] [PubMed] [Google Scholar]
- 50.Nikiforov YE, Seethala RR, Tallini G, Baloch ZW, Basolo F, et al. (2016) Nomenclature Revision for Encapsulated Follicular Variant of Papillary Thyroid Carcinoma: A Paradigm Shift to Reduce Overtreatment of Indolent Tumors. JAMA Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saji M, Xydas S, Westra WH, Liang CK, Clark DP, et al. (1999) Human telomerase reverse transcriptase (hTERT) gene expression in thyroid neoplasms. Clin Cancer Res 5: 1483–1489. [PubMed] [Google Scholar]
- 52.Rosen J, He M, Umbricht C, Alexander HR, Dackiw AP, et al. (2005) A six-gene model for differentiating benign from malignant thyroid tumors on the basis of gene expression. Surgery 138: 1050–1056; discussion 1056–1057. [DOI] [PubMed] [Google Scholar]
- 53.Takano T, Miyauchi A, Yoshida H, Kuma K, Amino N (2005) Decreased relative expression level of trefoil factor 3 mRNA to galectin-3 mRNA distinguishes thyroid follicular carcinoma from adenoma. Cancer Lett 219: 91–96. [DOI] [PubMed] [Google Scholar]
- 54.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, et al. (2014) The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26: 319–330. 10.1016/j.ccr.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurelac I, de Biase D, Calabrese C, Ceccarelli C, Ng CK, et al. (2015) High-resolution genomic profiling of thyroid lesions uncovers preferential copy number gains affecting mitochondrial biogenesis loci in the oncocytic variants. Am J Cancer Res 5: 1954–1971. [PMC free article] [PubMed] [Google Scholar]
- 56.Nikiforov YE, Biddinger PW, Thompson LDR (2012) Diagnostic Pathology and Molecular Genetics of the Thyroid, 2nd edition: Lippincott Williams & Wilkins. [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engstrom PG, Steijger T, Sipos B, Grant GR, Kahles A, et al. (2013) Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods 10: 1185–1191. 10.1038/nmeth.2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, et al. (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, et al. (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40: e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- 68.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, et al. (2009) (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A 106: 4519–4524. 10.1073/pnas.0900780106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The amino acid coordinate of Ribonuclease III domain and distribution of mutations in DICER1. (B) The expression level of DICER1 in normal, tumors with other mutations, and tumors with DICER1 mutations. (C) The distribution of mutation across TCGA cases. Each column represents individual specimen. Right matrix was omitted due to there is no overlapped mutation. (D) The amino acid coordinate and distribution of mutations of EIF1AX.
(PDF)
(A) PCR and Sanger sequencing results of mutations in driver candidates. Upper and lower panel represent results of matched normal and tumor tissues, respectively. Left and right panel represent results of forward and reverse sequences, respectively. (B) RT-PCR and Sanger sequencing result of novel PICALM–BRAF fusion gene.
(PDF)
(A) Aberrant overexpression of fusion genes. (B) FISH result of ETV6–NTRK3. The separated green and orange signals and green/orange fusion signals indicate rearranged gene. (C) Photomicrograph image of ALK IHC result. IHC showed strong cytoplasmic staining in periphery of tumor.
(PDF)
The result of K-means clustering via PCA on (A) All normal and tumor tissues using Ensembl gene set (Marked as histological subtype and driver gene). (B) All normal and tumor tissues using Ensembl gene set (Marked as LT and driver gene). (C) All tumors using Ensembl gene set (Marked as histological subtype and driver gene). (D) All tumors using Ensembl gene set (Marked as LT and driver gene). (E) Whole TCGA dataset using UCSC gene set. (F) Partial TCGA dataset (Two DICER1, four EIF1AX, four PAX8–PPARG, 15 H/K/NRAS, and 20 BRAF mutated samples were included) using UCSC gene set. Each cluster was represented by a 95.00% confidence ellipse.
(PDF)
The significantly enriched KEGG pathways are marked by–log (q-value).
(PDF)
The significantly enriched KEGG pathways are marked by–log (q-value).
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Sequenced paired-end reads are available in EBI European Nucleotide Archive database with accession number PRJEB11591 (Direct access: http://www.ebi.ac.uk/ena/data/view/PRJEB11591).
We submitted all the sequenced paired-end reads to EBI European Nucleotide Archive database with accession number PRJEB11591 (Direct access: http://www.ebi.ac.uk/ena/data/view/PRJEB11591).