Abstract
Genes in which germline mutations confer high or moderate increased risks of cancer are called cancer predisposition genes (CPG). Over 100 CPGs have been identified providing important scientific insights in many areas, particularly mechanisms of cancer causation. Moreover, clinical utilisation of CPGs has had substantial impact in diagnosis, optimised management and prevention of cancer. The recent transformative advances in DNA sequencing bring the promise of many more CPG discoveries and greater, broader clinical applications. However, there is also considerable potential for incorrect inferences and inappropriate clinical applications. Realising the promise of cancer predisposition genes for science and medicine will thus require careful navigation.
Genetic predisposition to cancer has been recognised for centuries, initially through observation of unusual familial clusterings of cancer. In 1866 neuroanatomist Paul Broca published one of the earliest reports, detailing a striking history of breast cancer in fifteen members of his wife’s family1. Broca, controversially for the time, proposed this was evidence of hereditary predisposition to cancer. Fifty years later, Theodor Boveri published his visionary theory that somatic acquisition of ‘particular, incorrect chromosome combinations’ underlie cancer. His paper was equally prophetic about inherited predisposition to cancer, predicting it could result from ‘weakened resistance against the action of factors that stimulate cell division’2. In 1971, Knudson’s mathematical modelling of the epidemiology of retinoblastoma suggested a ‘two-hit’ model whereby both alleles of a specific gene were required to be inactivated for retinoblastoma to occur, thus echoing Boveri’s predictions3. In 1987 the retinoblastoma predisposition gene, RB1, was discovered and, as predicted, in hereditary cases one allele was mutated in the germline with the second allele inactivated somatically4.
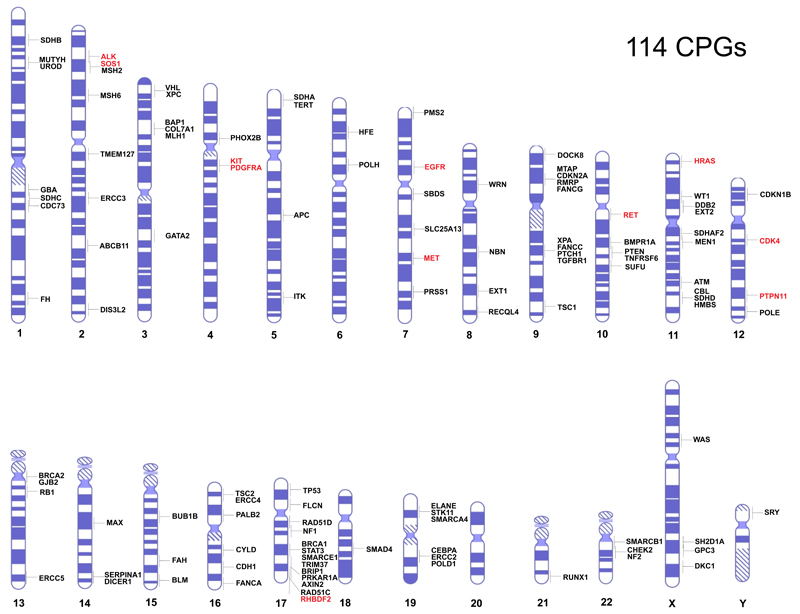
There is no definitive definition of a cancer predisposition gene (CPG). For the purposes of this review I have restricted inclusion to genes in which rare mutations confer high or moderate risks of cancer (>2 fold relative risks) and at least 5% of individuals with relevant mutations develop cancer. For the majority of genes both the risks and penetrance are considerably higher than these minimum criteria. Common variants conferring very small increases in risk discovered through genome-wide association studies are not included within this definition. Such variants are important components of the genetic architecture of cancer and are addressed in several other reviews5–7. Through extensive literature and database evaluations I identified 114 CPGs which form the basis of this review (Fig 1, Supplementary Table 1).
Figure 1. Chromosomal location of 114 Cancer Predisposition Genes.
Gain-of-function mutations predispose to cancer in genes in red text. Loss-of-function mutations predispose to cancer in genes in black text.
Traditionally cancer predisposition reviews have focussed on select cancers and/or sets of genes. In an era of whole genome sequencing, information about all known CPGs is increasingly desirable in both research and clinical practice. Here, I have aspired to integrate knowledge from three decades of research to provide a distillation of key CPG characteristics. I also discuss the exciting prospects and potential pitfalls of future discoveries and clinical applications.
Discovery of cancer predisposition genes
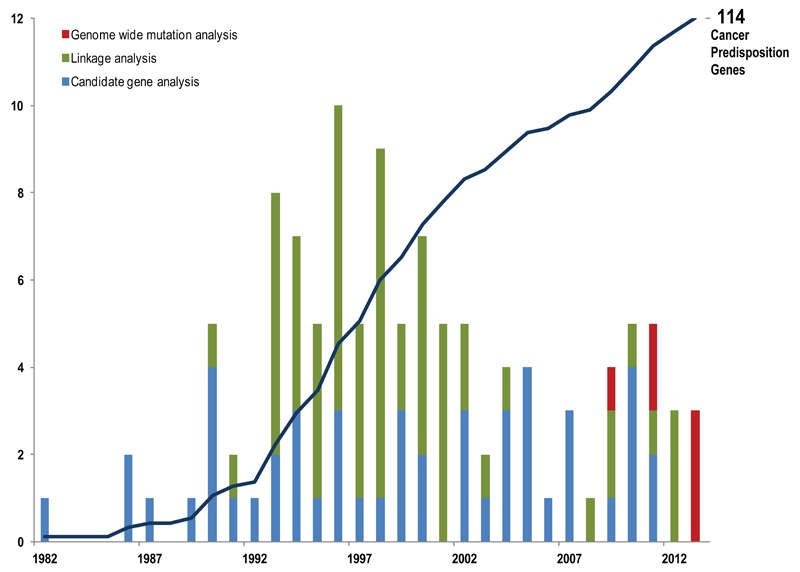
The 114 CPGs were discovered over the last thirty years through multiple different strategies (Fig 2). Since 1990 at least one new CPG has been identified each year, peaking in 1996 when ten CPGs were reported8–16. Genome-wide linkage analysis, an agnostic approach that allows tracking of disease-associated genomic markers in high-penetrance familial clusters has been the most successful strategy, yielding 59 CPGs, mostly in the 1990s when the methodology became routine. More recently, next-generation sequencing is leading to a new crop of CPGs being discovered through genome-wide mutational analyses such as exome and genome sequencing17,18.
Figure 2. Timeline of Cancer Predisposition Gene Discovery.
The cumulative total of CPGs discovered since 1982 is shown by the solid line. The number of CPGs discovered each year and the discovery method used is shown in the graph. See text for fuller explanation of the different methods.
The remaining genes were identified through various candidate-based strategies. Large numbers of candidate CPGs have been proposed and investigated. For the majority, no association with cancer predisposition has been found. This perceived failure led to candidate-based approaches falling from favour. However, certain strategies have proved very successful both as stand-alone discovery methods and in facilitating linkage studies. Candidates pursued as surrogates of cancer predisposition for example distinctive cellular phenotypes such as defective DNA repair, mosaic aneuploidies or telomere shortening have contributed to the discovery of many CPGs9,10,19,20. Genetic pathway candidates, i.e. genes selected because they function in similar pathways to known CPGs, has also yielded new predisposition genes, particularly in colorectal, breast, ovarian and endocrine cancers21–28. Candidate genes chosen because they are somatically mutated in cancers have, perhaps surprisingly, led to the identification of only twelve CPGs29–40.
Overlap of somatic and germline cancer genes
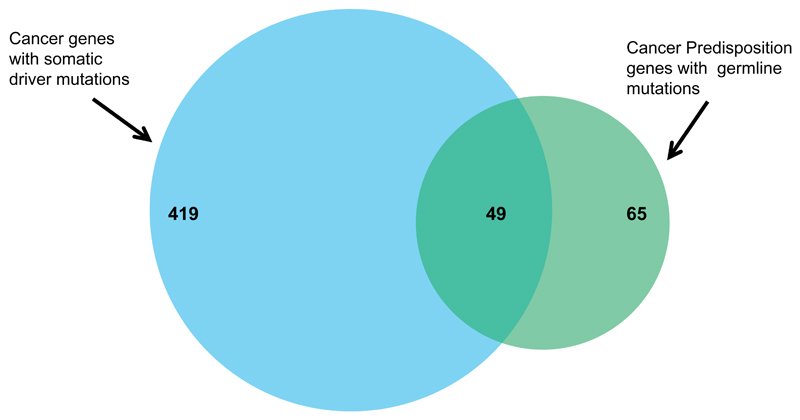
It is interesting and instructive to consider the overlap between the known germline and tumor mutated cancer genes. Currently, the COSMIC database includes 468 genes somatically mutated in cancers41. Of these, 49 are also known to be CPGs. Conversely, 65 of the 114 CPGs are known to be somatically mutated. These data imply that 10% of somatically mutated cancer genes also confer susceptibility to cancer when mutated in the germline, but that 40% of germline mutated cancer predisposition genes can also contribute to oncogenesis when mutations occur only in tumors (Fig 3). This apparent discrepancy is, at least in part, an artefact of different research approaches; it is common for the frequency of somatic CPG mutations to be investigated but it is unusual for somatically mutated genes to be evaluated for their role in cancer predisposition. The latter has been exacerbated by cancer genome sequencing studies in which a key filtering step is removal of variants present in normal tissue to focus attention on the potential cancer-driving mutations present only in the tumor42. It is thus highly likely that there is an underestimate of the overlap between somatic cancer driver mutations and germline cancer predisposing mutations and mutual interrogation of such genes could prove a useful approach for identification of new cancer genes.
Figure 3. Overlap between somatically mutated cancer genes and cancer predisposition genes.
468 genes with somatic driver mutations in cancers are recorded in COSMIC of which 49 are also included within the 114 cancer predisposition genes.
Overlap of high and low penetrance cancer associated variants
One notable absentee in the successful CPG discovery strategies is identification through genome-wide association study candidature. There are 391 known common variants that confer small increased risks of cancer43 (http://www.genome.gov/gwastudies/). The underlying causal gene/mechanism has only been identified for a small minority, but none have been shown to be sentinels of rare, higher penetrance mutations in new CPGs. Furthermore, only 15 SNPs are within known CPGs and none are associated with cancers that occur in carriers of rare, penetrant mutations. For example, ATM D1853N, (the only exonic cancer GWAS variant in a CPG), confers a protective effect of melanoma but it is not associated with the cancers that occur in biallelic or monoallelic ATM mutation carriers44,45. Similarly, rs78378222 alters the polyadenylation signal of TP53 and is associated with various cancers, but not those that typically occur in Li Fraumeni syndrome, which is due to germline TP53 mutations46. Multiple variants in the vicinity of TERT have been associated with several different cancers, but again not those that occur in dyskeratosis congenita, a recessive condition due to germline exonic TERT mutations 47–49. These data suggest the mechanisms underlying the association with cancer of rare, high penetrance alleles and common, low penetrance alleles are largely distinct. This differs from several other common, complex conditions which show considerable overlap between these components of the genetic architecture50,51.
Characteristics of cancer predisposition genes
The 114 genes are located throughout the genome with little evidence of chromosomal clustering (Fig 1).
Inheritance and mechanisms of oncogenesis
The inheritance pattern of cancer predisposition is varied; autosomal dominant for 65 CPGs, autosomal recessive for 28, X-linked for four and one is Y-linked. 16 genes cause phenotypes in both monoallelic and biallelic mutation carriers, i.e. they cause autosomal dominant and autosomal recessive conditions. For half of these the recessive condition is a more severe manifestation of the dominant condition. For example, biallelic BRCA2, PALB2, MLH1, MSH2, MSH6, PMS2 mutation carriers have high risks of childhood cancer whereas monoallelic mutation carriers have increased risks of adult cancers52. For other genes in which monoallelic and biallelic mutations cause clinical phenotypes, such as FH and SDHA, cancer has not been reported in biallelic mutation carriers. This is likely due to early mortality from other manifestations.
The great majority of CPGs act as tumor suppressor genes with mutations that abrogate their function promoting oncogenesis. Only eleven genes predispose to cancer through gain-of-function mutations. Several of these, such as RET, MET, KIT and ALK, encode kinases which are rendered constitutively active by cancer predisposing mutations53. There is much more diversity in the types, functions and mechanisms of oncogenesis of CPGs that are inactivated to increase the risk of cancer. Many are classic tumor suppressor genes, requiring both alleles to be inactivated, but haploinsufficiency and dominant-negative mechanisms also occur. For some genes different mutations operate through different mechanisms and lead to distinct phenotypes as described below54,55. For many genes, the clinical phenotypes and cancer risks associated with CPG mutations are also influenced by other factors, both genetic and non-genetic.
Functions of cancer predisposition genes
CPG research has directly resulted in fundamental insights into basic biological pathways and gene function. Indeed, some centrally important genes were first isolated because germline mutations within them predispose to cancer, as is transparent in their names (BRCA1, RB1, NF1 etc) which reflect the clinical phenotypes that led to their identification. The functions of these genes were only subsequently elucidated, often directly because of research into their role in cancer predisposition.
CPGs have a very broad range of different functions. Many are ubiquitously expressed and participate in fundamental processes such as DNA repair and cell cycle regulation. One of the enduring conundrums of cancer predisposition is why, and how, perturbation of universal cellular functions can cause exquisitely specific cancer phenotypes. However, some genes have organ-specific functions that are transparently related to the cancers with which they are associated. For example, mutations in SLC25A13, ABCB11, FAH, HMBS and UROD all lead to hepatic overload, liver cirrhosis and hence an increased the risk of hepatocellular carcinoma56.
Meaningful evaluation of the 114 CPGs for functional associations is currently precluded because so many were identified due to their functional relationships with known CPGs. Nonetheless, some noteworthy functional networks are emerging as important in cancer predisposition, in addition to well-recognised pathways such as DNA repair. Amongst these are the SWI/SNF chromatin remodelling pathway which has been particularly linked to rhabdoid tumors and meningiomas, the succinate dehydrogenase enzyme complex which is associated with phaeochromocytoma and paragangliomas and the PI3kinase/mTOR signalling pathway which has links with several CPGs including TSC1, TSC2, PTEN, LKB1, FLCN, HRAS, TMEM127 and hence is associated with a diverse cancers57–59.
Cancer predisposition gene phenotypes
Cancer phenotypes
It is currently estimated that ~3% of cancers are due to CPG mutations, which is >300,000 cancers per year worldwide. This is an underestimate as the contribution of known genes has been poorly characterised and not all genes have been identified. The contribution to individual cancers is highly variable. The highest attribution is to childhood embryonal tumors such as retinoblastoma and pleuropulmonary blastoma which are often caused by germline mutations in RB1 and DICER1 respectively29,60. This simplicity is not applicable to all childhood cancers; the embryonal kidney cancer, Wilms tumor, is associated with several CPGs and other predisposition mechanisms, which together account for <5% of cases61,62. At the other end of the spectrum, known CPGs make a very small contribution to some adult cancers, such as prostate and lung cancer. However, germline CPG mutations in multiple genes predispose to other adult cancers such as breast, colorectal, melanoma and ovarian cancer. For some the overall contribution of CPGs is sizeable with ~15% of ovarian cancer, ~20% of medullary thyroid cancer and >30% of phaeochromocytoma due to CPG mutations63–65.
Some CPGs preferentially predispose to specific histological subtypes of a cancer. For example BRCA1 is particularly associated with triple-negative breast cancer and serous ovarian cancer, whereas CDH1 is particularly associated with lobular breast cancer and diffuse gastric cancers66,67. The genomic profiles of cancers arising in individuals with germline CPG mutations can also be distinctive; chromothripsis, which describes localised chromosomal shattering, is striking in medulloblastomas that occur in TP53 germline mutation carriers and cancers in BRCA1/BRCA2 mutation carriers have a characteristic mutational signature that include substantial numbers of deletions with overlapping microhomology at the breakpoint junctions68,69.
Non-cancer phenotypes
Clinical phenotypes in addition to cancer often occur in individuals with CPG mutations with 87 CPGs being associated with non-cancer clinical features. These are often more discriminating and more common than cancer, and can be critical to clinical diagnosis of the underlying cancer syndrome. The spectrum of additional clinical features is very broad. Skin manifestations are most frequent and can be specific to the relevant CPG. They include hypopigmented and/or hyperpigmented areas, freckling, rashes, blistering, hypertrophy, skin tags, nodules and/or lumps. Neurological, dysmorphic and skeletal manifestations also occur, but are usually non-specific features such as microcephaly, macrocephaly, short stature and/or developmental delay. The proportion of CPGs associated with non-cancer clinical phenotypes is likely an over-estimate of the true proportion, as identification of genes that result in a readily clinically recognisable phenotype is inevitably more tractable.
Genotype-phenotype associations
One of the illuminating outcomes of CPG research has been increased knowledge about the diversity of mutational mechanisms and their relationship with phenotype. Even superficially straightforward associations can mask profound complexity. For example, gain-of-function germline HRAS missense mutations cause enhanced MAPK and P13 kinase signalling similar to somatic mutations70. However, the spectrum of germline and somatic mutations differs, and germline HRAS mutations not only predispose to cancer, they cause a multisystem disorder called Costello syndrome71. This condition includes distinctive facial dysmorphism and a wide range of cardiac, dermatological, musculoskeletal and developmental abnormalities. The role of HRAS in these processes and why HRAS mutations lead to such a complex phenotype is unknown.
The genotype-phenotype relationships of some CPGs are extraordinarily intricate and hint at deep uncharted complexities in gene function. Most WT1 mutations predispose to Wilms tumor, genitourinary abnormalities and renal dysfunction, the severity of which is influenced by the mutational type. However, intronic mutations that alter the relative abundance of WT1 isoforms cause a distinct condition, called Frasier syndrome, which includes gonadoblastoma rather than Wilms tumor, a focal-segmental nephropathy and severe gonadal dysgenesis which can manifest as complete sex reversal61.
TERT, which encodes telomerase, is another example. Recently, activating promoter mutations that result in increased telomerase expression were shown to predispose to melanoma72. Monoallelic and biallelic, primarily missense TERT mutations cause dyskeratosis congenita, which is characterised by various physical abnormalities, pulmonary fibrosis, bone marrow failure and increased incidence of acute myelogenous leukemia and squamous carcinomas of the head and neck and anogenital region48. Furthermore, various common SNPs in the vicinity of TERT confer small risks of several cancers, including breast, colorectal, testicular and prostate cancer47,49. The mechanisms underlying the diversity of TERT phenotypic associations are unknown.
TGFBR1, which encodes a transmembrane serine/threonine kinase receptor, is one of the most extreme examples of genotype-phenotype diversity. Missense mutations in the kinase domain cause marfanoid vasculopathies with no increased risk of cancer. Truncating mutations in the same kinase domain, or missense mutations in the extracellular ligand-binding domain, cause a highly unusual condition called multiple self-healing squamous epithelioma. Affected individuals develop squamous carcinoma-like locally invasive skin tumors that grow rapidly for a few weeks then spontaneously regress and scar73.
Cancer predisposition gene cancer risks
There is also deep and widely underappreciated complexity in the risks of cancer conferred by CPG mutations. A specific CPG mutation can confer different risks of different cancers. Different CPG mutations can confer different risks of a particular cancer. A specific CPG mutation can even confer different risks of a particular cancer in different contexts. The BRCA2 gene illustrates all these scenarios. Loss-of-function BRCA2 mutations confer substantial increased lifetime risks of breast and ovarian cancer but only small increased lifetime risks of prostate and pancreatic cancer74. However not all mutations confer the same risk, despite the great majority being protein truncating mutations predicted to result in nonsense-mediated RNA decay and thus to be functionally equivalent. BRCA2 loss-of-function mutations in the central part of the gene confer significantly higher relative risks of ovarian cancer compared to breast cancer than mutations at either end75. The mechanistic basis for this highly unusual pattern remains unknown. The degree of family history also impacts on the risk of cancer of BRCA2 mutations. The lifetime breast cancer risk of female BRCA2 mutation carriers with a strong family history is ~80% but is only ~45% for relatives of unselected breast cancer cases76,77. This reflects, at least in part, additional modifying factors within familial clusters that increase the cancer risk. Some genetic and non-genetic modifying factors of BRCA2 and BRCA1 cancer risks have already been identified, though it is likely there is still much to be discovered.78–81.
TP53 is another gene that has been known to predispose to cancer for over 20 years and yet our knowledge of the associated cancer risks is still lamentably incomplete. All germline TP53 mutations are typically assumed to be highly penetrant, but the widely quoted cancer lifetime risks are derived from small series of highly selected cases82. In fact there is strong evidence of high variability in the types and risks of cancer associated with different TP53 mutations. This is exemplified by TP53 R337H which confers a modest 10% risk of adrenocortical cancer and is not associated with increased risks of other classic Li Fraumeni cancers such as breast cancer or sarcoma83.
Clinical utility of cancer predisposition genes
The identification of CPGs has had substantial clinical impact. Indeed cancer is one of the foremost diseases in which such discoveries have transformed medical care in multiple areas, including cancer prevention.
Diagnosis and patient management
The benefits of determining if a cancer is due to a germline CPG mutation are incontrovertible. As such, CPG testing has become standard for many genes, albeit typically only in highly selected cases. From the patient perspective, simply having a better understanding of why their cancer occurred is usually highly valued. It also provides important information that can aid diagnosis and management, for instance whether to have conservative or radical surgery. Radiotherapy and chemotherapy may be also altered. For example, platinum based therapies are not standard treatment for breast cancer but can have utility in BRCA carriers84,85. Conversely, temozolomide is unlikely to be of benefit and may actually promote neoplastic progression in MSH6 mutation carriers86,87. Identifying an underlying CPG mutation also provides important prognostic information; survival is significantly better for BRCA2 mutation-positive ovarian cancer patients but significantly worse for BRCA2 mutation-positive prostate cancer patients88,89. The likelihood of recurrence, a new primary and/or a second malignancy can all be increased in CPG mutation carriers who require ongoing review and consideration of tailored surveillance and/or risk-reducing interventions. Management of non-cancer associated problems can also be important, for example certain WT1 mutations result in insidious renal dysfunction which requires monitoring and early intervention.
Targeted therapies
There is intense activity in developing tailored therapies for cancer and strategies targeting CPGs and their constituent pathways have been amongst the most innovative and fruitful. The rationale is to harness knowledge of the underlying cause of cancer to identify tumor-specific vulnerabilities that can be therapeutically exploited. The simplest model is in cancers caused by gain-of-function mutations which can be directly downregulated by inhibitors such as imatinib (KIT, PDGFRA), vandetanib (RET) and foretinib (MET)90–92. Trying to switch on genes that have been mutationally inactivated is more challenging. A direct approach of using compounds that ‘read-through’ stop codons is showing some promise, as is gene therapy93,94. Inhibiting a pathway member that is upregulated as a result of the CPG mutation has also had success. For example, everolimus, an mTOR inhibitor, is now approved for treatment of astrocytomas in tuberous sclerosis and vismodegib, which inhibits the hedgehog pathway, has shown responses in basal-cell nevus syndrome patients with PTCH1 mutations95,96. Perhaps, the most innovative approach has been through inducing synthetic lethality. PARP inhibitors, which cause lethality in BRCA deficient tumor cells but not normal cells with monoallelic mutations exemplify this approach, which is now being pursued for other CPGs97–99. Currently, these therapies are still largely being evaluated in research studies and clinical trials, but there is optimism that identification of CPG mutations will increasingly lead to personalised management for cancer patients.
Screening and prevention
An important benefit of CPG testing is in providing information about cancer risks for relatives. One of the unusual characteristics of CPGs is their capacity to serve as a biomarker of future disease. Identifying a CPG mutation can provide a window of opportunity to implement surveillance and/or risk-reducing measures that mitigate or prevent cancer. The type of screening is naturally determined by the type of cancer but most often involves imaging to detect a lesion before it presents clinically. Sometimes a biochemical marker of risk can be measured, such as catecholamines or calcitonin in individuals at-risk of phaeochromocytoma or thyroid cancer respectively100. The presumption is that if a cancer is detected early, treatment and survival will be improved, though this has rarely been proven and for some cancers the available evidence does not suggest benefit101. Prevention usually involves surgical removal of the at-risk tissue and is necessarily reserved for non-essential organs in individuals at very high-risk, such as the stomach in CDH1 mutation carriers, the thyroid in RET mutation carriers and the colon in APC mutation carriers100,102,103. Chemoprevention is an attractive strategy but to date there have been few applications. A notable exception is mismatch repair gene mutation carriers in whom the risk of colorectal cancer is significantly reduced by daily aspirin104. Of equal value, though commanding much less fanfare, is the use of CPG mutation testing in identifying people without a familial CPG mutation. Such individuals are released from anxiety for themselves and their offspring, and do not require costly interventions.
Pitfalls in CPG research and clinical practice
The study of CPGs has led to tremendous scientific and medical advancements of broad and lasting impact. However, the field has been hampered by incorrect interpretations of genetic data, which can have substantial negative consequences.
The first major problem is the incorrect classification of a gene as a CPG. There are, unfortunately, dozens of genes in widely-used databases such as OMIM and HGMD that are designated CPGs but for which the evidence is at best uncertain. For the majority, interpretation of available data strongly suggests the gene does not confer high or moderate risks of cancer. There are various reasons for these misclassifications. Until recently, the extent of human coding variation was poorly appreciated leading to over-estimation of the likely causal link between the presence of a gene variant and cancer in an individual. This problem is actually increasing with exome sequencing studies, with many researchers failing to appreciate that rare coding variation, including putative deleterious mutations, are collectively common105,106. The most widespread misconception is that absence from controls of a specific rare mutation identified in cancer cases provides evidence of causality, when it merely provides additional evidence that it is rare.
Over-extrapolation of concepts and data is a pervasive problem in CPG research and manifests in many ways. Firstly, it is often presumed that if a gene mutation causes one cancer that any other cancer that occurs in a mutation carrier is also likely attributable to that gene, whereas frequently it will be coincidental because cancer is very common. Secondly, it is frequently incorrectly assumed that because one mutation class (e.g. truncating mutations) predisposes to cancer that variants in other classes (e.g. missense mutations) are also causative, but many will be rare, innocuous variants. Thirdly, it is commonly assumed that if some genes in a pathway are CPGs that variants in other gene members of that pathway are de facto likely to predispose to cancer. Fifthly, it is widely believed that cancer risks of CPG mutations are constant and can be extrapolated from one context to another, whereas many factors can influence the clinical expression of a CPG mutation, as outlined above. Finally, it is often incorrectly assumed that if a variant is shown to have some kind of functional impact, this proves it is pathogenic. CPGs have multiple complex functions and the relationship between functional aberrations and clinical phenotype is typically unclear or unknown. There are virtually no CPG functional mutational assays that have been validated as robust tests of clinical pathogenicity. Thus although functional data can provide supportive evidence for pathogenicity it can very rarely serve as a substitute for robust genetic evidence.
The extent to which these presumptions lead to incorrect scientific inferences and inappropriate clinical management depends on the specific CPG and scenario. However, significant and unacceptable negative impacts can result, including unwarranted surgery in healthy individuals.
Future opportunities and challenges
The future for CPG research is very bright both in terms of scientific discovery and clinical translation. Strong evidence from multiple sources indicates that more CPGs remain to be discovered. Exome and genome sequencing are ideally suited to their identification, though standards to ensure consistent, robust designation as a CPG are required. For familial and syndromic cancer conditions, exome sequencing methods developed for Mendelian disorders will likely be successful, and are already yielding new genes17,18. As with other common complex conditions, identification of non-syndromic genes will remain challenging, at least until it is possible to sequence, analyse and interpret data from many thousands of individuals. Innovative sample and analytical prioritisation strategies, in the spirit of those used so successfully in the past will thus likely have high utility over the next few years106.
It is also important to recognise that in this review I have focussed on germline gene mutations with high/intermediate risks of cancer. Other components of the genetic architecture of cancer predisposition, such as common variants with small effects are also important and the interplay of different genes and variants is a topical field that will likely reveal new, clinically relevant insights78,81,107. It is also increasingly apparent that many other mechanisms are likely to play a role. One emerging area is the role of mosaic mutations, particularly in individuals with multiple cancers. Genetic and epigenetic cancer predisposing post-zygotic events have been identified, for example mosaic HIF2A mutations in individuals with paraganglioma and H19 hypermethylation in children with bilateral Wilms tumor108,109. More recently, mosaic PPM1D mutations associated with increased risks of ovarian and breast cancer have been reported, though the mechanism of cancer association is currently unclear110. Thus, although a considerable proportion of genetic predisposition to cancer likely resides in CPGs, the genetic architecture of cancer predisposition also includes other components, many of which may be undiscovered.
Further opportunities to use CPGs to improve management of cancer patients should be vigorously pursued. This will lead to optimised, personalised care for mutation carriers and will likely provide insights of broader relevance to cancer, as exemplified by countless CPG-based discoveries of the past. The rarity of CPG mutations impedes research and improved networks and registries of mutation carriers would greatly enhance the field. Routine integration of germline CPG testing into clinical trials will be invaluable, as will better collaborative links between somatic and germline cancer research. Probably the most important goal, which would facilitate all the above, is to increase availability of CPG testing to cancer patients. Next-generation sequencing makes large-scale, high-throughput CPG testing possible and affordable, but the clinical infrastructure needed to appropriately deliver such testing requires development. Various initiatives are seeking to achieve this, such as the UK Mainstreaming Cancer Genetics programme (www.mcgprogramme.com).
It is imperative that comprehensive evaluation of known CPGs in large patient and population series is performed so their cancer risks, clinical phenotypes, genotype-phenotype associations, genetic and non-genetic modifying factors and contribution to cancer can be clarified. Large-scale, international, integrated molecular and clinical databases and analysis will greatly facilitate these endeavours. Enthusiasm for feedback of incidental findings is best tempered until these data are available. Recently the American College of Medical Genetics issued a policy statement recommending incidental findings in 24 CPGs should be returned, irrespective of age or specific consent111. This has stimulated intense debate about possible ethical and legal ramifications. However, scant attention has been given to the arguably more pressing concern of our insufficient knowledge about the clinical consequences of mutations identified opportunistically. As discussed above, there is evidence to suggest the impact of incidental mutations may differ substantially from mutations detected in individuals with a clinical phenotype and thus more curation and more caution are required.
That being said, the impact of CPG mutations in the general population is of high interest and has potential to provide health benefits and opportunities for cancer prevention. It is often assumed cancer surveillance is of intrinsic value and should automatically be instituted in CPG mutation carriers. However, for most surveillance programmes there is little or no actual evidence of an improvement in outcome. The lack of proven efficacy and the potential risks of screening, such as overdiagnosis, misdiagnosis, false positives and false negatives are rarely discussed. The low frequency of CPG conditions makes randomised clinical surveillance trials challenging. It also leads to the misguided impression that ad-hoc screening in individual CPG families is a trivial burden. In fact, instituting decades of surveillance to relatives in a single family can be a very considerable financial outlay. To implement at population level, which may insidiously occur as genome sequencing becomes routine, could spiral into sizeable strains on the capacity and purse of health services. This is particularly likely if individuals with rare variants of unproven pathogenicity are (inappropriately) included in enhanced surveillance programmes, as is currently often the case. To ensure consistent, appropriate, affordable management of at-risk individuals there needs to be a grass-roots move away from reflex interventions to application and adherence to the accepted criteria of effective screening tests112. In parallel, we need to invest energy in developing carefully considered and evaluated strategies that maximise the benefits of identifying people at increased risk of cancer.
Supplementary Material
Supplementary Table 1. Cancer Predisposition Genes
The genes included all have proven association with cancer and confer RR>2 and at least 5% of individuals with the relevant mutations develop cancer. Inevitably, some subjective and/or pragmatic decisions, particularly for rare CPGs were required. For example, Fanconi anaemia genes were only included if individuals with cancer have been reported. It is quite likely that mutations in genes that cause the rarer subtypes do predispose to cancer but that insufficient individuals with the condition are known and/or individuals have died from other causes before cancer developed. It is also quite possible that, despite my best efforts, there are genes that should be included but I have missed. If you believe other genes should be included in this list please contact Rahmanlab@icr.ac.uk. I am grateful for any input. A web-based searchable database of CPGs that will be kept up-to-date is in development and will be available on the Institute of Cancer Research, London website.
Acknowledgements
I am very grateful to many colleagues with whom I have discussed discovery, characterisation and clinical translation of cancer predisposition genes over the last 15 years in particular Mike Stratton, Helen Hanson and Clare Turnbull. I am indebted to Ann Strydom for editorial assistance, Sandra Hanks for construction of Figure 1 and Shazia Mahamdallie, Bianca De Souza, Clare Turnbull and Elise Ruark for input into Supplementary Table 1.
References
- 1.Broca P. Trait, des tumeurs. 1866 [Google Scholar]
- 2.Boveri T. Zur frage der entstehung maligner tumoren. Gustav Fischer; 1914. [Google Scholar]
- 3.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fung YK, et al. Structural evidence for the authenticity of the human retinoblastoma gene. Science. 1987;236:1657–1661. doi: 10.1126/science.2885916. [DOI] [PubMed] [Google Scholar]
- 5.Varghese JS, Easton DF. Genome-wide association studies in common cancers--what have we learnt? Curr Opin Genet Dev. 2010;20:201–209. doi: 10.1016/j.gde.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Chang CQ, et al. A systematic review of cancer GWAS and candidate gene meta-analyses reveals limited overlap but similar effect sizes. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.161. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadler ZK, Gallagher DJ, Thom P, Offit K. Genome-wide association studies of cancer: principles and potential utility. Oncology (Williston Park) 2010;24:629–637. [PubMed] [Google Scholar]
- 8.Zuo L, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 9.Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J Biol Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 10.Sijbers AM, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 11.Stickens D, et al. The EXT2 multiple exostoses gene defines a family of putative tumour suppressor genes. Nat Genet. 1996;14:25–32. doi: 10.1038/ng0996-25. [DOI] [PubMed] [Google Scholar]
- 12.Pilia G, et al. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 13.Feder JN, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 14.Whitcomb DC, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 15.Yu CE, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RL, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 17.Comino-Mendez I, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet. 2011;43:663–667. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 18.Smith MJ, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45:295–298. doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
- 19.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 20.Armanios M, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci U S A. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolaides NC, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 22.Miyaki M, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 23.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 24.Meijers-Heijboer H, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 25.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 26.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao HX, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loveday C, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann DR. RB1 gene mutations in retinoblastoma. Hum Mutat. 1999;14:283–288. doi: 10.1002/(SICI)1098-1004(199910)14:4<283::AID-HUMU2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 30.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 31.Huff V, et al. Evidence for WT1 as a Wilms tumor (WT) gene: intragenic germinal deletion in bilateral WT. Am J Hum Genet. 1991;48:997–1003. [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida T, et al. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet. 1998;19:323–324. doi: 10.1038/1209. [DOI] [PubMed] [Google Scholar]
- 33.Sevenet N, et al. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor MD, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 35.Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med. 2004;351:2403–2407. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 36.Chompret A, et al. PDGFRA germline mutation in a family with multiple cases of gastrointestinal stromal tumor. Gastroenterology. 2004;126:318–321. doi: 10.1053/j.gastro.2003.10.079. [DOI] [PubMed] [Google Scholar]
- 37.Bell DW, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 38.Niemeyer CM, et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42:794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiesner T, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 41.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10:Unit 10 11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hudson TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrett JH, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao LB, et al. The association between ATM D1853N polymorphism and breast cancer susceptibility: a meta-analysis. J Exp Clin Cancer Res. 2010;29:117. doi: 10.1186/1756-9966-29-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stacey SN, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson ND, Bertuch AA. Dyskeratosis congenita as a disorder of telomere maintenance. Mutat Res. 2012;730:43–51. doi: 10.1016/j.mrfmmm.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mocellin S, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104:840–854. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahman N, Scott RH. Cancer genes associated with phenotypes in monoallelic and biallelic mutation carriers: new lessons from old players. Hum Mol Genet. 2007;16 Spec No 1:R60–66. doi: 10.1093/hmg/ddm026. [DOI] [PubMed] [Google Scholar]
- 53.Dixit A, et al. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS One. 2009;4:e7485. doi: 10.1371/journal.pone.0007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;11:111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villanueva A, Newell P, Hoshida Y. Inherited hepatocellular carcinoma. Best practice & research Clinical gastroenterology. 2010;24:725–734. doi: 10.1016/j.bpg.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 57.Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion. 2010;10:393–401. doi: 10.1016/j.mito.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santen GW, Kriek M, van Attikum H. SWI/SNF complex in disorder: SWItching from malignancies to intellectual disability. Epigenetics. 2012;7:1219–1224. doi: 10.4161/epi.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheppard K, Kinross KM, Solomon B, Pearson RB, Phillips WA. Targeting PI3 kinase/AKT/mTOR signaling in cancer. Critical reviews in oncogenesis. 2012;17:69–95. doi: 10.1615/critrevoncog.v17.i1.60. [DOI] [PubMed] [Google Scholar]
- 60.Slade I, et al. DICER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet. 2011;48:273–278. doi: 10.1136/jmg.2010.083790. [DOI] [PubMed] [Google Scholar]
- 61.Scott RH, Stiller CA, Walker L, Rahman N. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet. 2006;43:705–715. doi: 10.1136/jmg.2006.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott RH, et al. Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat Genet. 2008;40:1329–1334. doi: 10.1038/ng.243. [DOI] [PubMed] [Google Scholar]
- 63.Gayther SA, Pharoah PD. The inherited genetics of ovarian and endometrial cancer. Curr Opin Genet Dev. 2010;20:231–238. doi: 10.1016/j.gde.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pacini F, Castagna MG, Cipri C, Schlumberger M. Medullary thyroid carcinoma. Clin Oncol (R Coll Radiol) 2010;22:475–485. doi: 10.1016/j.clon.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Jafri M, Maher ER. The genetics of phaeochromocytoma: using clinical features to guide genetic testing. European journal of endocrinology / European Federation of Endocrine Societies. 2012;166:151–158. doi: 10.1530/EJE-11-0497. [DOI] [PubMed] [Google Scholar]
- 66.Mavaddat N, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2011 doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benusiglio PR, et al. CDH1 germline mutations and the hereditary diffuse gastric and lobular breast cancer syndrome: a multicentre study. J Med Genet. 2013;50:486–489. doi: 10.1136/jmedgenet-2012-101472. [DOI] [PubMed] [Google Scholar]
- 68.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aoki Y, et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 71.Hafner C, Groesser L. Mosaic RASopathies. Cell Cycle. 2013;12:43–50. doi: 10.4161/cc.23108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 73.Goudie DR, et al. Multiple self-healing squamous epithelioma is caused by a disease-specific spectrum of mutations in TGFBR1. Nat Genet. 2011;43:365–369. doi: 10.1038/ng.780. [DOI] [PubMed] [Google Scholar]
- 74.Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 75.Thompson D, Easton D. Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet. 2001;68:410–419. doi: 10.1086/318181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ford D, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antoniou A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Antoniou AC, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Couch FJ, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaudet MM, et al. Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS Genet. 2010;6:e1001183. doi: 10.1371/journal.pgen.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moorman PG, et al. Evaluation of established breast cancer risk factors as modifiers of BRCA1 or BRCA2: a multi-center case-only analysis. Breast Cancer Res Treat. 2010;124:441–451. doi: 10.1007/s10549-010-0842-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chompret A, et al. P53 germline mutations in childhood cancers and cancer risk for carrier individuals. Br J Cancer. 2000;82:1932–1937. doi: 10.1054/bjoc.2000.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Figueiredo BC, et al. Penetrance of adrenocortical tumours associated with the germline TP53 R337H mutation. J Med Genet. 2006;43:91–96. doi: 10.1136/jmg.2004.030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Byrski T, et al. Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer. Breast Cancer Res. 2012;14:R110. doi: 10.1186/bcr3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turner NC, Tutt AN. Platinum chemotherapy for BRCA1-related breast cancer: do we need more evidence? Breast Cancer Res. 2012;14:115. doi: 10.1186/bcr3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scott RH, et al. Medulloblastoma, acute myelocytic leukemia and colonic carcinomas in a child with biallelic MSH6 mutations. Nat Clin Pract Oncol. 2007;4:130–134. doi: 10.1038/ncponc0719. [DOI] [PubMed] [Google Scholar]
- 88.Vencken PM, et al. Outcome of BRCA1- compared with BRCA2-associated ovarian cancer: a nationwide study in the Netherlands. Ann Oncol. 2013;24:2036–2042. doi: 10.1093/annonc/mdt068. [DOI] [PubMed] [Google Scholar]
- 89.Castro E, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bachet JB, et al. Diagnosis, prognosis and treatment of patients with gastrointestinal stromal tumour (GIST) and germline mutation of KIT exon 13. Eur J Cancer. 2013 doi: 10.1016/j.ejca.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 91.Logan TF. Foretinib (XL880): c-MET inhibitor with activity in papillary renal cell cancer. Curr Oncol Rep. 2013;15:83–90. doi: 10.1007/s11912-013-0299-3. [DOI] [PubMed] [Google Scholar]
- 92.Wells SA, Jr, et al. Vandetanib for the treatment of patients with locally advanced or metastatic hereditary medullary thyroid cancer. J Clin Oncol. 2010;28:767–772. doi: 10.1200/JCO.2009.23.6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bordeira-Carrico R, Pego AP, Santos M, Oliveira C. Cancer syndromes and therapy by stop-codon readthrough. Trends in molecular medicine. 2012;18:667–678. doi: 10.1016/j.molmed.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 94.Aiuti A, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jozwiak S, Stein K, Kotulska K. Everolimus (RAD001): first systemic treatment for subependymal giant cell astrocytoma associated with tuberous sclerosis complex. Future Oncol. 2012;8:1515–1523. doi: 10.2217/fon.12.146. [DOI] [PubMed] [Google Scholar]
- 96.Tang JY, et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med. 2012;366:2180–2188. doi: 10.1056/NEJMoa1113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 98.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 99.Brough R, Frankum JR, Costa-Cabral S, Lord CJ, Ashworth A. Searching for synthetic lethality in cancer. Curr Opin Genet Dev. 2011;21:34–41. doi: 10.1016/j.gde.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 100.Wells SA, Jr, Pacini F, Robinson BG, Santoro M. Multiple endocrine neoplasia type 2 and familial medullary thyroid carcinoma: an update. J Clin Endocrinol Metab. 2013;98:3149–3164. doi: 10.1210/jc.2013-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reade CJ, Riva JJ, Busse JW, Goldsmith CH, Elit L. Risks and benefits of screening asymptomatic women for ovarian cancer: A systematic review and meta-analysis. Gynecol Oncol. 2013;130:674–681. doi: 10.1016/j.ygyno.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 102.Rozen P, Macrae F. Familial adenomatous polyposis: The practical applications of clinical and molecular screening. Fam Cancer. 2006;5:227–235. doi: 10.1007/s10689-005-5674-2. [DOI] [PubMed] [Google Scholar]
- 103.Seevaratnam R, et al. A systematic review of the indications for genetic testing and prophylactic gastrectomy among patients with hereditary diffuse gastric cancer. Gastric Cancer. 2012;15(Suppl1):S153–163. doi: 10.1007/s10120-011-0116-3. [DOI] [PubMed] [Google Scholar]
- 104.Burn J, Mathers JC, Bishop DT. Chemoprevention in Lynch syndrome. Fam Cancer. 2013 doi: 10.1007/s10689-013-9650-y. [DOI] [PubMed] [Google Scholar]
- 105.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Snape K, et al. Predisposition gene identification in common cancers by exome sequencing: insights from familial breast cancer. Breast Cancer Res Treat. 2012;134:429–433. doi: 10.1007/s10549-012-2057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Turnbull C, et al. Gene-gene interactions in breast cancer susceptibility. Hum Mol Genet. 2012;21:958–962. doi: 10.1093/hmg/ddr525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhuang Z, et al. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367:922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scott RH, et al. Stratification of Wilms tumor by genetic and epigenetic analysis. Oncotarget. 2012;3:327–335. doi: 10.18632/oncotarget.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruark E, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wilson JMG, Jungner G. Vol. Public Health Papers No. 34. WORLD HEALTH ORGANIZATION; Geneva: 1968. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.